Abstract
The majority of pharmacogenomic (PGx) studies have been conducted on European ancestry populations, thereby excluding minority populations and impeding the discovery and translation of African American–specific genetic variation into precision medicine. Without accounting for variants found in African Americans, clinical recommendations based solely on genetic biomarkers found in European populations could result in misclassification of drug response in African American patients. To address these challenges, we formed the Transdisciplinary Collaborative Center (TCC), African American Cardiovascular Pharmacogenetic Consortium (ACCOuNT), to discover novel genetic variants in African Americans related to clinically actionable cardiovascular phenotypes and to incorporate African American–specific sequence variations into clinical recommendations at the point of care. The TCC consists of two research projects focused on discovery and translation of genetic findings and four cores that support the projects. In addition, the largest repository of PGx information on African Americans is being established as well as lasting infrastructure that can be utilized to spur continued research in this understudied population.
A remarkable journey in genetic research began in 1990 when scientists sought to map the human genome. It culminated in mapping 3 billion base pairs and identifying ~ 20,500 human genes in what has been a remarkable achievement in genomic research.1 These discoveries spawned a massive wave of new areas of research in genetics, including the field of pharmacogenomics (PGx), which is aimed at identifying genetic variations such as single‐nucleotide polymorphisms (SNPs) that influence interindividual differences in drug response and adverse events. Its application promises to enable targeted drug administration, improve therapeutic outcomes, and inform drug development. Pharmacogenomic insights have improved our understanding of the underlying pathways and mechanisms behind adverse drug reactions, which account for ~ 100,000 deaths per year around the United States and markedly increase healthcare costs.2 With scans of human genetic variation and the consequent deluge of genomic information readily available,3, 4 the promise of personalized medicine is tantalizingly close.
Medical centers around the country have begun adopting clinical implementation of well‐established PGx findings,5 with up‐to‐date recommendations from the Clinical Pharmacogenetics Implementation Consortium (CPIC)6 on how available genetic test results can be used to optimize drug therapy. Unfortunately, even though many of these medical centers have substantial numbers of minority patients, they have been largely excluded from this revolution in medicine. This is only going to become more of an issue over time because the Veterans Administration projects that by 2037 around 32% of its patient population will be minorities, with over 16% of their patients being African American.7 The US census projects that by 2060, 25.7% of the population will be non white, and 17.9% of the population will be African American.8, 9
Our Transdisciplinary Collaborative Center (TCC) is focused on addressing this gap in precision medicine by undertaking research projects centering on DNA variant discovery in African Americans (Discovery Project) and clinical translational of these findings, which will assess the impact of genetic biomarkers on drug selection, dosing, and clinical outcomes (Translational Project). We have incorporated patient‐centered approaches to each of these initiatives and are working within African American communities on the design, recruitment, and dissemination of this important work. Figure 1 illustrates how each of the TCC components works synergistically with one another.
Figure 1.
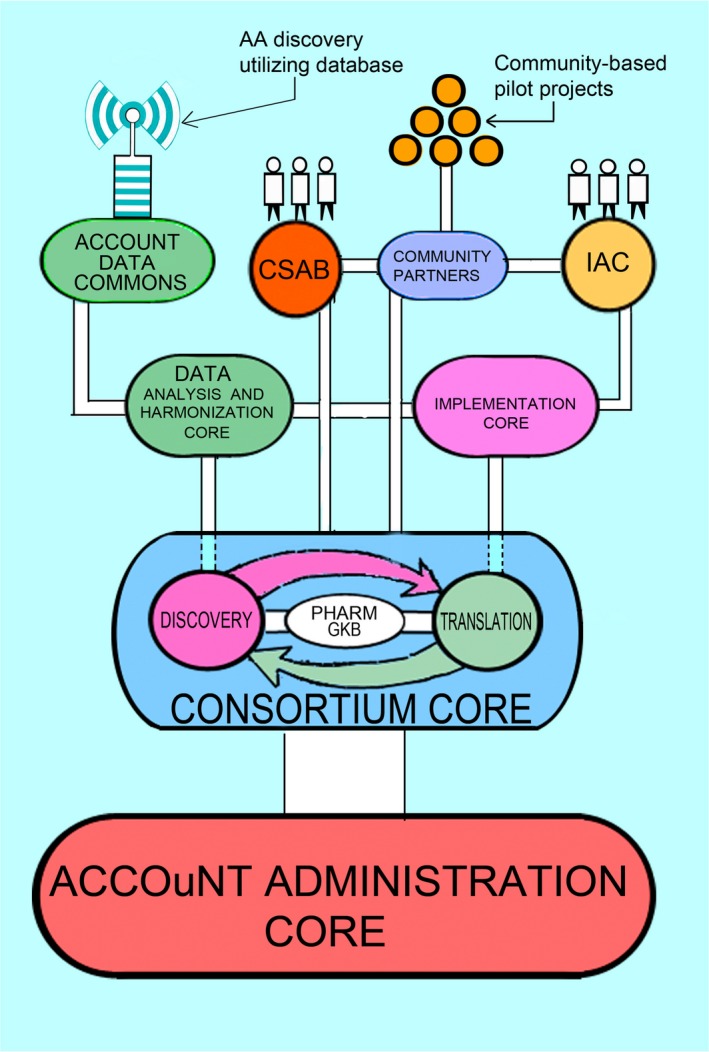
