Abstract
Structure-function studies of the cytochrome b6f complex, the central hetero-oligomeric membrane protein complex in the electron transport chain of oxygenic photosynthesis, which formed the basis for a high resolution (2.5 Å) crystallographic solution of the complex, are described. Structure-function differences between the structure of subunits of the bc complexes, b6f, and bc1 from mitochondria and photosynthetic bacteria, which are often assumed to function identically, are discussed. Major differences which suggest that quinone-dependent electron transport pathways can vary in b6f and bc1 complexes are: (a) an additional c-type heme, cn, and bound single copies of chlorophyll a and β-carotene in the b6f complex; and (b) a cyclic electron transport pathway that encompasses the b6f and PSI reaction center complexes.
The importance of including lipid in crystallization of the cytochrome complex, or with any hetero-oligomeric membrane protein complex, is emphasized, and consequences to structure-function of b6f being a lipo-protein complex discussed, including intra-protein dielectric heterogeneity and resultant pathways of trans-membrane electron transport. The role of the b6f complex in trans-membrane signal transduction from reductant generated on the p-side of the electron transport chain to the regulation of light energy to the two photosystems by trans-side phosphorylation of the light harvesting chlorophyll protein is presented. Regarding structure aspects relevant to plastoquinol-quinone entrance-egress: (i) modification of the p-side channel for plastoquinone access to the iron-sulfur protein would change the rate-limiting step in electron transport; (ii) the narrow niche for entry of plastoquinol into b6f from the PSII reaction center complex would seem to require close proximity between the complexes.
Keywords: cytochrome b6f, bc1 complexes, cytochrome f, crystal structure, heme cn, lipoprotein, plastoquinone, rate-limiting step, super-complex, trans-membrane signaling
1. Introduction.
The background of this perspective on structure-function of the cytochrome b6f complex, shown in the context of the oxygenic photosynthetic electron transport chain (Fig. 1), can be traced to studies described in references (Bendall, 1982; Boardman and Anderson 1967; Duysens 1955; Levine et al. 1966) the latter reference to the location of the rate limiting step in the chain, oxidation of plastoquinol coupled to reduction of cytochrome f. This rate-limiting step was associated with proton (H+) translocation across the thylakoid membrane and generation of the trans-membrane proton electrochemical potential gradient across the electro-negative (n) and -positive (p) sides of the photosynthetic membrane. This mechanism for membrane energy transduction and its application to mitochondrial membranes was described at that time (Mitchell 1966), and also applied to oxygenic photosynthetic membrane energy transduction (Jagendorf and Uribe 1966). The initiation of crystallographic analysis of membrane proteins in our laboratory at Purdue University was facilitated by studies at the Max Planck Inst, für Biophysik (Frankfurt) with H. Michel, G. Fritzsch, and H. Schägger.
Fig. 1.
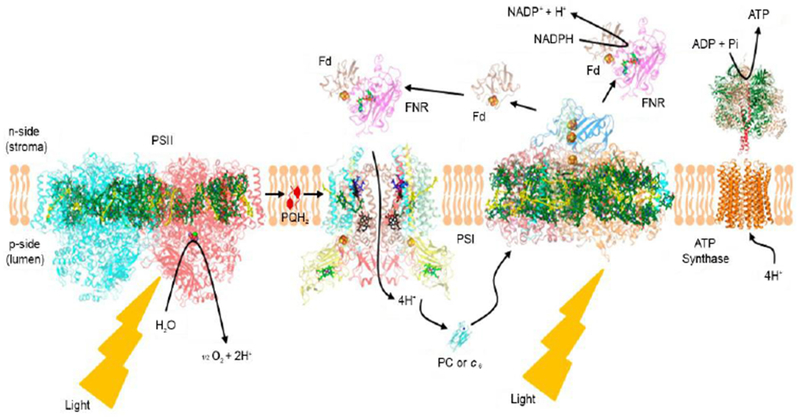
Electron transport chain associated with oxygenic photosynthesis; embedded in a lipidic membrane approximately 30 Å in width; p, n, positive, negative sides of the proton trans-membrane electrochemical gradient established by redox-linked proton translocation.
The high resolution structure of the cytochrome b6f electron transfer/proton translocating complex is of interest, not only because of insight provided on its function in the linear and cyclic electron transport pathways of oxygenic photosynthesis, and the role of the protein structure in determining the rate limiting step in electron transport step (Cramer and Hasan 2016; Hasan and Cramer 2012; Hasan and Cramer 2014; Hasan et al. 2013a). In addition, its structure is a member of the set of fewer than 500 hetero-oligomeric integral membrane proteins solved to a resolution < 2.5 Å in the Protein Data Bank (PDB), in which more than 1.2 × 105 structures of soluble proteins are stored presently. Also, see White (2018).
