Abstract
Thyroid hormone (TH) is a key regulator of transcriptional homeostasis in the heart. While hypothyroidism is known to result in adverse cardiac effects, the molecular mechanisms that modulate TH signaling are not completely understood. Mediator is a multiprotein complex that coordinates signal-dependent transcription factors with the basal transcriptional machinery to regulate gene expression. Mediator complex protein, Med13, represses numerous thyroid receptor (TR) response genes in the heart. Further, cardiac-specific overexpression of Med13 in mice that were treated with propylthiouracil (PTU), an inhibitor of the biosynthesis of the active TH, triiodothyronine (T3), resulted in resistance to PTU-dependent decreases in cardiac contractility. Therefore, these studies aimed to determine if Med13 is necessary for the cardiac response to hypothyroidism. Here we demonstrate that Med13 expression is induced in the hearts of mice with hypothyroidism. To elucidate the role of Med13 in regulating gene transcription in response to TH signaling in cardiac tissue, we utilized an unbiased RNA sequencing approach to define the TH-dependent alterations in gene expression in wild-type mice or those with a cardiac-specific deletion in Med13 (Med13cKO). Mice were fed a diet of PTU to induce a hypothyroid state or normal chow for either 4 or 16 weeks, and an additional group of mice on a PTU diet were treated acutely with T3 to re-establish a euthyroid state. Echocardiography revealed that wild-type mice had a decreased heart rate in response to PTU with a trend toward a reduced cardiac ejection fraction. Notably, cardiomyocyte-specific deletion of Med13 exacerbated cardiac dysfunction. Collectively, these studies reveal cardiac transcriptional pathways regulated in response to hypothyroidism and re-establishment of a euthyroid state and define molecular pathways that are regulated by Med13 in response to TH signaling.
Keywords: Hypothyroidism, heart failure, Mediator, transcription
Introduction
Increasing evidence from both clinical and animal studies associate low thyroid hormone (TH) levels with accelerated progression to heart failure with a poor prognosis [1–9]. Conversely, high levels of TH induce molecular and cellular alterations in the heart that can result in tachyarrhythmias and ventricular dysfunction [10–12]. In cardiac tissue, TH predominantly regulates the transcription of genes involved in maintaining proper cardiac structure and function, although the exact mechanisms by which this occurs are unclear. Accordingly, hypothyroidism due to decreased thyroid activity, results in detrimental transcriptional alterations that ultimately promote cardiac atrophy, impaired diastolic and systolic function, and reductions in cardiac output [13, 14]. Clinically, the use of the active TH, triiodothyronine (T3), or T3 analogs successfully restores patients to a euthyroid state and improves cardiac output [15–17]. However, an increase in ischemic heart disease is observed in patients under the age of 65 being treated with L-thyroxine, a synthetic form of inactive TH (T4) commonly used to treat hypothyroidism [18, 19]. Thus, the correlation between thyroid status and heart failure warrants further studies of transcriptional regulation in cardiac tissue in the context of hypothyroidism and TH replacement therapy to develop potential novel therapeutics.
One of the predominant roles of TH in the heart is to regulate transcription mediated by the interaction of T3 with its nuclear receptor, thyroid hormone receptor (TR) [12, 20]. TRs remain bound to thyroid response elements (TREs) in a ligand-independent manner. When unbound, TRs repress the transcription of positively-regulated target genes by interacting with transcriptional corepressors [21, 22]. However, once bound, TR heterodimerizes with retinoid X receptor (RXR) and undergoes conformational alterations that promote its interaction with transcriptional coactivators such as the Mediator complex [23–25]. In the cardiac myocyte, TH regulates expression of both structural and functional genes involved in the preservation of cardiac function, including alpha myosin heavy chain (α-MHC, Myh6) and sarcoplasmic/endoplasmic reticulum calcium ATPase 2a (Serca2a) [26–32]. In addition, unbound TR suppresses expression of certain genes, including β-MHC (Myh7) and phospholamban (Pln) [27, 33]. Despite the known regulation of key heart failure genes in the hypothyroid state resulting in disruption of the structural integrity of the cardiac myocyte, calcium handling dysfunction and reduced cardiac contractility, relatively few genes regulated by TH have been defined in the adult heart.
The Mediator complex is a transcriptional coactivator that was initially identified through ligand-dependent interactions with TRα in HeLa cells [23, 24, 34]. This complex consists of approximately 30 proteins that function to regulate gene expression by coordinating the actions of signal-dependent transcription factors with the basal transcriptional machinery [35]. Interactions between Mediator subunits and TR or other nuclear receptors confer specificity in the regulation of gene programs, which can be modified by alterations in the composition of the Mediator complex. For example, Mediator complex contains a kinase submodule consisting of Mediator subunit 13 (Med13), Med12, cyclin-dependent kinase 8 (Cdk8), and cyclin C that associates and dissociates with the core complex to alter Mediator-dependent transcriptional regulation [36–40]. Consistent with the ability of the Mediator complex to interact with nuclear receptors, we recently reported that cardiac-specific deletion of Med1 and cardiac-specific overexpression of Cdk8 alters cardiac metabolism, which is mediated in part through the regulation of nuclear metabolic transcription factors [41, 42]. In addition, cardiac-specific deletion of Med12 or overexpression of Cdk8, were both reported to disrupt calcium homeostasis in the heart [41, 43]. These data demonstrate that the Mediator complex is central to integrating cardiac-specific signals with transcriptional regulation, suggesting that it plays a key role in regulating gene expression in cardiac tissue in response to TH. However, there is much that remains to be learned about how these molecular pathways converge to regulate TH signaling in the heart, particularly with respect to modulating existing therapies for improved outcomes in cardiac function.
Our previous studies identified Med13 as a component of the miR-208a pathway that regulates cardiac responses to hypothyroidism [44]. Mice with a genetic deletion of miR-208a fail to upregulate β-MHC in response to treatment with propylthiouracil (PTU), an inhibitor of the biosynthesis of the active TH, triiodothyronine (T3) [45]. Furthermore, miR-208a-deficient mice display phenotypes similar to mice in a hyperthyroid state, both of which display protection against pathological hypertrophy and fibrosis as well as a block in expression of β-MHC [45]. These findings suggest that miR-208a acts, at least in part, by regulating a common component of the stress-responsive and TH signaling pathways in the heart. Notably, Med13 was initially implicated in TR-signaling as a target of miR-208a. Further, cardiac-specific overexpression of Med13 (i.e., Med13 transgenic mice) regulated expression of a subset of TH-responsive genes in the heart, resulting in a decreased sensitivity to hypothyroidism-induced dysfunction in cardiac contractility [44]. Together, these data suggest that Med13 expression in the heart has a protective role during hypothyroidism by modulating the transcription of TH-responsive genes, which ultimately preserves cardiac function.
While the clinical use of T3 to combat hypothyroidism in the context of many diseases, including heart failure, has been investigated, the cardiac-specific transcriptional pathways that are regulated in hypothyroidism and by TH-responsive genes remain incompletely understood. The current study utilizes RNA-sequencing to define differential gene regulation in the context of hypothyroidism and re-establishment of a euthyroid state in a mouse model. Furthermore, we demonstrate that in response to hypothyroidism, cardiac-specific expression of Med13 is induced while cardiac-specific deletion of Med13 exacerbates cardiac dysfunction. Thus, our findings identify transcriptional pathways regulated in cardiac tissue in response to altered TH signaling providing potential mechanisms that can be targeted to reduce the negative consequences associated with hypothyroidism.
Methods
Animal model
Med13 mice containing LoxP sites flanking exons 7 and 8 (Med13fl/fl) were bred to α-MHC Cre mice (C57bl/6 background, Charles River) to generate mice harboring a cardiac-specific deletion in Med13 (Med13cKO mice) [44]. Mice were housed under standard 12-hour light/dark cycles and all treatments were approved by the University of Iowa Institutional Animal Care and Use Committee (protocols 6011627, 5101529). Med13cKO and Med13fl/fl Cre-negative littermate mice were placed on a normal chow (NC) diet or one containing 0.15% propylthiouracil (PTU) at 8 weeks of age for 4 or 16 weeks (Harlan Teklad cat# TD.95125). Mice treated with 3,3′,5-Triiodo-L-thyronine (T3) received daily intraperitoneal injections for 3 days prior to harvesting to achieve a dose of 1 μg/g body weight (PTU + T3) (Sigma Aldrich cat#T2877).
