Abstract
Background: Chronic kidney disease (CKD) is associated with a decreased intestinal barrier function, causing bacterial translocation over the intestinal wall and triggering a systemic inflammatory response. Butyrate, a short-chain fatty acid produced by certain bacterial strains, is considered instrumental to keep the intestinal barrier intact. There are indications that a decreased amount of these specific bacterial species is part of the cause of the decreased intestinal barrier function in CKD. The aim of this study is (i) to determine if Dutch patients with end-stage renal disease (ESRD) have a decreased amount of butyrate-producing species and butyrate-producing capacity and (ii) whether this correlates with systemic inflammation.
Methods: We used qPCR to evaluate the most abundant butyrate-producing species F. prauznitzii, E. rectale and Roseburia spp. and the BCoAT gene, which reflects the butyrogenic capacity of the intestinal microbiota. Fecal samples were collected from healthy kidney donors (n=15), preemptive renal transplant recipients (n=4) and dialysis patients (n=31). Markers of inflammation (CRP and IL-6) and intestinal permeability (D-lactate) were measured in plasma.
Results: Patients with ESRD did not have a significantly decreased amount F. prauznitzii, E. rectale and Roseburia spp. or the BCoAT gene. Neither was there a significant correlation with CRP, IL-6 or D-lactate. On the individual level, there were some patients with decreased BCoAT levels and increased levels of CRP, IL-6 and D-lactate.
Conclusions: Patients with ESRD do not have a decreased amount of the most abundant butyrate-producing species nor a decreased butyrate-producing capacity.
Keywords: renal failure, intestinal barrier function, butyrate, intestinal microbiota, inflammation
Introduction
Despite the improvements that have been made in treatment modalities for patients with chronic kidney disease (CKD), morbidity and mortality rates remain high. Cardiovascular disease is the leading cause of morbidity and mortality and accounts for 50% of deaths in CKD patients regardless of biological age.1 This (disproportional) rate of cardiovascular mortality is hypothesized to stem from the presence of specific nontraditional risk factors that accompany the loss of renal function, such as the alterations in the gut that are seen in CKD.2
Numerous studies have pointed out intestinal alterations in CKD, including changes of the microbes, a decreased intestinal barrier function and inflammation of the intestinal wall.3,4 This decreased intestinal barrier function may lead to bacterial translocation and endotoxemia, which is known to potentiate systemic inflammation and atherosclerosis.5 Systemic inflammation is associated with progression of CKD and is a mediator of a variety of complications in CKD such as anemia, cognitive impairment, insulin resistance and cardiovascular disease,4,6–8 which points out the importance of a healthy intestinal barrier. Microbes appear also to have essential functions for the maintenance of this intestinal barrier function. One of these functions is the production of butyrate.
Butyrate, a short-chain fatty acid, is an important energy source for the colonocytes. It is synthesized from butyryl-CoA by the enzymes butyrate kinase and butyryl-CA: acetate CoA-transferase (BCoAT). Since the majority of human colonic butyrate-producing species use BCoAT for the last step of butyrate formation, this is considered as the dominant system for butyrate production.9 Butyrate is known to promote cell differentiation, suppresses colonic inflammation and facilitate the transcription of Claudine-1, which enhances the tight-junction assembly.3,10 Therefore, butyrate might attenuate bacterial translocation and enhance the gut barrier function. It is found in little amounts in foods such as parmesan cheese and butter; nonetheless, intestinal levels are mainly dependent on bacterial fermentation of dietary fiber by specific bacterial species.11 Butyrate can also be administrated, which appeared to be beneficial in a variety of gastrointestinal conditions such as diverticulitis and ulcerative colitis.12,13
A decreased amount of butyrate could be part of the cause of the decreased intestinal barrier function and thereby the systemic inflammation and high rate of cardiovascular disease in CKD. Studies in the Chinese population have pointed out a decreased amount of specific butyrate-producing species in different stages of CKD (non-dialysis), which also appeared to be correlated with inflammatory markers and was even associated with the progression of CKD.14,15 Thus, butyrate could be an interesting potential therapeutic target to decrease the systemic inflammatory response. However, literature on butyrate levels in CKD is scarce; the current available evidence is only based on the previously mentioned Chinese studies and there are no data on the western CKD population, which hinders the translation to actual intervention studies. In addition to this, the actual ability from the intestinal microbiota to synthesize butyrate has never been evaluated in CKD.
With our study, we aim to:
Evaluate the presence of the most abundant butyrate-producing species in the Dutch population with end-stage renal disease (ESRD), which is the most severe form of CKD, and healthy controls.
Evaluate whether the abundance of these butyrate-producing species correlates with inflammatory markers and the intestinal barrier function.
Measure the actual capacity of the intestinal microbiota to produce butyrate in these patients.
Materials and methods
Patients and samples
We enrolled healthy kidney donors, preemptive renal transplant recipients (transplant before the need to start dialysis) and patients undergoing dialysis treatment. Participants had to be able to collect stool samples to be included in this study. Participants who had received antibiotic treatment within the past 6 months were excluded; otherwise, there were no exclusion criteria. This study was conducted according to the Declaration of Helsinki and all participants gave written informed consent prior to enrolment.
From the kidney donors and the preemptive renal transplant recipients, samples were obtained one day before the scheduled renal transplantation. From each participant, blood and fecal samples were obtained and stored in the Biobank Renal Diseases in the Amsterdam UMC. This Biobank has been approved by the Biobank Ethical Committee of the Amsterdam UMC. Participants were asked to collect the fecal samples at home or in the Amsterdam UMC. Samples were kept at 7°C and were transported to the hospital in a cooler bag, after which they were immediately stored at −80°C. In addition to this, participants were asked questions about the use of antibiotics, prebiotics and dietary habits.
After the initial analysis of our patient samples, we performed an additional analysis of fecal samples from 7 young healthy volunteers at two time points (0 and at 3 months) to analyze the stability of the measured bacterial species and the butyrate-producing capacity of the intestinal microbiota.
