Abstract
In nature, biological nanomaterials are synthesized under ambient conditions in a natural microscopic-sized laboratory, such as a cell. Biological molecules, such as peptides and proteins, undergo self-assembly processes in vivo and in vitro, and these monomers are assembled into various nanometer-scale structures at room temperature and atmospheric pressure. The self-assembled peptide nanostructures can be further organized to form nanowires, nanotubes, and nanoparticles via their molecular-recognition functions. The application of molecular self-assemblies of synthetic peptides as nanometer-scale building blocks in devices is robust, practical, and affordable due to their advantages of reproducibility, large-scale production ability, monodispersity, and simpler experimental methods. It is also beneficial that smart functionalities can be added at desired positions in peptide nanotubes through well-established chemical and peptide syntheses. These features of peptide-based nanotubes are the driving force for investigating and developing peptide nanotube assemblies for biological and non-biological applications.
Graphical Abstract
1. Introduction
There has been rapid progress in the development of nanomaterials, such as nanowires, nanotubes, nanoparticles, and nanocrystals, which show superior electronic, magnetic, optical, and mechanical properties as compared to their bulk counterparts.[1–9] These materials are extremely attractive as potential building blocks for various applications, for instance, electronics, sensors, catalysis, magnetic devices, drug delivery, tissue engineering, ion channels, and medical imaging, because their physical and chemical properties are tunable via control of their size and shape.[10–17] Recently, various inorganic and carbon-based nanomaterials, such as nanocrystals and nanotubes, have been used as building blocks to assemble nanometer-scale devices. Some of these devices show significant improvements compared to existing devices.[18–22] while these nanometer-scale devices are close to reaching industrial application, there are still problems to be overcome with these approaches. For example, inorganic and carbon-based nanotubes have been produced by expensive techniques like chemical vapor deposition (CVD); such instruments usually demand large laboratory space and manpower. The high-temperature requirements for nanomaterial syntheses also lead to large-scale water consumption and higher production costs. If nanomaterial production can be achieved under milder experimental conditions in a smaller laboratory, the development of nanomaterial-based devices may be accelerated. Another issue in nanodevice fabrication from nanomaterials is to address these building blocks to exact locations, so as to design complex device geometries such as integrated circuits.[23] External forces, such as microfluidics, magnetic fields, electric fields, and the Langmuir–Blodgett technique, have been applied to align inorganic/carbon nanotubes,2,24–29] but the development of simpler and more economical methods is desirable for large-scale production.
In nature, biological nanomaterials are synthesized under ambient conditions in a microscopic-sized laboratory such as a cell. Biological molecules, like peptides and proteins, undergo self-assembly processes in vivo and in vitro, and these monomers are assembled into various nanometer-scale structures at room temperature and atmospheric pressure.[30,31] Self-assembled peptide nanostructures can be further organized into nanowires, nanotubes, or nanoparticles via their molecular recognition function.[32,33] This smart recognition function can also address the biological nanomaterials to exact locations on substrates where their complementary recognition groups are marked.[32–37] Therefore, the bionanotechnology approach seems to have many advantages for nanodevice development compared to existing technologies, except for the feasibility of certain physical properties, such as electronic and mechanical properties (although later in this review, it is shown that these problems can be overcome by peptide-regulated biomineralization of bionanomaterials under ambient conditions). These features of peptide-based nanotubes are, therefore, the driving force for the investigation and development of peptide nanotube assemblies for biological and non-biological applications. Here, we define peptide-based nanotubes as structures that possess a well-defined hollow cylinder with a diameter range of 0.5–500 nm and an aspect ratio greater than five.
While the concept of using biological nanomaterials, such as peptide nanotubes, as building blocks for device fabrication is relatively new, the self-assembly of peptides has been investigated for a long time in basic scientific studies carried out in molecular aggregation and medical fields.[30] Supramolecular self-assembly has been studied extensively for decades as it has been widely observed that many molecules aggregate via non-covalent interactions, such as hydrogen bonding and hydrophobic interactions. Therefore, the control of aggregated structures has quickly become a popular area of research for chemists. Since peptides and proteins are efficiently assembled into exact shapes with certain functionalities in biological systems by means of their smart recognition functions, they have been studied as model systems for advanced supra-molecular self-assemblies. For example, fibers spun by spiders show exceptional mechanical properties, due to their microstructure, relative to any synthetic nanofibers, and various artificial nanofibers have been self-assembled by using spider fibers as model systems.[38] Another motivation to study peptide/protein aggregation is to understand the mechanism for prion and neurodegenerative diseases, such as Alzheimer’s and Parkinson’s diseases; the peptides aggregate to form amyloid fibrils, natural biological nanotubes, that cause various neurological diseases (Fig. 1).[39–41] Inhibition of amyloid fibril formation has been studied systematically by analyzing the self-assembly of various synthetic peptides whose structures resemble those of amyloid-forming cellular polypeptides.[42,43]
Figure 1.

Proposed mechanism for the self-assembly of prion trimers to form an amyloid fibril [41,95]. (Reprinted with permission from [41]; copyright 2004, American Chemical Society.)
The application of molecular self-assemblies to nanometer-scale building blocks in devices is robust, practical, and affordable due to their advantages of reproducibility, large-scale production, monodispersity, and simpler experimental procedures. Recent studies showed that the amino acid sequence controls the peptide-assembled structures and their physical properties,[44–48] which is advantageous for applying self-assembled peptides as building blocks in various devices. Peptide nanotubes also maintain smart functionalities, such as molecular recognition and biomineralization, after self-assembling into the nanotube form; this is extremely useful in practical applications.[49–51] It should be noted that smart functionalities can be added at desired positions in peptides through well-established chemical and peptide syntheses. This feature enables decoration of the inside and outside walls of peptide nanotubes with different functional groups, which opens up various new applications.[52] As many peptide nanotubes are soluble in aqueous solution (i.e., peptide nanotubes do not aggregate in water), the assembly of peptide nanotubes via molecular recognition is relatively straightforward in solution. Peptide nanotubes seem to be unstable in harsh environments; however, their structures are fairly rigid because they are assembled via a large number of three-dimensional (3D) hydrogen bonds. For example, the melting point of some peptide nanotubes reaches 235 °C, which is suitable for the operating conditions of the nanodevices described above.[53]
In this review, we would like to introduce peptide nanotubes assembled from i) linear peptide monomers and ii) non-linear peptide monomers. In Section 4, we highlight recent advances in metal and semiconductor nanotube fabrication by using peptide nanotubes as templates. Recently, a significant number of approaches for templating peptide nanotubes have been published because metal-coated nanotubes are not only important building blocks for micro-electronics but are also fundamentally interesting model systems to understand biomineralization in nature. In Section 5, applications of peptide nanotubes in the fields of microelectronics and bioengineering are described. It should be noted that the hollow structure of nanotubes allows the advantage of functionalizing and/or templating various molecules inside the nanotubes. This type of structure can be applied, for example, in materials synthesis, functional channels, heterogeneous nanotube alloys, and waveguides, as explained in Section 5. Since this field is still in the early stages of development, there are more reports of peptide nanotube synthesis than reports of their applications. However, promising results of their use in applications are beginning to appear, as summarized in Section 5.