Protein expression
Following a NC, PTU, or PTU + T3 diet for 4 or 16 weeks, hearts were harvested, rinsed in cold phosphate buffered saline, and flash frozen. Cardiac ventricles were isolated and homogenized in RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% Na-deoxycholate, 0.1% SDS; Thermo) containing cOmplete Mini protease inhibitors (Roche). Protein lysate concentrations were quantified by BCA assays, and 40 μg of protein was separated by 7.5% SDS-PAGE gels. Proteins were transferred to 0.45 μm nitrocellulose (Biorad) at 35 V for 16 h at 4° C and stained with ponceauS. Blots were blocked for 1 h at room temperature in 3% Bovine Serum Albumin/1% Polyvinylpyrrolidone and incubated overnight at 4°C with primary antibodies to detect Med12 (Cell Signaling cat#4529, 1:1000), Med13 (Bethyl cat#A301–277A, 1:500), CDK8 (Santa Cruz cat#sc-1521, 1:1000), Med1 (Bethyl cat#A300–793A, 1:1000), and GAPDH (Cell signaling cat#2118, 1:3000). Blots were incubated for 2 h with HRP-conjugated secondary antibody and developed with ECL reagents (GE healthcare) using chemiluminescent-sensitive X-ray film (Fisher Scientific).
RNA analysis
RNA was extracted from flash frozen, pulverized ventricles using TRIzol reagent (Invitrogen). cDNA was synthesized with SuperScript III First-Strand Synthesis using random hexamers (Thermo Fisher) and qPCR was performed using 25 ng cDNA per reaction with iTaq SYBR Green reagents (Biorad). qPCR primers were used to detect expression of Med13 (Forward 5’-atccatcaagtgcctgcttg-3’, Reverse 5’-ggactgaggatcaactgtttgga-3’), Med1 (Forward 5’-acgagggacgaggaagttg-3’, Reverse 5’-tctggttaaattttgcatggag-3’), Med12 (Forward 5’-cacatcgactgctggacaat-3’, Reverse 5’-tggtccattggtctaaattcttg-3’), CDK8 (Forward 5’-tgccgacatagaaattccag-3’, Reverse 5’-cacttacgggcacgtctaca-3’), Myh6 (Forward 5’-acattcttcaggattctctg-3’, Reverse 5’-ctccttgtcatcaggcac-3’), Myh7 (Forward 5’- ttccttacttgctaccctc-3’, Reverse 5’-cttctcagacttccgcag-3’), Serca2a (Forward 5’-tgatcctcatggatgagacg, Reverse 5’-ccacatcacacagtgagttgg-3’), Thrsp (Forward 5’-tcggggtcttcatcagtctt-3’, Reverse 5’-gcggaaataccaggaaatga-3’), and 18S rRNA (Forward 5’-gccgctagaggtgaaattctt-3’, Reverse 5’-ctttcgctctggtccgtctt-3’). Gene expression was determined using the ΔΔCT method with normalization to 18S rRNA.
RNA sequencing
RNA quality was assessed by Qubit analysis and RNA integrity numbers greater than eight were accepted for RNA sequencing (RNAseq) library preparation. The University of Iowa Institute of Human Genetics, Genomics Division generated RNA libraries of 150 bp polyA-enriched RNA. RNAseq was performed on a HiSeq 4000 genome sequencing platform (Illumina). Sequencing results were uploaded and analyzed with BaseSpace (Illumina). Sequences were trimmed to 125 bp using the Fastq toolkit and aligned to Mus musculus mm10 genome using Bowtie2. Transcripts were assembled and differential gene expression determined using Cufflinks Assembly and DE (Version 2.1.0). Ingenuity Pathway Analysis (IPA; QIAGEN) was used to determine significantly enriched pathways, which were filtered using cutoffs of >1.5 fragments per kilobase per million reads, >1.5 fold changes in gene expression, and a false discovery rate <0.05. EaSeq software124 v1.05 (easeq.net, University of Copenhagen; Copenhagen, Denmark) was utilized to generate genes regulated for heatmap analysis using cutoffs of ≥2 fold change, false discovery rate <0.01, and FPKM ≥0.5. Data have been deposited into GEO (accession GSE124117).
Echocardiography
Two-dimensional echocardiography using a VisualSonics Vevo 2100 ultrasound was performed by the University of Iowa Cardiology animal phenotyping core laboratory. Long- and short-axis views were performed on unsedated mice. Serial echocardiographs were performed on mice at 4 weeks, 8 weeks, 12 weeks, and 16 weeks. Measurements were performed in a blinded manner with respect to treatment and genotype.
Histology
Hearts were preserved in 4% paraformaldehyde and embedded in paraffin. Sectioning (5 μm) and staining was performed by the University of Iowa Central Microscopy Core Facility. Four chamber sections were stained with hemotoxylin and eosin for morphological analysis or Masson’s Trichrome to observe fibrosis. Histology was performed on three to four hearts per group. Pictures were taken using a Nikon Eclipse Ti-S microscope.
Statistics
Results are expressed as mean ± SEM. Student’s t-tests were performed for studies of two groups. For studies containing greater than two groups, two-way ANOVA was used followed by Tukey’s multiple comparison tests. Significance was accepted at p<0.05.
Results
Expression of Mediator complex in mouse cardiac tissue in response to four weeks of hypothyroidism
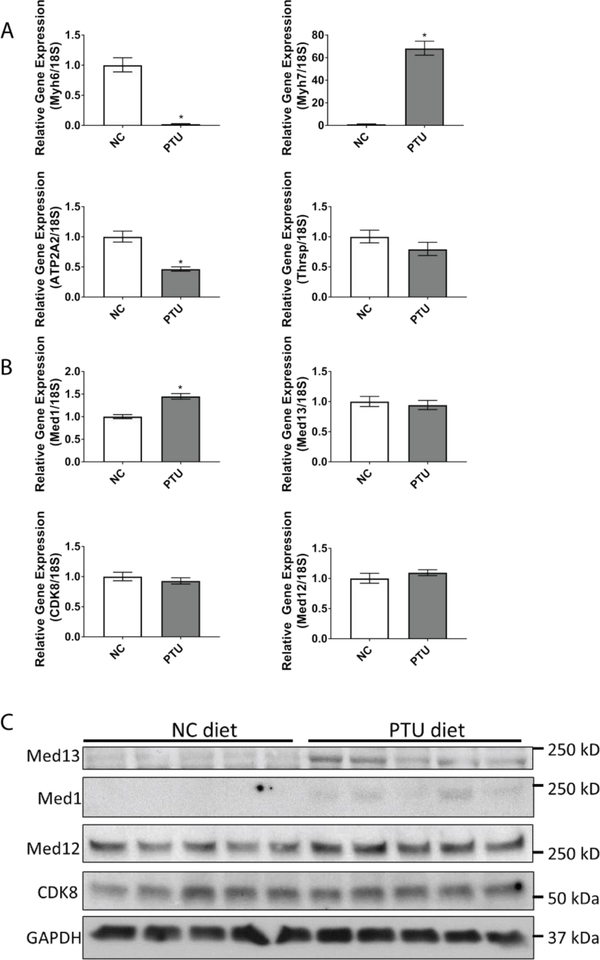
To determine the effects of hypothyroidism on the expression of TH-responsive genes, we first confirmed that a hypothyroid state was induced in mice treated with PTU for four weeks. As expected, PTU treatment resulted in a significant decline in the TH responsive genes Myh6 and Atp2a and an increase in Myh7 expression, indicative of a hypothyroid state (Fig. 1a) [26–31, 46]. Mediator complex is recruited to TH responsive genes by TRs to form the pre-initiation complex and regulate gene expression. Therefore, we assessed cardiac expression of select Mediator subunits in C57bl/6 mice that were fed a PTU diet for four weeks in comparison to mice on a normal chow (NC) diet. The mRNA expression of Cdk8, Med12, and Med13 was unaltered in response to PTU; however, Med1 mRNA expression was significantly increased by 1.45 fold (Fig. 1b). Consistent with RNA expression, Western blot analysis revealed that Med12 and Cdk8 expression were unchanged whereas Med1 expression was induced in mice fed a PTU diet for four weeks. Notably, protein expression of cardiac Med13 also was induced in mice fed a PTU diet for four weeks (Fig. 1c), suggesting that Med13 has a role in regulating responses to thyroid hormone signaling in the heart, as suggested by previously published studies [44, 45]. Taken together, these data suggest that in the context of a hypothyroid state, the regulation of Med13 expression occurs post-translationally.