DNA extraction
DNA was extracted from approximately 200 mg feces by using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany). Samples were kept frozen until DNA extraction by manufacturer manual for extraction of bacterial DNA was started. After extraction, DNA concentration was determined by using the NanoDrop photospectrometer (model ND2000, Thermo Fisher, Waltham, MA USA), after which the DNA was stored at 4°C until further processing.
qPCR
We used qPCR to quantify the total amount of bacterial DNA (16S), the three most abundant butyrate-producing species: F. prausnitzii, E. rectale, Roseburia spp. and the BCoAT gene (reflecting the ability of the intestinal microbiota to produce butyrate) in the fecal samples of each of the participants. For each of these outcome measures, a qPCR was set up by using the standard curve method for absolute quantification. Primers, listed in supplemental Table S1, were synthesized by Sigma Aldrich. Amplification of the product for standard curve design was done at the BioRad 48 well PCR machine (S1000 thermal cycler, Bio-Rad, Hercules, CA, USA) qPCR assays were performed in a 96-well optical plate on the Step One Plus Cycler (StepOnePlus, Applied Biosystems, Foster City, CA, USA). All assays were carried out in triplicate. The reaction mixture consisted of 2 ng DNA, 5 µM of each primer, SensiFast SYBR No ROX Kit (BioLine, London, UK) SybrGreen and PCR Grade water (Roche, Basel, Switzerland). The copy number of target DNA was determined by serially diluting standards running on the same plate. Data was analyzed using StepOne Software 2.1 (Applied Biosystems, Foster City, CA, USA).
Table S1.
Primers used in qPCR
| Target Bacteria | Primer | Sequence (5ʹto 3ʹ) | Product (bp) | Reference |
|---|---|---|---|---|
| Universal Bacteria | Uni-F | GTGSTGCAYGGYYGTCGTCA | 147 | [1] |
| Uni-R | ACGTCRTCCMCNCCTTCCTC | |||
| F. Prausnitzii | FPR-2F | GGAGGAAGAAGGTCTTCGG | 248 | [2] |
| Fprau645mR | AATTCCGCCTACCTCTGCACT | |||
| E. Rectale | Eubrect-F | AAGGGAAGCAACGCTGTGAA | 200 | [3] |
| Eubrect-R | TCGGTTAGGTCACTGGCTTC | |||
| Roseburia spp | RosF | TACTGCATTGGAAACTGTCG | 230 | [4] |
| RosR | CGGCACCGAAGAGCAAT | |||
| BCOAT gene | BCoATscrF BCoATscrR |
GCIGAICATTTCACITGGAAYWSITGGCAYATG CCTGCCTTTGCAATRTCIACRAANGC |
530 | [5] |
Biochemical analysis
CRP, IL-6 (inflammatory markers) and D-lactate (reflecting the intestinal barrier function) were measured in plasma. An automatic and validated immunoturbidimetric assay was performed to determine CRP levels by using a Roche Cobas C8000 with a c702 module (Roche Diagnostics, Risch-Rotkreuz, Zwitserland). IL-6 was measured by using an ELISA assay. Plasma samples were measured in duplicates by using the Sanquin PeliKine human IL6 ELISA with additional reagents from the PeliKine Toolset (Sanquin, Amsterdam, the Netherlands). Absorption was determined by ELISA plate reader (BioRad, model 680). Quantification of the D form of lactic acid in the plasma samples was performed by using the highly sensitive method of ultra-performance liquid chromatography-electrospray ionization-tandem mass spectrometry, as described before.16 The detection limit of this assay was 0.06 mmol/L, and D-lactate values below 0.06 are displayed as 0. In healthy subjects, D-lactate levels are below this detection limit.
Statistical analysis
All data analysis was conducted using IBM SPSS Statistics 23; graphs and figures were created using Graphpad Prism 7. The assumption of normality of each of the variable was assessed both visually and trough normality tests, of which the skewness, kurtosis and Shapiro–Wilk test were performed. For positive right-skewed variables, a log transformation was checked for improvement. We used parametric tests in case of normal distributed variables or nonparametric tests in case of a persisting non-normal distribution or categorical variables.
ESRD vs healthy donors
A one-tailed independent sample T-test or the Mann–Whitney U test was used to compare copy numbers of each of the bacterial species, the BCoAT gene, CRP, IL-6 and D-lactate levels between the dialysis group and the healthy kidney donors. Due to the small number of preemptive renal transplant recipients, no statistical comparison could be made between the preemptive renal transplant recipients and the other study groups.
(II) Correlation with inflammatory markers
Depending on a normal or non-normal distribution, the Pearson (r) or the Spearman (ρ) coefficient was calculated for each of the butyrate-producing species, the BCoAT gene and CRP, IL-6 and D-lactate levels to evaluate a possible correlation. In this analysis, all study participants were included (n=50). Sensitivity analysis was performed for the dialysis group. Since in this analysis multiple correlations were tested, p-values <0.007 were considered as statistically significant after Bonferroni correction.17
In an additional analysis, both the correlation (Pearson or Spearman) between each of the butyrate-producing species and the BCoAT gene were also evaluated, as well as the correlation between CRP, IL-6 and D-lactate.
(III) Stability over time
The Wilcoxon signed rank test was used to compare the copy numbers of each of the butyrate-producing species and the BCoAT gene between the two time points.
Ethics approval and informed consent
All participants gave informed consent for participation in the Biobank Renal Diseases. This Biobank has been approved by the Biobank Ethical Committee of the Amsterdam UMC.