The African American Cardiovascular Pharmacogenetic Consortium (ACCOuNT) Transdisciplinary Collaborative Center (TCC). Although both the discovery and translational projects are independent efforts, the overall interactions of these projects within ACCOuNT will help to develop a pipeline from which newly discovered single‐nucleotide polymorphisms (SNPs) in the Discovery Project and SNPs curated via PharmGKB will be investigated within the Translational Project. Data gathered within the Translational Project will inform which drugs are relevant candidates for investigation within the Discovery Project. Results of the Translational Project will be evaluated by the Implementation Advisory Council (IAC), which will collaborate with regional community partners for implementation planning and pilot project development. The ACCOuNT Data Commons will house information generated via the Discovery Project will become a resource for the greater PGx community for discovery and validation of African American findings. All efforts will have oversight of our Community and Stakeholder Advisory Board (CSAB) within the Consortium Core, which has launched community‐based pilot projects. All projects and cores are supported by the infrastructure within the Administrative Core, which provides the foundation of the TCC.
Our centers in Chicago, IL, and Washington, DC, are optimally located because of their large African American communities. Census data show that Chicago, IL, and Washington, DC, have African American populations of 30.9% and 47.1%, respectively, well above the national average of 13.4%.10 Research projects within the African American Cardiovascular Pharmacogenetic Consortium (ACCOuNT) were chosen because of the disproportional risk of cardiovascular disease in African Americans, the lack of comprehensive PGx studies related to treatment of this disease in African Americans, the existing history of collaboration between many of the TCC institutions, and the community engagement opportunities that exist in both Chicago, IL, and Washington, DC, which will allow for greater inclusion of African Americans in research (Table 1). The following two projects represent complementary aspects of precision medicine, which focus on interactions among biological, genetic, and contextual predictors of drug response.
Table 1.
Institutions in the ACCOuNT consortium
| Institution | Key investigators | Expertise | Discovery project | Translational project | Implementation core | Consortium core | Data analysis and harmonization core |
|---|---|---|---|---|---|---|---|
| Northwestern University | Minoli Perera, Kevin O'Leary, Marc Rosenman | Clinical cardiovascular medicine, PGx, statistical genetics | X | X | X | X | |
| The University of Chicago | David Meltzer, Robert Grossman, Peter O'Donnell, Doriane Miller | , health outcomes research in minority populations, bioinformatics, community connections | X | X | X | X | X |
| University of Illinois at Chicago | Edith Nutescu, William Galanter, Shane Borkowski | Clinical cardiovascular medicine, PGx, health outcomes | X | X | |||
| The George Washington University | Travis O'Brien, Norman Lee, April Barbour | Clinical cardiovascular medicine, platelet splicing, PGx | X | ||||
| Shenandoah University | Art Harralson | X | |||||
| Washington DC VA Medical Center | Matthew Tuck | Clinical cardiovascular medicine, PGx | X | ||||
| Stanford University | Teri Klein, Li Gong | , bioinformatics, PharmGKB, biomedical knowledge management | X | X |
ACCOuNT, African American Cardiovascular Pharmacogenetic Consortium.
This table illustrates the variety of scientific organizations in ACCOuNT. Each institution has been chosen for their recognized expertise as well as ongoing collaboration with other investigators in the area.
Discovery Project
In this project, we hypothesize that through whole‐genome genotyping and transcriptome analysis focused on African Americans, we will identify novel population‐specific predictive biomarkers of cardiovascular drug response that can be rapidly moved into translational outcome studies. We aim to recruit ~ 1,200 subjects to both the warfarin and the novel oral anticoagulant (NOAC) studies and 500 subjects to the clopidogrel study. We will use tools, such as genotyping arrays and RNA‐sequencing (targeted to those with extreme phenotypes, such as bleeding or nonresponse) to discover novel sequence variation and its effect on gene expression in the African American population. We have also been collecting a sample for cryopreservation of peripheral blood mononuclear cells, which can be converted to induce pluripotent stem cells for future studies to advance precision medicine in African Americans.