2. Peptide Nanotubes from Linear Peptide Monomers
Syntheses of peptide nanotubes and their applications have just started to be explored; however, the study of peptide aggregation related to neurological diseases has a long history. For example, soluble cellular proteins undergo a self-assembly process that leads to the formation of well-ordered tubular deposits in prion and neurodegenerative diseases, such as Alzheimer’s and Parkinson’s diseases (Fig. 1).[39,40,42] Nanotube formation from genetically modified peptide monomers has been studied extensively to understand which amino acid residues and sequences are responsible for tubular aggregation.[43,54,55] Information about the peptide sequences responsible for aggregation is utilized to prevent problematic nanotube formation in diseases. However, in the field of materials engineering this knowledge can be applied to design peptide nanotubes. For example, synthetic polypeptides were designed to fold into a nanotube structure by means of a peptide combinatorial library. These “de-novo” designs of polypeptides consist of polar amino acid residues (His, Lys, Asn, Asp, Gln, Glu) and non-polar residues (Leu, Ile, Val, Phe, Met), and their sequence determines the nanotube dimensions (Fig. 2, left).[48] This approach demonstrates that peptide nanotube formation can be regulated by amino acid sequences as shown in Figure 2 (right).
Figure 2.

Helical nanotube formation from de-novo peptides (left) and a transmission electron microscopy (TEM) image of this nanotube (right) [48]. (Reprinted with permission from [48]; copyright 2002, National Academy of Sciences.)
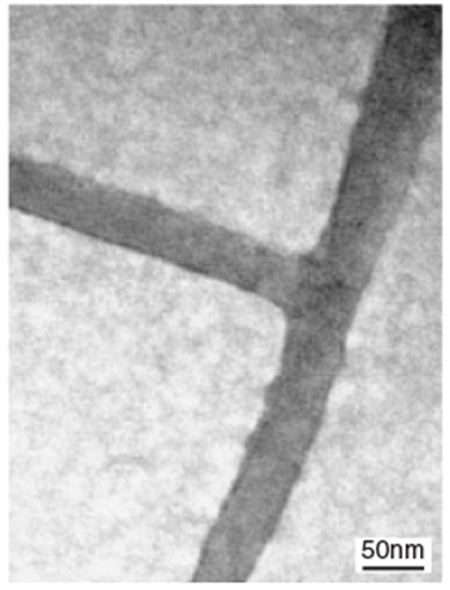
Recently, de-novo designs of polypeptides were also applied to form linear and nonlinear peptide nanotubes.[56] The peptide monomers contained sticky-end residues (Fig. 3a), which could selectively bind complementary residues. Therefore, the position of the sticky ends in the linear peptide monomers determined the assembled structures. For example, when the stiff and straight peptide monomers (peptides ABABAB and CDCDCD in Fig. 3b) incorporated the sticky ends, as shown in Figure 3b, they were assembled into straight peptide nanotubes (Fig. 3b, right). However, kinked and wavy peptide nanotubes (Fig. 3c, right) could be formed when a flexible peptide monomer (peptide CC in Fig. 3c) was mixed with a straight peptide monomer, AA. When a T-shaped peptide monomer, CDC (Fig. 3d), was incubated with the straight peptide monomer AB, these peptides assembled into branched peptide nanotubes (Fig. 3d, right).[57] The advantage of this technique is that by placing the sticky ends at proper residue positions the sequence of peptide monomers leads to predictable nanotube structures. De-novo polypeptides were also assembled into peptide nanotubes by using highly directional metal–ligand interactions instead of sticky-end interactions.[58] When polypeptide monomers were incubated with Re ions, the ions linked the monomers, and they were assembled into nanotubes, as shown in Figure 3e. The length of this peptide nanotube was controlled by the number of linking ions.
Figure 3.

Schematic principles and TEM images of morphology-controlled peptide nanotube assemblies, a) The sequences of standard self-assembling fibers. Block A complements block D, and block B complements block C. This dimerization creates the sticky ends in the nanotubes [56]. b) Straight nanotube formation (scale bar = 50 nm in TEM image) [56], c) kinked nanotube formation (scale bar = 100 nm in TEM image) [56], and d) branched nanotube formation (scale bar = 50 nm in TEM image) [57], e) Peptide assemblies mediated via an octahedral rhenium complex between peptide monomers (shown as bars) [58]. (Reprinted with permission from [56–58]; [56] copyright 2003, Nature Publishing Croup, [58] copyright 2004, the Royal Society of Chemistry.)
Another approach to produce peptide nanotubes is to use simpler organic molecules that are known to self-assemble to form nanotubes. If peptide motifs can be incorporated into these organic monomers, the modified monomers may still be assembled into nanotubes. For example, self-assemblies of amphiphilic molecules (i.e., molecules with charged head groups and non-polar tail groups) have been studied for decades because they are known to mimic biological membrane structures. These amphiphile monomers self-assemble by hydrophilic/hydrophobic interactions between the amphiphiles and surfactants. Experimental conditions to form nanotubes can be determined from phase diagrams of these assemblies, which have already been well-established.[59] Therefore, it was a natural progression to produce peptide nanotubes from peptide-derivatized amphiphile monomers. This system adds a new parameter to the control of self-assembled structures by controlling hydrogen bonds between amino acid residues,[60–62] which may make peptide amphiphile-assembled nanotubes more sensitive to pH, because the protonation of amino acid residues changes the strength of intermolecular hydrogen bonding.[63] In addition to hydrogen bonding, the hydrophobicity of peptide monomers also becomes an important factor in determining the dimensions of peptide nanotubes.[64]
Figure 4 illustrates how linear peptide monomers self-assemble into nanotubes.[65] The peptide amphiphile monomers in Figure 4a are assembled to orient the hydrophobic alkyl chain towards the core and the amino acid residues to the outer surface. While this assembled nanotube structure (Fig. 4b) is similar to a micelle, the protonation state of amino acids still plays an important role in organizing this structure; this is supported by the observation that the appearance of nanotubes depends on the pH of the solution.[66] The functionalization of peptide nanotubes can be achieved by introducing the functional groups into monomers via chemical synthesis. If some functional groups are incorporated into the head groups, those groups appear on the outer surfaces of nanotubes. For example, a certain peptide sequence for cell adhesion was incorporated into the head group of a monomer in order to anchor the peptide nanotubes onto cells.[65] It should be noted that recently amino acid residues were sequenced to form head and tail groups resembling amphiphile molecules, and some of these polypeptides with particular sequences self-assembled into peptide nanotubes, as was observed for the amphiphile nanotubes.[44]
Figure 4.

a) Illustration of IKVAV (Ile-Lys-Val-Ala-Val)-containing peptide amphiphile molecules and their assembly into peptide nanotubes, b) Scanning electron microscopy (SEM) image of an IKVAV peptide nanotube network formed by adding cell media to the peptide monomer aqueous solution. c) Neuron progenitor cells encapsulated in the IKVAV peptide nanotube network for 7 days. A large degree of neurite outgrowth was typically observed [118]. (Reprinted with permission from [118]; copyright 2004, American Association for the Advancement of Science.)
Another type of linear peptide monomer with two amide head groups connected by a hydrocarbon tail group (called a bola-amphiphile) shows pH dependence in the assembled structures. For example, a peptide bola-amphiphile, bis(N-α-amido-glycylglycine)-l,7-heptane dicarboxylate, assembled into a nanotube at a pH of 5 via 3D hydrogen bonds between amide and carboxylic acid groups.[53,67,68] Functionalization of this type of peptide nanotube can be achieved by anchoring functional molecules onto the nanotube surfaces via non-covalent bonding.[49] An interesting characteristic of such peptide nanotubes is that their free amide groups, which are not involved in nanotube self-assembly, can capture and template biological molecules, such as DNA, synthetic peptides, proteins, porphyrins, and azobenzenes, via hydrogen bonding.[36,49,50,69] For example, this feature of bola-amphiphile peptide nanotubes allowed them to be functionalized with antibodies, which could smartly navigate the nanotubes to antigen-marked surfaces.[33] These nanotubes could also be functionalized with synthetic peptides, known to mineralize particular metals/semiconductors, and this functionalization made the peptide nanotubes smart templates for growing metals and semiconductors in controlled morphologies that can tune the electric and magnetic properties of the nanotubes.[17,70–73] This templating feature distinguishes bola-amphiphile peptide nanotubes from other peptide nanotubes, because the functionalization of the nanotube does not affect its structure and stability.
