Abstract
Purpose
N6-methyladenosine (m6A), the most abundant mRNA modification in mammals, is involved in various biological processes. KIAA1429 is an important methyltransferase participating in m6A modification. However, the role of KIAA1429 in hepatocellular carcinoma (HCC) is still not well understood. Here, we aimed to investigate the function of KIAA1429 and its corresponding regulation mechanisms in HCC.
Patients and methods
HCC-related genes were analyzed by clinical and expression data of HCC patients in The Cancer Genome Atlas (TCGA) database. Expression of KIAA1429 was verified by quantitative reverse-transcription PCR, and interference efficiency was obtained using small interfering RNA (siRNA). Cell proliferation, migration, and invasion were assessed by cell counting kit-8 and transwell assays, and the m6A modification was detected by methylated RNA immunoprecipitation-PCR (MeRIP-PCR).
Results
We found a difference in the expression of KIAA1429 between HCC and normal hepatic tissues by analyzing data from the TCGA database. Comparing HCC cell lines (HepG2, Huh-7, HepG2.2.15) with normal hepatic cells (HL-7702), we observed an identically significant difference in KIAA1429 expression. KIAA1429 significantly enhanced proliferation, migration, and invasion of HepG2 cells. Moreover, Kyoto Encyclopedia of Genes and Genomes functional enrichment analysis and correlation analysis revealed a significant negative correlation between KIAA1429 and ID2. In the subsequent MeRIP-PCR assay, downregulation of KIAA1429 inhibited m6A modification of ID2 mRNA.
Conclusion
KIAA1429 facilitated migration and invasion of HCC by inhibiting ID2 via upregulating m6A modification of ID2 mRNA.
Keywords: N6-methyladenosine, methyltransferases, tumor metastasis, invasion
Introduction
Hepatocellular carcinoma (HCC), followed by lung and stomach cancer, is one of the greatest challenges in medicine. In China, it is the most common malignant tumor and the primary cause of death in men with cancer.1 Worldwide, HCC is the sixth most common malignant tumor, and accounted for 5.7% of the new cases of cancer in 2002.2 It is estimated that over 700,000 people die every year from this cancer.3 The main cause for the manifestation of HCC is chronic liver inflammation caused by hepatitis B virus, hepatitis C virus, alcohol abuse, or hemochromatosis.4 The situation is particularly severe in Asia.5 With the progress in science and technology, treatments for cancer, such as surgical resection, radiotherapy, and chemotherapy, have been developing continuously. However, high postoperative recurrence and low 5-year survival rate are problems that still exist.6 The poor prognosis can largely be attributed to the dissemination of tumor cells7 and the invasion of vein, especially the portal vein.4,8 Therefore, it is of great significance to study the invasion and transfer mechanism of HCC.
As the most abundant modification of eukaryotic mRNA,9 N6-methyladenosine (m6A) has been shown to play an important role in regulating mRNA splicing, translation, and stability.10 Additionally, m6A affects the development of a variety of human diseases, including tumors.11 There are three types of enzymes that control m6A modifications: “writers” (methyltransferases, which include METTL3, METTL14, WTAP, and KIAA1429), “erasers” (demethylases, which include ALKBH5 and FTO), and “readers” (YTH domain containing RNA binding proteins and heterogeneous nuclear ribonucleoprotein).12 Previous literature reported that FTO regulates the progression of cervical squamous cell carcinoma (CSCC) through mRNA demethylation,13 overexpression of YTHDF1 is associated with poor prognosis of HCC,11 and METTL14 suppresses the metastatic potential of HCC.7
In this study, we aimed to investigate the functions and related mechanism of KIAA1429 in HCC. Expression of KIAA1429 in HCC and in corresponding normal tissues and cells was compared via The Cancer Genome Atlas (TCGA) data analysis and quantitative PCR. Afterward, we knocked down KIAA1429 to study its effect on cell proliferation, migration, and invasion. Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis and correlation analysis were performed to explore the relevant pathways and molecules. Furthermore, methylated RNA immunoprecipitation-PCR (MeRIP-PCR) assay was used to carry out methylation of KIAA1429.
