Abstract
In 1936, John Joseph Bittner identified mouse mammary tumor virus (MMTV), a milk transmitted beta retrovirus, a form of single-stranded positive-sense RNA virus. A retrovirus inserts a copy of its genome into the DNA of a host cell, thus altering the cell's genome. In the current analysis, we searched for MMTV sequences within the human genome. To compare the MMTV genome to the human genome, we used BLAT, the Blast-Like Alignment Tool of the UCSC Genome Browser. BLAT can align a user sequence of 25 bases or more to the genome. 60 MMTV sequences were in the human genome. Of 56 sequences from the MMTV POL gene, 36 POL sequences were from the same part of the gene, beginning at viral nucleotide 4800 but of different lengths. 8 viral sequences began at nucleotide ~3430 of the POL gene. Four viral sequences were from GAGdUTPase, encoded by the MMTV PRO gene. Deoxyuridine 5′-triphosphate nucleotidohydrolase (dUTPase) is an enzyme present in several major retroviral families. In MMTV dUTPase may be essential for viral replication. Since BLAT identified no MMTV envelope (env) sequence in the human genome, the env sequences from breast tumors and normal breast tissue found in other studies may have come from an MMTV infection. However, no one is certain how MMTV could enter human cells, since the cells do not have a cellular receptor for MMTV, as do mouse cells.
Keywords: Mouse mammary tumor virus, Breast cancer, The UCSC Genome Browser, The Cancer Genome Atlas
Breast cancer is the most common form of cancer in women. The cause of sporadic cases is usually difficult to ascertain. A minority, about 30%, are related to a gene.
An infectious origin is sometimes inferred, since 16% of all human cancer can be attributed to a micro-organism. Among the viruses that might be related to breast cancer are papillomaviruses, herpes viruses and retroviruses. The mouse mammary tumor virus (MMTV), especially, has always been a prime suspect (Gannon et al., 2018; Lawson et al., 2018).
In 1936, John Joseph Bittner identified MMTV, a milk transmitted beta retrovirus, a form of single-stranded positive-sense RNA virus. A retrovirus inserts a copy of its genome into the DNA of a host cell, thus altering the cell's genome.
MMTV is a lentivirus (lente-, Latin for “slow”), a genus of retroviruses that cause chronic, lethal diseases with long incubation periods in humans and other mammals. The best known lentivirus is the Human Immunodeficiency Virus (HIV). Lentiviruses can become endogenous, integrating their genome into the host germline genome. The virus or part thereof is then inherited by the host's offspring. In the current analysis, we searched for MMTV sequences within the entire human genome.
We utilized the UCSC Genome Browser, an on-line genome browser at the University of California, Santa Cruz (UCSC). The browser is an interactive website offering access to genome sequence data from a variety of vertebrate and invertebrate species and major model organisms, integrated with a large collection of aligned annotations. The graphical viewer is optimized to support fast interactive performance and is an open-source, web-based tool suite built on top of a MySQL database for rapid visualization, examination, and querying of data at many levels. The Genome Browser Database, browsing tools, downloadable data files, and documentation are all accessible on the UCSC Genome Bioinformatics website (https://genome.ucsc.edu) (Kuhn et al., 2013).
To compare the MMTV genome to the human genome, we used BLAT, the Blast-Like Alignment Tool of the UCSC Genome Browser (Kuhn et al., 2013). BLAT can align a user sequence of 25 bases or more to the genome. Because some level of mismatch is tolerated, cross-species alignments may be performed provided the species have not diverged too far from each other; this capability allowed comparison of the MMTV genome to the human genome. BLAT calculates a percent identity score to indicate differences between sequences without a perfect match (i.e. without 100% identity). The differences include mismatches and gaps (Bhagwat et al., 2012). The MMTV sequence we analyzed with BLAT was FASTA Mouse mammary tumor virus, complete genome, NCBI Reference Sequence: NC_001503.1.
Because of the genetic basis for breast cancer (Shiovitz and Korde, 2015), we used cBioportal to access data in The Cancer Genome Atlas (TCGA). cBioPortal for Cancer Genomics provides visualization, analysis and download of large-scale cancer genomics data sets (http://www.cbioportal.org). The Cancer Genome Atlas is a project, begun in 2005, to catalog genetic mutations responsible for cancer, using genome sequencing and bioinformatics. We used the KM curve plotting tool in the UCSC Xena Browser to evaluate survival (https://xenab-rowser.net). UCSC Xena allows users to explore functional genomic data sets for correlations between genomic and/or phenotypic variables.
60 MMTV sequences BLAT identified in the human genome are listed in Table 1. The viral sequences are of three groups, but of variable length.