Recent studies of amyloid fibrils revealed that the stacking of aromatic amino acid residues may play a key role in amyloid fibril formation.[74] Based on this information, peptide nanotubes assembled from short aromatic peptide monomers have been reported, and well-ordered tubular structures with high directionality were observed due to the attractive forces of the aromatic moieties and the restricted geometry of the monomer conformation.[75] This peptide monomer was extended by adding amino acid residues to the core recognition motif of the diphenylalanine element, and the resulting peptide monomers could still be assembled to form nanotubes. A similar approach was also taken to assemble peptide nanotubes from the critical KLVFFAE region of the β-amyloid peptide.[55] This short synthetic peptide was first assembled to form a sheet-like structure and then folded into a helical ribbon form. The helical ribbon was further twisted to form the nanotube structure. It should be noted that this type of transformation, from sheet to nanotube, was also observed in bola-amphiphile-peptide nanotubes, aromatic-peptide nanotubes, and polymer nanotubes.[67,76,77]
Peptide nanotubes have been produced from various linear peptide monomers.[78] Whereas morphology changes of peptide nanotubes, via changes in peptide chain length, functional groups in the chain, and pH of the solution, have been studied systematically,[79–81] the assembly of structures into nanotubes cannot be predicted easily. For example, a change in pH will alter charge distributions and intermolecular hydrogen bonds between peptide monomers that determine the assembled structures, however, it will also influence the individual peptide monomer conformation via intramolecular charge interactions and/or intramolecular hydrogen bonds.[66] Since self-assembled nanotube dimensions are also sensitive to peptide monomer conformations, changes in the assembled structure as a function of pH cannot be solely predicted from interactions between the peptide monomers. For example, peptide nanotubes formed from peptide amphiphiles with ten hydrocarbons in the hydrophobic chain aligned in parallel in solution to form bundles of nanotubes, while peptide nanotubes from peptide amphiphiles with sixteen hydrocarbons in the hydrophobic chain were shorter and aggregated with a random orientation in solution.[66] Therefore, the mechanical, chemical, and physical properties of peptide nanotubes are very sensitive to the chemical structure of the peptide monomers. However, it may be advantageous that peptide nanotube structures are flexible with experimental conditions, and that the formation of peptide nanotubes with monodisperse sizes is very reproducible via molecular self-assembly under fixed nanotube growth conditions.[82] In fact, the sensitivity of changes in assembled structure to experimental conditions may turn out to be an advantage for the fabrication of exotic structures by controlling growth conditions. For example, peptide monomers, which self-assemble into microspheres at pH 8 and nanotubes at pH 7, can be assembled into a structure intermediate between the two (i.e., a dumbbell) by switching the pH between spherical growth and tubular growth conditions.[83] This is an example of how the sequential adjustment of experimental conditions during the self-assembly of peptide monomers alters the growth process, producing exotic, nanotube-based structures such as bridges, dumbbells, and multi-branched structures.
3. Peptide Nanotubes from Nonlinear Peptide Monomers
In 1974, De Santis and co-workers proposed that peptide nanotubes could self-assemble by stacking cyclic peptide monomers;[84] later, Ghadiri and co-workers demonstrated nanotube assembly from cyclic peptides.[85] A typical mechanism for the self-assembly of cyclic peptide monomers is shown in Figure 5a. Peptide rings are stacked through backbone–backbone hydrogen bonds between neighboring amide groups to form peptide nanotubes. In this example, the octa-peptide sequence of cyclo[−(l-Gln-d-Ala-l-Glu-l-Ala)2−] was necessary to prevent subunit association through columbic repulsion in basic aqueous solution, by alternating the stereochemistry of the amino acids. Generally, this type of peptide nanotube can be assembled from cyclic peptide monomers with alternating d,l-α-amino acids, β-amino acids, alternating α,β-amino acids, alternating α,γ-amino acids, δ-amino acids, and oligoureasd.[85–93] Recently, the heterocyclic alternation between α- and ε-amino acids in a cyclic peptide was also demonstrated to form an open-ended, hollow tubular structured.[52] New designs of peptide sequence protocols can be identified systematically by developing a cyclic-peptide library based on automated computer analyses of mass spectra.[94]
Figure 5.

a) Schematic representation of the formation of peptide nanotubes from cyclic peptide monomers [124]. b) Illustration of a single porous column layer assembled from dendritic peptides [95,98]. c) Illustration of the peptide nanotube assembled by stacking the layers in (b) [95,98]. d) Illustration of peptide nanotube assembly from leucine-zipper dendrimers, EZ (NH2-AQALEKELQALEKELQALEWELQALE-KELSGSGCCOOH) and KZ (NH2-AQALKKKLQALKKKLQALK-WKLQALKKKLSGSGCCOOH) [96]. (Reprinted with permission from [96,98,124]; copyrights 1994 and 2004, American Chemical Society.)
Recently, dendritic peptide monomers have also been reported to form peptide nanotubes. For example, dendritic peptides self-assemble to form cyclic structures by hydrogen bonding the narrow ends of the peptides and spreading out of the dendritic parts (Fig. 5b); these cyclic dendritic peptides stack to form peptide nanotubes via hydrogen bonding, as shown in Figure 5c.[95] In this structure, the diameter of the peptide nanotube could be changed from 1 to 24 Å by changing the amino acid composition. Another example of a dendritic peptide nanotube is one assembled from a dendritic peptide monomer consisting of a zero-generation poly(aminoamine) (PAMAM) core (circles in Fig. 5d) and pH-sensitive leucine zipper peptides, EZ (NH2-AQALEKELQALEKELQALE-WELQALE-KELSGSGC-COOH) and KZ (NH2-AQALKKKLQALKKKLQALK-WKLQALKKKLSGSGC-COOH), shown by ribbons in Figure 5d.[96] In solution, these peptides were zipped-up dendritic monomers, and the dendron cores linked them to extend to the nanotube form.
One of the characteristic features of cyclic peptide nanotubes is their precise diameter control, determined by chain length, side-chain size, peptide monomer bond angle, and stereochemistry of the amino acids. The proper number and sequence of the amino acids define the surface properties inside and outside the nanotubes. This feature enables cyclic peptide nanotubes to possess distinctive properties on the inner and outer surfaces. As pointed out in Section 1, hydrogen-bond-driven self-assemblies are very sensitive to external conditions, such as pH and solvent, because the protonation states of peptide monomers influence intermolecular hydrogen bonds in self-assembled peptide structures. Since stacking of peptide monomers by means of hydrogen bonds produces nanotubes in only one direction, rather than three, the optimization of experimental conditions to grow nanotubes from cyclic peptide monomers may be more straightforward than that for nanotubes from linear peptide monomers. While the amino acid sequences of the cyclic peptide monomers control the internal diameter of the peptide nanotubes, nanotubes derived from cyclic peptides are limited to a narrow range of sizes. To produce larger-diameter peptide nanotubes from cyclic peptide monomers, the ring diameters of the cyclic peptide monomers need to be larger. However, cyclic d,l-peptide monomers can only be sequenced between 9 and 13 Å in diameter to maintain a stable cyclic structure, and nanotubes need to be bundled to obtain larger nanotube diameters.[97] To achieve a wide range of peptide-nanotube sizes, it may be more advantageous to assemble them from linear peptide monomers. Another limitation of the monomer structure of cyclic peptides is that the amino acid stereochemistries must be alternated. However, it should be noted that dendritic peptide monomers can avoid this problem because the self-assembly of dendritic monomers can occur with any combination of amino acid residues.[98] For example, this feature allowed dendritic-peptide-based nanotubes to form a hydrophobic channel regardless of amino acid sequences, and these hydrophobic channels mediated proton transport in the same way as the pore formed by the natural protein, gramicidin.[95]
4. Metal/Semiconductor Nanotube Formation Using Peptide Nanotubes as Templates
Recently, a large number of reports about templating peptide nanotubes with metals and semiconductors have been published because metal/semiconductor nanotubes have been recognized as important building blocks for microelectronics.[99] Since peptides with particular sequences in nature are known to biomineralize specific types of metals and semiconductors,[100–105] peptide nanotubes with these sequences have the potential to be excellent templates for metal/semiconductor nanotubes. In nature, the tobacco mosaic virus (TMV) is a stable viron nanotube 18 nm in diameter and assembled from protein monomers.[103] The repeated polypeptide subunits in the TMV nanotube offer nucleation sites to produce highly crystalline semiconductor and metal coatings such as CdS, PbS, silica, iron oxides, and Cu.[103,106] The use of peptide nanotubes as templates also has the advantage of producing materials that cannot be created by synthetic methods. For example, Se nanotubes synthesized via a traditional solution-phase method possessed a trigonal structure, whereas the protein cytochrome, c3, extracted from the bacteria Desulfovibrio vulgaris grew monoclinic Se nanotubes.[107,108] It should be noted that biomineralization with peptides can generally be accomplished under much more moderate experimental conditions, such as room temperature and atmospheric pressure, as compared to synthetic methods, such as CVD.[109] For industrial application, these features are very important to fabricate exotic nanotubes, which are difficult and expensive to produce with existing technologies.