Fig. 1.
Effects of hypothyroidism on cardiac gene expression and expression of the Mediator complex. WT mice were fed normal chow (NC) or a PTU diet (PTU) for four weeks. Subsequently, mRNA and protein lysates were obtained from the ventricles for expression analysis. A-B. qRT-PCR analysis of the indicated TH responsive genes (A) or components of the Mediator Complex (B). Values are relative to 18S rRNA. Bars represent mean ± SEM. C. Western blot to assess protein expression of components in the Mediator Complex. N=5 mice/treatment. *p<0.05, students T-test.
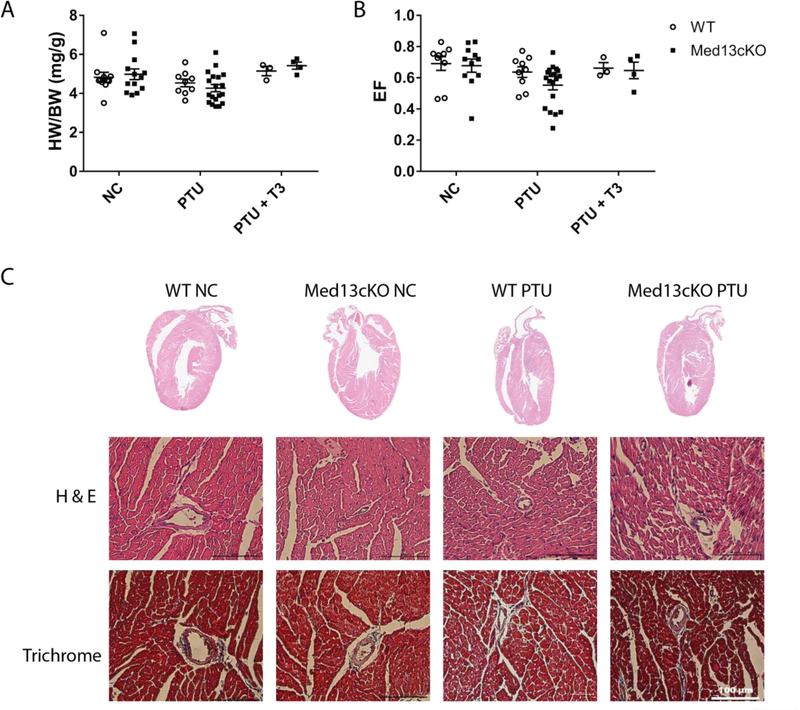
To investigate the short-term structural and functional consequence of altering the levels of TH on the heart, we placed Med13fl/fl (i.e., WT) and Med13fl/fl;αMHC Cre (i.e., Med13cKO) mice on a PTU diet or a NC diet for four weeks. Additionally, re-establishment of a euthyroid state with administration of short term T3 treatment following four weeks of PTU diet (PTU + T3) was performed to investigate TH-dependent transcriptional alterations regulated by Med13. We also assessed cardiac function and performed histological analysis on mice that were placed on a NC, PTU, or PTU + T3 diet. No significant changes in cardiac size, as measured by the ratio of heart weight to body weight (HW/BW), or contractile function, as represented by cardiac ejection fraction (EF), were observed between WT or Med13cKO mice in either treatment group (Fig. 2A, 2B). As expected, Med13cKO mice fed a PTU diet displayed a trend toward reduced cardiac output in part due to significant reductions in heart rate (Table 1). Similar to previous findings, short-term T3 replacement therapy resulted in a subtle, but not significant, increase in cardiac size demonstrating rapid correction from a hypothyroid state (Fig. 2A). In WT or Med13cKO mice fed a normal chow or a PTU diet for four weeks, histological analysis of the four-chamber view of hearts by hematoxylin and eosin staining revealed normal cardiac architecture, and trichrome staining revealed no significant fibrosis following establishment of short-term hypothyroidism in either WT or Med13cKO mice (Figure 2C). Therefore, short-term hypothyroidism was not sufficient to produce alterations in structure or function of the heart.
Fig. 2.
Loss of cardiac Med13 results in minimal alterations in cardiac function following 4 weeks of PTU. WT or Med13cKO mice were fed normal chow (NC) or a PTU diet for four weeks or a PTU diet for four weeks followed by T3 treatment at a dose of 1 μg/g body weight for three days (PTU + T3). A-B. Ratio of heart weight to body weight (HW/BW) or ejection fraction (EF) in mice of the indicated genotype in each group. n=3–19. Data are mean ± SEM. C. Whole heart hematoxylin and eosin (H&E) stain (top) or trichrome stain at 40x magnification (bottom) in mice of the indicated genotype that received either normal chow (NC) or a PTU diet (PTU). Statistics performed using Two-way ANOVA with Tukey’s multiple comparisons test.
Table 1.
Echocardiographic parameters from mice of each genotype fed a diet of PTU or NC for four weeks, or fed a diet of PTU for four weeks followed by T3 treatment. n=3-19 per treatment group. Data represent mean ± SEM. Statistics performed using Two-way ANOVA with Tukey’s multiple comparisons test.
| HW (mg) | BW (g) | HW/BW (mg/g) | HR (bpm) | EDV (μl) | ESV (μl) | CO (mcL/m) | EF (SV/EDV) | |
|---|---|---|---|---|---|---|---|---|
| WT NC | 129.29 ± 5.54 | 26.82 ± 0.81 | 4.81 ± 0.074 | 639.89 ± 40.00 | 38.46 ± 4.46 | 12.70 ± 3.15 | 16575.05 ± 1926.83 | 0.69 ± .044 |
| Med13cKO NC | 128.25 ± 3.56 | 29.92 ± 1.35 | 4.32 ± 0.12 | 639.36 ± 35.17 | 43.92 ± 6.12 | 16.39 ± 5.01 | 17679.82 ± 1691.59 | 0.68 ± .042 |
| WT PTU | 91.98 ± 2.36 | 25.20 ± 0.90 | 3.68 ± 0.14 | 575.00 ± 24.96 | 34.25 ± 4.43 | 13.42 ± 3.04 | 11943.50 ± 1066.04 | 0.64 ± .034 |
| Med13cKO PTU | 94.77 ± 2.29 | 25.69 ± 0.51 | 3.70 ± 0.095 | 528.79 ± 13.82* | 38.93 ± 2.54 | 18.42 ± 2.42 | 10933.2 ± 732.43* | 0.55 ± .030 |
| WT PTU + T3 | 113.47 ± 1.02 | 22.14 ± 1.03 | 5.14 ± 0.24 | 532 ± 15.28 | 30.34 ± 5.56 | 10.22 ± 2.29 | 10668.42 ± 1993.03 | 0.66 ± 0.036 |
| Med13cKO PTU + T3 | 124.00 ± 3.60 | 22.99 ± 1.37 | 5.43 ± 0.18 | 540.5 ± 16.08 | 34.11 ± 1.03 | 12.00 ± 1.83 | 11945.94 ± 1170.60 | 0.65 ± 0.053 |
p<0.05 in comparison to NC control of same genotype.