Table 1.
60 MMTV sequences BLAT identified in the human genome. The viral sequences are of approximately three varieties. Four are from GAGdUTPase. 8 begin at viral nucleotide ~3430 of the POL gene. 48 begin at viral nucleotide ~4800 of the POL gene.
| VIRAL SEQ |
Human |
Viral | Sequence | MMTV | |||||
|---|---|---|---|---|---|---|---|---|---|
| Size | Identity | Chromosome | Gene | Strand | Start | End | Start | End | Gene |
| 253 | 64.5% | 6q22.31 | + + | 121365109 | 121365361 | 3084 | 3336 | GAGdUTPase | |
| 253 | 64.1% | 11q14.3 | − + | 92681138 | 92681390 | 3084 | 3336 | GAGdUTPase | |
| 49 | 91.9% | 11p12 | − + | 43188209 | 43188291 | 3084 | 3166 | GAGdUTPase | |
| 197 | 67.6% | 8p11.1 | − + | 43553754 | 43553977 | 3230 | 3453 | GAGdUTPase | |
| 56 | 91.1% | 4q22.1 | − + | 88326156 | 88326406 | 3434 | 3684 | POL | |
| 65 | 92.4% | 11q12.1 | − + | 56096830 | 56097083 | 3434 | 3687 | POL | |
| 254 | 57.1% | 17q21.31 | − + | 41406291 | 41406544 | 3434 | 3687 | POL | |
| 25 | 96.0% | 19q13.41 | ZNF578 | − + | 52979141 | 52979165 | 3434 | 3458 | POL |
| 65 | 90.8% | Xq22.1 | − + | 100441118 | 100441371 | 3434 | 3687 | POL | |
| 20 | 100.0% | 6 qbl hap6 | HLA-DRB1 | − + | 3763979 | 3763998 | 3440 | 3459 | POL |
| 44 | 86.4% | 11q22.1 | − + | 101087161 | 101087204 | 3620 | 3663 | POL | |
| 21 | 100.0% | 19p12 | − + | 20687696 | 20687716 | 3643 | 3663 | POL | |
| 98 | 75.6% | 1q25.2 | − + | 178525196 | 178525383 | 4797 | 4992 | POL | |
| 72 | 84.8% | 3p12.3 | + + | 75605430 | 75605727 | 4799 | 5097 | POL | |
| 297 | 66.0% | 1q23.3 | CD48 | + + | 160666454 | 160666750 | 4800 | 5097 | POL |
| 298 | 66.5% | 1q22 | BC041646 | − + | 155599516 | 155599813 | 4800 | 5097 | POL |
| 104 | 83.7% | 1p36.21 | + + | 13684991 | 13685276 | 4800 | 5085 | POL | |
| 82 | 83.0% | 1p22.2 | − + | 89553867 | 89553948 | 4800 | 4881 | POL | |
| 298 | 66.8% | 3q12.3 | + + | 101416506 | 101416803 | 4800 | 5097 | POL | |
| 220 | 70.0% | 5p13.1 | + + | 40109055 | 40109621 | 4800 | 5354 | POL | |
| 199 | 69.9% | 6p11.2 | + + | 57626420 | 57626705 | 4800 | 5085 | POL | |
| 41 | 85.4% | 6q14.3 | − + | 87209196 | 87209236 | 4800 | 4840 | POL | |
| 286 | 65.8% | 8q24.3 | − + | 146250053 | 146250338 | 4800 | 5085 | POL | |
| 199 | 71.9% | 8q11.1 | − + | 47179713 | 47179998 | 4800 | 5085 | POL | |
| 286 | 66.8% | 9q34.11 | CCBL1 | + + | 131616161 | 131616446 | 4800 | 5085 | POL |
| 110 | 84.6% | 10q24.2 | ABCC2 | − + | 101582566 | 101582864 | 4800 | 5097 | POL |
| 298 | 66.8% | 11q22.1 | + + | 101571611 | 101571908 | 4800 | 5097 | POL | |
| 286 | 63.3% | 15q21.3 | + + | 55182155 | 55182440 | 4800 | 5085 | POL | |
| 110 | 75.5% | 19q13.12 | − + | 36065731 | 36066011 | 4800 | 5097 | POL | |
| 286 | 64.0% | 22q11.23 | + + | 23885809 | 23886094 | 4800 | 5085 | POL | |