If peptide nanotubes incorporate certain mineralizing peptide motifs at proper locations on the nanotube surfaces, efficient coating of the nanotubes with metal/semiconductor materials with controlled morphologies should be obtained, just as is observed in nature. For example, peptide nanotubes assembled from synthetic peptide monomers with a diphenylalanine aromatic core could grow Ag nanotubes inside their cavities, and then the enzymatic degradation of the peptide nanotubes extracted the Ag nanotubes (Fig. 6a).[75] In Figure 6b, a Ag nanotube with a darker contrast in the TEM image is observed inside the peptide nanotube, while the removal of the peptide nanotube mold exposes the Ag nanotubes, as shown in Figures 7c,d.
Figure 6.

a) Schematic diagram of the process to cast silver nanotubes within peptide nanotubes. b) TEM image of a peptide nanotube filled with a silver nanotube, formed by the reduction of silver ions within the peptide tube. c,d) TEM images of silver nanotubes after the peptide nanotubes were removed by the addition of the proteinase K enzyme [75]. (Reprinted with permission from [75]; copyright 2003, American Association for the Advancement of Science.)
Figure 7.

a) TEM image (left) and size distribution (right) of Cu nanocrystals grown on a nanotube at pH 6. The inset shows the TEM image at higher magnification. b) TEM image (left) and size distribution (right) of Cu nanocrystals grown on a nanotube at pH 8. The inset shows the TEM image at higher magnification. c) Proposed structure of the Cu nanocrystal–HG12 peptide complex on the bionanotube. d) UV-vis spectra of bionanotubes coated with 10 nm diameter Cu nanocrystals, grown in a pH 6 solution (i), and those coated with 30 nm diameter Cu nanocrystals, grown in a pH 8 solution (ii) [71]. e) Cu2S nanocrystal growth on a bionanotube coated with the HG12 peptide. Scale bar = 100 nm. (Reprinted with permission from [71]; copyright 2003, National Academy of Sciences.)
While the syntheses of metal nanotubes in the peptide-nanotube templates are robust and useful, nanocrystal coatings on peptide nanotubes have the potential to control the physical properties of the resulting peptide nanotubes, because the electronic structure of the nanocrystal-coated peptide nanotubes is very sensitive to the size, shape, and density of the nanocrystals that form the coating. In other words, the conductivity, magnetism, and bandgap of coated peptide nanotubes may become tunable by controlling the nanocrystal coatings. To grow uniform and isotropic nanocrystals on the peptide-nanotube sidewalls, a new biological approach was recently developed by immobilizing sequenced “mineralizing peptides” on the nanotubes.[50] For example, peptide nanotubes self-assembled from small bolaamphiphile peptide monomers, bis(N-α-amido-glycylglycine)-1,7-heptane dicarboxylate, were shown to be an excellent template for this purpose.[67,79] These template nanotubes can immobilize the “mineralizing peptides” at free amide sites on their sidewalls via hydrogen bonding, by incubating the “mineralizing peptides” in the nanotube solution for one day.[50,70] After the reduction of ions on the nanotubes, highly crystalline nanocrystals were grown uniformly and with a high-density coverage on the nanotubes. The biological recognition of these peptides toward specific elements led to efficient coating of the nanotubes, and various sequences of “mineralizing peptides” were demonstrated to grow a variety of nanocrystals on the nanotubes.[17,50,70–72] In addition, the conformations and charge distributions of the “mineralizing peptides” were sensitive to the pH and ion concentrations of the growth solutions; changes of peptide conformations and charges could alter the size, packing density, and shape of the nanocrystals coating the peptide nanotubes. For example, when the peptide, Ala-His-His-Ala-His-His-Ala-Ala-Asp (HRE), was immobilized on the nanotubes and used to grow Au nanocrystals on the sidewalls, the packing density of the Au nanocrystals (average diameter of 6 nm) was controllable as a function of pH.[50] Growth solutions with a higher pH increased the packing density due to an increase in the number of nucleation sites on the peptides. When Cu nanocrystals were mineralized on nanotubes incorporating the peptide, His-Gly-Gly-Gly-His-Gly-His-Gly-Gly-Gly-His-Gly (HG12), the diameter of the Cu nanocrystals on the nanotubes could be varied between 10 and 30 nm by altering the conformation of HG12 by changing the pH (Figs. 7a,8b).[71] In the higher pH range, HG12 peptides were more aggregated on the nanotube, which generated more spaces to grow Cu nanocrystals and induced the growth of larger Cu nanocrystals (Fig. 7c). The size of the Cu nanocrystals on the nanotubes influenced the electronic structure of the peptide nanotubes, as shown in Figure 7d; this observation indicates that the bandgaps of the resulting nanotubes will be tunable by controlling the size of the nanocrystals grown on the nanotubes. The HG12 peptide was also successful in growing Cu2S semiconductor nanocrystals on nanotubes in a CuCl2/Na2S solution; the size of the Cu2S nanocrystals were controlled in the diameter range 14–40 nm (Fig. 7e). When the size of Ni nanocrystals on peptide nanotubes was controlled in the same manner, the magnetic properties of the resulting nanotubes were affected by the Ni nanocrystal size.[73] Shape control of Ag nanocrystals on nanotubes was demonstrated using the AG4 peptide, Asn-Pro-Ser-Ser-Leu-Phe-Arg-Tyr-Leu-Pro-Ser-Asp, as the “mineralizing peptide”.[17] This peptide sequence was found to recognize and affect the Ag nanocrystal growth kinetics on the (111) face via the combinatorial phage display peptide library.[45] When the AG4 peptide was sequenced and incorporated onto the nanotube surface, the biomineralization of Ag ions on the nanotubes led to an isotropic coating of hexagonal-shaped Ag nanocrystals, as the (111) face of Ag crystals is hexagonal. The TEM images of Figures 8a,b show the different sizes of hexagonal-shaped Ag nanocrystals grown on the nanotubes as a function of the growth period. The plasmon band of the hexagonal-shaped Ag nanocrystals on the nanotubes red-shifted as the size of the nanocrystals increased (Fig. 8c); the UV-vis spectra indicate the distinctive electronic structure of hexagonal-shaped Ag nanocrystals as compared to spherical Ag nanocrystals of a similar size,[15] which adds another dimension to the control of the band gap and conductivity of these nanotubes. It should be noted that nanocrystals could be coated only inside the nanotubes when the peptide and ion concentrations were optimized.[70]
Figure 8.

