Genome-wide association studies (GWASs) have identified numerous variants associated with cardiovascular traits and diseases, but progress in heart failure has been limited. Since many GWAS variants appear to affect cis gene expression, we asked whether there was overlap between variants associated with cardiac structure or function and expression quantitative trait locus (eQTL) variants in the human left ventricle (LV). Our human studies were approved by institutional review boards, and all participants provided written informed consent. Animal studies and procedures were performed in accordance with institutional guidelines.
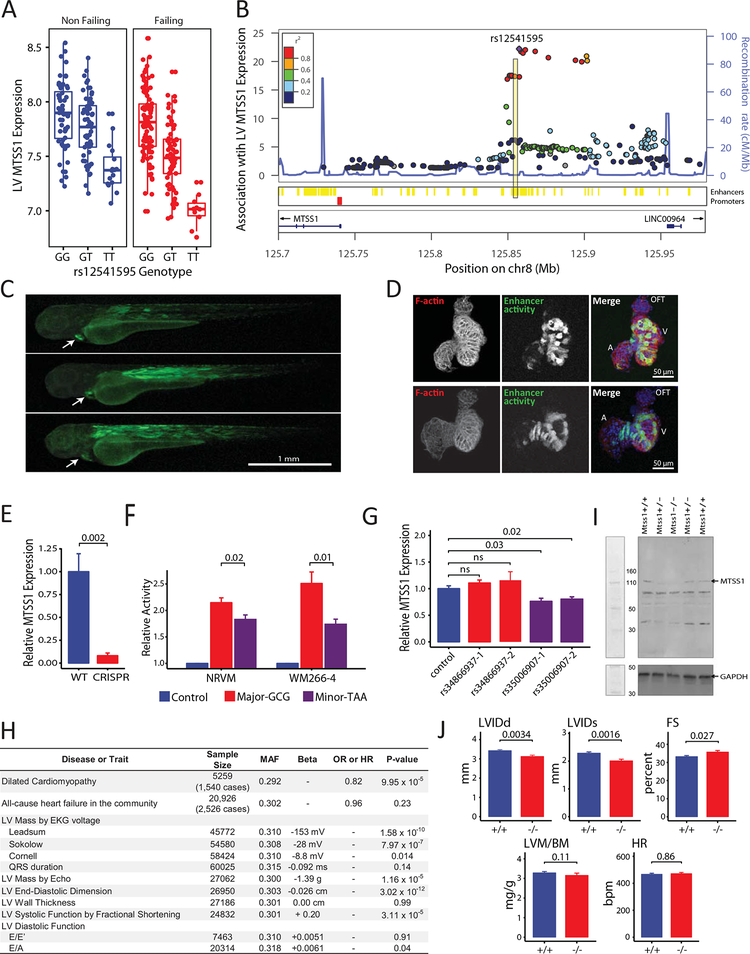
A lead variant, rs12541595, in the MTSS1 locus stood out for its associations with LV end-diastolic dimension (LVEDD, P=2×10–13) in EchoGen1 and MTSS1 expression (P=6×10–23) in 313 LV samples from individuals of European descent in the Myocardial Applied Genomics Network (MAGNet) consortium1 (Figure [A]). We also compared the association between rs12541595 and MTSS1 expression across multiple human tissues in the Genotype-Tissue Expression project,2 and the association was strongest in the left ventricle (P=1×10–28, n=272). Across these studies, the rs12541595 minor allele (T) is associated with both reduced LVEDD and reduced MTSS1 expression.
Figure 1. Interrogation of the MTSS1 locus.