Cardiac gene expression in the context of hypothyroidism
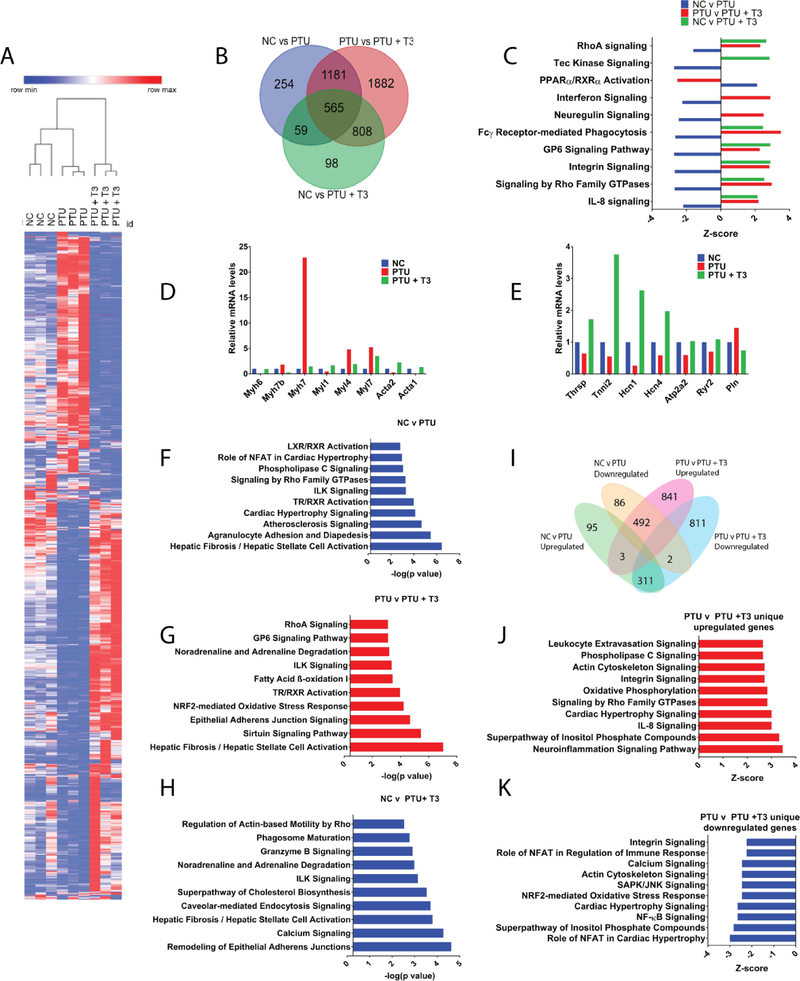
TH binds to TRs to modify gene expression [12, 20], although how transcriptional regulation by TR is accomplished is incompletely understood. TH-dependent gene expression can be activated or inhibited in the presence of free TH depending on the type of TRE that is involved, the isoform of the TR that is being used, and the transcriptional cofactors that are present [22, 23, 47–49]. Therefore, to identify TH-regulated cardiac transcriptional networks that precede cardiac dysfunction, we performed RNA sequencing using wildtype mice fed a NC or PTU diet for four weeks. We also performed RNA sequencing in mice fed a PTU diet for four weeks followed by short-term T3 replacement therapy in order to identify transcriptional targets of T3. Differentially expressed genes among the three groups were visualized by heatmap analysis, which demonstrated distinct subsets of genes were either induced or repressed in response to hypothyroidism and that this effect was largely reversed following re-establishment of a euthyroid state (Fig. 3A). Venn diagram analysis was performed to visualize the overlap in gene expression changes between the three groups (NC vs PTU, PTU vs PTU+T3, NC vs PTU+T3). This identified 2059 genes that were differentially expressed following four weeks of a PTU diet whereas treatment with T3 following consumption of a PTU diet resulted in 4436 differentially regulated genes. In addition, this analysis revealed altered expression of a common set of 565 genes in all treatment groups (Fig. 3B). Thus, a subset of the cardiac transcriptional network is regulated by T3.
Fig. 3.
Hypothyroidism and TH replacement therapy alter cardiac transcription. RNA-sequencing was performed from RNA obtained from ventricles of WT mice fed a normal chow (NC) or PTU diet for four weeks or a PTU diet for four weeks followed by T3 treatment for three days at a dose of 1 μg/g body weight (PTU + T3). A. Heat map displaying genes that are differentially regulated between each treatment group. The heatmap contains 1235 total genes with a fold change ≥2, a false discovery rate < 0.01, and minimum FPKM of 0.5. The heatmap was generated using Morpheus GENE-E software (Broad Institute). Each lane represents a single mouse. B. Venn diagram demonstrating the overlap of genes that were significantly differentially regulated in WT mice in each treatment group. C. Ingenuity pathway analysis (IPA) canonical pathway analysis demonstrating pathways regulated in WT mice in each treatment group as determined by z-score. D. Relative gene expression of structural genes identified by IPA in the Rho signaling and integrin signaling pathways. E. Relative gene expression of known TH-regulated genes F,G,H. IPA canonical pathway analysis displaying pathways significantly regulated for NC v PTU (F), PTU v PTU + T3 (G), and NC v PTU + T3 (H). I. Venn diagram analysis displaying genes that are differentially upregulated and downregulated with FPKM values ≥1.5 and ≥ 1.5 fold change in both NC v PTU and PTU v PTU + T3. J. IPA canonical pathway analysis of genes upregulated between PTU v PTU + T3 groups. K. IPA canonical pathway analysis of genes downregulated in PTU v PTU + T3 groups. n = 3 for each group. For analysis with IPA, FPKM cutoffs of 1.5, fold change of ≥1.5, and false discovery rate < 0.05 were utilized for significantly differentially regulated genes.
To further delineate alterations in gene expression between the different conditions, we utilized the Ingenuity Pathway Analysis (IPA) tool. Consistent with heatmap analysis, IPA canonical pathway analysis revealed that many genes were dynamically regulated in a hypothyroid state but were reversed upon re-establishment of a euthyroid state (Fig. 3C). This analysis largely represents expected pathways (e.g. TR/RXR activation), including those that are the result of structural alterations in cardiomyocytes such as Rho signaling and integrin signaling. Genes regulated in these pathways include alpha actin (Acta1, Acta2), myosin heavy chains (Myh7, Myh7b), and Myosin light chains (Myl1, Myl4, Myl7). Furthermore, known TH-regulated genes that were downregulated in expression in a hypothyroid state, including thyroid hormone responsive (Thrsp), Serca2a (Atp2a2), hyperpolarization cyclic nucleotide (HCN1 and HCN4), ryanodine receptor (Ryr2), Troponin I (Tnni2), and phospholamban (Pln), were reversed with T3 administration (Fig. 3D, E) [12, 28–31, 33, 46, 50–52]. The IPA canonical pathway analysis also revealed pathways known to be regulated in response to hypothyroidism, including atherosclerosis signaling, cardiac hypertrophy signaling, and pathways involved in cardiac calcium handling (Fig. 3F). In addition, hepatic fibrosis is the most significantly enriched pathway that is regulated by both hypothyroidism and the re-establishment of euthyroid state (Fig. 3F, 3G). Notably, this pathway contained numerous genes involved in cardiac fibrosis, including collagens, which were downregulated in mice on a PTU diet and reversed in response to treatment with T3. In addition, comparison of gene expression changes in mice fed a NC diet relative to those fed a PTU diet and treated with T3 revealed significant alterations in pathways involved in calcium signaling, cardiomyocyte structure, cellular morphology and adhesion, and fibrosis (Fig. 3H).
To further explore the relationship between differentially expressed genes in response to a PTU diet or a PTU diet followed by T3 treatment, IPA analysis was performed on genes that were upregulated or downregulated in the NC v PTU dataset and the PTU v PTU + T3 dataset. Specifically, we analyzed genes that were significantly differentially regulated, as determined by those with a fragments per kilobase million (FPKM) of at least 1.5 that demonstrated a 1.5 fold change in gene expression within each set of comparisons. This analysis revealed that genes that were differentially regulated in response to hypothyroidism were largely reversed upon short-term T3 treatment (Fig. 3I). For example, of the 409 genes that were significantly upregulated in mice fed a PTU diet relative to those fed a NC diet, only three genes were similarly upregulated in PTU + T3 mice, whereas 311 of those genes were downregulated following treatment with T3. Likewise, of the 580 genes downregulated in response to a PTU diet relative to those fed a NC diet, only two of these genes were similarly downregulated in mice fed a PTU + T3 diet relative to those that were fed a PTU diet, whereas 492 genes of the 580 upregulated in response to a PTU diet were reversed upon treatment with T3 (PTU vs PTU + T3) (Fig. 3I). These data demonstrate that genes regulated in a hypothyroid state are largely reversed, and not exacerbated, following T3 replacement therapy.
This analysis further revealed that the majority of genes regulated in response to acute T3 treatment were not altered by PTU alone. To better identify the gene programs specifically regulated within the PTU + T3 group, IPA was performed on genes that were either uniquely upregulated or downregulated in mice fed a PTU diet that also received T3 relative to mice fed a PTU diet. Canonical pathway analysis revealed that genes involved in numerous pathways that promote cardiac hypertrophy were upregulated in mice that were fed a PTU diet and also received treatment with T3 in comparison to PTU mice (n=841; Fig. 3J). In addition, numerous inflammatory pathways including the neuroinflammation signaling pathway, dendritic cell maturation, IL-8 signaling, leukocyte extravasation signaling, and interferon signaling were upregulated in this group. Genes that were uniquely downregulated in mice fed a PTU + T3 diet relative to those that were fed a PTU diet (n=811) were involved in cardiac hypertrophy and inflammation, including Mef2a and Nfatc4, which are known to be involved in pathological cardiac hypertrophy (Fig. 3K). These data demonstrate that while treatment with T3 largely reverses altered gene expression caused by hypothyroidism, it also alters the expression of additional genes that may contribute toward the adverse effects of hyperthyroidism on the heart.