TEM images of hexagonal-shaped Ag nanocrystals on bionanotubes: a) after 14 h in the pH 7 growth solution (scale bar = 100 nm), the inset shows a higher-magnification TEM image (scale bar = 15 nm); b) after 72 h in the pH 7 growth solution (scale bar = 50 nm). c) UV-vis spectra of hexagonal-shaped Ag nanocrystals on bionanotubes; the nanocrystal diameters were 5 nm (top), 14 nm (middle), and 50 nm (bottom) [17]. (Reprinted with permission from [17]; copyright 2003, American Chemical Society.)
5. Applications of Peptide Nanotubes
While the self-assembling nature of peptide nanotubes offers interesting prospects, peptide nanotubes may not display ideal physical properties for particular applications. Thus, what makes peptide nanotubes special as compared to other nanotubes? We believe that their flexibility in functionality and their molecular-recognition properties distinguish them from other nanotubes. In nature, peptide monomers usually possess certain functions, and these functions are retained after their assembly into peptide nanotubes. For example, a positioning function can be added to nanotubes by the use of immobilizing polypeptides that can recognize and attach to the surface of their complementary ligand. Mineralizing functions can be added by incorporating peptides whose molecular recognition and biomineralization abilities allow them to selectively mineralize various metals and semiconductors; this behavior is routinely observed in living systems.[47,110] Here we summarize examples of peptide-nanotube applications where they use smart peptide functions.
5.1. Microelectronics
While various nanomaterials have been used as building blocks to construct nano-devices,[4,5,19–22,99] more reproducible and simpler methods to address them to precise positions are desirable. For example, improved methods are necessary to place nanotubes in electrical circuits in order to interconnect appropriate electronic components.
There are several techniques to align nanotubes by using external electric fields, magnetic fields, microfluidics, and by the Langmuir–Blodgett technique.[2,24–29] However, when multiple types of nanotubes are required to be aligned in more complex device configurations, these methods may not be sufficient. For example, consider fabricating a nanodevice whose configuration has two different types of nanotubes crossing perpendicular to each other (Fig. 9a). In this configuration, when one nanotube is metallic and the other nanotube is semiconducting, this nanodevice will function as a switch in memory circuits for reading and writing information.[111] Even though these device elements can be switched between well-defined on and off states by transiently charging the coated nanotubes to produce attractive and repulsive electrostatic forces, precisely aligning two or more types of nanotubes in desired directions is not an easy task.
Figure 9.

a) Illustration of proposed scheme to assemble multiple protein nanotubes into device configurations via biological recognition between the protein nanotubes and complementary protein-patterned surfaces (SAM = self-assembled monolayer), b) Illustration and SEM image of a streptavidin-functionalized nanotube immobilized onto a complementary biotin–SAM/Au substrate [32]. (Reprinted with permission from [32]; copyright 2001, American Chemical Society.)
When circuit elements and connecting wires are functionalized with biomolecular recognition of complementary connecting units, we can mimic biological systems, in which organic/in-organic nanoscale building blocks are routinely and precisely turned into complex structures for biological function with almost perfect reproducibility. In theory, antibody-coated peptide nanotubes, which can recognize and selectively bind to a well-defined region of a complementary antigen-patterned substrate, can be used as building blocks to assemble 3D nanoscale architectures by placing them at uniquely defined positions. For example, in Figure 9a, an antibody 1 nanotube can attach to antigen 1 areas, while an antibody 2 nanotube recognizes and immobilizes on antigen 2 surfaces. By this approach, the immobilization of antibody-coated peptide nanotubes onto antigen self-assembled monolayers (SAMs) will be programmed in aqueous solution without the need for complicated multistep fabrication procedures. To demonstrate the feasibility of this scheme, streptavidin-coated nanotubes were dispersed on biotin-incorporating SAMs (biotin–SAMs) in solution, and their selective immobilization onto the biotin–SAMs was observed after one day.[32] Figure 9b shows that the streptavidin-functionalized peptide nanotubes were positioned to interconnect patterned, complementary biotin–SAM/Au surfaces via biomolecular recognition.
Antibody-nanotube assembly on substrates can also be scaled up to form arrays. To scale up the assembly, an array of antigens was written on alkylthiol-SAM-coated Au substrates via nanografting, by using the tip of an atomic force microscoped,[112] and antigens were immobilized onto the shaved regions of the alkylthiol SAMs. Then, antibody nanotubes, produced by templating antibodies on peptide nanotubes, were selectively attached onto the antigen regions (Fig. 10a). Figure 10b shows atomic force microscopy (AFM) images of an example where peptide nanotubes (100 nm diameter) coated with anti-mouse immunoglobulin G (IgG) were selectively aligned and attached onto mouse-IgG-patterned areas.[33]
Figure 10.

a) Schematic diagram of antibody nanotube assembly on complementary antigen substrates via biological recognition: i) self-assembly of alkylthiol monolayers on Au substrates; ii) shaving trenches on the alkylthiol SAM using the tip of an atomic force microscope (nanoshaving); iii) deposition of antigens in the shaved trenches (nanografting); iv) location-specific immobilization of the antibody nanotube onto the complementary antigen regions via biological recognition, b) AFM images of anti-mouse-IgG-coated nanotubes immobilized on mouse-IgG-deposited regions (400 nm ×2 μm) (left) and the section analysis (right) along the white dotted line in the AFM image. Scale bars: 1 μm in the main AFM image and 300 nm in the inset [33]. (Reprinted with permission from [33]; copyright 2004, American Chemical Society.)
While the more complex configurations of logic gates, such as OR, AND, and NOR, have been fabricated by using microfluidics,[2,113] simple and reproducible fabrication techniques for nanotube alignment may be practical for larger scale industrial production. Peptide nanotubes may be alternative building blocks for microelectronics because functionalization with antibodies allows biomolecular-recognition-driven nanotube alignment, and the metal/semiconductor coatings on peptide nanotubes are quite controllable, as described in the previous section. There are various ways to accomplish multifunctionalization of peptide nanotubes. For example, recognition antibodies can be attached to the ends of the nanotubes while mineralizing peptides are coated on their sidewalls.[51] Metal/semiconductor materials can also be placed inside the nanotubes while recognition antibodies are attached to the outside of the nanotubes.[70]
It should be noted that recent computational studies show that some peptide nanotubes may not be insulating even without any metallic coating. For example, the highest occupied molecular orbital (HOMO) to the lowest unoccupied molecular orbital (LUMO) gap of the cyclic d,l-octapeptide nanotube was computed to be approximately 5 eV,[114] and these nanotubes may be conductive due to delocalization of electrons and holes through hydrogen bonds between the peptide rings.[115] As theory suggests that the electronic band-edge states are very sensitive to the strength of intermolecular hydrogen bonds along the nanotube axis.[116] optimized structures of peptide nanotubes may possess adequate electronic transport properties to build microelectronic devices.