This project aims at identifying and validating new genetic associations in drug phenotypes that have been well studied in the European Ancestry populations. This is vital work that will allow us to answer the question, “Are the known European predictors of drug response adequate to predict drug response in African Americans, or are new population‐specific predictors needed to guide therapy in African Americans?” To answer this question, we must conduct population‐specific discovery studies as opposed to testing established European findings in African Americans. We have chosen to begin with cardiovascular PGx phenotypes of clopidogrel response, bleeding risk with warfarin, and NOACs (see inclusion/exclusion criteria for the discovery cohorts in Tables 2 and 3). For each of these phenotypes, we are recruiting a prospective cohort of African Americans with the corresponding clinical and demographic data to test for genetic association in a genome‐wide manner. All participants are self‐identified as African Americans, and we will further assess ancestry thorough genomic methods (such as principal component analysis or ancestry estimation). As has been established in our previous work, we will correct for population substructure and population outliers as appropriate.11 We are collecting a whole‐blood transcriptome from each patient in which splice variant discovery and expression quantitative trait loci studies can be performed. Additionally, because the discovery project will also collect adverse events while on therapy, such as thrombosis, bleeding, and major adverse cardiac events, we will be able to determine if transcriptomic differences between cases and controls exist. The integrative approach of linking genomes to transcriptome and clinical data has proven to be a powerful tool in elucidating potentially functional SNPs and possible biological mechanisms for the associated SNPs (reviewed in ref. 12). Our project also incorporates social and demographic factors that help to explain drug response (e.g., geocodes for patient environment interaction with response), which may be independent of genetics. These factors will be important to both physicians and patients in order to choose the most appropriate medication for treatment of disease.
Table 2.
Inclusion criteria for the discovery study
| Warfarin arm | Clopidogrel arm | NOAC arm |
|---|---|---|
| Both new starts and currently on warfarin | Clopidogrel 75 mg daily | Taking 15 mg/day of rivaroxaban (Xarelto) or taking 5 mg/day of apixaban (Eliquis) or taking 30 mg/day of edoxaban (Savaysa, Lixiana) |
| Expected to remain on warfarin for ≥ 3 months |
Has been on treatment for at least 14 days Expected to remain on treatment for ≥ 30 days |
Has been on treatment for at least 3 days Expected to remain on standard daily dose for ≥ 30 days |
| Primary indication (at least one): venous thromboembolism, atrial fibrillation, mechanical valve replacement | Primary indication (at least one): history of myocardial infarction, coronary stent placement (drug‐eluting and nondrug‐eluting stents), history of stroke, aspirin intolerance, including hypersensitivity to aspirin or aspirin resistance | Primary indications (at least one): nonvalvular atrial fibrillation, venous thromboembolism treatment, venous thromboembolism prophylaxis |
NOAC, novel oral anticoagulant.
Self‐described as African American.
Age > 18 years.
Starting or on treatment with the selected agents.
Speak and understand English.
Table 3.
Exclusion criteria for the discovery study
| Hemoglobin < 7.0 g/dL at enrollment |
| Life expectancy < 8 weeks |
| Active cancer diagnosis |
| Inability or unwillingness to comply with study procedures or study medications |
| History of significant bleeding (defined as requiring hospitalization) within 6 months prior to enrollment and/or prior to starting anticoagulant or antiplatelet therapy |
| History of any significant gastrointestinal, intracranial, pulmonary, or urogenital bleeding, ongoing peptic ulcer disease or ongoing or acute gastritis, neurospinal disease or spinal surgery (NOAC arm only) within 2 years prior to enrollment or beginning therapy |
| Contraindications to anticoagulant therapy |
| Platelet count < 100,000 mm3 |
| Concurrent or history of alcohol or drug abuse within 1 year prior to enrollment or while on therapy |
| History of hypersensitivity to study drug |
| Concomitant dual antiplatelet therapy daily (however, patients may be on low‐dose aspirin) or daily NSAIDs |
NOAC, novel oral anticoagulant; NSAIDs, nonsteroidal anti‐inflammatory drugs.
Phenotypes
For all arms of the genomic studies, we will review the electronic medical record (EMR) as well as recontact subjects 6 months after enrollment to assess bleeding events. The primary phenotypes will be clinically relevant bleeding as defined by the International Society on Thrombosis and Hemostasis. The two categories include non major bleeding in nonsurgical patients or clinically relevant nonmajor bleeds (see definitions below).
Major bleeding in nonsurgical patient
Fatal bleeding
Symptomatic bleeding in critical area or organ, such as intracranial, intraspinal, intraocular, retroperitoneal, intraarticular or pericardial, or intramuscular with compartment syndrome.
Clinically relevant nonmajor bleeding
Hospital admission.
Physician‐guided medical or surgical treatment for bleeding.
A change in antithrombotic therapy (including interruption or discontinuation of drug).
Given that African Americans incur venous thromboembolism at a higher rate than other populations, we will also collect data on embolic events that occur while on either warfarin or NOACs (see definitions below).