Results
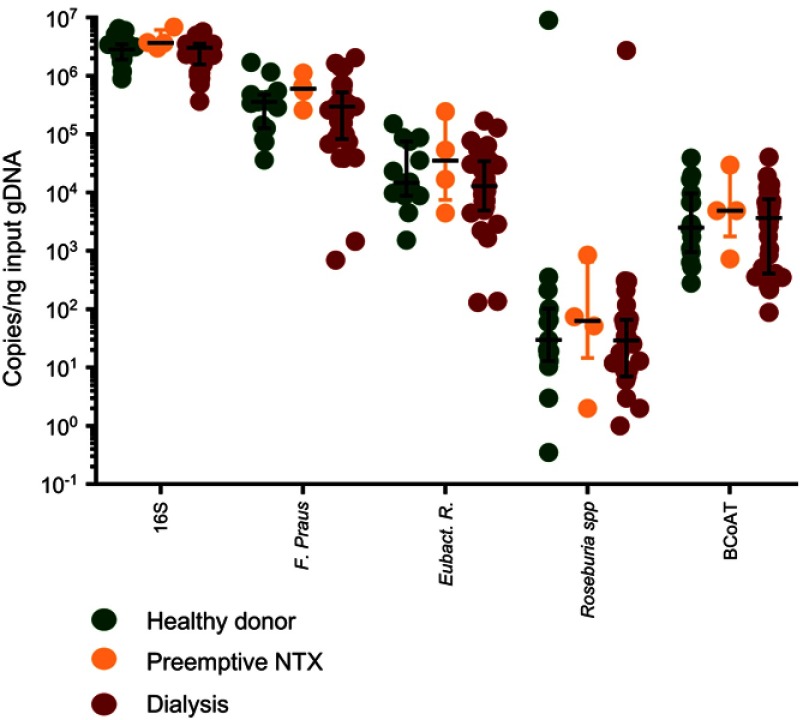
We included 15 healthy kidney donors, 4 preemptive renal transplant recipients and 31 dialysis patients. Demographic characteristics are displayed in Table 1. Copy numbers of the total amount of bacterial DNA (16S), each of the butyrate-producing species and the BCoAT gene are expressed per ng input DNA and are displayed in Figure 1. Median copy numbers/ng input DNA for each group and for each of the bacterial species and the BCoAT gene are displayed in Table 2. There were no significant differences between the groups for either of the butyrate-producing species or considering the butyrate-producing capacity.
Table 1.
Baseline characteristics
| Donors (n=15) | Preemptive (n=4) | Dialysis (n=31) | |
|---|---|---|---|
| Male sex, n (%) | 53.3% | 100% | 67.8% |
| Age, mean (SD) | 62.1 (7.6) | 51.8 (12.3) | 55.0 (15.5) |
| Type of renal disease - Glomerulopathy - Reno vascular - ADPKD - Anatomical abnormalities - Other |
- |
100% |
28.9% 36.8% 10.5% 10.5% 13.2% |
| Time on dialysis (months), mean (SD) | - | - | 49.3 (55.0) |
| Ethnicity (%) -Caucasian -African -Mediterranean -Asian -Surinamese |
93.3% 6.7% 0% 0% 0% |
75% 0% 0% 25% 0% |
45.2% 32.3% 9.7% 6.5% 6.5% |
| History of CVD (%) | 0% | 0% | 58% |
| Diabetes (%) | 0% | 25% | 16% |
| Vegetarian diet (%) | 7.7% | 0% | 0% |
| Probiotic use (%) | 14.3% | 0% | 18.5% |
| PPI use (%) | 13.3% | 50% | 58% |
| Phosphate binders (%) | 0% | 100% | 90% |
| Potassium binders (%) | 0% | 25% | 16% |
Abbreviations: SD, standard deviation; ADPKD, autosomal dominant polycystic kidney disease; CVD, cardiovascular disease; PPI, proton pump inhibitor.
Figure 1.
Butyrate-producing species and the butyrate-producing capacity in healthy donors, preemptive renal transplant recipients and dialysis patients. Copy numbers/nanogram input gDNA of the total amount of bacterial DNA (16S), the three most abundant butyrate-producing species and the BCoAT gene (median + IQR).
Abbreviation: NTX = kidney transplant recipient.
Table 2.
Butyrate-producing species, inflammatory markers and the intestinal barrier function
| 16S copies/ng input gDNA | F. prausnitzii copies/ng input gDNA | E rectale copies/ng input gDNA | Roseburia spp. copies/ng input gDNA | BCoAT copies/ng input gDNA | CRP mg/l | IL-6 pg/ml | D-lactate mmol/l | |
|---|---|---|---|---|---|---|---|---|
| Healthy controls (n=15) | 2,828,150 (1,904,217–3,465,626) | 356,986 (127,028–478,816) | 14,625 (8845 −75 283) | 30 (13–102) | 2493 (949–9609) | 0.8 (0.5–2.2) | 1.4 (1.3–1.6) | <0.1 (0–0) |
| Preemptive (n=4) | 3,680,962 (3,166,966–6,10,362) | 600,164 (331,419–1,003,237) | 35,026 (7532–196,945) | 63 (15–656) | 4862 (1761–23,368) | 0.9 (0.1–35.8) | 2.5 (1.5–11.5) | <0.1 (0–0.1) |
| Dialysis group (n=31) | 3,027,599 (1 576,133–3,544,898) | 297,402 (82,416–524,638) | 12,896 (4928–34,306) | 29 (7–66) | 3656 (410–7684) | 2.2 (0.9–4.2)* | 2.7 (1.9–3.8)* | <0.1 (0–0.13)* |
Notes: Median (Interquartile range) copy numbers 16S, the measured butyrate-producing species and the BCoAT gene and levels of inflammatory markers and D-lactate. For comparison between the healthy controls and dialysis group, either the one-tailed independent sample T-test or the Mann–Whitney U test was used: *p<0.05.