5.2. Bioengineering
In nature, biomineralization of self-assembled fibrils routinely produces various composite materials. For example, collagen fibrils are formed in bone tissues by self-assembled collagen triple helices, and hydroxyapatite crystals grow in these fibrils with the hierarchical organization of their c axes along the long axes of the fibrils.[117] Peptide nanotubes were shown to mimic the collagen fibril by growing inorganic hydroxyapatite crystals.[65] In this peptide monomer, the phosphoserine residue was incorporated into the peptide sequence to nucleate and grow calcium phosphate crystals along the c axis of the scaffold, while the amino acid sequence (Arg-Gly-Asp) was also in the head group of the peptide to mediate cell adhesion. Cystein amino acids were in the monomer sequence to stabilize the nanotubes by forming disulfide bonds with adjacent monomers. Recently, when the peptide epitope IKVAV, known to promote neurite sprouting and to direct neurite growth, was incorporated into the head group of a peptide amphiphile monomer, the modified monomers self-assembled into a 3D network of nanotubes in vitro (Fig. 4b).[118] This peptide-nanotube scaffold with IKVAV could induce very rapid differentiation of cells into neurons, while discouraging the development of astrocytes. Figure 4c shows the neural progenitor cells, encapsulated in the peptide nanotube network, that differentiated into neurons after 1–7 days. This system will be widely applicable in tissue engineering because peptide nanotubes can be assembled in vivo by simple injection of peptide monomer solutions into tissues. Another advantage is that peptide nanotubes can be customized to incorporate peptide sequences for specific cell responses. The ability of peptide nanotubes to target specific cells also makes them attractive for drug-delivery applications.
As discussed in the previous section, inner surfaces of peptide nanotubes can be designed to have distinct properties from the outer surfaces. For example, this feature allows one to fabricate peptide nanotubes with hydrophobic outer surfaces and hydrophilic inner surfaces. This type of nanotube can be inserted into cell membranes and can mimic the ion channels in membranes.[119,120] When these peptide nanotubes, which incorporated molecules with biological recognition in the capping groups, were inserted into membranes, the resultant membranes were able to function as biosensors.[97] In this modified membrane, the conductivity change due to the binding between capped antibodies and antigens was used as a probe to determine the antigen concentration.[121,122] When cyclic d,l-α-peptide nanotubes were designed to target bacterial membranes, their insertion in the bacterial membranes also led to antibacterial activity by increasing the membrane permeability.[123] It should be noted that combinatorial split-and-pool syntheses of cyclic peptides were recently used for rapid identification of cyclic-peptide libraries based on automated computer analyses of mass spectra for antimicrobial screening.[94]
6. Conclusions
The field of device fabrication based on peptide nanotubes is still very young. At this stage, research efforts to develop peptide nanotubes as exotic nanomaterials have been more active; however, steady progress is being made into organizing peptide nanotubes in certain device configurations. The bio-compatibility of peptide nanotubes also makes them valuable for applications as biosensors and biomolecular filters. The physical properties of peptide nanotubes may not be ideal for certain applications, however, the functionalization of peptide nanotubes may enable them to acquire the desired physical properties. As explained in Section 5, functionalization can be achieved via peptide-monomer modification or the non-covalent attachment of functional molecules to the peptide nanotubes. It should also be noted that the development of improved analytical tools for nanoscale materials is essential in this field. Nanomaterial synthesis and nanodevice fabrication attract significant attention and publicity, however, the reliable and reproducible measurement of properties, such as conductivity, magnetism, mechanical strength, band gap, and chemical structure, is crucial for developing peptide-nanotube-based assemblies into realistic workable devices.
Biographies

Hiroshi Matsui is an Associate Professor in the Department of Chemistry at the City University of New York (CUNY), the Graduate Center and Hunter College. After completion of his B.S. degree in Chemistry at Sophia University (Japan), he received his M.S. degree in Materials Science and Engineering from Stanford University in 1992. He obtained his Ph.D. degree in Chemistry from Purdue University in 1996. He was a postdoctoral associate in the Department of Chemistry at Columbia University from 1996 to 1998. He came to CUNY from the University of Central Florida in 2001.

Xueyun Gao was born in Shandong (China) in 1970. He received his B. S. degree in Chemistry from Zhengzhou Institute of Light Industry and his Ph.D. degree in Condensed Matter Physics from the Institute of Solid State Physics at the Chinese Academy of Sciences, under the supervision of Prof. Lide Zhang. He joined Professor Hiroshi Matsui’s group as a research associate in 2004. His research interests are nanostructured materials and their properties.
Footnotes
This work was supported by the U.S. Department of Energy (DE-FG-02-01ER45935), the National Science Foundation CARRER Award (ECS-0103430), and partially supported by a NIGMS MBRS SCORE grant (2-S06-GM60654).
References
- [1].Cui Y, Wei QQ, Park HK, Lieber CM, Science 2001, 293, 1289. [DOI] [PubMed] [Google Scholar]
- [2].Huang Y, Duan X, Cui Y, Lauhon LJ, Kim KH, Lieber CM, Science 2001, 294, 1313. [DOI] [PubMed] [Google Scholar]
- [3].Diehl MR, Yaliraki SN, Beckman RA, Barahona M, Heath JR, Angew. Chem. Int. Ed 2001, 41, 353. [DOI] [PubMed] [Google Scholar]
- [4].Collins PG, Arnold MS, Avouris P, Science 2001, 292, 706. [DOI] [PubMed] [Google Scholar]
- [5].Bachtold A, Hadley P, Nakanishi T, Dekker C, Science 2001, 294, 1317. [DOI] [PubMed] [Google Scholar]