Patients and methods
Clinical samples and cell lines
HCC specimens and the corresponding normal tissues used in this study were obtained from eleven patients with HCC with informed consent. Demographics and clinical characteristics of the eleven HCC patients are shown in Table 1. Human HCC cell lines HepG2, Huh-7, and HepG2.2.15 and normal liver cell line HL-7702 were purchased from the cell bank of the Chinese Academy of Sciences. Cells were cultured in DMEM (Corning, Shanghai, China) containing 10% FBS (Gibco, Waltham, MA, USA) at 37°C and 5% CO2. The study was approved by the First Affiliated Hospital of Nanchang University, and all the participants signed written informed consents. The study was conducted in accordance with the ethical guidelines of the World Medical Association’s Declaration of Helsinki.
Table 1.
Demographics and clinical characteristics of HCC specimens
| Terms | All (n=11) |
|---|---|
| Demographic variables | |
| Age, years | 55 (51–69) |
| Sex, male | 8 (72.7) |
| Etiology, HBV/HCV/alcohol/others | 7/1/2/1 (63.6/9.1/18.2/9.1) |
| Previous HCC treatment history | 3 (27.2) |
| Tumor variables | |
| Size of tumor, <5 cm/≥5 cm | 5/6 (45.5/54.5) |
| Number of tumors, single/multiple | 4/7 (36.4/63.6) |
| T stage, 1–2/3–4 | 5/6 (40.5/59.5) |
| N stage, 0/1 | 10/1 (96.2/3.8) |
| M stage, 0/1 | 9/2 (93.7/6.3) |
| Modified UICC stage, I–II/III–IV | 4/7 (40.5/59.5) |
| Clinical variables | |
| ESR, mm/h | 15.5 (6.15–22.75) |
| CRP, mg/dL | 0.30 (0.22–0.48) |
| Platelet count, 109/L | 152 (97–185) |
| Prothrombin time, INR | 1.11 (1.06–1.14) |
| Aspartate aminotransferase, IU/L | 43.00 (37–48) |
| Alanine aminotransferase, IU/L | 34.00 (24–39) |
| Total bilirubin, mg/dL | 0.77 (0.48–1.14) |
| Serum albumin, g/dL | 3.7 (3.3–4.1) |
| Creatinine, mg/dL | 0.92 (0.75–1.03) |
| Alpha-fetoprotein, ng/mL | 27.6 (6.3–1,512) |
| LC | 5 (45.5) |
| Child-Pugh class, A/B | 3/2 (60/40) |
| Presence of ascites | 0 |
Notes: Demographic variables and clinical variables terms are presented as average (min–max); tumor variables and LC terms are presented as number (%).
Abbreviations: CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; INR, international normalized ratio; LC, liver cirrhosis; UICC, Union for International Cancer Control.
RNA extraction and qRT-PCR
Total RNA of tissue and cells was extracted with Trizol (Thermo Fisher Scientific) and all reagents used in the experiment were RNase-free. The concentration and quality of the extracted RNA were determined by OD260/OD280 ratio, and the integrity of RNA was observed using agarose gel electrophoresis. cDNA was synthesized using a commercial reverse-transcription kit made by Thermo Bio-Analysis. Actin was used as internal reference in quantitative reverse-transcription PCR (qRT-PCR) and the result was analyzed by 2−ΔΔct method. Primer sequences are listed in Table 2.
Table 2.
Primer and siRNA sequences
| Name | Sequence |
|---|---|
| Actin | F 5′-AGCACAGAGCCTCGCCTTTG-3′ |
| R 5′-CTTCTGACCCATGCCCACCA-3′ | |
| KIAA1429 ID2 |
F 5′-CGATAACTTGATGACCCCAGAA-3′ |
| R 5′-ATAACGGCAAGATTCCATTTC-3′ | |
| F 5′-CACGGACATCAGCATCCTGT-3′ | |
| R 5′-CCCAAATGCCATTTATTTAGC C-3′ | |
| siKIAA1429-1 | F 5′-GAGGATGATCGACGAACAGTA-3′ |
| siKIAA1429-2 | R 5′-AAGGCTTATTAACCTCCTAGA-3′ |
Abbreviations: F, forward; R, reverse.