Embolic events
Stroke (ischemic, hemorrhagic, or unspecified)—sudden focal neurological deficit in the distribution of a single brain artery that persisted beyond 24 hours and was not due to another identifiable cause.
Systemic embolism—abrupt vascular insufficiency associated with clinical or radiological evidence of arterial occlusion in the absence of another likely mechanism, such as atherosclerosis.
Myocardial infarction—typical symptoms and cardiac biomarker elevation above the upper limit of normal, new pathological Q waves, or ST segment elevations in at least two contiguous echocardiogram leads, or confirmation at autopsy.
Platelet function testing is not routinely conducted prior to administration of clopidogrel; therefore, all tests of platelet function will be done at 14–30 days after beginning therapy. Studies in European populations have shown that the predictive CYP2C19*2 SNP was associated with postdose Adenosine diphosphate‐stimulated platelet aggregation. Therefore, the magnitude of on‐clopidogrel platelet reactivity in whole blood, expressed as platelet reactivity units, will be measured with the VerifyNow (Accumetrics, San Diego, CA) point‐of‐care assay, similar to previous studies in Europeans.
Healthy platelet subproject
Given that there is no information on population‐specific splice variants in platelets (a clinically relevant tissue), a subproject was established to investigate potential African American–specific splicing variants in platelets. We will seek to establish differences in alternative transcript abundance and novel transcripts from platelets between African American and European American healthy volunteers. Relatively nothing is known regarding population differences in gene expression in platelets or the role of alternative transcripts. Platelets play an instrumental role in the early stages of thrombus formation and atherogenesis in coronary artery disease.13, 14 Inhibiting platelet aggregation is a primary goal in the treatment, as is secondary prevention, of ischemic events following acute coronary syndromes as well as percutaneous coronary intervention using dual antiplatelet therapy.15 These results will be valuable for both future functional validation studies as well as studies aimed at elucidating platelet‐specific splice variants associated with cardiovascular disease risk and therapeutic outcome. These data will be made publicly available through the ACCOuNT Data Commons.
Account Data Commons
An important component of the Data Commons effort will be the dissemination of the genomics, transcriptomic, and clinical information into a publicly available searchable resource, which, in turn, will fuel the discovery of predictive biomarkers in African Americans by the larger scientific community. Data such as these (e.g., imputed genotypes obtained from genotyping arrays and RNA‐seq normalized gene expression from blood) have never been integrated in a way that would allow any investigator to determine whether specific SNPs (anywhere in the genome) may show evidence for association to the expression of a gene of interest in African Americans. Furthermore, we have incorporated publicly available Gene Expression Omnibus gene expression data from African Americans into the Data Commons so that analysis can be conducted across several study cohorts. This is a highly valuable addition to our own data as most public data in African Americans are sparse and, hence, a resource in which all available data can be used together will be a powerful tool for discovery and translational efforts.
Challenges and strategies
One current concern is the ability to recruit a sufficient numberof subjects for all phenotypes proposed. Because of our large collaborative network, no one site will carry the burden of recruitment. Each site has the infrastructure to facilitate recruitment. Furthermore, many of these sites have contributed significantly to previous large‐scale efforts, such as the African American genome‐wide association study in warfarin dose.11 To facilitate recruitment, we are building education materials to help educate physicians and patients about precision medicine and the gap in discovery that remains for African Americans. An excellent example of this can be seen in the community partnership with Taking Effective Action, which produced an educational film to introduce African American patients to the benefits of participation in genomics research.16 This partnership was made possible through our Consortium Core.
Translational Project
This project aims to deliver PGx to healthcare personnel (including physicians, pharmacists, physicians’ assistants, etc.) at the point of patient encounter. The interactive Genomic Prescribing System (GPS) was developed to provide decision support by translating patients’genotypic results into individualized PGx summaries.5 Patients are pre‐emptively genotyped so that these summaries allow their healthcare provider to determine if any results impact drugs they are considering prescribing. The GPS provides no raw genotypes but rather a clinical interpretation that the provider can read in 30 seconds or less. This system was designed to deliver guidance that is harmonious with current CPIC guidelines.17 The translational project has customized the GPS to deliver African American findings when appropriate. Our test case is warfarin PGx (ClinicalTrials.gov record: NCT03225820), in which a recent genome‐wide association study identified an African American variant associated with lower warfarin dose requirement.11 This variant was subsequently added to the newest CPIC guidelines regarding the use of genomic information to guide therapy in warfarin.18 The GPS delivers warfarin PGx recommendations based on patient‐specific genotype and clinical data. This warfarin decision support tool was built primarily by using algorithms that were largely developed in white patients previously published by Gage et al.19 Randomized prospective trials, however, have called into question the utility of these algorithms in nonwhite patients and, specifically, there are data to suggest potential harm to African Americans if genetic information is included in these dosing algorithms for African American patients.20 In fact, because of this, many institutions that have implemented warfarin PGx only use clinical factors to guide dosing—omitting PGx data for safety reasons (personal communication, Dr Perera). In contrast, for white patients, both PGx and clinical data are used.