- [6].Murray CB, Kagan CR, Bawendi MG, Annu. Rev. Mater. Sci 2000, 30, 545. [Google Scholar]
- [7].Alivisatos AP, Science 1996, 271, 933. [Google Scholar]
- [8].Hamad-Schifferli K Schwartz JJ, Santos AT, Zhang S, Jacobson JM, Nature 2002, 415, 152. [DOI] [PubMed] [Google Scholar]
- [9].Johnson JC, Choi HJ, Knutsen KP, Schaller RD, Yang PD, Saykally RJ, Nat. Mater 2002, 1, 106. [DOI] [PubMed] [Google Scholar]
- [10].Wang ZL, Adv. Mater 2003, 15, 432. [Google Scholar]
- [11].Sun Y, Xia Y, Science 2002, 298, 2176. [DOI] [PubMed] [Google Scholar]
- [12].Gou LF, Murphy CJ, Nano Lett 2003, 3, 231. [Google Scholar]
- [13].Pinna N, Weiss K, Urban J, Pileni MP, Adv. Mater 2001, 13, 261. [Google Scholar]
- [14].Ahmadi TS, Wang ZL, Green TC, Henglein A, El-Sayed MA, Science 1996, 272, 1924. [DOI] [PubMed] [Google Scholar]
- [15].Jin RC, Cao YW, Mirkin CA, Kelly KL, Schatz GC, Zheng JG, Science 2001, 294, 1901. [DOI] [PubMed] [Google Scholar]
- [16].Puntes VF, Scher EC, Alivisatos AP, J. Am. Chem. Soc 2000, 122, 12700. [Google Scholar]
- [17].Yu L, Banerjee IA, Matsui H, J. Am. Chem. Soc 2003, 125, 14837. [DOI] [PubMed] [Google Scholar]
- [18].Zhong ZH, Wang DL, Cui Y, Bockrath MW, Lieber CM, Science 2003, 302, 1377. [DOI] [PubMed] [Google Scholar]
- [19].Chen RJ, Bangsaruntip S, Drouvalakis KA, Kam NWS, Shim M, Li YM, Kim W, Utz PJ, Dai HJ, Proc. Natl. Acad. Sci USA 2003, 100, 4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fennimore AM, Yuzvinsky TD, Han WQ, Fuhrer MS, Cumings J, Zettl A, Nature 2003, 424, 408. [DOI] [PubMed] [Google Scholar]
- [21].Modi A, Koratkar N, Lass E, Wei BQ, Ajayan PM, Nature 2003, 424, 171. [DOI] [PubMed] [Google Scholar]
- [22].Melosh NA, Boukai A, Diana F, Gerardot B, Badolato A, Petroff PM, Heath JR, Science 2003, 300, 112. [DOI] [PubMed] [Google Scholar]
- [23].Rao SG, Huang L, Setyawan W, Hong SH, Nature 2003, 425, 36. [DOI] [PubMed] [Google Scholar]
- [24].Jin S, Whang DM, McAlpine MC, Friedman RS, Wu Y, Lieber CM, Nano Lett 2004, 4, 915. [Google Scholar]
- [25].Long DP, Lazorcik JL, Shashidhar R, Adv. Mater 2004, 16, 814. [Google Scholar]
- [26].Smith PA, C. D. Nordquist, T. N. Jackson, T. S. Mayer, B. R. Martin, J. Mbindyo, T. E. Mallouk, Appl. Phys. Lett 2000, 77, 1399. [Google Scholar]
- [27].Chung JY, Lee KH, Lee JH, Ruoff RS, Langmuir 2004, 20, 3011. [DOI] [PubMed] [Google Scholar]
- [28].Lay MD, Novak JP, Snow ES, Nano Lett 2004, 4, 603. [Google Scholar]
- [29].Huang L, Wind SJ, O’Brien SP, Nano Lett 2003, 3, 299. [Google Scholar]
- [30].Greig LM, Philp D, Chem. Soc. Rev 2001, 30, 287. [Google Scholar]
- [31].Niemeyer CM, Angew. Chem. Int. Ed 2001, 40, 4128. [DOI] [PubMed] [Google Scholar]
- [32].Matsui H, Porrata P, Douberly GEJ, Nano Lett 2001, 1, 461. [Google Scholar]
- [33].Nuraje N, Banerjee IA, MacCuspie RI, Yu L, Matsui H, J. Am. Chem. Soc 2004, 126, 8088. [DOI] [PubMed] [Google Scholar]
- [34].Mbindyo JKN, Reiss BD, Martin ER, Keating CD, Natan MJ, Mallouk TE, Adv. Mater 2001, 13, 249. [Google Scholar]
- [35].Braun E, Eichen Y, Sivan U, Ben-Yoseph G, Nature 1998, 391, 775. [DOI] [PubMed] [Google Scholar]
- [36].Banerjee IA, Yu L, Matsui H, J. Am. Chem. Soc 2003, 125, 9542. [DOI] [PubMed] [Google Scholar]
- [37].Gyorvary E, Schroedter A, Talapin DV, Weller H, Pum D, Sleytr UB, J. Nanosci. Nanotechnol 2004, 4, 115. [DOI] [PubMed] [Google Scholar]
- [38].Kaplan DL, Nat. Biotech 2002, 20, 239. [DOI] [PubMed] [Google Scholar]
- [39].Harper JD, Lansbury PT, Annu. Rev. Biochem 1997, 66, 385. [DOI] [PubMed] [Google Scholar]
- [40].Sipe JD, Cohen AS, J. Struct. Biol 2000, 130, 88. [DOI] [PubMed] [Google Scholar]
- [41].Borman S, Chem. Eng. News 2004, 82 (31), 9. [Google Scholar]
- [42].Gazit E, Drugs Future 2004, 29, 1. [Google Scholar]
- [43].Legname G, Baskakov IV, Nguyen HOB, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB, Science 2004, 305, 673. [DOI] [PubMed] [Google Scholar]
- [44].Vauthey S, Santoso S, Gong HY, Watson N, Zhang SG, Proc. Natl. Acad. Sci. USA 2002, 99, 5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Naik RR, Stringer SJ, Agarwal G, Jones SE, Stone MO, Nat. Mater 2002, 1, 169. [DOI] [PubMed] [Google Scholar]
- [46].Sarikaya M, Tamerler C, Jen AKY, Schulten K, Nat. Mater 2003, 2, 577. [DOI] [PubMed] [Google Scholar]
- [47].Whitling JM, Spreitzer G, Wright DW, Adv. Mater 2000, 12, 1377. [Google Scholar]
- [48].Wang WX, Hecht MH, Proc. Natl. Acad. Sci. USA 2002, 99, 2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Douberly GJ, Pan S, Walters D, Matsui H, J. Phys. Chem. B 2001, 105, 7612. [Google Scholar]
- [50].Djalali R, Chen Y-F, Matsui H, J. Am. Chem. Soc 2002, 124, 13 660. [DOI] [PubMed] [Google Scholar]
- [51].Banerjee IA, Matsui H, Nano Lett 2003, 3, 283. [Google Scholar]
- [52].Horne WS, Stout CD, Ghadiri MR, J. Am. Chem. Soc 2003, 125, 9372. [DOI] [PubMed] [Google Scholar]
- [53].Kogiso M, Masuda M, Shimizu T, Supramol. Chem 1998, 9, 183. [Google Scholar]
- [54].Scheibel T, Bloom J, Lindquist SL, Proc. Natl. Acad. Sci. USA 2004, 101, 2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lu K, Jacob J, Thiyagarajan P, Conticello VP, Lynn DG, J. Am. Chem. Soc 2003, 125, 6391. [DOI] [PubMed] [Google Scholar]
- [56].Ryadnov MG, Woolfson DN, Nat. Mater 2003, 2, 329. [DOI] [PubMed] [Google Scholar]
- [57].Ryadnov MG, Woolfson DN, Angew. Chem. Int. Ed 2003, 42, 3021. [DOI] [PubMed] [Google Scholar]
- [58].Tsurkan MV, Ogawa MY, Chem. Commun 2004, 2. [DOI] [PubMed] [Google Scholar]
- [59].Fuhrhop J-H, Koning J, Membranes and Molecular Assemblies: The Synkinetic Approach, Royal Society of Chemistry, Cambridge: 1994. [Google Scholar]
- [60].Nakashima N, Asakuma S, Kunitake T, J. Am. Chem. Soc 1985, 107, 509. [Google Scholar]
- [61].Fuhrhop J-H, Helfrich W, Chem. Rev 1993, 93, 1565. [Google Scholar]
- [62].Shimizu T, Hato M, Biochim. Biophys. Acta 1993, 1147, 50. [DOI] [PubMed] [Google Scholar]
- [63].Matsuzawa Y, Kogiso M, Matsumoto M, Shimizu T, Shimada K, Itakura M, Kinugasa S, Adv. Mater 2003, 15, 1417. [Google Scholar]
- [64].Privalov PL, Gills SJ, Pure Appl. Chem 1989, 61, 1097. [Google Scholar]
- [65].Hartgerink JD, Beniash E, Stupp SI, Science 2001, 294, 1684. [DOI] [PubMed] [Google Scholar]