Bioinformatics analysis
GEPIA (http://gepia.cancer-pku.cn/detail.php) was used to analyze the data from TCGA database in order to determine the long non-coding RNA associated with HCC. Moreover, the DAVID tool (National Institute of Allergy and Infectious Diseases, Bethesda, MD, USA) was used to analyze KEGG pathways and for enrichment of gene ontology terms.
SiRNA transfection
HepG2 cells were seeded at a density of 3 × 105 cells per well in six-well plates and grown for 24 hours in complete media until the fusion rate reached 80%–90%. For transient knockdown, cells were rinsed with fresh media and incubated with siRNA (GenePharma) and Lipofectamine 2000 (Thermo Fisher Scientific) for 24 hours, and were ready for subsequent experiments. The sequences of siRNA are listed in Table 2.
Western blot
Protein samples were prepared by lysing cells in RIPA lysis buffer and collecting the supernatant after centrifuging. Total protein level was measured using BCA assay kit (Pierce Biotechnology, Inc., Rockford, IL, USA). Next, the same quantity of protein from each sample was run on 10% SDS-PAGE and transferred to polyvinylidene fluoride membranes (MD Millipore, Billerica, MA, USA). Membranes were blocked with TBST solution (0.1% Tween 20) with 5% skim milk and incubated with mouse anti-ID2 (1:1,000, ab85990; Abcam, Cambridge, UK) and anti-GAPDH antibodies (1:1,000, ab9482; Abcam) for 12 hours at 4°C. Membranes were washed and incubated with horseradish peroxidase-conjugated goat anti-mouse IgG (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) for 2 hours. Protein bands were revealed using enhanced chemiluminescence reagent (Thermo Fisher Scientific) and visualized via a ChemiDoc MP system (Bio-Rad, Hercules, CA, USA).
CCK-8 assay
Cells were seeded in a 96-well plate (3599; Corning) at a density of 2 × 103 cells per well. Each well contained six repetitions and sterile water was added to marginal wells. After culturing for 0, 24, 48, 72, or 96 hours, CCK-8 reagent (Beyotime) was added and OD was measured at 450 nm.
Transwell assay
In the migration experiment, 500 µL of serum-free DMEM (Corning), which contained 1 × 105 cells, was loaded into the upper chambers (Corning) of the transwell device. Next, 700 µL of DMEM with 10% FBS was loaded into the lower chamber. After 24 hours, cells on the underside of the membrane were stained and counted under a microscope in three random high-power fields. In the invasion experiment, chambers precoated with matrix adhesive (Corning) were used and the experiments described above were repeated.
MeRIP-PCR
HepG2 cells transfected with siRNAs against KIAA1429 and control cells were irradiated with ultraviolet light at 254 nm, 400 mJ/cm2 (Stratagene Stratalinker), and lysed by disruptive sonication with RIP lysis buffer (MagnaRIP Kit; EMD Millipore) at 4°C. Immunoprecipitations were performed using an anti-m6A antibody (202003; Synaptic Systems) at 4°C, overnight. After washes, RNA was extracted with phenol:chloroform isoamyl alcohol and subjected to qRT-PCR using primers for ID2 mRNA, as listed in Table 2.
Statistical analysis
Data were analyzed with SPSS 19.0 (SPSS Inc., Chicago, IL, USA) and presented as mean ± SD. Student’s t-test was used for comparison between two groups and two-way ANOVA was used for comparison between more than two groups. P<0.05 was considered as statistically significant.