The first solved crystal structure of such a membrane protein complex was that of the reaction center complex from the purple photosynthetic bacterium, Rhodopseudomonas viridis (Deisenhofer and Michel 1989). In addition to providing further understanding of the function of the b6f complex, the structure studies summarized here also describe the role in the structure, and presumably in function, of integral lipid bound in the protein complex (Hasan et al. 2013; Hasan and Cramer 2014a). It is now realized that lipids not only provide the intra-membrane bilayer environment for membrane proteins, but also participate in the internal structure of hetero-oligomeric membrane proteins (Barrera et al. 2013; Valiyaveetil et al. 2002) such as the b6f complex (Zhang et al. 2003). The position and general functions of the b6f complex in the linear electron transport chain that functions to transfer electrons from water to NADP+ in oxygenic photosynthesis is shown (Fig. 1). It should be noted that this figure is somewhat simplified because it does not show the presence of “super-complexes” formed between the distinct electron transfer complexes, e.g., that between the b6f and PSI complexes (Iwai et al. 2010), as well as those between reaction centers and light harvesting complexes, which are not discussed here.
2. Initial approach to the crystal structure of the b6f complex.
Purification of the p-side soluble domains of (i) cytochrome f (Cramer et al. 1994) and (ii) the high potential Rieske* (Rieske et al. 1964) [2Fe-2S] protein (Zhang et al. 1996) led to crystal structures obtained In collaboration with S. Martinez in J. L. Smith’s lab. Crystal structures were obtained of the p-side soluble domains of plant (Martinez et al. 1992, 1994, 1996) and cyanobacterial cytochrome f (Carrell 1999), the latter in collaboration with the group of Derek Bendall. After isolation of a 139-residue C-terminal fragment of the Rieske [2Fe-2S] protein (Zhang et al. 1996), a 1.83 A structure was obtained of this lumen-side domain (Carrell et al. 1997). These structures led to an understanding via a collaboration with Prof. Lev Krishtalik of the electrostatic forces that mediate the interaction between the positively charged region around the cytochrome f heme and a complementary negatively charged region on the surface of the plastocyanin electron acceptor (Soriano et al. 1996, 1997, 1998). An unexpected result was that the cyt f heme on the p-side of the membrane is linked to an internal chain of 5 water molecules (Martinez et al. 1996). It was proposed that this water chain, whose interruption was found to impair electron transport (Ponamarev et al. 1998; Sainz et al. 2000), might function in a mechanism of H+ translocation via a water-supported ‘proton wire.’ It was not possible, however, to determine the function of this H2O chain in the p-side domain of the cytochrome f structure, and this aspect of the cytochrome f structure remains an enigma.
Concerning redox-linked dynamics of the p-side domains of the Rieske protein and cytochrome f, the flash-induced oxidation of cytochrome f, the chloroplast analogue of cytochrome c1, was found to be inhibited reversibly by increased lumenal viscosity, as was the subsequent reduction of hemes b6 and f, both rates varying inversely with lumenal viscosity over a range of 1- 10 centipoise (Heimann et al., 2000). The kinetic data suggested that upon initiation of electron transfer by a light flash, cytochrome b6 reduction requires movement of reduced ISP from an initial position predominantly proximal to cytochrome f, apparently favored by the reduced ISP, to the quinol binding site at which the oxidant-induced reduction of cytochrome b6 hemes are initiated. Subsequent reduction of cytochrome f requires the additional movement of the ISP back to a site proximal to cytochrome f. The dependence of the cytochrome redox reaction on ambient viscosity implies that ‘tethered diffusion’ of the ISP could be part of the rate limitation for charge transfer through the b6f complex (see section 11.1 below).
3. Differences between b6f and bc1 complexes.
A common origin or significant overlap in evolution of the b-cytochrome component of b6f and bc1 complexes is implied by the similarity of amino acid sequences and, to an even greater extent, correspondence of hydropathy functions that describe the ca. 20 residue membrane spanning domains of the b-cytochrome component of (i) the 8 trans-membrane helices (TMH) of the bc1 complex, and (ii) the combined 7 TMH of the b-cytochrome (4 TMH) and ‘subunit IV’ (3 TMH) components of the b6f complex (Widger et al. 1984).
In contrast, regarding cyt f and c1 subunits, except for the single TMH that binds the cytochrome f or c1 subunit to the complex and the membrane, the structures of the cyt f and cyt c1 subunits are different in the respective cytochrome b6f and bc1 complexes (Figs. 2A, B vs. Figs. 2C, D). The only conserved sequence element is the Cys-X-Y-Cys-His sequence, which is responsible for covalent binding of the c-type heme. The p-side heme binding domain of cyt f and c1, respectively, has a very different secondary structure in b6f and bc1 complexes, predominantly β and α, respectively (Figs. 2C, D). Both cyt f and cyt c1 have a relatively positive redox potential, +370 mV and +260 mV, respectively, sufficient to provide an effective oxidant for ISP. Along with the structure dissimilarity, the evolutionary origin of cytochrome f relative to cytochrome c1 in the bc1 complex of mitochondria or photosynthetic bacteria is unclear (Martinez et al. 1994). Structure differences between cyt f and cyt c1 are discussed below (section 4) in the context of other differences in structure or subunit composition between bc1 and b6f complexes that suggest the possibility of differences in function. More perspective on issues of molecular evolution involving the bc complexes can be found in (Nitschke et al., 2010; Kao and Hunte 2014; Esser et al, 2015; Dibrova et al, 2013; Mulkidjanian et al., 2007)
Fig. 2.