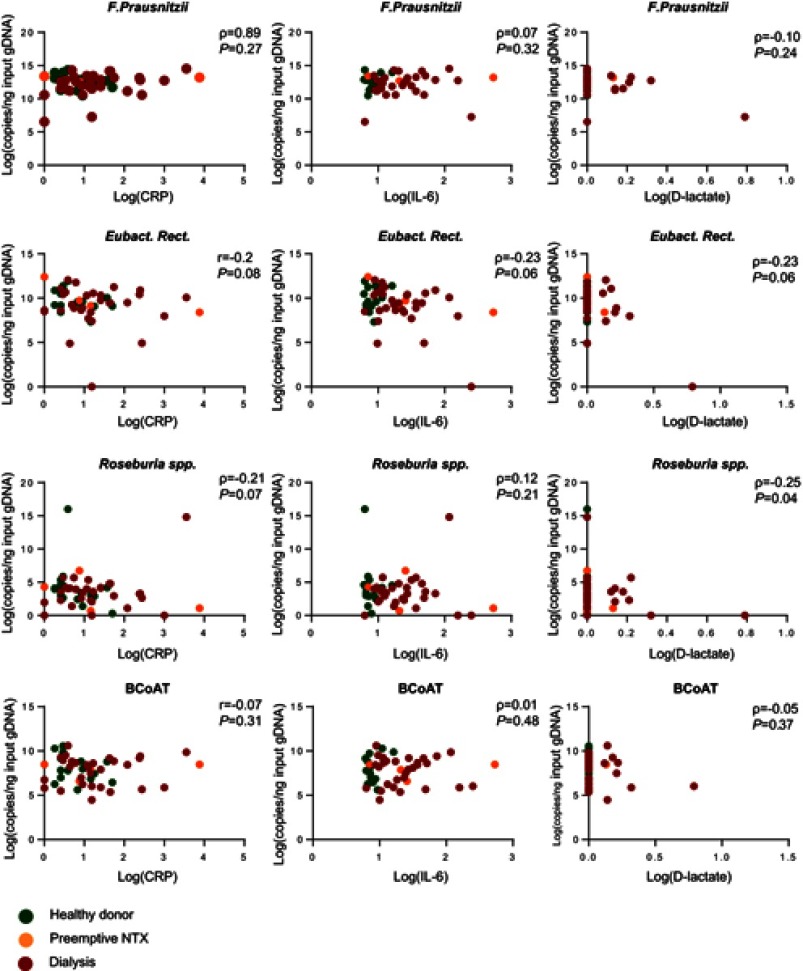
As expected, CRP, IL-6 and D-lactate were elevated in the dialysis group compared to the healthy kidney donors (p<0.05; p<0.001; p<0.05, respectively). There was no statistically significant correlation between either of the butyrate-producing species or the BCoAT gene and CRP, IL-6 or D-lactate (Table 3; Figure 2). The weak negative correlation between Roseburia spp. and D-lactate (ρ=−0.25, p=0.04) was not significant after Bonferroni correction.
Table 3.
Correlation between butyrate-producing species, the butyrate-producing capacity, inflammatory markers and the intestinal barrier function
| Total group (n=50) | Dialysis patients (n=31) | |||||
|---|---|---|---|---|---|---|
| CRP | IL-6 | D-lactate | CRP | IL-6 | D-lactate | |
| F. prausnitzii |
ρ=0.89 p=0.27 |
ρ=0.07 p=0.32 |
ρ=−0.10 p=0.24 |
ρ=0.34 p=0.03* |
ρ=0.23 p=0.10 |
ρ=−0.12 p=0.25 |
| E. rectale |
r=−0.20 p=0.08 |
ρ=−0.23 p=0.06 |
ρ=−0.23 p=0.06 |
r=−0.02 p=0.46 |
ρ=−0.2 p=0.14 |
ρ=−0.18 p=0.16 |
| Roseburia spp. |
ρ= −0.21 p=0.07 |
ρ=0.12 p=0.21 |
ρ=−0.25 p=0.04* |
ρ=0.01 p=0.48 |
ρ=−0.05 p=0.39 |
ρ=−0.21 p=0.13 |
| BCoAT |
r=−0.07 p=0.31 |
ρ=0.01 p=0.48 |
ρ=−0.05 p=0.37 |
r=0.06 p=0.38 |
ρ=0.02 p=0.47 |
ρ=−0.06 p=0.38 |
Abbreviations: r, Pearson coefficient; ρ, Spearman coefficient. *p<0.05. None of the parameters were statistically significantly different after Bonferroni correction.
Figure 2.
Correlation between butyrate-producing species, the BCoAT gene, inflammatory markers and D-lactate.
Abbreviations: r, Pearson coefficient; ρ, Spearman coefficient; log, natural logarithm +1; NTX, kidney transplant recipient.
Sensitivity analysis for only the dialysis group did reveal a positive correlation between F. prausnitzii, and CRP (ρ=0.34, p=0.03); however, this was not significant after Bonferroni correction (Table 3). There were 7 dialysis patients with values below the 25th percentile of the BCoAT gene. These patients did not share any specific patient characteristics (Table 4). The patients with elevated levels of D-lactate did not share specific patients characteristics either (Table S2).
Table 4.
Baseline characteristics of patients with a low butyrogenic capacity
| Age years | M/F | Ethn | Renal disease | CVD | DM | Time on dialysis months | Veg diet | PPI | PO4 binder | K binder | F. prausnitzii copies/ng input gDNA | E. rectale copies/ng input gDNA | Roseburia spp. copies/ng input gDNA | BCoAT copies/ng input gDNA | CRPmg/L | IL-6 pg/mL | D-lactate mmol/L |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 38 | M | Black | Reno-vascular | Yes | No | 12 | No | Yes | Yes | Yes | 99,178 | 1646 | 7 | 89 | 2.3 | 1.7 | 0.2 |
| 63 | M | Black | Glomer-ulopathy | Yes | No | 36 | No | Yes | Yes | No | 297,402 | 5540 | 9 | 243 | 0.5 | 1.8 | <0.1 |
| 62 | M | Asian | FSGS | No | No | 10 | No | Yes | Yes | No | 349,171 | 2883 | 0 | 353 | 19 | 8.0 | 0.4 |
| 73 | M | Black | Reno-vascular | Yes | No | 24 | No | No | Yes | No | 145,251 | 17,381 | 61 | 360 | 3.1 | 3.4 | <0.1 |
| 66 | F | Cauc | Hyper-oxaluria | No | No | 112 | No | No | Yes | No | 1450 | 0 | 0 | 409 | 2.3 | 10.1 | 1.2 |
| 40 | M | Cauc | Dense deposit disease | No | No | 7 | No | Yes | Yes | No | 692 | 4928 | 0 | 338 | <0.3 | 1.22 | <0.1 |
| 28 | M | Asian | Glomerulo-pathy | No | No | 96 | No | No | No | No | 1,417,213 | 12,896 | 118 | 217 | 4.2 | 2.7 | <0.1 |
Note: Characteristics of patients with low (<25th percentile) values of the BCoAT gene.
Abbreviations: M, male; F, female, Ethn, ethnicity; FSGS, focal segmental glomerulosclerosis; veg diet, vegetarian diet; CVD, cardiovascular disease; DM, diabetes mellitus; PPI, proton pump inhibitor; P04, phosphate; K, potassium; Cauc, Caucasian.
Table S2.