Results
KIAA1429 is elevated in HCC and associated with prognosis
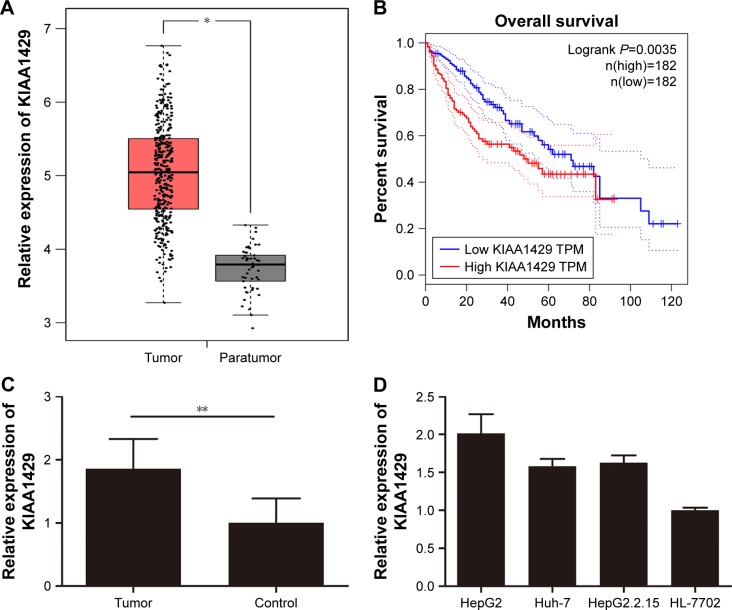
First, we analyzed data from TCGA, which included 371 liver cancer tissues and 50 normal liver tissues. Result revealed that mRNA level of KIAA1429 in liver tumor was significantly higher compared to that in normal liver tissues (Figure 1A). Analysis of survival data showed that high expression of KIAA1429 was significantly correlated with poor overall survival (P=0.0035, Figure 1B). Furthermore, qRT-PCR analysis of KIAA1429 mRNA showed that HCC tissues expressed significantly increased mRNA level of KIAA1429 compared to that of the corresponding normal tissues (Figure 1C). Similarly, the mRNA expression level of KIAA1429 in three human HCC cell lines (HepG2, Huh-7, and HepG2.2.15) was dramatically elevated compared to that in the normal hepatic cell lines (HL-7702) (Figure 1D).
Figure 1.
KIAA1429 is elevated in HCC and is associated with prognosis.
Notes: (A) KIAA1429 levels in 371 liver cancer tissues and 50 normal liver tissues from TCGA database. (B) Survival data of patients with high or low levels of KIAA1429. (C) KIAA1429 mRNA levels in eleven pairs of HCC tissues and normal tissues. (D) KIAA1429 levels in different cell lines. n=3, *P<0.05, **P<0.01, Student’s t-test.
Abbreviations: HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas; TPM, trans per million.
KIAA1429 promotes cell proliferation in HCC
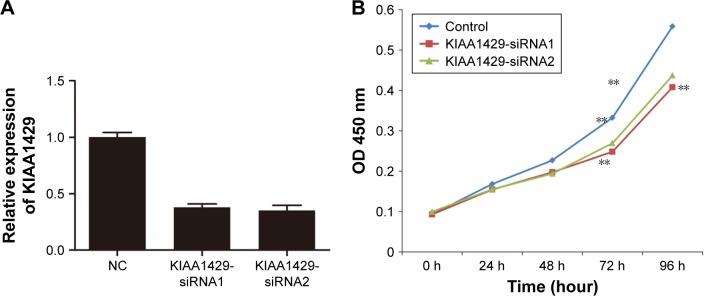
To elucidate the function of KIAA1429 in HCC, HepG2 cell line, which expressed high level of KIAA1429, was selected for further confirmatory studies. Knockdown of KIAA1429 in cells transfected with siRNA targeting KIAA1429 was confirmed by qRT-PCR (Figure 2A). CCK-8 assay revealed that underexpression of KIAA1429 decreased proliferation of HCC cells (Figure 2B).