| 269 | 67.0% | Xq28 | − + | 153840065 | 153840351 | 4800 | 5085 | POL | |
| 105 | 78.1% | 7p12.3 | + + | 48182602 | 48183141 | 4801 | 5352 | POL | |
| 225 | 63.2% | 1q21.3 | GOLPH3L | + + | 150629956 | 150630587 | 4802 | 5370 | POL |
| 56 | 78.6% | 2q11.2 | MRPL30 | − + | 99889556 | 99889611 | 4802 | 4857 | POL |
| 113 | 78.8% | 3q25.1 | + + | 151422866 | 151423419 | 4802 | 5357 | POL | |
| 191 | 62.9% | 3q13.11 | + + | 102932546 | 102932736 | 4802 | 4992 | POL | |
| 191 | 64.4% | 4q35.1 | − + | 185967894 | 185968084 | 4802 | 4992 | POL | |
| 191 | 62.4% | 4q21.3 | MAPK10 | + + | 87386270 | 87386460 | 4802 | 4992 | POL |
| 24 | 95.9% | 4q13.3 | + + | 74233739 | 74233762 | 4802 | 4825 | POL | |
| 128 | 78.2% | 7p12.3 | − + | 46079431 | 46079992 | 4802 | 5359 | POL | |
| 94 | 74.5% | 8q24.23 | − + | 137922224 | 137922413 | 4802 | 4992 | POL | |
| 214 | 66.4% | 17q11.2 | + + | 29963846 | 29964402 | 4802 | 5359 | POL | |
| 229 | 66.0% | 17q11.2 | + + | 26026606 | 26027172 | 4802 | 5359 | POL | |
| 234 | 68.9% | 19p13.2 | ZNF433 | + + | 12136441 | 12137033 | 4802 | 5385 | POL |
| 191 | 63.9% | 19p13.2 | + + | 9569541 | 9569731 | 4802 | 4992 | POL | |
| 191 | 62.9% | 19p12 | − + | 21851294 | 21851484 | 4802 | 4992 | POL | |
| 45 | 95.6% | 19q13.41 | − + | 53254794 | 53254985 | 4802 | 4992 | POL | |
| 269 | 56.9% | Yq11.223 | − + | 26164973 | 26165241 | 4808 | 5076 | POL | |
| 250 | 60.4% | 4q21.23 | − + | 86047722 | 86047976 | 4829 | 5085 | POL | |
| 274 | 62.5% | 5p14.3 | − + | 18783631 | 18784154 | 4829 | 5353 | POL | |
| 30 | 93.4% | Xq28 | + + | 148806315 | 148806344 | 4829 | 4858 | POL | |
| 267 | 65.6% | 5p13.3 | − + | 30490099 | 30490365 | 4831 | 5097 | POL | |
| 159 | 66.1% | Yp11.2 | − + | 8984533 | 8985051 | 4847 | 5359 | POL | |
| 56 | 82.2% | 1q32.3 | − + | 213200478 | 213200890 | 4955 | 5359 | POL | |
| 56 | 82.2% | 1q21.3 | NUP210L | + + | 154024314 | 154024721 | 4955 | 5354 | POL |
| 57 | 80.8% | 14q21.1 | − + | 42777950 | 42778368 | 4955 | 5359 | POL | |
| 40 | 92.5% | 5q22.3 | − + | 114373450 | 114373842 | 4976 | 5359 | POL | |
| 29 | 89.7% | 1q24.1 | FMO9P | − + | 166577918 | 166577946 | 5057 | 5085 | POL |
| 28 | 92.9% | 7p14.1 | + + | 38202294 | 38202321 | 5057 | 5084 | POL | |
| 29 | 96.6% | Yp11.2 | TBL1Y | − + | 6829855 | 6829884 | 5057 | 5085 | POL |
Of 56 sequences from the MMTV POL gene, 36 POL sequences were from the same part of the gene, beginning at viral nucleotide 4800 but of different lengths.
8 viral sequences began at nucleotide ~3430 of the POL gene.
Four viral sequences were from GAGdUTPase, encoded by the MMTV PRO gene. Deoxyuridine 5′-triphosphate nucleotidohydrolase (dUTPase) is an enzyme present in several major retroviral families. In MMTV dUTPase may be essential for viral replication (Hizi and Herzig, 2015).