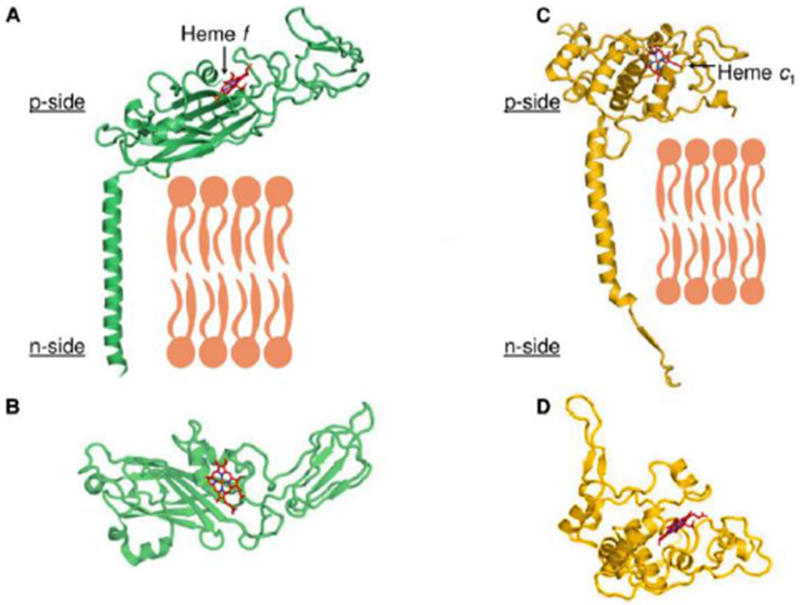
High potential electron acceptor of cytochrome bc complexes: cyt f of the b6f complex (17-19) (left) and cyt c1 of bc1 complex (right). (A, B) Cyt f subunit of b6f complex attached to the complex and membrane through one TMH. Cyt f subunit binds high potential c-type heme (/heme f, red/blue sticks) in the p-side β-sheet extrinsic domain (panel B). (C, D) Cyt c1 subunit of bc1 complex also bound to complex through one TMH (C). In contrast to cyt f, p-side extrinsic domain of cyt c1, with c-type heme bound covalently (red/blue sticks), mostly α-helical (D). B, D panels rotated 90° about horizontal axis relative to (A, B). Derived from original drawing by S. Saif Hasan.
4. Approach to the crystal structure of the b6f complex; problems and strategies.
Initial studies that formed the basis of efforts toward crystallization of the b6f complex were: (i) the plant (spinach) complex was found to be a dimer with a molecular weight, including lipid, of approximately 2.3 × 105 (Huang et al. 1994), and to contain (a) one integral chlorophyll a molecule, with a stoichiometry of one per monomer, a ligand whose presence was confirmed in the crystal structures, and which subsequently received substantial study (Dashdorj et al. 2005; Kim et al. 2005; Yan et al. 2008). Initially thought to be complementary to the single chlorophyll molecule, a single β-carotene was found in the complex (Zhang et al., 1999). Partly because significant light energy transfer between the chlorophyll and β-carotene could not be demonstrated Dashdorj et al., 2005), the function of the single chlorophyll in the complex still remains enigmatic except for the unexpected finding that it is not the porphyrin ring that presently has a defined function. Rather, the chlorophyll phytyl chain seems to have a function, that of gating the passage of plastoquinol/quinone within the complex (Hasan et al., 2014b).
Because thermophilic cyanobacteria had been used in the initial successes in crystallization of oxygenic photosynthetic hetero-oligomeric membrane proteins (Jordan et al. 2001; Ferreira et al. 2004; Zouni et al. 2001), we chose initially to utilize such an organism, Mastigocladus laminosus (Kurisu et al. 2003), as the protein source. When applied to the purification of the b6f complex, however, an unexpected result was proteolysis, resulting inevitably in poor crystal quality, which was refractory toward inhibition by protease inhibitors, which was mostly a consequence of long times required for crystallization (Baniulis et al., 2011; Zhang et al., 2005). The latter problem, which had to be solved before crystals displaying reasonable diffraction could be obtained, followed the realization that a key component, lipid, was missing from the crystallization mix.
Confirmation of the composition of the b6f complex was obtained through an analysis by electrospray ionization mass spectrometry. Size-exclusion and reverse-phase separations allowed accurate measurement of their molecular masses, those for the cytochrome f and b6 subunits purified from spinach being 31,935 and 24,887 Da, respectively (Whitelegge et al. 2002).