Cardiac Med13-dependent regulation of thyroid hormone signaling
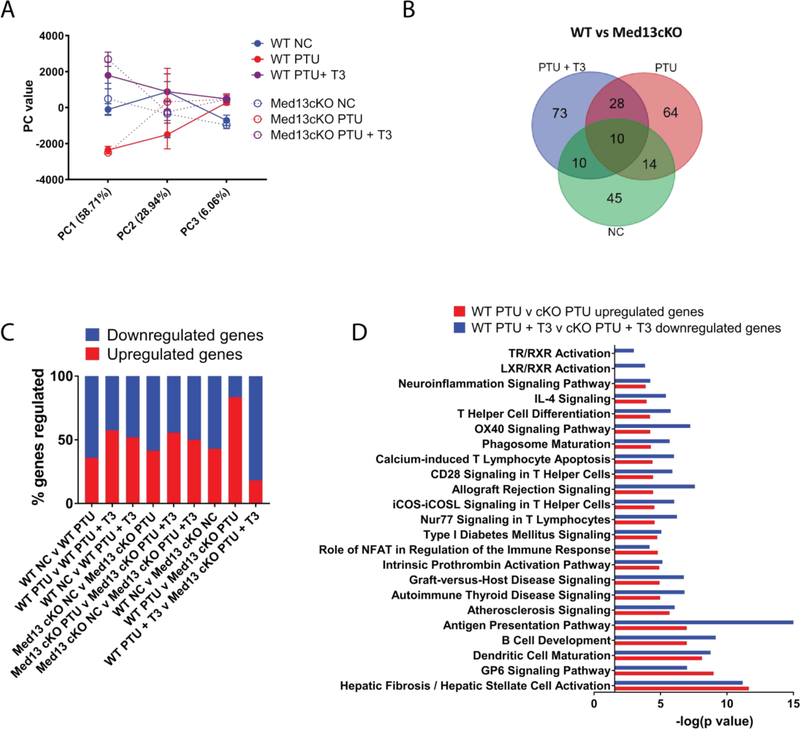
To understand the role of Med13-dependent transcription in TH signaling, Med13cKO mice and WT littermate controls were placed on a PTU diet for four weeks to assess differences in gene expression that occur due to hypothyroidism but that precede cardiac dysfunction. In addition, mice on a PTU diet were also compared to a group of mice that were fed a PTU diet and also received T3 treatment in order to re-establish a euthyroid state, as described above. Ventricles from WT and Med13cKO mice in each group were obtained for analysis by RNA sequencing. Principle component analysis revealed that the greatest variance in gene expression was due to the effects of diet, as demonstrated by PC1 (58.71%) whereas genotype specific differences also account for some variation in gene expression, as explained by PC2 (28.94%) (Fig. 4A). In total, there were 244 genes that were differentially regulated in Med13cko mice compared to WT mice in all treatment conditions (Fig. 4B). A similar proportion of genes were upregulated or downregulated in WT or Med13cKO mice when comparing euthyroid and hypothyroid states and upon re-establishment of a euthyroid state. However, these differences became skewed upon deletion of Med13 (Fig. 4C, last two bars). Specifically, in mice fed a PTU diet for four weeks, the loss of cardiac-specific Med13 expression resulted in an increased percentage of genes that were upregulated in comparison to WT mice fed a PTU diet, suggesting that Med13 acts to transcriptionally inhibit gene expression in a hypothyroid state where TR is unbound. In contrast, T3 replacement therapy increased the percentage of genes that were downregulated in Med13cko mice compared WT mice suggesting that Med13 acts as an activator of gene expression during T3 replacement therapy where TR is ligand-bound (Fig. 4C).
Fig. 4.
Cardiac-specific deletion of Med13 results in transcriptional alterations in the heart. RNA-seq was performed from RNA obtained from ventricles of WT or Med13cKO mice fed NC or PTU diet for four weeks or a PTU diet for four weeks followed by T3 treatment for three days (PTU + T3). A. Principle component analysis generated by BaseSpace that compares genes that are significantly differentially regulated in each treatment group. B. Venn diagram analysis displaying the overlap of significantly differentially expressed genes in Med13cKO mice in each treatment group. C. Percentage of genes upregulated or downregulated in each group. D. IPA canonical pathway analysis comparing genes that are specifically upregulated in WT v Med13cKO mice that were fed a PTU diet and genes that are downregulated in WT v Med13cKO mice that were fed a PTU diet and received T3 treatment. n = 3/treatment group. For analysis with IPA, FPKM cutoffs of 1.5 and fold change of ≥1.5 were utilized for significantly differentially regulated genes.
Because genotype-specific alterations in the percentage of genes regulated in hypothyroidism and re-establishment of a euthyroid state were observed, IPA was performed on genes upregulated in Med13cKO PTU hearts compared to WT PTU hearts and genes downregulated in Med13cKO PTU + T3 hearts compared to WT PTU + T3 hearts. This analysis revealed that in mice fed a PTU diet, genes involved in inflammatory and fibrotic pathways were upregulated in Med13cKO mice compared to WT mice (Fig. 4D). In addition, genes that were downregulated in Med13cKO mice that were fed a PTU diet and treated with T3 compared to WT mice in the same conditions were enriched in inflammatory and fibrotic pathways. These results suggest that induction of Med13 in hypothyroidism may have a role in repressing pro-inflammatory and pro-fibrotic transcriptional programs that are reversed upon treatment with T3.
Cardiac dysfunction is exacerbated in the absence of Med13
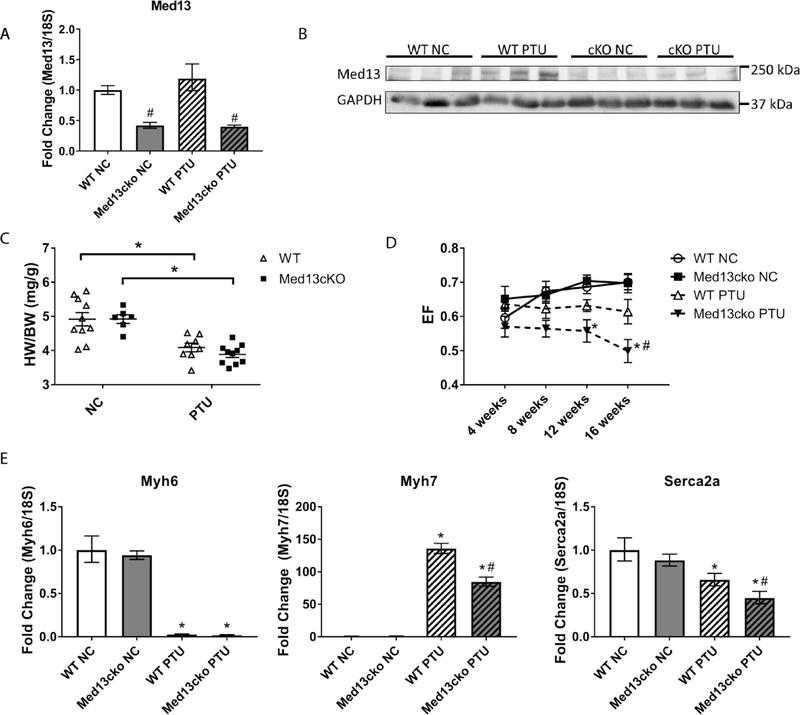
The induction of Med13 in response to PTU (Fig. 1C) suggests it has a role in transcriptional responses to TH in the heart. To assess the long-term effects of cardiac-specific deletion of Med13 in hypothyroidism, mice were placed on a PTU diet for 16 weeks and compared to mice given a NC diet. Cardiac-specific expression of Med13 was assessed for mice of each genotype and on each diet at the end of the 16-week period. A significant reduction in Med13 mRNA expression (an approximate two-fold reduction) was observed in Med13cKO mice in comparison to WT mice, irrespective of diet (Fig. 5A). It is likely that the remaining Med13 expression that is observed can be attributed to its expression in cardiac fibroblasts. Of note, cardiac-specific mRNA expression of Med13 failed to be induced even after mice were in a hypothyroid state for 16 weeks. Similar to the increased protein expression of Med13 observed in mice given a PTU diet for four weeks, WT mice that received a PTU diet for 16 weeks showed increases in Med13 protein expression compared to WT mice fed a NC diet. However, Med13 protein expression in Med13cKO mice was blunted in response to a hypothyroid state, suggesting that Med13 expression is largely induced in the cardiomyocyte fraction of the heart in response to TH levels (Fig. 5B).