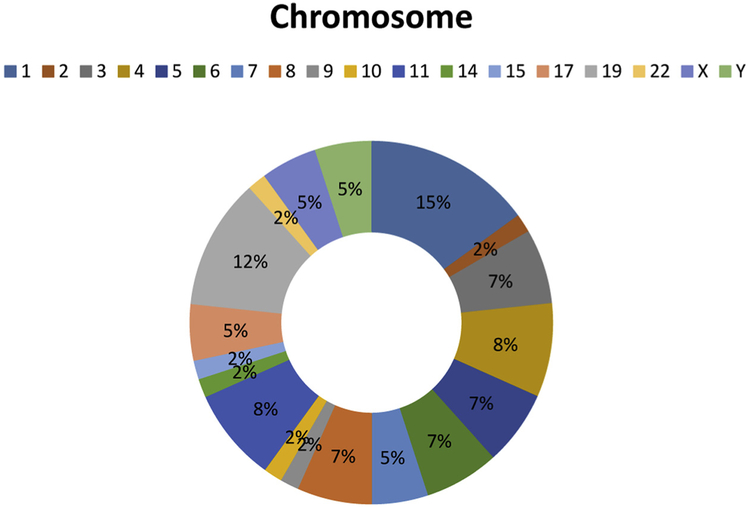
Distribution of MMTV sequences in the human genome by chromosome is shown in Fig. 1. Most of the sequences (15%) are within chromosome 1. Next was chromosome 19 (12%). Chromosome 3 had 7% of MMTV sequences and chromosome 11 had 8%. No sequence was from the MMTV env (envelope) gene. Moreover, BLAT did not identify ENV sequences in the baboon genome, bonobo genome, chimpanzee genome, gorilla genome, or green monkey genome.
Fig. 1.
Distribution of MMTV sequences in the human genome by chromosome. The largest percentage of sequences (15%) is within chromosome 1. Chromosome 3 had 7% and chromosome 11 had 8%, chromosome 19 12%.
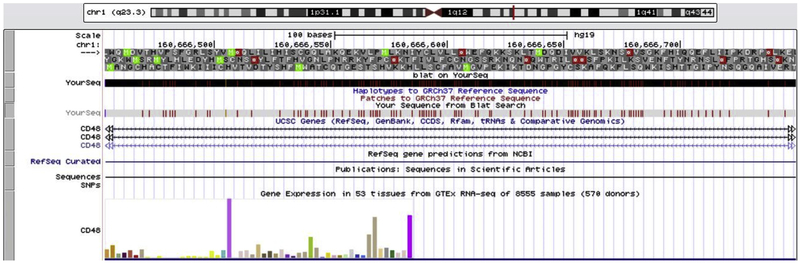
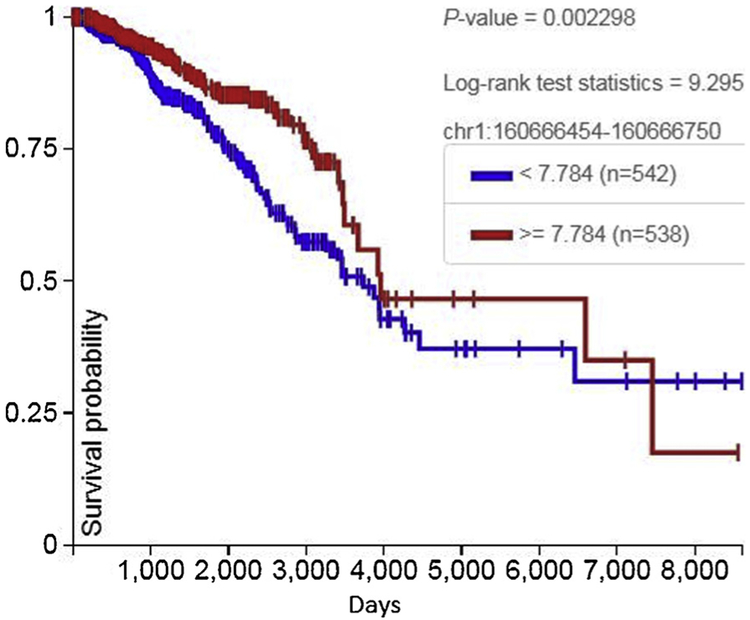
The 297 BP MMTV POL gene sequence in an intronic region of the human CD48 gene is shown in the UCSC genome browser (Fig. 2). Increased RNA expression of the sequence is associated with increased survival (Fig. 3).
Fig. 2.
The 297 BP MMTV POL gene sequence in an intronic region of the human CD48 gene is shown in the UCSC genome browser. Red: Genome and MMTV sequence have different bases at this position. Green: The query sequence appears to have a polyA tail that is not aligned to the genome.
Fig. 3.
Increased RNA expression of the 297 BP MMTV POL gene sequence in an intronic region of the human CD48 gene is associated with increased overall survival.
MMTV sequences BLAT identified in the human genome were in intronic segments of these genes:
CD48 (1q23.3). The CD48 molecule is a glycosyl-phosphatidyl-in-ositol (GPI)-anchored cell-surface protein of the CD2 family of molecules (Elishmereni and Levi-Schaffer, 2011). In breast cancer patients, CD48 expression is a favorable sign (Fig. 2).