- [66].Hartgerink JD, Beniash E, Stupp SI, Proc. Natl. Acad. Sci. USA 2001, 99, 5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Matsui H, Gologan B, J. Phys. Chem. B 2000, 104, 3383. [Google Scholar]
- [68].Shimizu T, Kogiso M, Masuda M, J. Am. Chem. Soc 1997, 119, 6209. [Google Scholar]
- [69].Matsui H, MacCuspie R, Nano Lett 2001, 1, 671. [Google Scholar]
- [70].Djalali R, Chen Y-F, Matsui H, J. Am. Chem. Soc 2003, 125, 5873. [DOI] [PubMed] [Google Scholar]
- [71].Banerjee IA, Yu L, Matsui H, Proc. Natl. Acad. Sci. USA 2003, 100, 14678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Yu L, Banerjee IA, Matsui H, J. Mater. Chem 2004, 14, 739. [Google Scholar]
- [73].Yu L, Banerjee IA, Shima M, Rajan K, Matsui H, Adv. Mater 2004, 16, 709. [Google Scholar]
- [74].Gazit E, FASEB J 2002, 16, 77. [DOI] [PubMed] [Google Scholar]
- [75].Reches M, Gazit E, Science 2003, 300, 625. [DOI] [PubMed] [Google Scholar]
- [76].Reches M, Gazit E, Nano Lett 2004, 4, 581. [DOI] [PubMed] [Google Scholar]
- [77].Schlaad H, Krasia T, Antonietti M, J. Am. Chem. Soc 2004, 126, 11 307. [DOI] [PubMed] [Google Scholar]
- [78].Vauthey S, Santoso S, Gong H, Watson N, Zhang S, Proc. Natl. Acad. Sci. USA 2002, 99, 5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kogiso M, Ohnishi S, Yase K, Masuda M, Shimizu T, Langmuir 1998, 14, 4978. [Google Scholar]
- [80].Kogiso M, Okada Y, Hanada T, Yase K, Shimizu T, Biochim. Biophys. Acta 2000, 1475, 346. [DOI] [PubMed] [Google Scholar]
- [81].Shimizu T, Iwaura R, Masuda M, Hanada T, Yase K, J. Am. Chem. Soc 2001, 123, 5947. [DOI] [PubMed] [Google Scholar]
- [82].Whitesides GM, Sci. Am 1995, 273 (3), 146. [Google Scholar]
- [83].Matsui H, Holtman C, Nano Lett 2002, 2, 887. [Google Scholar]
- [84].De Santis P, Morosetti S, Rizzo R, Macromolecules 1974, 7, 52.4837982 [Google Scholar]
- [85].Ghadiri MR, Granja JR, Milligan RA, McRee DE, Khazanovich N, Nature 1993, 366, 324. [DOI] [PubMed] [Google Scholar]
- [86].Hartgerink JD, Granja JR, Milligan RA, Ghadiri MR, J. Am. Chem. Soc 1996, 118, 43 [Google Scholar]
- [87].Seebach LD, Matthews JL, Meden A, Wessels T, Naerlocher C, McCusker B, Helv. Chim. Acta 1997, 80, 173. [Google Scholar]
- [88].Clark TD, Buriak JM, Kobayashi K, Isler MP, McRee DE, Ghadiri M,R, J. Am. Chem. Soc 1998, 120, 8949. [Google Scholar]
- [89].Karle IL, Handa BK, Hassall CH, Acta Crystallogr., Sect. B 1975, 31, 555. [Google Scholar]
- [90].Amorin M, Castedo L, Granja JR, J. Am. Chem. Soc 2003, 125, 2844. [DOI] [PubMed] [Google Scholar]
- [91].Gauthier D, Baillargeon P, Drouin M, Dory YL, Angew. Chem. Int. Ed 2001, 40, 4635. [DOI] [PubMed] [Google Scholar]
- [92].Semetey V, Didierjean C, Briand JP, Aubry A, Guichard G, Angew. Chem. Int. Ed 2002, 41, 1895. [DOI] [PubMed] [Google Scholar]
- [93].Ranganathan D, Lakshmi C, Karle IL, J. Am. Chem. Soc 1999, 121, 6103. [Google Scholar]
- [94].Redman JE, Wilcoxen KM, Ghadiri MR, J. Comb. Chem 2002, 5, 33. [DOI] [PubMed] [Google Scholar]
- [95].Percec V, Dulcey AE, Balagurusamy VSK, Miura Y, Smidrkal J, Peterca M, Nummelin S, Edlund U, Hudson SD, Heiney PA, Hu DA, Magonov SN, Vinogradov SA, Nature 2004, 430, 764. [DOI] [PubMed] [Google Scholar]
- [96].Zhou M, Bentley D, Ghosh I, J. Am. Chem. Soc 2004, 126, 734. [DOI] [PubMed] [Google Scholar]
- [97].Bong DT, Clark TD, Granja JR, Ghadiri MR, Angew. Chem. Int. Ed 2001, 40, 988. [PubMed] [Google Scholar]
- [98].Rouhi M, Chem. Eng. News 2004, 82 (33), 4. [Google Scholar]
- [99].Lieber CM, MRS Bull 2003, 28, 486. [Google Scholar]
- [100].Field M, Smith CJ, Awshalom DD, Mayes EL, Davis SA, Mann S, Appl. Phys. Lett 1998, 73, 1739. [Google Scholar]
- [101].Behrens S, Rahn K, Habicht W, Bohm K-J, Rosner H, Dinjus E, Unger E, Adv. Mater 2002, 14, 1621. [Google Scholar]
- [102].Seeman NC, Belcher AM, Proc. Natl. Acad. Sci. USA 2002, 99, 6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Shenton W, Douglas T, Young M, Stubbs G, Mann S, Adv. Mater 1999, 11, 253. [Google Scholar]
- [104].Douglas T, Young M, Adv. Mater 1999, 11, 679. [Google Scholar]
- [105].Wang Q, Kaltgrad E, Lin TW, Johnson JE, Finn MG, Chem. Biol 2002, 9, 805. [DOI] [PubMed] [Google Scholar]
- [106].Demir M, Stowell MHB, Nanotechnology 2002, 13, 541. [Google Scholar]
- [107].Gates B, Mayers B, Cattle B, Xia YN, Adv. Funct. Mater 2002, 12, 219. [Google Scholar]
- [108].Abdelouas A, Gong WL, Lutze W, Shelnutt JA, Franco R, Moura I, Chem. Mater 2000, 12, 1510. [Google Scholar]
- [109].Sumerel JL, Yang WJ, Kisailus D, Weaver JC, Choi JH, Morse DE, Chem. Mater 2003, 15, 4804. [Google Scholar]
- [110].Slocik JM, Moore JT, Wright DW, Nano Lett 2002, 2, 169. [Google Scholar]
- [111].Rueckes T, Kim K, Joselevich E, Tseng GY, Cheung C-L, Lieber CM, Science 2000, 289, 94. [DOI] [PubMed] [Google Scholar]
- [112].Liu G-Y, Xu S, Qian Y, Acc. Chem. Res 2000, 33, 457. [DOI] [PubMed] [Google Scholar]
- [113].Javey A, Wang Q, Ural A, Li YM, Dai HJ, Nano Lett 2002, 2, 929. [Google Scholar]
- [114].Lewis JP, Pawley NH, Snakey OF, J. Phys. Chem. B 1997, 101, 10576. [Google Scholar]
- [115].Fukusaki K, Takeda K, Shiraishi K, J. Phys. Soc. Jpn 1997, 66, 3387. [Google Scholar]
- [116].Okamoto H, Kasahara M, Takeda K, Shiraishi K, Pept. Sci 1999, 36, 67. [Google Scholar]
- [117].Traub W, Weiner HD, Proc. Natl. Acad. Sci. USA 1989, 86, 9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI, Science 2004, 303, 1352. [DOI] [PubMed] [Google Scholar]
- [119].Ghadiri MR, Adv. Mater 1995, 7, 675. [Google Scholar]
- [120].Engels M, Bashford D, Ghadiri MR, J. Am. Chem. Soc 1995, 117, 9151. [Google Scholar]
- [121].Braha O, Walker B, Cheley S, Kasianowicz JJ, Song L, Gouaux JE, Bayley H, Chem. Biol 1997, 4, 497. [DOI] [PubMed] [Google Scholar]
- [122].Gu L-Q, Braha O, Conlan S, Cheley S, Bayley H, Nature 1999, 398, 686. [DOI] [PubMed] [Google Scholar]
- [123].Fernandez-Lopez S, Kim HS, Choi EC, Delgado M, Granja JR, Khasanov A, Kraehenbuehl K, Long G, Weinberger DA,Wilcoxen K, Ghadiri MR, Nature 2001, 412, 452. [DOI] [PubMed] [Google Scholar]
- [124].Khazanovich N, Granja JR, McRee DE, Milligan RA, Ghadiri MR, J. Am. Chem. Soc 1994, 116, 6011. [Google Scholar]