Fig. 5.
Med13 is induced by and is important for cardiac responses to hypothyroidism. WT and Med13cKO mice were fed a normal chow (NC) diet or a PTU diet for 16 weeks. A. Ventricular Med13 mRNA expression in WT and Med13cKO mice after 16 weeks on the indicated diet. n =5–6/group B. Ventricular Med13 protein expression in WT and Med13cKO mice after 16 weeks on the indicated diet. C. Heart weight to body weight ratio (HW/BW; mg/g) after four weeks on the indicated diet. n =6–10/group. D. Ejection fraction of WT or Med13cKO mice, as determined by serial echocardiogram at 4 weeks, 8 weeks, 12 weeks, and 16 weeks following the start of the indicated diet. n =6–10/group. E. Ventricular mRNA expression of known TH-regulated, hypertrophic markers in WT and Med13cKO mice following 16 weeks on the indicated diet. n =5–6/group. *p < .05 diet-specific comparison. # < 0.05 genotype-specific comparison, Two way ANOVA, Tukey’s multiple comparisons test. Data represent mean ± SEM.
Following PTU diet for 16 weeks, WT and Med13cKO mice were sacrificed and HW/BW ratios were measured. As expected, mice fed a PTU diet demonstrated a reduced HW/BW ratio in comparison to mice fed a NC diet within each respective genotype (Fig. 5C). However, no genotype-specific differences were observed in HW/BW due to diet. To determine if a hypothyroid state impacted cardiac function over time in either WT or Med13cKO mice, serial echocardiograms were performed on mice fed a NC diet or a PTU diet at 4 weeks, 8 weeks, 12 weeks, and 16 weeks (Fig. 5D). WT mice fed a PTU diet had a lower, although not significant, EF relative to WT mice fed a NC diet. In contrast, Med13cKO mice fed a PTU diet displayed a significant reduction in EF in comparison to Med13cKO mice fed a NC diet as early as 12 weeks after starting the PTU diet. Furthermore, Med13cKO mice that were fed a PTU diet displayed a significant reduction in EF in comparison to WT mice fed a PTU diet 16 weeks after the start of the diet, suggesting that cardiac-specific expression of Med13 is required to preserve cardiac function in a hypothyroid state. This genotype-specific reduction in EF occurred without significant differences in heart rate suggesting altered cardiac contractility (Table 2).
Table 2.
Echocardiographic parameters of mice for each genotype fed a diet of PTU or NC for 16 weeks. n=6-10/group.
| HW (mg) | BW (g) | HW/BW (mg/g) | HR (bpm) | EDV (μl) | ESV (μl) | CO (mcL/m) | EF (SV/EDV) | ||
|---|---|---|---|---|---|---|---|---|---|
| 4 weeks | WT NC | 742.0 ± 7.49 | 27.79 ± 2.65 | 10.93 ± 0.99 | 12503.33 ± 1515.01 | 0.60 ± 0.031 | |||
| Med13cko NC | 689.4 ± 26.10# | 29.80 ± 2.75 | 10.26 ± 1.11 | 13445.29 ± 1723.1 | 0.64 ± 0.036 | ||||
| WT PTU | 525.0 ± 13.51* | 26.42 ± 1.58 | 9.70 ± 0.89 | 8777.47 ± 752.52 | 0.64 ± 0.016 | ||||
| Med13cko PTU | 514.2 ± 5.40* | 32.06 ± 2.39 | 14.02 ± 1.61 | 9272.38 ± 738.63 | 0.57 ± .030 | ||||
| 8 weeks | WT NC | 702.3 ± 12.82 | 29.77 ± 2.28 | 9.73 ± 1.10 | 14118.47 ± 1249.53 | 0.67 ± 0.030 | |||
| Med13cko NC | 707.13 ± 12.89 | 31.26 ± 3.24 | 10.59 ± 1.01 | 14688.72 ± 1852.89 | 0.65 ± 0.019 | ||||
| WT PTU | 500.88 ± 7.53* | 26.29 ± 2.73 | 9.88 ± 1.14 | 8212.77 ± 969.90* | 0.62 ± 0.028 | ||||
| Med13cko PTU | 492.8 ± 6.59* | 34.33 ± 3.94 | 15.32 ± 2.02 | 9303.36 ± 786.23* | 0.56 ± 0.025 | ||||
| 12 weeks | WT NC | 730.6 ± 9.56 | 29.67 ± 2.32 | 9.25 ± 0.86 | 14886.66 ± 1267.93 | 0.69 ± 0.019 | |||
| Med13cko NC | 705.57 ± 28.96 | 41.95 ± 4.82# | 12.00 ± 1.50 | 20927.91 ± 2411.78# | 0.72 ± 0.018 | ||||
| WT PTU | 492.75 ± 5.93* | 31.04 ± 3.82 | 11.48 ± 1.59 | 9589.73 ± 1157.46 | 0.63 ± 0.018 | ||||
| Med13cko PTU | 468.8 ± 8.55* | 34.82 ± 2.37 | 15.79 ± 2.02 | 8925.00 ± 638.49* | 0.56 ± 0.033* | ||||
| 16 weeks | WT NC | 60.79 ± 3.18 | 33.09 ± 1.36 | 4.92 ± 0.20 | 738.9 ± 12.58 | 36.89 ± 4.82 | 11.52 ± 2.42 | 18587.85 ± 1883.87 | 0.70 ± 0.022 |
| Med13cko NC | 156.67 ± 5.84 | 31.93 ± 1.51 | 4.92 ± 0.13 | 719.0 ± 9.66 | 48.74 ± 7.02 | 14.42 ± 1.79 | 24760.22 ± 4196.61# | 0.70 ± .028 | |
| WT PTU | 98.84 ± 2.71 | 24.21 ± 0.41 | 4.09 ± 0.13 | 482.5 ± 7.74* | 35.45 ± 2.39 | 13.60 ± 1.40 | 10514.33 ± 957.52* | 0.61 ± 0.036 | |
| Med13cko PTU | 94.46 ± 2.72 | 24.33 ± 0.62 | 3.89 ± 0.091 | 447.9 ± 11.79* | 42.49 ± 2.28 | 21.40 ± 1.88*# | 9432.30 ± 803.03* | 0.50 ± .034*# |
p<0.05 diet-specific comparison.
p<0.05 genotype-specific comparison
Two way ANOVA, Tukey’s multiple comparisons test. Data represent mean ± SEM.
To determine the role of Med13 in preserving cardiac function following induction of a hypothyroid state for 16 weeks, we assessed expression of hypertrophy markers known to be regulated by TH. As expected, Myh6 was reduced in mice fed PTU for 16 weeks, although no significant differences were observed between genotypes (Fig. 5E). In contrast, expression of both Myh7 and Serca2a was significantly reduced in Med13cKO mice on a PTU diet compared to WT mice on a PTU diet. These alterations in gene expression are necessary to preserve cardiac function in response to a hypothyroid state and may, in part, account for the reduced cardiac function observed in a hypothyroid state in Med13cKO mice.
Discussion
Dynamic transcriptional responses occur due to altered TH levels in the heart [12, 20]. Transcriptional regulation of genes that are necessary for maintaining cardiac contractility is dependent on TH; however, the global regulation of cardiac gene expression in hypothyroidism and upon re-establishment of a euthyroid state is incompletely defined. Here, we elucidate transcriptional networks that are regulated by a short-term (four weeks) hypothyroid state in comparison to those affected by rapid correction to a euthyroid state. Transcriptional analysis was performed prior to any detectable alterations in cardiac function and revealed novel genes that are differentially expressed due to TH. Similar to previous studies, short-term replacement with T3 following a four-week PTU diet resulted in slight increases in HW/BW and contractile function demonstrating the rapid correction from a hypothyroid state. Further, genetic deletion of Med13 in cardiomyocytes alters the expression of a subset of genes that are differentially expressed due to altered TH levels (i.e., on a PTU diet), which exacerbates the detrimental cardiac response to hypothyroidism.