Figure 2.
KIAA1429 promotes cell proliferation in HCC.
Notes: (A) KIAA1429 levels in negative control and in cells transfected with siRNA targeting KIAA1429. (B) CCK-8 assays of negative control and KIAA1429 siRNA transfected cells (**P<0.01).
Abbreviations: HCC, hepatocellular carcinoma; CCK, cell counting kit; NC, negative control.
KIAA1429 increases cell migration and invasion in HCC
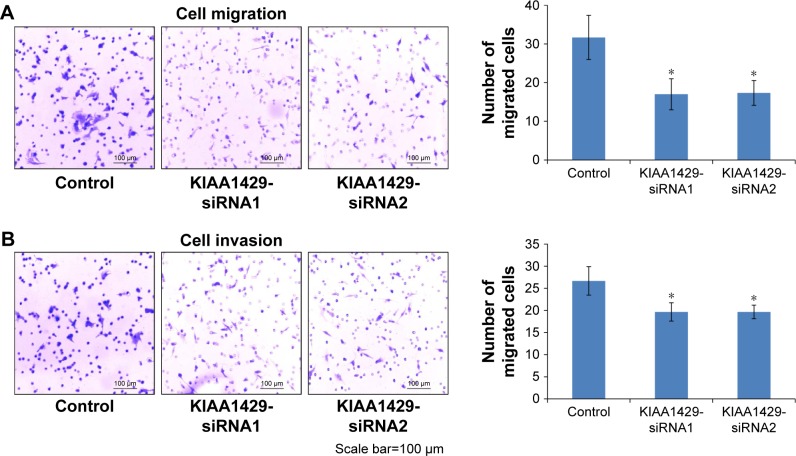
To study the effect of KIAA1429 on cell migration and invasion, transwell assays were performed in HepG2 cells. Knockdown of KIAA1429 led to significant inhibition of cell migration in HepG2 cells (Figure 3A and B). Similarly, invasive ability of KIAA1429 siRNA transfected cells was significantly decreased as compared to that of control cells (Figure 3A and B).
Figure 3.
KIAA1429 increases cell migration and invasion in HCC.
Notes: (A) Transwell migration of HepG2 cells transfected with siRNAs against KIAA1429. (B) Transwell invasion assays of HepG2 cells transfected with siRNAs against KIAA1429. Data of migration/invasion are shown in the histogram on the right (*P<0.05).
Abbreviation: HCC, hepatocellular carcinoma.
Downregulation of KIAA1429 increases ID2 level
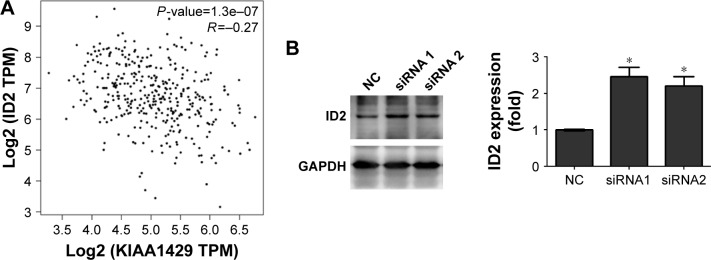
To obtain insights into the molecular mechanisms through which KIAA1429 controls hepatoma cell functions, we analyzed the genes that are differentially expressed and contain alterative m6A deposition after the interference with KIAA1429 expression in Gene Expression Omnibus database (GSE102493). Three pathways were found in subsequent KEGG functional enrichment analysis. We further analyzed three genes of the TGF-β signaling pathway in TCGA database and found a significant negative correlation between KIAA1429 and ID2, which was reported to regulate metastatic potentials of HCC in a previous study (Figure 4A).6 The result of Western blot also showed that silencing of KIAA1429 significantly increased the protein levels of ID2 (Figure 4B).
Figure 4.
Downregulation of KIAA1429 increases ID2 level.
Notes: (A) Correlation analysis between KIAA1429 and ID2 using TCGA database. (B) Protein levels of ID2 in knockdown and control cells (*P<0.05).