We are currently using the GPS within the inpatient Hospitalist service at the University of Chicago and have also begun implementing it at other ACCOuNT sites within Chicago with the goal of prospectively assessing prescribing/treatment changes and relevant outcomes in African American patients who receive GPS‐guided therapy vs. those who receive standard of care (nongenotype‐guided therapy). Given that both healthcare personnel and patients are enrolled in this study, we can also assess the uptake and attitude of multiple members of the healthcare team on using this type of intervention in health management.
Through the Consortium Core of ACCOuNT, we are conducting our translational efforts with a patient‐centered approach that incorporates the input of community partners, frontline physicians, and African American patients, who will provide input on relevant clinical phenotypes in this population. We anticipate that these studies will reveal attitudes of both healthcare personnel and patients to the GPS technology, as well as the benefit to both patients and the healthcare system in terms of clinical outcomes and quality of care received. This project aims to create a large repository of hospitalized African American patients who receive GPS‐guided therapy in which we can: (i) determine highly utilized medications that would benefit from discovery studies and (ii) initiate new translational efforts in drugs in which novel SNPs that predict response are found in the future. The project aims to recruit two cohorts of patients: (i) a feasibility cohort of frequently hospitalized patients (Figure 2 a) and (ii) a warfarin cohort (Figure 2 b). In this way, we create an on‐going pipeline from discovery to translation and ultimately to implementation.
Figure 2.
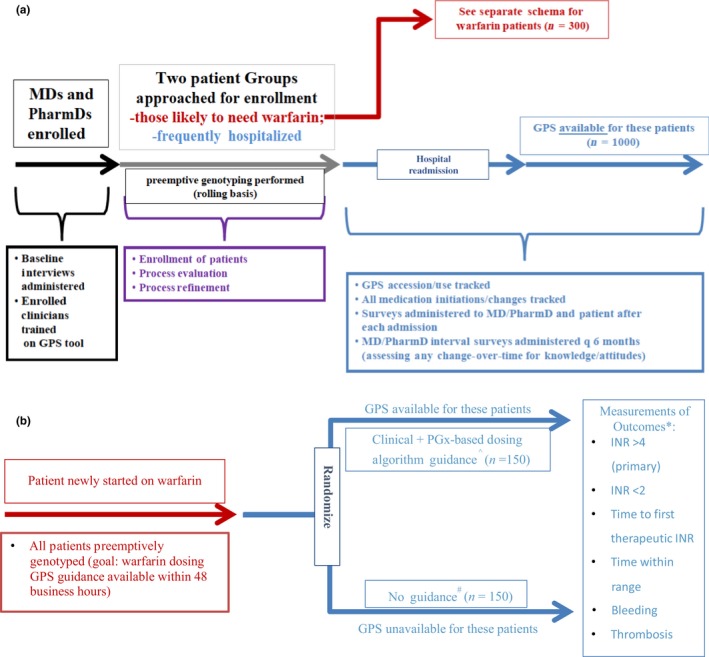
Translational Project Study Schemas. Overall scheme of planned recruitment within the Translational Project. (a) The hospitalized African American cohort, which will receive Genomic Prescribing System (GPS)‐guided therapy on their next readmission, (b) shows the randomization of African American patients newly started on warfarin who will either receive pharmacogenomics (PGx)‐based therapy or standard of care with assessment of outcome collect 3 months after dose initiation.^Via GPS; *Includes phone follow‐up at 14 and 28 days after warfarin initiation if patient is already discharged, plus medical record review (including International Normalized Ratios (INRs)) for the first 3 months; time within range will include allowance for variability by (±) 0.2 for the INR to be counted within range; #physicians and pharmacists are permitted to seek standard available decision support/guidance/dosing algorithms if they wish, but specific guidance from this study will be unavailable.
Across clinical situations, there are choices of different drugs or drug doses that can be given to the patient. Except for situations in which a serious toxicity could be avoided using PGx, we believe that these situations of “clinical drug equipoise” (because of how widely they occur) could be the most influenced by the availability of PGx information. In these “drug choice/dose option” situations, the clinician traditionally will make a choice of one of the medications over the others based upon some combination of clinical factors specific to his or her patient (i.e., allergies; experiences with one or several of the drugs in question), drug costs, availability (formulary influences), or, commonly, based simply upon habit. We have hypothesized that PGx could offer additional, useful information for informing these types of clinical prescribing dilemmas. A recent paper highlights clear examples of prescribing equipoises that can be informed by PGx specifically for cardiovascular medications.21
Our approach to assess the therapeutic and clinical outcomes of population‐specific prescribing will again begin with African American inpatients initiating warfarin and later expand to other cohorts: For the healthcare professionals who have consented to use the GPS to determine its impact on prescribing practice, we will “track” the pharmacogenetic Web portal for the following: (i) did the provider view the patient's results during the admission? (ii) If so, did the provider follow the secondary link summarizing the sources of evidence implicating that genetic result with a given drug (information that will encapsulate the number, size, design, magnitude, and statistical significance of the published studies on that drug‐gene pair). Among those viewing the dual layers of the above information, we will then determine (through querying the patient, the provider, and the medication records) whether a new medication was started at that admission or whether a previously prescribed prescription was changed (either the dose or the drug itself) at a given encounter.