5. An awakening; it’s the lipid!
It was finally realized that the existence of ‘boundary lipid,’ known from studies on mitochondrial membrane proteins, implied that we should examine the effect of lipid on the rate and quality of crystallization. Dr. H. Zhang, with admirable experimental intuition, added dioleoyl-phosphatidyl choline (DOPC) to the crystallization well at a stoichiometry of 10 equivalents per cytochrome f. Whereas it had previously taken weeks or months to obtain crystals of meaningful size, prominent reddish-brown crystals appeared overnight! The implied general requirement of lipid for crystallization of the b6f complex was described (Zhang et al. 2003). The generality of such a requirement for crystallization and implied proper conformation of integral membrane proteins could have been anticipated from the precedent already set in studies on other hetero-oligomeric membrane proteins, for example, the yeast cytochrome bc1 complex (Palsdottir and Hunte 2004) and the trans-membrane potassium (K+) channel (Valiyaveetil et al., 2002). The resulting structure of the cyanobacterial b6f complex, obtained in the presence of added lipid as described above from M. laminosus, is shown in ribbon format (Fig. 3). A higher resolution 2.5 Å structure, obtained from the cyanobacterium Nostoc sp. PCC 7120 (Hasan and Cramer 2014), showed 23 potential lipid binding sites (i. e., sites occupied by lipids, fatty acids, or detergent molecules) per monomer. The ubiquitous presence of interstitial lipid in oligomeric integral membrane proteins is now well established (Barrera et al. 2013).
Fig. 3.
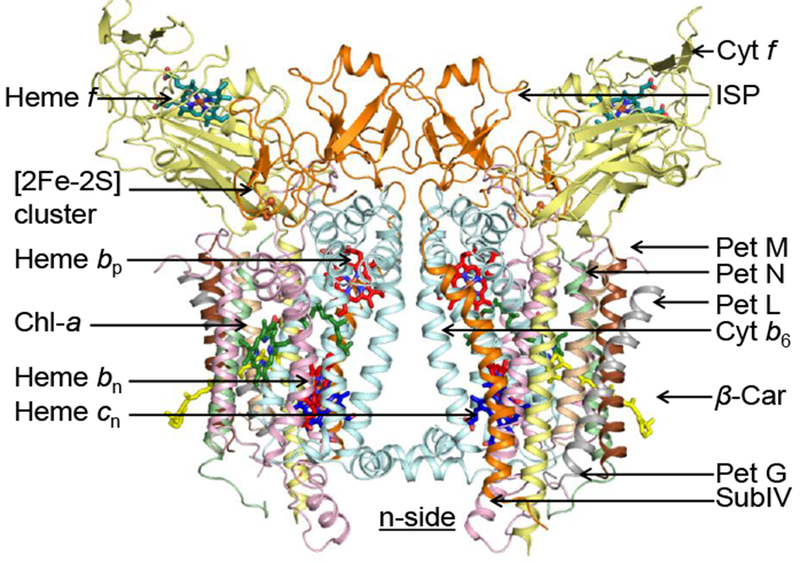
Ribbon diagram of cyto b6f complex [Nostoc sp. PCC 7120 (4), PDB 40GQ, a symmetric dimer (spinach b6f, 268 kDa including redox co-factors, determined by mass spectrometry (pers. comm., C. Bechera), resolution, 2.5 Å, bound lipids; one bound FNR molecule); C2 symmetry; 13 TMH per monomer including 4 small (3.4 – 4.1 kDa Pet M, N, L, and G ‘picket fence’ subunits in each monomer), 7 prosthe-tic groups [5 redox (4 hemes: f, bn, bp, cn) 1 FeS cluster], 1 chl a (33), 1 β - carotene protruding 11 Å from the complex (Zhang et al., 1999). 23 lipid binding sites per monomer. Derived from original drawing by S. Saif Hasan.
6. Publication of the first b6f crystal structures – trans-atlantic diplomacy.
Good communication and diplomatic relations existed between the laboratory of J.-L. Popot at the Institut de Biologie Physico-Chemie in Paris, France, where a crystal structure was sought from the green alga, C. reinhardtii, and our laboratory at Purdue University where we had the same goal using a thermophilic cyanobacterial (M. laminosus) source. In the summer of 2003, the two labs presented their respective findings on the structure of the b6f complex at two different Gordon Research Conferences in New Hampshire, U. S. A. Oral Communication was arranged on this topic between the two Conferences using a cell phone and a unique transmission apparatus. The communication was difficult, but was improved if one leaned against the base of a tall Church on the site of one of the Conferences, the Church structure apparently providing an unanticipated antenna function! After the Conference, the Paris and West Lafayette groups met in the Holiday Inn near Boston’s Logan Airport and coordinated submission of the two studies, to be submitted to Science and Nature. respectively. Both studies were published in late 2003 (Kurisu et al. 2003; Stroebel et al. 2003)