RNA sequencing analysis revealed a subset of differentially regulated genes that were distinct in both a euthyroid state and hypothyroid state in WT mice. Canonical pathway analysis of these genes revealed enrichment of pathways known to be dysregulated in hypothyroidism. Specifically, genes belonging to categories indicative of structural remodeling, including RhoA signaling and integrin signaling, were regulated in a hypothyroid state and upon re-establishment of a euthyroid state. Genes in these pathways are known TH-regulated genes including Acta1, Acta2, and Myh6/7. In addition, TH regulates numerous metabolic processes and RNA sequencing analysis revealed dysregulation of oxidative phosphorylation, cholesterol signaling, as well as pathways that maintain glucose homeostasis. Alterations in these pathways could potentially be due to stoichiometric alterations in RXR since it heterodimerizes with Pparα, Vitamin D receptors (VDR), and LXR to regulate metabolic processes. Notably, mice on a PTU diet for four weeks had reduced expression of genes involved in cardiac fibrosis, which was reversed upon treatment with T3. However, no differences in collagen deposition were observed by histology in these mice compared to mice on a NC diet. Therefore, it is possible that longer exposure to PTU would induce pro-fibrotic transcriptional programs.
Previous studies in rats demonstrated that correcting hypothyroidism with T3 replacement therapy resulted in an exacerbation of genes that are related to the development of cardiac fibrosis and inflammation [53]. In these studies, hypothyroidism was induced by treating rats for 6 weeks with PTU, followed by its reversal with T3 treatment for 7 days. PTU reportedly induced pro-inflammatory and pro-fibrotic transcriptional responses. Expression of specific gene markers, including Ccl2 and Ctgf, were exacerbated following reversion to a euthyroid state. In contrast, our findings demonstrate that expression of Ccl2 and Ctgf are reduced following treatment with T3. The differences observed could be associated with the treatment timelines, which were sufficient to induce apparent fibrosis by histological analysis in the former study and are in contrast to the findings presented here.
Med13 has been implicated in TH signaling, thus we further assessed its role in TH signaling in both a hypothyroid state and upon re-establishment of a euthyroid state. Our findings demonstrate that Med13 is induced in response to hypothyroidism. Furthermore, we demonstrate that cardiac-specific deletion of Med13 results in impaired cardiac function with a concomitant reduction of TH-regulated genes that are involved in preserving cardiac contractility following long-term PTU treatment. In support of this finding, cardiac-specific overexpression of Med13 was previously reported to preserve cardiac function in a hypothyroid state. In this context, Med13 overexpressing mice placed on a PTU diet maintained Myh7 expression, which we observed to be significantly reduced in Med13cKO mice in comparison to WT littermates. In addition, Serca2a expression was significantly reduced in Med13cKO mice fed a PTU diet in comparison to WT mice fed a PTU diet. Recently, Med12, a component of the Mediator kinase submodule, was found to positively regulate calcium-handling genes, including Serca2a. Therefore, it is possible that deletion of Med13 disrupts the Mediator complex kinase submodule resulting in diminished Serca2a expression and cardiac dysfunction.
To identify transcriptional programs that are regulated by Med13 in a hypothyroid state and upon re-establishment of a euthyroid state, we performed RNA sequencing analysis prior to the development of cardiac dysfunction in WT and Med13cKO mice fed a NC diet or a PTU diet for four weeks. Notably, our results suggest that cardiac-specific Med13 acts as a repressor in a hypothyroid state whereas it acts to activate transcription upon the re-establishment of euthyroid state. The fact that our RNA sequencing analysis did not identify strong differential gene expression programs in response to hypothyroidism or upon re-establishment of a euthyroid state in Med13cKO mice was somewhat surprising, although recent reports have suggested that Med13 and it’s paralogue, Med13 like (Med13L) are redundant in the genes they regulate at the transcriptional level [54]. Therefore, it is possible that deletion of Med13 is not sufficient to alter global gene regulation due to the presence of Med13L.
The transcriptional effects of hypothyroidism that are observed in mice fed a PTU diet provide insight into alterations that may occur in humans who are suffering from heart disease. Specifically, several studies have associated heart disease with low serum TH levels which may in part drive cardiac fetal transcriptional reprograming [1–5, 8, 12, 20]. Therefore, TH replacement therapy to maintain a euthyroid state has been proposed as a potential therapeutic for heart failure. Altogether, we have identified cardiac-specific transcriptional programs that are regulated in response to both a hypothyroid state and the re-establishment of a euthyroid state. Our work furthers our understanding of the transcriptional regulation mediated by Med13 and highlights the importance of transcriptional cofactors in modifying the transcriptome. In the future, it will be important to define the dynamics of TH signaling during development and in the complex interplay between TR and cofactors that are involved in heart disease.
Acknowledgements
This work was supported by the NIH/NHLBI R01 HL125436 (CEG). RNA sequencing library generation and data were obtained at the Genomics Division of the Iowa Institute of Human Genetics through the support of the University of Iowa Institute on Human Genetics which is supported, in part, by the University of Iowa Carver College of Medicine. Histological services were performed at the University of Iowa Central Microscopy Research Facility, which is supported by the Vice President for Research & Economic Development, the Holden Comprehensive Cancer Center, and the Carver College of Medicine. Echocardiography services were performed by the University of Iowa Cardiovascular Imaging Core using the Vevo 2100 system which was purchased using NIH Grants S10 OD019941–01 and S10 RR026293–01. We would like to thank Jennifer Barr for critically editing.
Footnotes
Conflicts of interest: The authors have declared that no conflicts of interest exist.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Selmer C, et al. , Subclinical and overt thyroid dysfunction and risk of all-cause mortality and cardiovascular events: a large population study. J Clin Endocrinol Metab, 2014. 99(7): p. 2372–82. [DOI] [PubMed] [Google Scholar]
- 2.Wang W, et al. , Free Triiodothyronine Level Correlates with Myocardial Injury and Prognosis in Idiopathic Dilated Cardiomyopathy: Evidence from Cardiac MRI and SPECT/PET Imaging. Sci Rep, 2016. 6: p. 39811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gencer B, et al. , Subclinical thyroid dysfunction and the risk of heart failure events: an individual participant data analysis from 6 prospective cohorts. Circulation, 2012. 126(9): p. 1040–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nanchen D, et al. , Subclinical thyroid dysfunction and the risk of heart failure in older persons at high cardiovascular risk. J Clin Endocrinol Metab, 2012. 97(3): p. 852–61. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell JE, et al. , Thyroid function in heart failure and impact on mortality. JACC Heart Fail, 2013. 1(1): p. 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fruhwald FM, et al. , Subclinical thyroid disorders in patients with dilated cardiomyopathy. Cardiology, 1997. 88(2): p. 156–9. [DOI] [PubMed] [Google Scholar]
- 7.Iervasi G, et al. , Low-T3 syndrome: a strong prognostic predictor of death in patients with heart disease. Circulation, 2003. 107(5): p. 708–13. [DOI] [PubMed] [Google Scholar]
- 8.Tang YD, et al. , Low thyroid function leads to cardiac atrophy with chamber dilatation, impaired myocardial blood flow, loss of arterioles, and severe systolic dysfunction. Circulation, 2005. 112(20): p. 3122–30. [DOI] [PubMed] [Google Scholar]
- 9.Mourouzis I, et al. , Thyroid hormone improves the mechanical performance of the post-infarcted diabetic myocardium: a response associated with up-regulation of Akt/mTOR and AMPK activation. Metabolism, 2013. 62(10): p. 1387–93. [DOI] [PubMed] [Google Scholar]
- 10.Freedberg AS, Papp JG, and Williams EM, The effect of altered thyroid state on atrial intracellular potentials. J Physiol, 1970. 207(2): p. 357–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnsdorf MF and Childers RW, Atrial electrophysiology in experimental hyperthyroidism in rabbits. Circ Res, 1970. 26(5): p. 575–81. [DOI] [PubMed] [Google Scholar]