HLA-DRB1 (chr 6qbl hap6). HLA-DRB1 has a key function in the immune system by presenting peptides derived from extracellular proteins. In breast cancer HLA-DRB1 may represent a protective allele (Chaudhuri et al., 2000) (RefSeq Summary NM_002124)
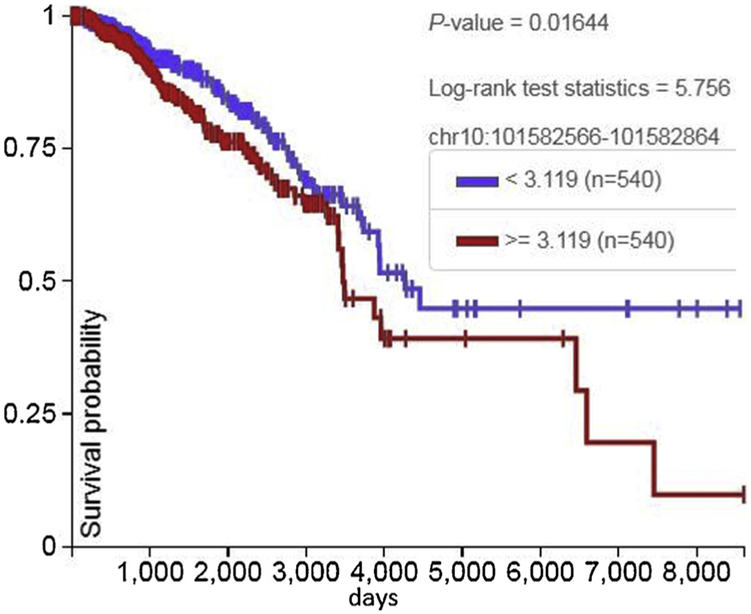
ABCC2 (MRP2, cMOAT) (chr 10q24.2). ABC proteins transport various molecules across extra- and intra-cellular membranes and belong to the MRP subfamily, involved in multi-drug resistance. ABCC2 is localized in the nuclear envelope of breast carcinoma cells and increased expression correlates with poor clinical outcome (Fig. 4) (Maciejczyk et al., 2012).
MAPK10 (chr 4q21.3) The protein encoded by this gene is a member of the MAP kinase family. MAP kinases act as integration points for multiple biochemical signals, and thus are involved in a wide variety of cellular processes, including proliferation, differentiation, transcription regulation and development (RefSeq Summary NM_138980). A breast cancer susceptibility locus has recently been identified on chr 4q.21 (Hamdi et al., 2016).
MRPL30 (chr 2q11.2) Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within mitochondria. (RefSeq Summary NR_028356)
TBL1Y (chr Yp11.2) This gene is highly similar to TBL1X gene in nucleotide sequence and protein sequence, but the TBL1X gene is located on chromosome X and TBL1Y is on chromosome Y, and therefore would be unrelated to female breast cancer.
GOLPH3L (chr 1q21.3) The Golgi complex plays a key role in the sorting and modification of proteins exported from the endoplasmic reticulum. GOLPH3L is associated with poor prognosis of patients with epithelial ovarian cancer and may be an independent prognostic factor (Feng et al., 2015).
NUP210L (chr 1q21.3) Homo sapiens nucleoporin 210 kDa-like (NUP210L), transcript variant 1, mRNA. Chromosome 1q21.3 amplification is a trackable biomarker and actionable target for breast cancer recurrence (Goh et al., 2017).
ZNF578 (chr 19q13.41) Homo sapiens zinc finger protein 578 (ZNF578), mRNA. A polymorphism at 19q13.41 predicts breast cancer survival after endocrine treatment (Khan et al., 2015).
TheZNF433 (chr 19p13.2) Human zinc finger protein may be a susceptibility gene for multiple sclerosis (Nischwitz et al., 2010). High-resolution 19p13.2-13.3 allelotyping of breast carcinomas demonstrates frequent loss of heterozygosity (Yang et al., 2004).
FMO9P (chr 1q24.1). Chr 1q24.1-2 has been found amplified in the JIMTI breast cancer cell line (Jonsson et al., 2007).
Fig. 4.
Increased RNA expression of the 110 BP MMTV POL gene sequence in an intronic region of the human ABCC2 gene is associated with diminished overall survival.
8 percent of DNA in the human genome comes from human endogenous retroviruses (HERV), and some human diseases have been attributed to this DNA. HERV sequences have occasionally been adopted by the human body to serve a useful purpose, such as in the placenta, where they may safeguard fetal-maternal tolerance (Kurth and Bannert, 2010). Some HERV sequences have oncogenic properties; yet treating cancer cells with methyltransferase inhibitors increases HERV RNA and DNA. Like infectious agents, the HERV-derived nucleic acids in the cytoplasm activate innate immune responses that produce tumor cell apoptosis (Bannert et al., 2018; Wildschutte et al., 2016).