7. Crystal structure of the cyanobacterial cytochrome b6f complex.
A ribbon diagram shows the atomic structure of the C2 symmetric structure of the cyanobacterial (Nostoc) b6f complex at 2.5 Å resolution (PDB showing the two b hemes, bp and bn, traversing the ~ 30 Å hydrophobic core (Fig. 3). The two other redox centers shown are the p- side 2Fe-2S cluster at the p-side interface of the left-side monomer, and the cyt f heme at the distal end of the p-side cyt f subunit that protrudes from the membrane. That the 2Fe-2S center and cytochrome f are seen only in the left-side monomer demonstrates C2 symmetry of the dimer.
The monomeric unit of the homo-dimeric b6f complex from M. laminosus (Fig. 3) contains eight polypeptide subunits, including the four, petA (cyt f), petB (cyt b6), petC (Rieske ISP), and petD (subunit IV, corresponding to the C-terminal half of the cytochrome b subunit of the bc1 complex, with molecular weights = 30.9, 24.7, 19.3, and 17.5 kDa, respectively, in spinach thylakoid membranes (Baniulis et al., 2009; Cramer and Hasan, 2016). These subunits bind or coordinate their five tightly bound metallo-redox prosthetic groups, four of which, hemes f, bp, bn, and cn, (defined c1 in the C. reinhardtii structure) and the 2Fe-2S ‘Rieske’ iron-sulfur protein (ISP) form the redox core of the complex conserved in all cytochrome bc (i. e., b6f, bc1 complexes. These include the electron transport proteins that function in mitochondrial respiration, oxygenic photosynthetic algae, and anoxygenic photosynthetic bacteria. The bound prosthetic groups are the hemes bn and bp (non-covalently bound b hemes on the n and p sides of the membrane, respectively; cf., Fig. 1). At the trans-membrane outside periphery of the conserved core in each monomer are four short hydrophobic membrane spanning peptides, Pet M, N, L, and G (Fig. 3) with molecular weights in M. laminosus of 3.8, 3.3, 3.5, and 4.1 kDa, respectively, which form a hydrophobic ‘picket fence’ on the periphery of each monomer, a structural feature that is possibly unique in the family of integral membrane proteins.
7.1. Domain-swapped ISP protein.
In both b6f and bc1 complexes the Rieske ISP is ‘domain-swapped,’ with the ISP C-terminal β-sheet extrinsic domain containing the redox-active [2Fe-2S] cluster residing on the p-side of each monomer in the dimeric complex, and connected to the other monomer of the b6f complex through an N-terminal trans-membrane α-helix of the ISP. Analysis of the function and structure of the b6f complex isolated from the cyanobacterium Fremyella diplosiphon SF33 shows that the domain-swapped ISP structure is necessary for function but not for maintenance of the dimeric structure (Agarwal et at. 2015). The latter function is proposed to be a consequence of the requirement that the anchoring helix of the ISP not perturb the heme organization or quinone channel in the conserved core of each monomer.
7.2. Heme cn
An important aspect of the structure found in both the cyanobacterial (Kurisu et al., 2003) and algal (Stroebel et al. 2003) structures, but emphasized more in the latter, is the presence of an additional c-type heme relative to the bc1 complex in mitochondria or photosynthetic bacteria, to which the notation heme c1 and heme x, respectively, was conferred in the studies on the C.reinhardtii alga and cyanobacterial structures (Fig. 4).
Fig. 4.
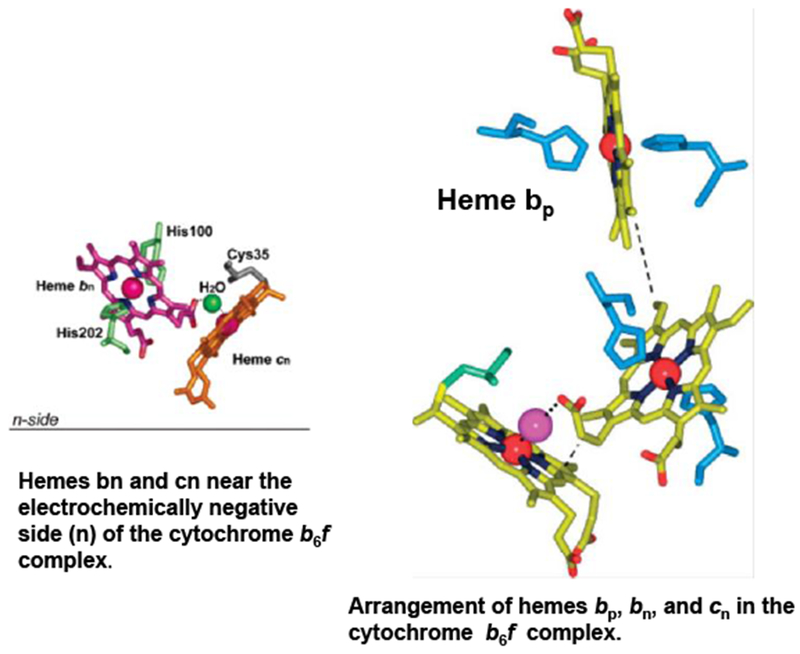
Trans-membrane heme ligation in the cytochrome b6f complex. The central Fe atom of heme bp and bn is axially ligated, respectively, by residues His86/His187 and His100/His202 which bridge the B and D trans-membrane helices of the cytochrome b subunit, Heme cn is covalently attached to the protein via Cys35. Heme cn is unique as it lacks an amino acid as axial ligand; central Fe-atom penta-coordinated. The only ligand of heme cn supplied by H2O or OH− ligand on heme bn side. Central Fe of heme cn separated by 4.0 Å from propionate oxygen of heme bn, resulting in electronic coupling, characterized by a high spin g =12 EPR signal (Zatsman et al. 2006; Baymann et al. 2007)) and an oxidase reaction with nitric oxide (Twigg et al. 2009).