Abbreviations: TCGA, The Cancer Genome Atlas; NC, negative control; TPM, trans per million.
Downregulation of KIAA1429 inhibits m6A modification of ID2 mRNA
To further delineate the action mechanism between KIAA1429 and ID2, we built a MeRIP-PCR assay. A significant inhibition of m6A modification was observed in KIAA1429 siRNA transfected cells (Figure 5), which suggests that KIAA1429 might methylate ID2 mRNA to regulate the expression of ID2.
Figure 5.
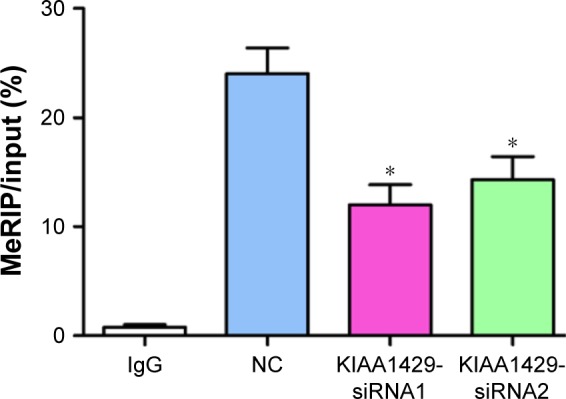
Downregulation of KIAA1429 inhibits m6A modification of ID2 mRNA.
Note: MeRIP-PCR revealed the m6A levels of ID2 mRNA in HepG2 cells transfected with siRNAs against KIAA1429 (*P<0.05).
Abbreviations: MeRIP-PCR, methylated RNA immunoprecipitation-PCR; NC, negative control.
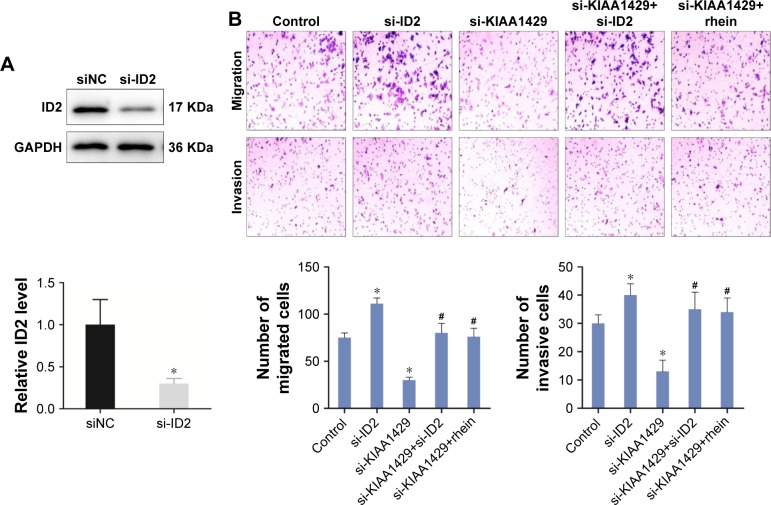
KIAA1429 regulates migration/invasion of HCC by altering m6A modification of ID2 mRNA
Then, we hypothesized that the alteration of m6A on ID2 mRNA is associated with the changes in cell function of KIAA1429 silenced cells. Interference of the expression of ID2 in HCC cells by siRNA was confirmed by Western blot (Figure 6A). Knockdown of ID2 led to a significant acceleration in cell migration and invasion capacities of HepG2 cells (Figure 6B). Interestingly, In Figure 3, we have mentioned that interfering with KIAA1429 expression can inhibit the migration and invasion of HCC cells. On this basis, interfering with ID2 expression was found to be able to reverse the effect of KIAA1429 on cell function, and KIAA1429 knockdown cells treated with rhein can restore the inhibitory effect of KIAA1429 knockdown on the migration and invasion of HCC (Figure 6B).
Figure 6.