Mouse mammary tumor virus (MMTV)-like elements have been identified in the genomes of pikas (Ochotona sp.), herbivorous smaller relatives of rabbits and hares. Lemos de Matos et al. found a nearly complete MMTV-like virus (Pika-BERV) in the American pika genome, which was estimated to have invaded the host genome around 3–7 million years ago, coinciding with the divergence time of American pikas from Asian pikas (Lemos de Matos et al., 2015).
Foley has analyzed the molecular phylogeny of lentiviruses and constructed phylogenetic trees of the POL and GAG genes. Foley reported that the protein-coding regions of the lentiviral genome are conserved to varying degrees, with GAG and POL being more conserved overall than TAT or ENV (Foley, 2000).
MMTV POL-Related Sequences have been reported in human DNA (Deen and Sweet, 1986). Similar MMTV-related sequences are present in normal breast tissue. The tissue sequences are over 90% homologous to the nucleotide sequence of the MMTV POL gene. In one study, no significant difference was found when comparing DNA from normal and tumor tissue (Moyret et al., 1992); and MMTV is not associated with neuroendocrine human breast cancers (Lawson et al., 2017).
By using a DNA fragment primarily encoding the POL region of the Syrian hamster intracisternal A particle (IAP; type A retrovirus) gene as a probe, human endogenous retrovirus genes, tentatively termed HERV-K genes, were cloned from a fetal human liver gene library. Typical HERV-K genes were 9.1 or 9.4 kb in length. By filter hybridization, the number of HERV-K genes was estimated to be approximately 50 copies per haploid human genome. The cloned mouse mammary tumor virus (type B) gene was found to hybridize with both the HERV-K and IAP genes to essentially the same extent (Ono, 1986; Ono et al., 1986). The 56 MMTV POL sequences we report here (Table 1) were considerably shorter, 20–298 BP, and correspond to only small sections of the MMTV genome, not the entire genome, as do the HERV-K genes.
As noted above, four MMTV viral sequences in the human genome were from GAGdUTPase, encoded by the MMTV PRO gene. During the replication of retroviruses, large numbers of Gag molecules must be generated to serve as precursors to the structural proteins of the virions. The enzymes encoded by the PRO and POL genes are, in most cases, needed in smaller numbers to carry out catalytic functions (Hughes and Varmus, 1999).
Whether MMTV or a related virus infects humans is controversial. MMTV DNA sequences have been detected inconsistently and serologic methods have varied widely. One study showed no MMTV-specific antibodies in 92 US women with breast cancer (Goedert et al., 2006). A second study could not confirm the presence of MMTV in human breast cancer patients (Perzova et al., 2017). Other studies have revealed antibodies (Nartey et al., 2017).
Yet MMTV-like env sequences (envelope protein sequences, also called HMTV sequences to denote their source) were found in 9 of 25 breast cancer specimens (36%). Among 25 non-cancerous breast biopsies of the same patients taken 1 to 11 years earlier, six contained MMTV-like sequences (24%). Five of the six were among the nine virally-associated breast cancers. In two pairs of specimens, benign and malignant, env sequences were 97% identical (Nartey et al., 2017).
Since BLAT identified no MMTV env sequence in the human genome, the env sequences may have come from an MMTV infection. However, no one is certain how MMTV could enter human cells, since the cells do not have a cellular receptor for MMTV, as do mouse cells (Brower, 2009).
As noted above (Fig. 1) the largest percentage of the MMTV sequences (15%) is within chromosome 1. Next was chromosome 19 (12%). Chromosome 3 had 7% of MMTV sequences and chromosome 11 had 8%. In one study, loss of heterozygosity (LOH) was observed in 30 chromosomal loci of primary breast tumors and 48 chromosomal loci of metastatic lesions. In metastatic lesions, incidence of LOH was highest on chromosome 19, followed by chromosomes 14, 3, and 11 (Li et al., 2014).
In sum, MMTV env sequences are not present in the human genome but are present in breast tumors and normal breast tissues. This finding suggests that MMTV infection may be related to human breast cancer. More evidence of causation is still needed.
Footnotes
Conflicts of interest
None.