Because of its proximity to the electrochemically negative ‘n’ side of the membrane, for purposes of teaching and discussion, it turned out to be helpful to use heme cn as the notation for this unique heme. It is proposed that the role of heme cn in trans-membrane electron transfer function is significant for structure and function, as the distance between the central Fe atom of heme cn and the propionate of heme bn is only about 4.0 Å, implying that electrons are shared between the two heme entities. This prediction from the structure was verified by EPR analysis of isolated b6f complex and demonstration that heme cn interacts with heme bn via an axial OH− or H2O bridged through an H-bond to a propionate oxygen of heme bn (Cramer et al. 2008; Zatsman et al. 2006; Baymann et al. 2007) It is noted that the structural coupling between hemes bn and cn, which can generate a pathway for two electron transfer in the bn-cn cytochrome component, suggests the possibility of a trans-membrane electron transfer pathway distinct from that described in the classical Q cycle (Mitchell 1966; Crofts et al. 2008).
8. Cytochrome b6f complex as a lipoprotein.
Lipid binding sites and properties were compared of the b6f (Fig. 5, left- and right-hand panels showing the distribution, respectively, of trans-membrane helices and lipids), and bc1 complexes that function in photosynthetic and respiratory membrane energy transduction. Comparison of lipid and detergent binding sites shows significant conservation of lipid positions: Seven lipid binding sites in the cyanobacterial b6f complex overlap three sites in the C. reinhardtii algal complex and four sites in the yeast mitochondrial bc1 complex, although the specific identity of the lipids is different between b6f and bc1 complexes, the b6f complex containing a unique sulfo-lipid,sulfoquinovosyl-diacylglycerol, as well as phosphatidyl-glycerol, phosphatidylcholine (PC), monogalactosyl-diacylglycerol, and digalactosyl-diacylglycerol. IN contrast, cardiolipin, phosphatidyl-ethanolamine (PE), and phosphatidic acid are the major lipid constituents in the yeast bc1 complex. The lipidic chlorophyll a and β-carotene (β-car) in cyanobacterial b6f, as well as eicosane in C. reinhardtii, are unique to the b6f complex. The lipid functions that could operate in the b6f complex are: (i) substitution of a trans-membrane helix by a lipid and chlorin ring, (ii) lipid and β-carotene connection of peripheral and core domains, (iii) stabilization of the iron-sulfur protein trans-membrane helix, (iii) n-side charge and polarity compensation; (iv) β-carotene-mediated super-complex formed with the PS I complex, and (v) stabilization of the domain-swapped TMH of the iron-sulfur protein.
Fig. 5.
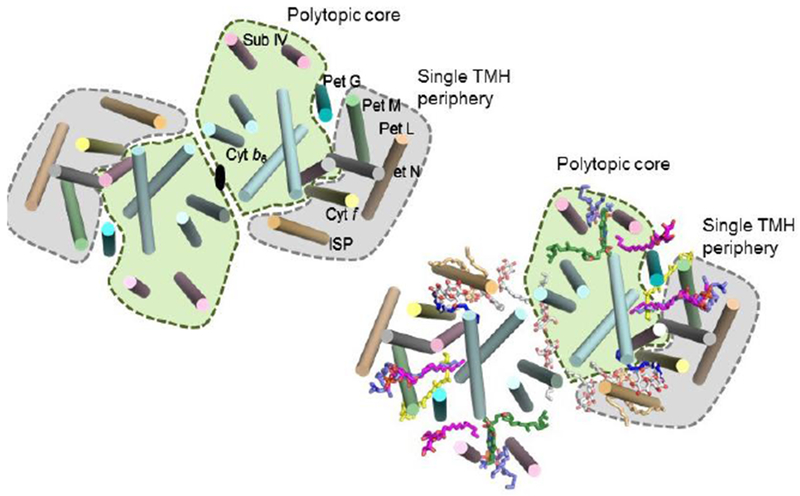
Lipid distribution in the b6f complex. View normal to the plane of the membrane of the b6f complex as a structural core (green) conserved in bc1 and b6f complexes, and external ‘hydrophobic picket fence,’ composed of small petM, N, L, and G peptides, a structural feature unique in b6f complexes. Annular lipids, par-ticularly a β-carotene which protrudes 11 Å from each side of the complex, may provide a connection for super-complex formation, with the photo-system reaction centers, or the LHCII kinase enzvme for trans-membrane signaling (Singh et al, 2016). Internal lipids could mediate (i) cross-linking to stabilize the domain-swapped iron-sulfur protein subunit (Agarwal et al., 2015), (ii) generate dielectric heterogeneity (section 9; Fig. 6) within inter-monomer and intra-monomer electron transfer pathways (Hasan et al. 2014a), and (iii) dimer stabilization through lipid-mediated inter-monomer interactions (Hasan et al. 2014a).