A, association of minor allele (T) at rs12541595 with MTSS1 expression in human LV (P=6×10–23 across all samples using an additive genetic model adjusting for presence/absence of heart failure; n=313). B, cis associations with LV MTSS1 expression (–log10 P-values using same model as in A; n=313). Variants within a putative enhancer (yellow) in linkage disequilibrium with rs12541595 shows strong associations. C, the enhancer region was cloned upstream of a green fluorescent protein reporter and transfected into zebrafish embryos to determine anatomical site of activity (green). Activity is specific to cardiac-muscle (white arrow) and skeletal-muscle (dorsal axis) 72 hour post fertilization. D, analysis of cardiac-restricted enhancer activity within excised zebrafish hearts demonstrates strong activity (green) in the ventricle (V) and limited activity in the atrium (A) and outflow tract (OFT). Hearts are counterstained for sarcomeric architecture (F-actin, red) and nuclei (blue). E, CRISPR-Cas9 enhancer deletion abolishes MTSS1 expression in WM266–4 cells (n=6 per group; Mann-Whitney U test P-value). F, Relative activity of major and minor enhancer haplotypes defined by rs12541595, rs35006907, and rs34866937. The minor enhancer haplotype, marked by the rs12541595 minor allele, shows reduced activity (n=6 per group; Mann-Whitney U test P-values). G, CRISPR interference targeting enhancer variants shows reduced activity with dCas9 positioned at rs35006907 using two different guide RNAs (n=3 per group; Mann-Whitney U test P-values). H, association of the MTSS1 rs12541595 minor allele (T) with cardiac traits in human populations (P-values by additive genetic models). I, immunoblot of murine myocardial protein extracts verifying successful knock-out of Mtss1. J, LV structure and function in Mtss1+/+ (n= 23; 8 female, 15 male) and Mtss1–/– (n=16; 10 female, 6 male) mice by echocardiography under isoflurane anesthesia (gender adjusted P-values). LVIDd, LV internal dimension in diastole; LVIDs, LV internal dimension in systole; FS, fractional shortening; LVM/BM, LV mass normalized to body mass; HR, heart rate.
To validate the cardiac-specific MTSS1 eQTL, we interrogated the genomic locus in detail in MAGNet (Figure [B]). The lead variant rs12541595 marks a cluster of variants that strongly influence LV MTSS1 expression and that reside in a predicted enhancer. We introduced a fluorescent reporter construct harboring the 1-kb predicted enhancer region into zebrafish embryos and verified strong cardiac- and myocyte-specific enhancer activity, predominating in the ventricle (Figure [C, D]). Deletion of the 1-kb region with CRISPR-Cas9 in WM266–4 melanoma cells, which are known to express MTSS1, reduced endogenous MTSS1 expression (Figure [E]). Resequencing of the enhancer in 145 MAGNet participants and examination of 1000Genomes data revealed 2 common enhancer variants (rs34866937 and rs35006907) that are in strong linkage disequilibrium (r2 > 0.97) with the lead variant rs12541595 in Europeans. Together, the rs12541595, rs35006907, and rs34866937 variants define a major haplotype (GCG, frequency 0.66) and a minor haplotype (TAA, frequency 0.33). We compared activity of both enhancer haplotypes using luciferase reporter constructs transfected into WM266–4 cells and neonatal rat ventricular myocytes. In both cell types, we observed decreased luciferase expression with the minor haplotype (Figure [F]), which is marked by the rs12541595 minor allele. Finally, we used CRISPR interference to assess for enhancer activity near rs35006907 or rs34866937; in WM266–4 cells and HEK 293T cells, both homozygous for the major haplotype, positioning of dCas9 at rs35006907 but not rs34866937 reduced MTSS1 expression (Figure [G]).
Given the relationship of the MTSS1 locus to cardiac structure, we assessed whether the lead variant rs12541595 was associated with all-cause heart failure or dilated cardiomyopathy, or with LV traits assessed by echocardiography and electrocardiography (Figure [H]).1, 3, 4 With a collection of 1,540 cases of dilated cardiomyopathy, we observed an 18% risk reduction per copy of the minor allele (P=1×10–4). Also for the minor allele, we observed a significant increase in LV systolic function by fractional shortening (P=3×10–5) and decrease in LV dimension and mass by several measures.