In particular, given extremely busy hospitalist care settings, will such clinicians choose to view patient‐specific PGx results when making prescribing decisions, just as they would consider more routine clinical results, like their patient's creatinine or hemoglobin level? Feasibility in this practice setting will be assessed by measuring the frequency of review of PGx results by participating hospitalists for study patients using the GPS. At least 1,000 patients being cared for by hospitalists will be enrolled in the study. The patients will be receiving care from preselected specialist physicians in this discipline. Patients from the outpatient setting who are admitted to the hospital will be included in this study, plus additional patients recruited specifically for this study. Given that we are studying “early‐adopter physicians” in this study (i.e., the physicians selected could inherently be among the more enthusiastic about the promise of PGx/precision medicine), we assume a likely bias toward utilization and, therefore, generously hypothesize that ≥ 50% of all patient hospitalizations evaluable in this study will incorporate an evaluation of genotype results via GPS. This study will have met its primary end point if the actual percentage is ≥ 50%. Providers will be reminded at the time of hospitalization which patients are study participants, but providers will not be required to access the GPS.
Analysis of warfarin response in the race‐specific prescribing study
As a secondary analysis we will examine clinical outcomes related to the delivery of GPS information for warfarin use. These predefined clinical outcomes will be assessed by medical record review during that patient's hospitalization and patient survey results collected by telephone at 14 and 28 days after discharge and review of the EMR or outside medical records for up to 3 months post‐drug initiation. Warfarin will have a pre‐identified adverse event or nonresponse dichotomized clinical outcome identified, which will be used to determine whether a patient exhibits a potentially preventable PGx “event.” The selected outcomes for warfarin include International Normalized Ratios (INRs; over 4 (primary), INRs below 2, time to first therapeutic INR, time within therapeutic INR range, and bleeding or thrombosis.
Challenges and strategies
One of the biggest challenges we face for warfarin PGx is to return genotyping results in a timely fashion to impact clinical care. Turnaround time for receiving, testing, and releasing the results of individual patients into the GPS will, therefore, be critical. To overcome this, we plan to also target enrollment of patients likely to require warfarin therapy in the near future, using bioinformatics algorithms that incorporate known clinical predictors of warfarin use. Northwestern, through its Electronic Medical Records and Genomics (eMERGE) project, has developed an algorithm to help determine which patients are most likely to require therapy and have deployed these algorithms to target PGx interventions within their hospital population.22
A second major challenge is the incorporation of the GPS within different EMRs and hospital settings within ACCOuNT, although this challenge is not unique to ACCOuNT. This challenge is currently being approached and tackled through customized solutions across many healthcare systems that are trying to incorporate genomic data and related clinical decision support within electronic workflows. The solutions that we develop and implement for ACCOuNT may, thus, serve as models for other such ongoing or future efforts if successful.
Account cores
Data analysis and harmonization core
It is a challenge today for most biomedical scientists to manage, analyze, and share with the research community large genomic data sets, especially those that contain protected health information that must be managed in a secure and compliant fashion. The ACCOuNT discovery and translational projects will generate genomics, transcriptomic, and clinical data on African American patients that will require extensive curation and quality control to yield usable data sets. Given that these data are being collected across multiple institutions, data quality, analysis, and harmonization efforts are needed to ensure that the data can be integrated into one comprehensive data set. Thus, the Data Analysis and Harmonization core has been established. This core will provide the needed computation infrastructure, data security pipelines, and genetic statistical expertise to accomplish our goals with the following aims:
Create common clinical data elements and phenotypes for projects, register participants, provide honest broker service, and coordinate sample collection, quality control of samples, and genotyping from all ACCOuNT sites;
Collect and integrate clinical data and molecular data of participants for all TCC activities, provide quality control, genomic and transcriptome analysis, and storage in an environment designed for the secure and compliant processing of controlled access data; and
Design and implement an African American portal and database of genomic and transcriptomic findings to support ongoing research within African American precision medicine.
Consortium core
In order for ACCOuNT to achieve its goals and to move the needle in African American PGx, we established the ACCOuNT Consortium Core. The core has engaged community leaders in the African American cardiovascular PGx research arena and has assumed responsibility for the administration of a pilot grant mechanism that stimulates new avenues of collaboration among community‐based participatory researchers in precision medicine. These pilots are unique in that they are projects, which are done in conjunction with community stakeholders.