KIAA1429 regulates migration/invasion of HCC by altering m6A modification of ID2 mRNA.
Notes: (A) ID2 levels in negative control and in cells transfected with siRNA targeting ID2 was confirmed by Western blot (top) and RT-PCR (bottom). (B) Transwell migration and invasion assays of HepG2 cells transfected with siRNA targeting ID2, siRNA-1 against KIAA1429, or siRNA-1 against KIAA1429 with rhein. Data of migration/invasion are shown in the histogram below. Rhein is an m6A eraser inhibitor. *P<0.05 (all groups vs control group), #P<0.05 (all groups vs si-KIAA1429 group).
Abbreviations: HCC, hepatocellular carcinoma; RT-PCR, reverse-transcription PCR; NC, negative control.
Discussion
In this study, we demonstrated that KIAA1429 contributed to the development of HCC via regulating mRNA methylation levels of ID2 in cell lines. At a cellular level, decrease of KIAA1429 significantly inhibited proliferation, migration, and invasion of HCC cells. From the perspective of mechanisms, processes mediated by KIAA1429 in HCC were at least partially carried out by ID2 via methylation. In more detail, KIAA1429 increased the m6A level of ID2 mRNA which subsequently reduced ID2 expression and promoted cell migration and invasion in HCC. ID2 was a dominant-negative antagonist of basic helix ± loop ± helix transcription factors and proteins of the retinoblastoma (Rb) family.14 It has been reported that ID2 was associated with the development of many diseases. Members of the ID gene family, including ID2, were known to be highly expressed in a number of different tumor types.15,16
The attention was first focused on KIAA1429 because of the discovery of its ortholog, which was shown to interact with Drosophila WTAP in the context of sex-specific splicing.17 Further research revealed that KIAA1429, as well as METTL3 and METTL14, was required for methylation in mammals.18 METTL3 and METTL14 regulate several biological process including cell division,19 neural development,20 and cerebellar development.21 In previous investigations, METTL3 was observed to be upregulated in liver cancer and was found to promote its progression;22 METTL14 was reported to suppress the metastatic potential of HCC and was downregulated in this type of cancer.7 Several studies have reported that KIAA1429, METTL3, METTL14, and WTAP are usually composed of a methyl-transferase complex.23,24 Since KIAA1429 is a methyltransferase similar to METTL3 and METTL14, we supposed that KIAA1429 might be abnormally expressed in HCC and might be associated with cell migration. In our study, this hypothesis was confirmed, and we showed that KIAA1429 plays an important role in HCC cells.
Among various modifications, m6A methylation, which participates in almost every step of RNA metabolism, is the most abundant.25 An increasing number of investigations suggest multiple functions for m6A RNA methylation in disease.26 FTO, a type of demethylase, regulates the chemo-radiotherapy resistance of CSCC via altering m6A level of β-catenin.13 Transcription of ID2 takes an obbligato effect in HCC and various regulations come true via ID2-mediated pathways. The metastatic potential of HCC was promoted by decreasing ID2 via altering the secretion of vascular endothelial growth factor.6 Similarly, in our study, KIAA1429 regulated migration and invasion of HCC by altering the m6A level of ID2.