References
- Bannert N, Hofmann H, Block A, Hohn O, 2018. HERVs new role in cancer: from accused perpetrators to cheerful protectors. Front Microbiol. 9, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat M, Young L, Robison RR, 2012. Using BLAT to find sequence similarity in closely related genomes. Curr. Protoc. Bioinformatics Chapter 10, Unit 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower V, 2009. Mouse mammary tumor virus: new tumor virus or just a rumor virus? J. Natl. Cancer Inst 101 (5), 293–295. [DOI] [PubMed] [Google Scholar]
- Chaudhuri S, Cariappa A, Tang M, Bell D, Haber DA, Isselbacher KJ, Finkelstein D, Forcione D, Pillai S, 2000. Genetic susceptibility to breast cancer: HLA DQB*03032 and HLA DRB1*11 may represent protective alleles. Proc. Natl. Acad. Sci U. S. A 97 (21), 11451–11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deen KC, Sweet RW, 1986. Murine mammary tumor virus pol-related sequences in human DNA: characterization and sequence comparison with the complete murine mammary tumor virus pol gene. J. Virol 57 (2), 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elishmereni M, Levi-Schaffer F, 2011. CD48: A co-stimulatory receptor of immunity. Int. J. Biochem. Cell Biol 43 (1), 25–28. [DOI] [PubMed] [Google Scholar]
- Feng Y, He F, Wu H, Huang H, Zhang L, Han X, Liu J, 2015. GOLPH3L is a novel prognostic biomarker for epithelial ovarian cancer. J. Cancer 6 (9), 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley BT, 2000. An overview of the molecular phylogeny of lentiviruses. HIV sequence compendium 2000: theoretical biology and biophysics. Los Alamos National Laboratory, Los Alamos, N. Mex, pp. 35–43. [Google Scholar]
- Gannon OM, Antonsson A, Bennett IC, Saunders NA, 2018. Viral infections and breast cancer - A current perspective. Cancer Lett. 420, 182–189. [DOI] [PubMed] [Google Scholar]
- Goedert JJ, Rabkin CS, Ross SR, 2006. Prevalence of serologic reactivity against four strains of mouse mammary tumour virus among US women with breast cancer. Br. J Cancer 94 (4), 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh JY, Feng M, Wang W, Oguz G, Yatim SMJM, Lee PL, Bao Y, Lim TH, Wang P, Tam WL, Kodahl AR, Lyng MB, Sarma S, Lin SY, Lezhava A, Yap YS, Lim AST, Hoon DSB, Ditzel HJ, Lee SC, Tan EY, Yu Q, 2017. Chromosome 1q21.3 amplification is a trackable biomarker and actionable target for breast cancer recurrence. Nat. Med 23 (11), 1319–1330. [DOI] [PubMed] [Google Scholar]
- Hamdi Y, Soucy P, Adoue V, Michailidou K, Canisius S, Lemacon A, Droit A, Andrulis IL, Anton-Culver H, Arndt V, Baynes C, Blomqvist C, Bogdanova NV, Bojesen SE, Bolla MK, Bonanni B, Borresen-Dale AL, Brand JS, Brauch H, Brenner H, Broeks A, Burwinkel B, Chang-Claude J, Couch FJ, Cox A, Cross SS, Czene K, Darabi H, Dennis J, Devilee P, Dork T, Dos-Santos-Silva I, Eriksson M, Fasching PA, Figueroa J, Flyger H, Garcia-Closas M, Giles GG, Goldberg MS, Gonzalez-Neira A, Grenaker-Alnaes G, Guenel P, Haeberle L, Haiman CA, Hamann U, Hallberg E, Hooning MJ, Hopper JL, Jakubowska A, Jones M, Kabisch M, Kataja V, Lambrechts D, Le ML, Lindblom A, Lubinski J, Mannermaa A, Maranian M, Margolin S, Marme F, Milne RL, Neuhausen SL, Nevanlinna H, Neven P, Olswold C, Peto J, Plaseska-Karanfilska D, Pylkas K, Radice P, Rudolph A, Sawyer EJ, Schmidt MK, Shu XO, Southey MC, Swerdlow A, Tollenaar RA, Tomlinson I, Torres D, Truong T, Vachon C, van den Ouweland AM, Wang Q, Winqvist R, Zheng W, Benitez J, Chenevix-Trench G, Dunning AM, Pharoah PD, Kristensen V, Hall P, Easton DF, Pastinen T, Nord S, Simard J, 2016. Association of breast cancer risk with genetic variants showing differential allelic expression: Identification of a novel breast cancer susceptibility locus at 4q21. Oncotarget 7 (49), 80140–80163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizi A, Herzig E, 2015. dUTPase: the frequently overlooked enzyme encoded by many retroviruses. Retrovirology 12, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SH, Varmus HE, 1999. Retroviruses. CSHL Press. [Google Scholar]