- 12.Kahaly GJ and Dillmann WH, Thyroid hormone action in the heart. Endocr Rev, 2005. 26(5): p. 704–28. [DOI] [PubMed] [Google Scholar]
- 13.Montalvo D, et al. , Underlying mechanism of the contractile dysfunction in atrophied ventricular myocytes from a murine model of hypothyroidism. Cell Calcium, 2018. 72: p. 26–38. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z and Gerdes AM, Influence of hypothyroidism and the reversal of hypothyroidism on hemodynamics and cell size in the adult rat heart. J Mol Cell Cardiol, 1990. 22(12): p. 1339–48. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton MA, et al. , Safety and hemodynamic effects of intravenous triiodothyronine in advanced congestive heart failure. Am J Cardiol, 1998. 81(4): p. 443–7. [DOI] [PubMed] [Google Scholar]
- 16.Pingitore A, et al. , Acute effects of triiodothyronine (T3) replacement therapy in patients with chronic heart failure and low-T3 syndrome: a randomized, placebo-controlled study. J Clin Endocrinol Metab, 2008. 93(4): p. 1351–8. [DOI] [PubMed] [Google Scholar]
- 17.Amin A, et al. , Effects of triiodothyronine replacement therapy in patients with chronic stable heart failure and low-triiodothyronine syndrome: a randomized, double-blind, placebo-controlled study. ESC Heart Fail, 2015. 2(1): p. 5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flynn RW, et al. , Serum thyroid-stimulating hormone concentration and morbidity from cardiovascular disease and fractures in patients on long-term thyroxine therapy. J Clin Endocrinol Metab, 2010. 95(1): p. 186–93. [DOI] [PubMed] [Google Scholar]
- 19.Leese GP, et al. , Morbidity in patients on L-thyroxine: a comparison of those with a normal TSH to those with a suppressed TSH. Clin Endocrinol (Oxf), 1992. 37(6): p. 500–3. [DOI] [PubMed] [Google Scholar]
- 20.Dillmann WH, Cellular action of thyroid hormone on the heart. Thyroid, 2002. 12(6): p. 447–52. [DOI] [PubMed] [Google Scholar]
- 21.Brent GA, et al. , Thyroid hormone aporeceptor represses T3-inducible promoters and blocks activity of the retinoic acid receptor. New Biol, 1989. 1(3): p. 329–36. [PubMed] [Google Scholar]
- 22.Hu X and Lazar MA, Transcriptional repression by nuclear hormone receptors. Trends Endocrinol Metab, 2000. 11(1): p. 6–10. [DOI] [PubMed] [Google Scholar]
- 23.Ito M and Roeder RG, The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol Metab, 2001. 12(3): p. 127–34. [DOI] [PubMed] [Google Scholar]
- 24.Fondell JD, Ge H, and Roeder RG, Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci U S A, 1996. 93(16): p. 8329–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fondell JD, et al. , Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc Natl Acad Sci U S A, 1999. 96(5): p. 1959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsika RW, et al. , Thyroid hormone regulates expression of a transfected human alpha-myosin heavy-chain fusion gene in fetal rat heart cells. Proc Natl Acad Sci U S A, 1990. 87(1): p. 379–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lompre AM, Nadal-Ginard B, and Mahdavi V, Expression of the cardiac ventricular alpha- and beta-myosin heavy chain genes is developmentally and hormonally regulated. J Biol Chem, 1984. 259(10): p. 6437–46. [PubMed] [Google Scholar]
- 28.Rohrer D and Dillmann WH, Thyroid hormone markedly increases the mRNA coding for sarcoplasmic reticulum Ca2+-ATPase in the rat heart. J Biol Chem, 1988. 263(15): p. 6941–4. [PubMed] [Google Scholar]
- 29.Rohrer DK, Hartong R, and Dillmann WH, Influence of thyroid hormone and retinoic acid on slow sarcoplasmic reticulum Ca2+ ATPase and myosin heavy chain alpha gene expression in cardiac myocytes. Delineation of cis-active DNA elements that confer responsiveness to thyroid hormone but not to retinoic acid. J Biol Chem, 1991. 266(13): p. 8638–46. [PubMed] [Google Scholar]
- 30.Zarain-Herzberg A, et al. , Thyroid hormone receptor modulates the expression of the rabbit cardiac sarco (endo) plasmic reticulum Ca(2+)-ATPase gene. J Biol Chem, 1994. 269(2): p. 1460–7. [PubMed] [Google Scholar]
- 31.Hartong R, et al. , Delineation of three different thyroid hormone-response elements in promoter of rat sarcoplasmic reticulum Ca2+ATPase gene. Demonstration that retinoid X receptor binds 5’ to thyroid hormone receptor in response element 1. J Biol Chem, 1994. 269(17): p. 13021–9. [PubMed] [Google Scholar]
- 32.Trivieri MG, et al. , Cardiac-specific elevations in thyroid hormone enhance contractility and prevent pressure overload-induced cardiac dysfunction. Proc Natl Acad Sci U S A, 2006. 103(15): p. 6043–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belakavadi M, et al. , Repression of cardiac phospholamban gene expression is mediated by thyroid hormone receptor-{alpha}1 and involves targeted covalent histone modifications. Endocrinology, 2010. 151(6): p. 2946–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan CX, et al. , The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci U S A, 1998. 95(14): p. 7939–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taatjes DJ, The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci, 2010. 35(6): p. 315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeronimo C, et al. , Tail and Kinase Modules Differently Regulate Core Mediator Recruitment and Function In Vivo. Mol Cell, 2016. 64(3): p. 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrenko N, et al. , Mediator Undergoes a Compositional Change during Transcriptional Activation. Mol Cell, 2016. 64(3): p. 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elmlund H, et al. , The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc Natl Acad Sci U S A, 2006. 103(43): p. 15788–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dewey CM, et al. , Cardiac-Secreted Factors as Peripheral Metabolic Regulators and Potential Disease Biomarkers. J Am Heart Assoc, 2016. 5(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grueter CE, Mediator complex dependent regulation of cardiac development and disease. Genomics Proteomics Bioinformatics, 2013. 11(3): p. 151–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall DD, et al. , Ectopic expression of Cdk8 induces eccentric hypertrophy and heart failure. JCI Insight, 2017. 2(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spitler KM, et al. , Cardiac Med1 deletion promotes early lethality, cardiac remodeling, and transcriptional reprogramming. Am J Physiol Heart Circ Physiol, 2017. 312(4): p. H768–H780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baskin KK, et al. , MED12 regulates a transcriptional network of calcium-handling genes in the heart. JCI Insight, 2017. 2(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grueter CE, et al. , A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell, 2012. 149(3): p. 671–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Rooij E, et al. , Control of stress-dependent cardiac growth and gene expression by a microRNA. Science, 2007. 316(5824): p. 575–9. [DOI] [PubMed] [Google Scholar]
- 46.Arai M, et al. , Effect of thyroid hormone on the expression of mRNA encoding sarcoplasmic reticulum proteins. Circ Res, 1991. 69(2): p. 266–76. [DOI] [PubMed] [Google Scholar]
- 47.Yen PM, Physiological and molecular basis of thyroid hormone action. Physiol Rev, 2001. 81(3): p. 1097–142. [DOI] [PubMed] [Google Scholar]
- 48.Zhu XG, et al. , NCoR1 regulates thyroid hormone receptor isoform-dependent adipogenesis. Journal of Molecular Endocrinology, 2011. 46(3): p. 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiss RE, et al. , Mice deficient in the steroid receptor co-activator 1 (SRC-1) are resistant to thyroid hormone. EMBO J, 1999. 18(7): p. 1900–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jump DB, et al. , Rapid Effects of Triiodothyronine on Hepatic Gene-Expression - Hybridization Analysis of Tissue-Specific Triiodothyronine Regulation of Messenger Rnas14. Journal of Biological Chemistry, 1984. 259(5): p. 2789–2797. [PubMed] [Google Scholar]
- 51.Gloss B, et al. , Cardiac ion channel expression and contractile function in mice with deletion of thyroid hormone receptor alpha or beta. Endocrinology, 2001. 142(2): p. 544–50. [DOI] [PubMed] [Google Scholar]
- 52.Averyhart-Fullard V, et al. , Differential regulation of slow-skeletal and cardiac troponin I mRNA during development and by thyroid hormone in rat heart. J Mol Cell Cardiol, 1994. 26(5): p. 609–16. [DOI] [PubMed] [Google Scholar]
- 53.Hajje G, et al. , Hypothyroidism and its rapid correction alter cardiac remodeling. PLoS One, 2014. 9(10): p. e109753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuuluvainen E, et al. , Depletion of Mediator Kinase Module Subunits Represses Superenhancer-Associated Genes in Colon Cancer Cells. Mol Cell Biol, 2018. 38(11). [DOI] [PMC free article] [PubMed] [Google Scholar]