9. Lipids and dielectric heterogeneity
An additional consequence of the multiple lipid binding sites in the cytochrome complex can be linked to the heterogeneity of polarity (i. e., dielectric constants) in the complex (Fig. 6), which was inferred from an analysis of the time course of (a) electron storage in this pathway, utilizing simultaneous measurement of the kinetics of dithionite reduction of the b-type hemes, bn and bp on the two sides of the membrane, and (b) Soret band excitonically split circular dichroism (CD) spectra which result from heme-heme interactions (Palmer and Degli-Esposti, 1994). Information on heme-heme interactions was obtained from kiinetic analysis of the Soret band CD spectrum initiated by addition of reductant. CD analysis is used because the identity of the b-hemes (bn or bp) on the two sides of the membrane reduced through oxidant (plastoquinone)-induced b-heme reduction cannot be accurately determined through spectrophotometric absorbance analysis at room temperature because (a) their α-band wavelength (λ) maxima (λm) are very similar (Δλm < 1 nm) [Joliot and Joliot, 1988]; (b) only approximately 0.5 b heme is reduced by a saturating light flash, and it is not possible to determine whether it is heme bp or bn (Furbacher et al., 1989).
Fig. 6.
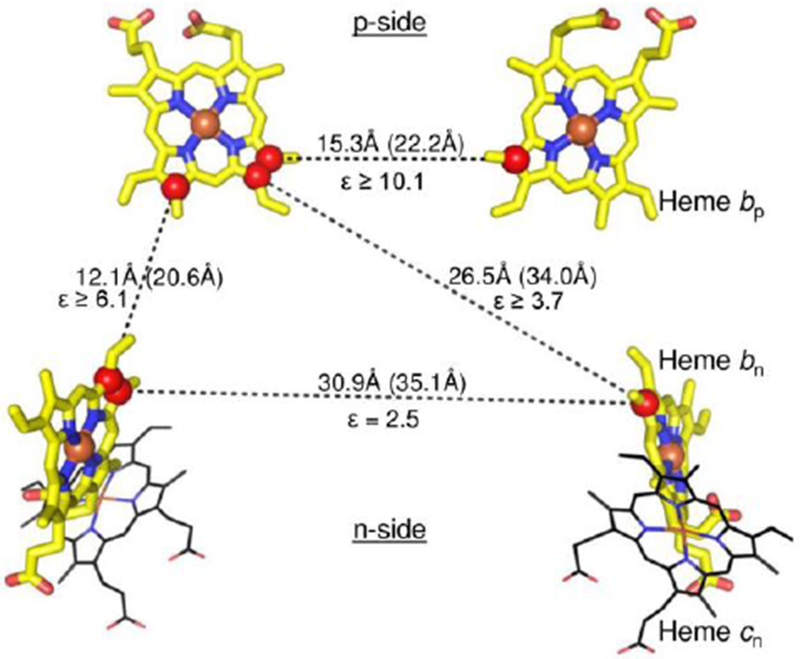
The Intra-membrane and inter-heme distance parameters and dielectric constants in the cytochrome b6f complex from the cyanobacterium Nostoc PCC 7120, showing the orientation of hemes bn, bp, and cn (Hasan et al. 2014a). Components of the [2Fe–2S] cluster of the Rieske iron–sulfur protein are depicted as spheres (Fe, brown; sulfur, yellow). Geometry of hemes within the trans-membrane domain of cytochrome b6f (PDB 40GQ). Edge–edge and center–center (Fe–Fe, in parentheses) distances are shown, along with the effective inter-heme dielectric constants.
Note: the dielectric constant varies from 2.5 (n-side) to at least 10 on the p-side.