Integrating these various observations, we hypothesized that reduced expression of MTSS1 in the mouse heart would result in favorable echocardiographic changes. We generated a Mtss1 knockout mouse with CRISPR-Cas9 (Figure [I]); the mice were viable and fertile. Focusing specifically on the heart, we assessed wild-type and knockout littermates at 8 weeks of age with echocardiography (Figure [J]). In the knockout mice, we observed significant decreases in LV end-diastolic dimension (P=0.0034) and LV end-systolic dimension (P=0.0016) as well as trends towards increased LV fractional shortening (P=0.027)—highly concordant with the human genetic findings.
MTSS1 is a member of the Inverse Bin-Amphiphysin-Rvs (I-BAR) protein family and interacts with the actin cytoskeleton and the cell membrane to regulate cell structure and intercellular junctions. MTSS1 is expressed in a variety of organs besides heart, and disruption of MTSS1 can lead to diverse effects. In kidney epithelium, MTSS1 co-localizes with F-actin and E-cadherin at adherens junctions and is induced in response to shear stress via MEF2, suggesting a role in stress-responsive tissue remodeling.5 We speculate that MTSS1 may play a similar role in cardiac myocytes and cardiac remodeling. Given our evidence that enhancer variants that reduce cardiac MTSS1 are cardioprotective, we suggest that therapeutic reduction of MTSS1 expression limited to the heart could represent a novel approach to address the burden of human heart failure.
Sources of Funding:
This work was supported by grants for the US National Institutes of Health (HL105993, HL088577, HL105993, HL092577, and HL128914), the Fondation Leducq (14CVD01), and the Winkelman Family Innovation Fund.
Disclosures: Dr. Ellinor receives grant support from Bayer AG to the Broad Institute for research focused on the development of therapeutics for cardiovascular disease.
Footnotes
Data Sharing
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
- 1.Wild PS, Felix JF, Schillert A, Teumer A, Chen MH, Leening MJG, Volker U, Grossmann V, Brody JA, Irvin MR, Shah SJ, Pramana S, Lieb W, Schmidt R, Stanton AV, Malzahn D, Smith AV, Sundstrom J, Minelli C, Ruggiero D, Lyytikainen LP, Tiller D, Smith JG, Monnereau C, Di Tullio MR, Musani SK, Morrison AC, Pers TH, Morley M, Kleber ME, Aragam J, Benjamin EJ, Bis JC, Bisping E, Broeckel U, Cheng S, Deckers JW, Del Greco MF, Edelmann F, Fornage M, Franke L, Friedrich N, Harris TB, Hofer E, Hofman A, Huang J, Hughes AD, Kahonen M, Investigators K, Kruppa J, Lackner KJ, Lannfelt L, Laskowski R, Launer LJ, Leosdottir M, Lin H, Lindgren CM, Loley C, MacRae CA, Mascalzoni D, Mayet J, Medenwald D, Morris AP, Muller C, Muller-Nurasyid M, Nappo S, Nilsson PM, Nuding S, Nutile T, Peters A, Pfeufer A, Pietzner D, Pramstaller PP, Raitakari OT, Rice KM, Rivadeneira F, Rotter JI, Ruohonen ST, Sacco RL, Samdarshi TE, Schmidt H, Sharp ASP, Shields DC, Sorice R, Sotoodehnia N, Stricker BH, Surendran P, Thom S, Toglhofer AM, Uitterlinden AG, Wachter R, Volzke H, Ziegler A, Munzel T, Marz W, Cappola TP, Hirschhorn JN, Mitchell GF, Smith NL, Fox ER, Dueker ND, Jaddoe VWV, Melander O, Russ M, Lehtimaki T, Ciullo M, Hicks AA, Lind L, Gudnason V, Pieske B, Barron AJ, Zweiker R, Schunkert H, Ingelsson E, Liu K, Arnett DK, Psaty BM, Blankenberg S, Larson MG, Felix SB, Franco OH, Zeller T, Vasan RS and Dorr M. Large-scale genome-wide analysis identifies genetic variants associated with cardiac structure and function. J Clin Invest. 