- Jonsson G, Staaf J, Olsson E, Heidenblad M, Vallon-Christersson J, Osoegawa K, de JP, Oredsson S, Ringner M, Hoglund M, Borg A, 2007. High-resolution genomic profiles of breast cancer cell lines assessed by tiling BAC array comparative genomic hybridization. Genes Chromosomes. Cancer 46 (6), 543–558. [DOI] [PubMed] [Google Scholar]
- Khan S, Fagerholm R, Rafiq S, Tapper W, Aittomaki K, Liu J, Blomqvist C, Eccles D, Nevanlinna H, 2015. Polymorphism at 19q13.41 predicts breast cancer survival specifically after endocrine therapy. Clin. Cancer Res. 21 (18), 4086–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Haussler D, Kent WJ, 2013. The UCSC genome browser and associated tools. Brief. Bioinform. 14 (2), 144–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth R, Bannert N, 2010. Beneficial and detrimental effects of human endogenous retroviruses. Int. J. Cancer 126 (2), 306–314. [DOI] [PubMed] [Google Scholar]
- Lawson JS, Mazzanti C, Civita P, Menicagli M, Ngan CC, Whitaker NJ, Hochman J, Braitbard O, Yosufi B, Glenn WK, 2018. Association of mouse mammary tumor virus with human breast cancer: histology immunohistochemistry and polymerase chain reaction analyses. Front. Oncol. 8, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson JS, Ngan CC, Glenn WK, Tran DD, 2017. Correction to: mouse mammary tumour virus (MMTV) and human breast cancer with neuroendocrine differentiation. Infect. Agent. Cancer 12, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos de Matos A, de Sousa-Pereira P, Lissovsky AA, van der Loo W, Melo-Ferreira J, Cui J, Esteves PJ, 2015. Endogenization of mouse mammary tumor virus (MMTV)-like elements in genomes of pikas (Ochotona sp.). Virus Res. 210, 22–26. [DOI] [PubMed] [Google Scholar]
- Li H, Yang B, Xing K, Yuan N, Wang B, Chen Z, He W, Zhou J, 2014. A preliminary study of the relationship between breast cancer metastasis and loss of heterozygosity by using exome sequencing. Sci. Rep. 4, 5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejczyk A, Jagoda E, Wysocka T, Matkowski R, Gyorffy B, Lage H, Surowiak P, 2012. ABCC2 (MRP2, cMOAT) localized in the nuclear envelope of breast carcinoma cells correlates with poor clinical outcome. Pathol. Oncol. Res 18 (2), 331–342. [DOI] [PubMed] [Google Scholar]
- Moyret C, Bernard D, Bignon Y, Dastugue B, Plagne R, Chollet P, Peters G, 1992. Presence of the mouse mammary-tumor virus (mmtv) pol gene in breast-cancer. Int. J. Oncol. 1 (4), 475–480. [DOI] [PubMed] [Google Scholar]
- Nartey T, Mazzanti CM, Melana S, Glenn WK, Bevilacqua G, Holland JF, Whitaker NJ, Lawson JS, Pogo BG, 2017. Mouse mammary tumor-like virus (MMTV) is present in human breast tissue before development of virally associated breast cancer. Infect. Agent. Cancer 12, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nischwitz S, Cepok S, Kroner A, Wolf C, Knop M, Muller-Sarnowski F, Pfister H, Roeske D, Rieckmann P, Hemmer B, Ising M, Uhr M, Bettecken T, Holsboer F, Muller-Myhsok B, Weber F, 2010. Evidence for VAV2 and ZNF433 as susceptibility genes for multiple sclerosis. J. Neuroimmunol 227 (1–2), 162–166. [DOI] [PubMed] [Google Scholar]
- Ono M, 1986. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. J. Virol 58 (3), 937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Yasunaga T, Miyata T, Ushikubo H, 1986. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J. Virol. 60 (2), 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perzova R, Abbott L, Benz P, Landas S, Khan S, Glaser J, Cunningham CK, Poiesz B, 2017. Is MMTV associated with human breast cancer? Maybe, but probably not. Virol. J 14 (1), 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiovitz S, Korde LA, 2015. Genetics of breast cancer: a topic in evolution. Ann. Oncol 26 (7), 1291–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildschutte JH, Williams ZH, Montesion M, Subramanian RP, Kidd JM, Coffin JM, 2016. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc. Natl. Acad. Sci U. S. A 113 (16), E2326–E2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TL, Su YR, Huang CS, Yu JC, Lo YL, Wu PE, Shen CY, 2004. High-resolution 19p13.2-13.3 allelotyping of breast carcinomas demonstrates frequent loss of heterozygosity. Genes Chromosomes. Cancer 41 (3), 250–256. [DOI] [PubMed] [Google Scholar]