Baseline characteristics of patients with a decreased intestinal barrier function
| Age years | M/F | Ethn | Renal disease | CVD | DM | Time on dialysis months | Veg diet | PPI | PO4-binder | K – binder | F. praus copies/ng input gDNA | Eubact. Rect. copies/ng input gDNA | Roseburia spp. copies/ng input gDNA | BCoAT copies/ng input gDNA | CRP mg/l | IL-6 pg/ml | D-lactate mmol/l |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 69 | M | Cauc | Glomerulo-pathy | No | No | N.A. | No | No | Yes | No | 547,389 | 4471 | 2 | 4862 | 47.3 | 14.3 | 0.14 |
| 62 | M | Asian | FSGS | No | No | 10 | No | Yes | Yes | No | 349,171 | 2883 | 0 | 353 | 19 | 8.0 | 0.38 |
| 40 | F | Cauc | Glomerulo-phaty | No | No | 24 | No | Yes | Yes | No | 258,156 | 4434 | 9 | 1778 | 2.2 | 3.0 | 0.23 |
| 39 | M | Black | Reno-vascular | Yes | No | 72 | No | No | No | No | 82,416 | 168,575 | 56 | 40,568 | 0.8 | 1.6 | 0.15 |
| 62 | M | Med | ADPKD | No | No | 48 | No | Yes | Yes | No | 709,905 | 38,521 | 34 | 5530 | 2.4 | 4.6 | 0.13 |
| 77 | F | Caus | Reno-vascular | Yes | No | 96 | No | Yes | Yes | No | 578,255 | 7150 | 298 | 5873 | 1.1 | 3.8 | 0.24 |
| 66 | M | Cauc | Hyper-oxaluria | No | No | 112 | No | No | Yes | No | 1450 | 0 | 0 | 409 | 2.3 | 10.1 | 1.2 |
| 61 | F | Cauc | ADPKD | No | No | 24 | No | No | Yes | Yes | 103,497 | 64,107 | 34 | 10,558 | 0.5 | 1.9 | 0.20 |
| 38 | M | Black | Reno-vascular | Yes | No | 12 | No | Yes | Yes | Yes | 99,178 | 1646 | 7 | 89 | 2.3 | 1.7 | 0.1 |
Note: Characteristics of patients with elevated D-lactate levels.
Abbreviations : M, male; F, female, Ethn, ethnicity; Cauc, Caucasian; FSGS, focal segmental glomerulosclerosis; ADPKD, autosomal dominant polycystic kidney disease; veg diet, vegetarian diet; CVD, cardiovascular disease; DM, diabetes mellitus; PPI, proton pump inhibitor; P04, phosphate; K, potassium; Med, Mediterranean; N.A., not applicable.
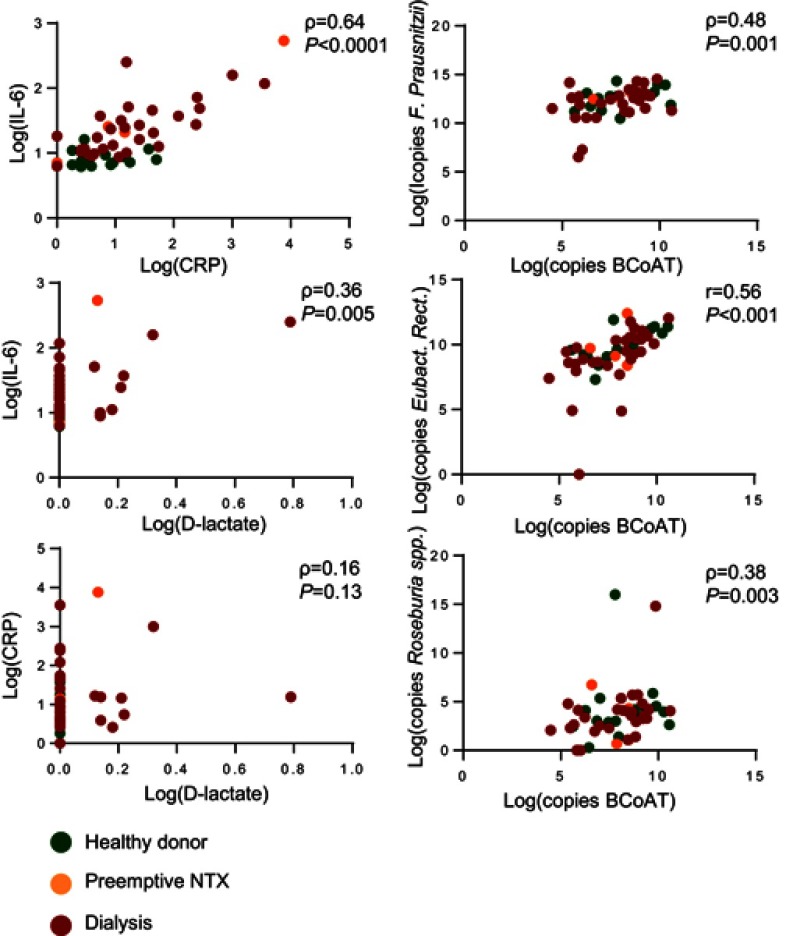
As expected there was a significant positive correlation between F. prausnitzii, E. rectale, Roseburia spp. and the BCoAT gene (ρ=0.48 p=0.001, r=0.56 p=<0.0001, ρ=0,38 p=0.003, respectively) and between CRP and IL-6 (ρ=0.64, p=<0.0001), as displayed in supplemental Figure 1. There was a weak positive correlation between D-lactate and IL-6 (ρ=0.36, p=0.005), but not between D-lactate and CRP (ρ=0.16, p=0.13) (Figure S1).
Figure S1.
Correlation between outcomes measures. The correlation between CRP, IL-6 and D-lactate and the correlation between each of the butyrate producing species and the BCoAT gene.
Abbreviations: r, Pearson coefficient; ρ, Spearman coefficient; log, natural logarithm +1; NTX, kidney transplant recipient.