2017;127:1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Harst P, van Setten J, Verweij N, Vogler G, Franke L, Maurano MT, Wang X, Mateo Leach I, Eijgelsheim M, Sotoodehnia N, Hayward C, Sorice R, Meirelles O, Lyytikainen LP, Polasek O, Tanaka T, Arking DE, Ulivi S, Trompet S, Muller-Nurasyid M, Smith AV, Dorr M, Kerr KF, Magnani JW, Del Greco MF, Zhang W, Nolte IM, Silva CT, Padmanabhan S, Tragante V, Esko T, Abecasis GR, Adriaens ME, Andersen K, Barnett P, Bis JC, Bodmer R, Buckley BM, Campbell H, Cannon MV, Chakravarti A, Chen LY, Delitala A, Devereux RB, Doevendans PA, Dominiczak AF, Ferrucci L, Ford I, Gieger C, Harris TB, Haugen E, Heinig M, Hernandez DG, Hillege HL, Hirschhorn JN, Hofman A, Hubner N, Hwang SJ, Iorio A, Kahonen M, Kellis M, Kolcic I, Kooner IK, Kooner JS, Kors JA, Lakatta EG, Lage K, Launer LJ, Levy D, Lundby A, Macfarlane PW, May D, Meitinger T, Metspalu A, Nappo S, Naitza S, Neph S, Nord AS, Nutile T, Okin PM, Olsen JV, Oostra BA, Penninger JM, Pennacchio LA, Pers TH, Perz S, Peters A, Pinto YM, Pfeufer A, Pilia MG, Pramstaller PP, Prins BP, Raitakari OT, Raychaudhuri S, Rice KM, Rossin EJ, Rotter JI, Schafer S, Schlessinger D, Schmidt CO, Sehmi J, Sillje HH, Sinagra G, Sinner MF, Slowikowski K, Soliman EZ, Spector TD, Spiering W, Stamatoyannopoulos JA, Stolk RP, Strauch K, Tan ST, Tarasov KV, Trinh B, Uitterlinden AG, van den Boogaard M, van Duijn CM, van Gilst WH, Viikari JS, Visscher PM, Vitart V, Volker U, Waldenberger M, Weichenberger CX, Westra HJ, Wijmenga C, Wolffenbuttel BH, Yang J, Bezzina CR, Munroe PB, Snieder H, Wright AF, Rudan I, Boyer LA, Asselbergs FW, van Veldhuisen DJ, Stricker BH, Psaty BM, Ciullo M, Sanna S, Lehtimaki T, Wilson JF, Bandinelli S, Alonso A, Gasparini P, Jukema JW, Kaab S, Gudnason V, Felix SB, Heckbert SR, de Boer RA, Newton-Cheh C, Hicks AA, Chambers JC, Jamshidi Y, Visel A, Christoffels VM, Isaacs A, Samani NJ and de Bakker PI. 52 Genetic Loci Influencing Myocardial Mass. J Am Coll Cardiol. 2016;68:1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith NL, Felix JF, Morrison AC, Demissie S, Glazer NL, Loehr LR, Cupples LA, Dehghan A, Lumley T, Rosamond WD, Lieb W, Rivadeneira F, Bis JC, Folsom AR, Benjamin EJ, Aulchenko YS, Haritunians T, Couper D, Murabito J, Wang YA, Stricker BH, Gottdiener JS, Chang PP, Wang TJ, Rice KM, Hofman A, Heckbert SR, Fox ER, O’Donnell CJ, Uitterlinden AG, Rotter JI, Willerson JT, Levy D, Vanduijn CM, Psaty BM, Witteman JC, Boerwinkle E and Vasan RS. The Association of Genome-Wide Variation with the Risk of Incident Heart Failure in Adults of European and African Ancestry: A Prospective Meta-Analysis from the CHARGE Consortium. Circ Cardiovasc Genet. 2010;3:256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia S, Li X, Johnson T, Seidel C, Wallace DP and Li R. Polycystin-dependent fluid flow sensing targets histone deacetylase 5 to prevent the development of renal cysts. Development. 2010;137:1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]