The Consortium Core has three essential roles that together will ensure smooth, coordinated, and productive operations of our TCC. First, it provides a focal point for interactions among the several academic, governmental, and community‐based participants in this program. Second, it facilitates technical coordination among the academic health centers conducting the research. Third, it administers a pilot grant mechanism that stimulates new avenues of collaboration among all participants, by funding pilot projects led by community stakeholders.
There are currently four funded pilot projects that are summarized below.
-
Taking Effective Action (TEA), African American Pharmacogenomics and Precision Medicine Learning Event (APPLES). Principal investigator (PI): Rev. Dr Gertie Hurley, MBA
Goals: Create brochures and an informational video for African American audiences about the benefits of genetic testing and to participate in learning events in the DC area to promote cardiovascular PGx education.
-
University of Pittsburgh, Pittsburg PA. PI: Amber Johnson MD
Goals: Increase provider and patient understanding of PGx in a federally qualified health center in Chicago, enhancing therapeutic decision making in African American patients by increasing awareness of ethnic‐specific genetic variants that can impact pharmacokinetics and pharmacodynamics, increasing recruitment into the All of Us precision medicine research program.
-
C.W. Williams Health Center, Mecklenburg County, NC. PIs: Debra Weeks (C.W. Williams, CEO), Dr Amina Abubakar, and Dr Olivia Bentley (Pharmacogenetic Center of Excellence founders a part of the Amity Group Foundation)
Goals: Provide education and training for medical professionals, community members, pharmacists, and patients in a nine‐county area in North Carolina in pharmacies, community meetings, and clinic visits.
-
Virginia Commonwealth University (VCU). PI: Elvin Price, PharmD, PhD
Goals: Introduce cardiovascular PGx testing and counseling to older adults living in subsidized housing Richmond, VA.
Implementation core
The purpose of the implementation is to assure that the discovery and translational findings of ACCOuNT are implemented into practice as promptly, effectively, and broadly as possible. Of course, a necessary precondition of successful implementation of PGx is the discovery of knowledge and the translation of that knowledge into practice. Moreover, because the conditions of practice vary over time and institutional contexts, broad implementation of potentially valuable interventions requires a careful appreciation of the range of contexts in which knowledge is implemented. We will approach this challenge of implementing ACCOuNT findings by applying tools of implementation science and by drawing on existing approaches to PGx implementation. Our approach to understanding implementation issues is informed theoretically by the Reach Efficacy Adoption Implementation Maintenance framework, which examines these as a function of the reach, efficacy, adoption, implementation, adoption, and maintenance of interventions.23 In applying this framework to address barriers in the implementation of population‐specific PGx for African Americans, we seek to reflect contextual factors that may be of particular importance to PGx implementation for African Americans in the United States. These include, for example, the geographic concentration of many African Americans in urban areas, the higher rates of poverty, and, hence, reliance on Medicaid, and the increased rate of distrust of the healthcare system among African Americans.24 These provide important context for our choice of approaches and partners in implementation. In addition, the Implementation Core will develop and disseminate curriculum, workshops, and collaborative interactions for patients, physicians, and policy makers on the use of population‐inclusive precision medicine in clinical care.
Conclusions
PGx is aimed at identifying genetic variations that influence interindividual differences in drug response and adverse events as well as widespread clinical relevance. Its application promises to enable targeted drug administration, improve therapeutic outcomes, inform drug development, and improve our understanding of adverse drug reactions.
Are sequence variations derived from investigating patients of European ancestry adequate to predict drug response in African Americans, or are new population‐specific predictors needed? We anticipate that these studies will reveal novel SNP associations and gene regulation pathways. By specifically interrogating genetic variation in African American patients as well as building a large cohort of African Americans receiving GPS‐guided therapy, we can help accelerate the implementation of genetic findings into clinical practice. We are focused on closing the gap in precision medicine by undertaking clinical translational initiatives that will assess the impact of genetic biomarkers on clinical outcomes. This model will establish a framework in which basic science discoveries can pass quickly into clinical translational studies.
Funding
This work was supported by grants from the National Institutes of Health (U54MD010723).
Conflict of Interest
P.H.O. is named as a coinventor on a pending patent application for a Genomic Prescribing System. The other authors declare that they have no competing interests.
Acknowledgments
The authors would like to gratefully thank Mr Edmund Perera for assistance in figure illustration preparation in this manuscript.