Additional longitudinal analysis in healthy volunteers
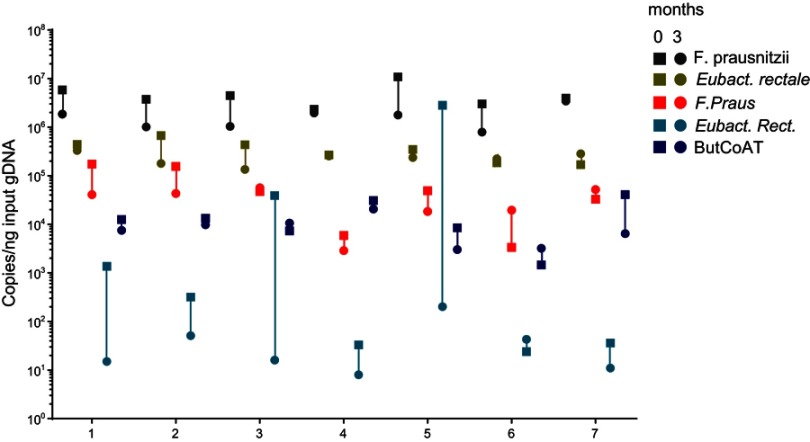
We evaluated 7 young and healthy individuals at two time points: 0 and 3 months. Table 5 shows the demographics of these healthy volunteers. Figure 3 shows the copy numbers of each of the bacterial species and the BCoAT gene at the two different time points.
Table 5.
Demographics healthy volunteers included in longitudinal analysis
| Healthy volunteers (n=7) | |
|---|---|
| Male sex, n (%) | 42% |
| Age, mean (SD) | 30 (3.42) |
| Ethnicity (%) -Caucasian |
100% |
| History of CVD (%) | 0% |
| Diabetes (%) | 0% |
| Vegetarian diet (%) | 14.3% |
| Use of PPI (%) | 0% |
| Antibiotics (%) <6 months | 0% |
Abbreviations: SD, standard deviation; CVD, cardiovascular disease; PPI, proton pump inhibitor.
Figure 3.
Longitudinal analysis of the most abundant butyrate-producing species and the BCoAT gene in healthy volunteers. Longitudinal analysis of 7 healthy volunteers, numbered 1–7. Copy numbers/nanogram input gDNA of the total amount of bacterial DNA (16S), the three most abundant butyrate-producing species and the BCoAT gene. Squares represent the first measured time point for each of the individuals, circles the second time point three months later.
Copy numbers of the BCoAT gene, F. prausnitzii and E. rectale were relatively stable, as there was no significant difference between the two time points, whilst copy numbers of Roseburia spp. did fluctuate (p=0.03).
Discussion
In our study, we found no difference in the amount of the measured butyrate-producing species between healthy kidney donors, preemptive renal transplant recipients and dialysis patients, neither was there a decreased capacity to produce butyrate in either of the patient groups. There was no correlation between the butyrate-producing species or the butyrate-producing capacity and inflammatory markers or D-lactate on a population level. As expected, dialysis patients did have increased levels of the inflammatory markers and D-lactate.
Our results of the Dutch ESRD (CKD stage V) patients are in contrast with the decreased amount of butyrate-producing species that has been reported in Chinese patients with different stages of CKD (stage I–V).14,15 There are several explanations for these conflicting results. Instead of per gram of isolated DNA, the results from the Chinese studies were expressed per gram of stool, an outcome measure that is easily influenced by factors such as the effectiveness of the DNA-isolation step, but also by the consistency of the stool sample, which especially in CKD patients can differ because of the different recommendations about restrictions in fluid intake. Besides stool consistency, the composition of the intestinal microbiota is known to be influenced by many factors such as dietary habits, ethnicity, smoking and medication (among others antibiotics and proton pump inhibitors).18–21 In this prospect, it is difficult to compare the Chinese population to the Dutch population as there are many differences considering environmental, behavioral and genetic factors, which may have influenced test results. Our study is, to the best of our knowledge, the first study evaluating butyrate-producing species in the western CKD population. It is unclear whether demographic characteristics explain the difference in test results between the Chinese and Dutch patient population.
The strength of our study is that this is the first time that the ability of the intestinal microbiota to produce butyrate has been evaluated in CKD patients. Quantification of the BCoAT gene has been pointed out as a valid tool to evaluate the butyrate-producing capacity of the intestinal microbiota and thus all butyrate-producing species.22 The additional analysis we performed in healthy volunteers also points out that the BCoAT gene is a robust outcome measure, as it appears to be relatively stable over time. Our results are reported in copy numbers per nanogram input gDNA, which is less easily influenced by technical issues than the expression per gram of feces. We used D-lactate to evaluate the intestinal barrier function. D-lactate is the metabolic product of bacteria, and in normal conditions, its serum level is very low, below our detection limit of 0.06 mmol/L.23,24 When the intestinal mucosa is damaged, the intestinal permeability increases and serum D-lactate levels will increase. Therefore, D-lactate is considered as a sensitive marker assessing the intestinal barrier function.25,26 To the best of our knowledge, D-lactate is currently the best available method to evaluate the intestinal barrier function in patients with CKD. There are no validated methods in CKD, and most other markers evaluating the intestinal barrier function are influenced by renal function.27 It is assumed that serum D-lactate levels are not influenced by renal function since studies have shown that D-lactate is metabolized in peritoneal dialysis patients without urinary production.28,29
Our study also has some limitations. Since we specifically focused on the intestinal butyrate production, we quantified the most abundant butyrate-producing species and the butyrogenic capacity of the intestinal microbiota. Quantitative PCR is a valid tool to measure actual copy numbers and to answer our research question. However, it is impossible to speculate about the composition of the intestinal microbiome in ESRD and the influence or competition with other bacterial species. In addition to this, all samples were collected at one time point, which we do consider as a limitation considering the interpretation of results from Roseburia spp. Furthermore, the included dialysis population was heterogeneous, from different ethnical backgrounds, with a wide variety of comorbidities and medication such as proton pump inhibitors. In the recent years, several studies have pointed out that a variety of medication, including these proton pump inhibitors, profoundly alter the intestinal microbiota.30 In addition to this, the most common comorbidities among dialysis patients, including diabetes and cardiovascular disease, are also associated with alterations in the intestinal microbiome.31,32 Other factors that might influence the intestinal microbiota are the dietary restrictions imposed to these patients and also the dialysis procedure itself. Intradialytic hypotension, a common complication in hemodialysis patients, is known to result in regional hypo-perfusion that can also affect the gut mucosa.33,34 It is unknown whether this disturbance of the homeostatic environment also affects the composition of the intestinal microbiome and butyrate-producing species. Accordingly, even though our patient population does truly reflect the general dialysis population, many factors may influence intestinal health in dialysis patients, and in this study, it is impossible to speculate about the influence of all the separate factors.