Conclusion
We found that KIAA1429 facilitated migration and invasion of HCC by inhibiting ID2 via upregulating m6A modification of ID2 mRNA. This result indicates that KIAA1249 may be a potential diagnostic and therapeutic target in HCC.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global Cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Bosch FX, Ribes J, Borràs J. Epidemiology of primary liver cancer. Semin Liver Dis. 1999;19(3):271–285. doi: 10.1055/s-2007-1007117. [DOI] [PubMed] [Google Scholar]
- 4.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31(4):339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 5.Chen M, Wei L, Law CT, et al. RNA N6-methyladenosine methyl-transferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 6.Tsunedomi R, Iizuka N, Tamesa T, et al. Decreased Id2 promotes metastatic potentials of hepatocellular carcinoma by altering secretion of vascular endothelial growth factor. Clin Cancer Res. 2008;14(4):1025–1031. doi: 10.1158/1078-0432.CCR-07-1116. [DOI] [PubMed] [Google Scholar]
- 7.Ma JZ, Yang F, Zhou CC, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65(2):529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 8.Vauthey J-N, Lauwers GY, Esnaola NF, et al. Simplified staging for hepatocellular carcinoma. J Clin Oncol. 2002;20(6):1527–1536. doi: 10.1200/JCO.2002.20.6.1527. [DOI] [PubMed] [Google Scholar]
- 9.Batista PJ. The RNA Modification N6-methyladenosine and its implications in human disease. Genomics Proteomics Bioinformatics. 2017;15(3):154–163. doi: 10.1016/j.gpb.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Z, Li J, Feng G, et al. MicroRNA-145 Modulates N6-Methyladenosine Levels by Targeting the 3′-Untranslated mRNA Region of the N6-Methyladenosine Binding YTH Domain Family 2 Protein. J Biol Chem. 2017;292(9):3614–3623. doi: 10.1074/jbc.M116.749689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao X, Chen Y, Mao Q, et al. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018;21(4):859–868. doi: 10.3233/CBM-170791. [DOI] [PubMed] [Google Scholar]
- 12.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15(5):293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 13.Zhou S, Bai ZL, Xia D, et al. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog. 2018;57(5):590–597. doi: 10.1002/mc.22782. [DOI] [PubMed] [Google Scholar]
- 14.Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407(6804):592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 15.Lasorella A, Benezra R, Iavarone A. The Id proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer. 2014;14(2):77–91. doi: 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- 16.Havrda MC, Paolella BR, Ran C, et al. Id2 mediates oligodendrocyte precursor cell maturation arrest and is tumorigenic in a PDGF-rich microenvironment. Cancer Res. 2014;74(6):1822–1832. doi: 10.1158/0008-5472.CAN-13-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ortega A, Niksic M, Bachi A, et al. Biochemical function of female-lethal (2)D/Wilms’ tumor suppressor-1-associated proteins in alternative pre-mRNA splicing. J Biol Chem. 2003;278(5):3040–3047. doi: 10.1074/jbc.M210737200. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz S, Mumbach MR, Jovanovic M, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014;8(1):284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi M, Ohsugi M, Sasako T, et al. The RNA methyltransferase complex of WTAP, METTL3, and METTL14 regulates mitotic clonal expansion in adipogenesis. Mol Cell Biol. 2018;38(16) doi: 10.1128/MCB.00116-18. 15. pii: e00116–e00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du K, Zhang L, Lee T, Sun T. M6a RNA methylation controls neural development and is involved in human diseases. Mol Neurobiol. 2019;56(3):1596–1606. doi: 10.1007/s12035-018-1138-1. [DOI] [PubMed] [Google Scholar]
- 21.Wang CX, Cui GS, Liu X, et al. METTL3-mediated M6a modification is required for cerebellar development. PLoS Biol. 2018;16(6):e2004880. doi: 10.1371/journal.pbio.2004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen M, Wei L, Law CT, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 23.Guo J, Tang HW, Li J, Perrimon N, Yan D. Xio is a component of the Drosophila sex determination pathway and RNA N6-methyladenosine methyltransferase complex. Proc Natl Acad Sci U S A. 2018;115(14):3674–3679. doi: 10.1073/pnas.1720945115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson M, Shah P, Cui YH, He YY. The role of dynamic m6A RNA methylation in photobiology. Photochem Photobiol. 2019;95(1):95–104. doi: 10.1111/php.12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai D, Wang H, Zhu L, Jin H, Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9(2):124. doi: 10.1038/s41419-017-0129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan Y, Ma P, Liu Y, Li W, Shu Y. Multiple functions of m6A RNA methylation in cancer. J Hematol Oncol. 2018;11(1):48. doi: 10.1186/s13045-018-0590-8. [DOI] [PMC free article] [PubMed] [Google Scholar]