References
- 1. Lander, E.S. et al Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 2. Lazarou, J. , Pomeranz, B.H. & Corey, P.N. Incidence of adverse drug reactions in hospitalized patients: a meta‐analysis of prospective studies. JAMA 279, 1200–1205 (1998). [DOI] [PubMed] [Google Scholar]
- 3. Venter, J.C. et al The sequence of the human genome. Science 291, 1304–1351 (2001). [DOI] [PubMed] [Google Scholar]
- 4. Gharani, N. et al The Coriell personalized medicine collaborative pharmacogenomics appraisal, evidence scoring and interpretation system. Genome Med. 5, 93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O'Donnell, P.H. et al The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin. Pharmacol. Ther. 92, 446–449 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Relling, M.V. & Klein, T.E. CPIC: clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. Clin. Pharmacol. Ther. 89, 464–467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. National Center for Veterans Analysis and Statistics [Internet]. <https://www.va.gov/vetdata/Veteran_Population.asp> Accessed August 8, 2018 (2016).
- 8. Bureau USC . 2014 National Population Projections Tables 2014 [updated May 9, 2017. The Population Projections Program produces projections of the United States resident population by age, sex, race, Hispanic origin, and nativity. The 2014 National Projections are based on the July 1, 3 population estimates, which are based on the 0 Census, and provide projections of the population for July 1 to July 1, 60. The projections were produced using a cohort‐component method and are based on assumptions about future births, deaths, and net international migration. The Census Bureau releases new national projections periodically] <https://www.census.gov/data/tables/2014/demo/popproj/2014-summary-tables.html>.
- 9. Bureau USC . QuickFacts: Chicago, Illinois 2017 [updated July 1, 2017. QuickFacts provides statistics for all states and counties, and for cities and towns with a population of 5000 or more]. <https://www.census.gov/quickfacts/chicagocityillinois>. Accessed 2017.
- 10. Bureau USC . QuickFacts: District of Columbia 2017 [updated July 1, 2017; cited 2017. QuickFacts provides statistics for all states and counties, and for cities and towns with a population of 5,000 or more]. <https://www.census.gov/quickfacts/fact/table/dc/PST045217#PST045217>.
- 11. Perera, M.A. et al Genetic variants associated with warfarin dose in African‐American individuals: a genome‐wide association study. Lancet 382, 790–796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cookson, W. , Liang, L. , Abecasis, G. , Moffatt, M. & Lathrop, M. Mapping complex disease traits with global gene expression. Nat. Rev. Genet. 10, 184–194 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang, Z.T. , Wang, Z. & Hu, Y.W. Possible roles of platelet‐derived microparticles in atherosclerosis. Atherosclerosis 248, 10–16 (2016). [DOI] [PubMed] [Google Scholar]
- 14. Freynhofer, M.K. , Bruno, V. , Wojta, J. & Huber, K. The role of platelets in athero‐thrombotic events. Curr. Pharm. Des. 18, 5197–5214 (2012). [DOI] [PubMed] [Google Scholar]
- 15. Nurden, A.T. Platelets, inflammation and tissue regeneration. Thromb. Haemost. 105 (suppl. 1), S13–S33 (2011). [DOI] [PubMed] [Google Scholar]
- 16. Taking Effective Action I. How Geonomics May Help You Receive a Better Prescription [mp4]. [culturally relevant interactive video tutorial on cardiovascular health, pharmacogenomics, precision medicine and African American involvement in medical research]. <https://precisionmedicine4all.com/research-projects/pilot-programs/> (2018)
- 17. O'Donnell, P.H. et al Adoption of a clinical pharmacogenomics implementation program during outpatient care–initial results of the University of Chicago “1,200 Patients Project”. Am. J. Med. Genet. C Semin. Med. Genet. 166C, 68–75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson, J.A. et al Clinical pharmacogenetics implementation consortium (CPIC) guideline for pharmacogenetics‐guided warfarin dosing: 2017 update. Clin. Pharmacol. Ther. 102, 397–404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gage, B.F. et al Effect of genotype‐guided warfarin dosing on clinical events and anticoagulation control among patients undergoing hip or knee arthroplasty: the GIFT randomized clinical trial. JAMA 318, 1115–1124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cavallari, L.H. & Perera, M.A. The future of warfarin pharmacogenetics in under‐represented minority groups. Future Cardiol. 8, 563–576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaufman, A.L. et al Evidence for clinical implementation of pharmacogenomics in cardiac drugs. Mayo Clin. Proc. 90, 716–729 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rasmussen‐Torvik, L.J. et al Design and anticipated outcomes of the eMERGE‐PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin. Pharmacol. Ther. 96, 482–489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Glasgow, R.E. , Vogt, T.M. & Boles, S.M. Evaluating the public health impact of health promotion interventions: the RE‐AIM framework. Am. J. Public Health 89, 1322–1327 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Halbert, C.H. , Armstrong, K. , Gandy, O.H. Jr & Shaker, L. Racial differences in trust in health care providers. Arch. Intern. Med. 166, 896–901 (2006). [DOI] [PubMed] [Google Scholar]