We included only 4 preemptive renal transplant recipients (ESRD not on dialysis). These patients did not start with renal replacement therapy yet, and thus had a less severe form of ESRD. As we did not find a significant difference between the dialysis patients and the healthy controls and results from these 4 patients were comparable to the dialysis patients, we do not expect that including more preemptive renal transplant recipients will change our results and conclusions.
Butyrate production
F. prausnitzii, E. rectale and Roseburia spp. are the butyrate-producing species with the highest relative abundance.35 Nevertheless, it is unknown what the exact contribution of each of these butyrate-producing species is to the total butyrate production. Our data suggest that whilst F. prausnitzii is by far the most abundant butyrate-producing species, E. rectale may deliver a greater contribution to the total butyrate production, as the correlation between the BCoAT gene and E. rectale was higher than the correlation between the BCoAT gene and F. prausnitzii. Thus, calculating the exact production of butyrate may be far more complex than the sum of the most abundant butyrate producing species.
Individual results
On a group level, no significant differences between healthy controls and ESRD patients in either of the outcome measures reflecting the intestinal butyrate production and no correlation between these outcome measures and the markers reflecting intestinal permeability or the systemic inflammatory response were found. On an individual level, there were patients with a decreased amount of the butyrate-producing species, a decreased capacity to produce butyrate and elevated levels of CRP, IL-6 and D-lactate. These patients did not share specific patient characteristics. Since in our patient group only a few patients had increased inflammatory markers and/or increased D-lactate levels, it is possible that our sample size was too small to detect a correlation between butyrate production and inflammatory markers and the intestinal barrier function.
Thus, our results point out that a decreased amount of butyrate may be a realistic concern in some but not all ESRD patients, but a clear subgroup cannot be defined. Additionally, not all dialysis patients appear to have a decreased intestinal barrier function and not all patients have increased inflammatory markers. Further research is necessary to determine which factors influence the intestinal butyrate production on the individual level but also whether butyrate administration may be a possible therapeutic target to prevent and treat the systemic inflammatory response specifically in these patients.
Conclusion
In conclusion, we did not find a significant difference in the amount of the three most abundant butyrate-producing species or in the butyrate-producing capacity between patients with ESRD and healthy kidney donors, neither was there a correlation with CRP, IL-6 or D-lactate. Among the dialysis patients, the variability was high and we did identify patients with low amounts of the butyrate-producing species, a low butyrogenic capacity and high inflammatory markers or D-lactate. Further research is warranted to determine which factors influence the intestinal butyrate production on individual level, especially in hemodialysis patients.
Acknowledgments
We would like to thank Nelly van der Bom for her help with processing of the samples and Marit van Sandwijk for her help with the inclusion of patients. Samples in this study have been stored in the Biobank Renal Diseases, this Biobank is funded by Astellas Pharma.
Abbreviation list
CKD, chronic kidney disease; ESRD, end-stage renal disease; SD, standard deviation; ADPKD, autosomal dominant polycystic kidney disease; CVD, cardiovascular disease; PPI, proton pump inhibitor; r, Pearson coefficient; ρ, Spearman coefficient; M, male; F, female, Ethn, ethnicity; FSGS, focal segmental glomerulosclerosis; veg diet, vegetarian diet; CVD, cardiovascular disease; DM, diabetes mellitus; PPI, proton pump inhibitor; P04, phosphate; K, potassium; Cauc, Caucasian.
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
Frederike J Bemelman reports grants from Astellas, during the conduct of the study. The authors report no other conflicts of interest in this work.
Supplementary materials
References
- 1.Maeda H, Fujimoto C, Haruki Y, et al. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol Med Microbiol. 2003;39(1):81–86. [DOI] [PubMed] [Google Scholar]
- 2.Ramirez-Farias C, Slezak K, Fuller Z, et al. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr. 2009;101(4):541–550. [DOI] [PubMed] [Google Scholar]
- 3.Balamurugan R, Rajendiran E, George S, et al. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. 2008;23(8 Pt 1):1298–1303. [DOI] [PubMed] [Google Scholar]
- 4.Larsen N, Vogensen FK, van den Berg FW, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5(2):e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louis P, Flint HJ. Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol. 2007;73(6):2009–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Shastri S, Sarnak MJ. Cardiovascular disease and CKD: core curriculum 2010. Am J Kidney Dis. 2010;56(2):399–417. doi: 10.1053/j.ajkd.2010.03.019 [DOI] [PubMed] [Google Scholar]
- 2.Jovanovich A, Isakova T, Stubbs J. Microbiome and cardiovascular disease in CKD. Clin J Am Soc Nephrol. 2018. doi: 10.2215/CJN.12691117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaziri ND. Effect of synbiotic therapy on gut-derived uremic toxins and the intestinal microbiome in patients with CKD. Clin J Am Soc Nephrol. 2016;11(2):199–201. doi: 10.2215/CJN.13631215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaziri ND, Goshtasbi N, Yuan J, et al. Uremic plasma impairs barrier function and depletes the tight junction protein constituents of intestinal epithelium. Am J Nephrol. 2012;36(5):438–443. doi: 10.1159/000343886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoll LL, Denning GM, Weintraub NL. Potential role of endotoxin as a proinflammatory mediator of atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24(12):2227–2236. doi: 10.1161/01.ATV.0000147534.69062.dc [DOI] [PubMed] [Google Scholar]
- 6.Stenvinkel P. Inflammation in end-stage renal disease: the hidden enemy. Nephrology (Carlton). 2006;11(1):36–41. doi: 10.1111/j.1440-1797.2006.00541.x [DOI] [PubMed] [Google Scholar]
- 7.Amdur RL, Feldman HI, Gupta J, et al. Inflammation and progression of CKD: the CRIC Study. Clin J Am Soc Nephrol. 2016;11(9):1546–1556. doi: 10.2215/CJN.13121215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nowak KL, Chonchol M. Does inflammation affect outcomes in dialysis patients? Semin Dial. 2018;31(4):388–397. doi: 10.1111/sdi.12686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louis P, Duncan SH, McCrae SI, Millar J, Jackson MS, Flint HJ. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J Bacteriol. 2004;186(7):2099–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7(3):189–200. doi: 10.1080/19490976.2015.1134082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stilling RM, van de Wouw M, Clarke G, Stanton C, Dinan TG, Cryan JF. The neuropharmacology of butyrate: the bread and butter of the microbiota-gut-brain axis? Neurochem Int. 2016;99:110–132. doi: 10.1016/j.neuint.2016.06.011 [DOI] [PubMed] [Google Scholar]
- 12.Hallert C, Bjorck I, Nyman M, Pousette A, Granno C, Svensson H. Increasing fecal butyrate in ulcerative colitis patients by diet: controlled pilot study. Inflamm Bowel Dis. 2003;9(2):116–121. [DOI] [PubMed] [Google Scholar]
- 13.Krokowicz L, Stojcev Z, Kaczmarek BF, et al. Microencapsulated sodium butyrate administered to patients with diverticulosis decreases incidence of diverticulitis–a prospective randomized study. Int J Colorectal Dis. 2014;29(3):387–393. doi: 10.1007/s00384-013-1807-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang S, Xie S, Lv D, et al. Alteration of the gut microbiota in Chinese population with chronic kidney disease. Sci Rep. 2017;7(1):2870. doi: 10.1038/s41598-017-02989-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang S, Xie S, Lv D, et al. A reduction in the butyrate producing species Roseburia spp. and Faecalibacterium prausnitzii is associated with chronic kidney disease progression. Antonie Van Leeuwenhoek. 2016;109(10):1389–1396. doi: 10.1007/s10482-016-0737-y [DOI] [PubMed] [Google Scholar]
- 16.Mason S, Reinecke CJ, Kulik W, van Cruchten A, Solomons R, van Furth AM. Cerebrospinal fluid in tuberculous meningitis exhibits only the L-enantiomer of lactic acid. BMC Infect Dis. 2016;16:251. doi: 10.1186/s12879-016-1987-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordi T, Khamis H. Simple solution to a common statistical problem: interpreting multiple tests. Clin Ther. 2004;26(5):780–786. [DOI] [PubMed] [Google Scholar]
- 18.Minalyan A, Gabrielyan L, Scott D, Jacobs J, Pisegna JR, Gastric T. Intestinal microbiome: role of proton pump inhibitors. Curr Gastroenterol Rep. 2017;19(8):42. doi: 10.1007/s11894-017-0577-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14(1):20–32. doi: 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016;65(1):57–62. doi: 10.1136/gutjnl-2015-309618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes V. Microbiome: cultural differences. Nature. 2012;492(7427):S14–S15. doi: 10.1038/492S14a [DOI] [PubMed] [Google Scholar]
- 22.Louis P, Flint HJ. Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol. 2007;73(6):2009–2012. doi: 10.1128/AEM.02561-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ewaschuk JB, Naylor JM, Zello GA. D-lactate in human and ruminant metabolism. J Nutr. 2005;135(7):1619–1625. doi: 10.1093/jn/135.7.1619 [DOI] [PubMed] [Google Scholar]
- 24.Brandt RB, Siegel SA, Waters MG, Bloch MH. Spectrophotometric assay for D-(-)-lactate in plasma. Anal Biochem. 1980;102(1):39–46. [DOI] [PubMed] [Google Scholar]
- 25.Tan SJ, Yu C, Yu Z, et al. High-fat enteral nutrition reduces intestinal mucosal barrier damage after peritoneal air exposure. J Surg Res. 2016;202(1):77–86. doi: 10.1016/j.jss.2015.12.010 [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Gong J, Wang W, et al. Are there any different effects of Bifidobacterium, Lactobacillus and Streptococcus on intestinal sensation, barrier function and intestinal immunity in PI-IBS mouse model? PLoS One. 2014;9(3):e90153. doi: 10.1371/journal.pone.0090153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terpstra ML, Singh R, Geerlings SE, Bemelman FJ. Measurement of the intestinal permeability in chronic kidney disease. World J Nephrol. 2016;5(4):378–388. doi: 10.5527/wjn.v5.i4.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yasuda T, Ozawa S, Shiba C, et al. D-lactate metabolism in patients with chronic renal failure undergoing CAPD. Nephron. 1993;63(4):416–422. doi: 10.1159/000187245 [DOI] [PubMed] [Google Scholar]
- 29.Adeva-Andany M, Lopez-Ojen M, Funcasta-Calderon R, et al. Comprehensive review on lactate metabolism in human health. Mitochondrion. 2014;17:76–100. doi: 10.1016/j.mito.2014.05.007 [DOI] [PubMed] [Google Scholar]
- 30.Imhann F, Vich Vila A, Bonder MJ, et al. The influence of proton pump inhibitors and other commonly used medication on the gut microbiota. Gut Microbes. 2017;8(4):351–358. doi: 10.1080/19490976.2017.1284732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tilg H, Moschen AR. Microbiota and diabetes: an evolving relationship. Gut. 2014;63(9):1513–1521. doi: 10.1136/gutjnl-2014-306928 [DOI] [PubMed] [Google Scholar]
- 32.Ahmadmehrabi S, Tang WHW. Gut microbiome and its role in cardiovascular diseases. Curr Opin Cardiol. 2017;32(6):761–766. doi: 10.1097/HCO.0000000000000445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou JA, Kalantar-Zadeh K, Mathew AT. A brief review of intradialytic hypotension with a focus on survival. Semin Dial. 2017;30(6):473–480. doi: 10.1111/sdi.12627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Sande FM, Dekker MJ, Leunissen KML, Kooman JP. Novel insights into the pathogenesis and prevention of intradialytic hypotension. Blood Purif. 2018;45(1–3):230–235. doi: 10.1159/000485160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dillon SM, Kibbie J, Lee EJ, et al. Low abundance of colonic butyrate-producing bacteria in HIV infection is associated with microbial translocation and immune activation. AIDS. 2017;31(4):511–521. doi: 10.1097/QAD.0000000000001366 [DOI] [PMC free article] [PubMed] [Google Scholar]