The Soret band CD signal is dominated by excitonic interaction between the intra-monomer b-hemes, bn and bp, on the electrochemically negative and positive sides of the complex. Kinetic data imply that the most probable pathway for transfer of the two electrons needed for quinone oxidation-reduction utilizes this intra-monomer heme pair, contradicting the expectation based on heme redox potentials and thermodynamics, that the two higher potential hemes bn on different monomers would be preferentially reduced. Energetically preferred intra-monomer electron storage of electrons on the intra-monomer b-hemes requires heterogeneity of inter-heme dielectric constants. Relative to the medium separating the two higher potential hemes bn, a relatively large dielectric constant must exist between the intra-monomer b-hemes, allowing a smaller electrostatic repulsion between the reduced hemes. Thus, heterogeneity of intra-protein dielectric constants is an additional structure-function parameter of membrane protein complexes (Hasan et al., 2014a). The sequence of b-heme reduction in the b6f complex does not follow the prescription of the Q-cycle mechanism. The latter mechanism was, however, found to apply to the bacterial bc1 complex where the difference in redox potentials between n- and p-side hemes is substantially larger (Bhaduri et al., 2016).
10. Plastoquinol-quinone passage in p-side channel of b6f complex, binding site of quinone analog inhibitors, e.g., tridecyl-stigmatellin.
Quinol oxidation on the electro-chemically positive, p-side interface (Qp site) of the complex occurs at the end of a narrow quinol/quinone entry/exit portal, 11 Å long in the b6f complex (Hasan & Cramer, 2014; PDB 4OGQ) , which connects to the protein-deficient space between the two monomers (Fig. 7). Superoxide, which has multiple signaling extra-organelle signaling functions, is a product of the p-side quinol oxidation, and is generated at a rate approximately 1% of that of non-cyclic electron transport (Baniulis et al., 2013). Although the trans-membrane core and the chemistry of quinone redox reactions are conserved in cytochrome bc complexes, the rate of superoxide generation is an order of magnitude greater in the b6f complex relative to bc1. It was subsequently found that amino acid residues near the p-side of the b6f complex and the unique single chlorophyll molecule are oxidatively modified (Taylor et al., 2018).
Figure 7.
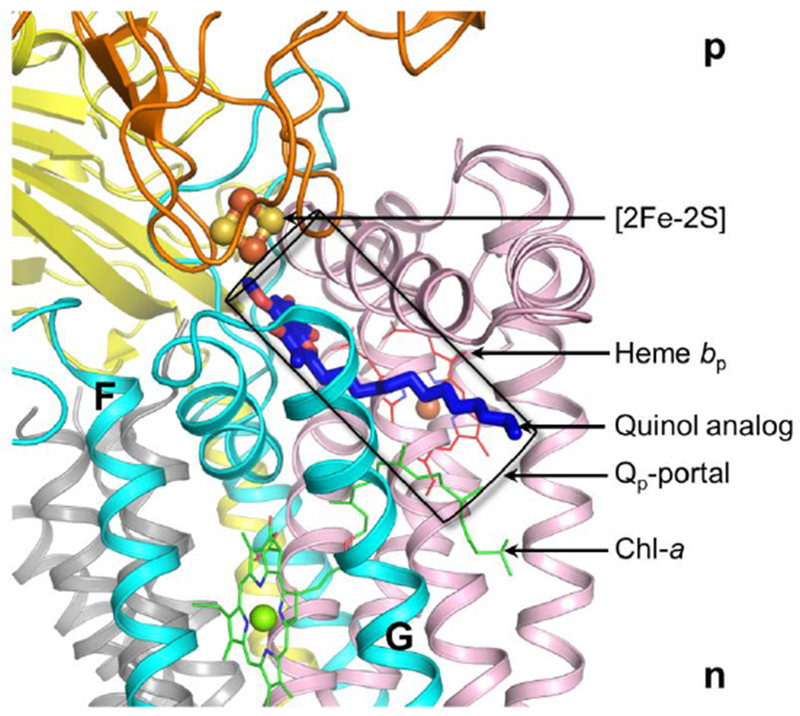
p-side quinone analog inhibitor, tridecyl-stigmatellin, bound in an 11 Å long channel (in purple) terminating proximal to the p-side 2Fe-2S cluster, the electron-proton acceptor of plastohydroquinone, PQH2, the Qp-portal in the cytochrome b6f complex for p-side quinol/quinone traffic. As inferred from the position of the bound TDS molecule, the quinol within the Qp-portal interacts with the [2Fe-2S] cluster that is covalently linked to the ISP extrinsic domain, which is shown as orange ribbons. A chlorophyll molecule (Chl-a), inserted between F and G helices of subunit IV (cyan), using its phytyl tail, functions as a gate for quinol/quinone traffic in the Qp-portal (Hasan and Cramer, 2012). Drawing by S. Saif Hasan.
The portal leading to the Qp site, can be occupied by quinone analog/inhibitors such as tridecyl-stigmatellin (TDS) in the Qp portal of the b6f complex (Fig. 7). A unique structural feature of the b6f complex near the p-side plastoquinone (Qp) binding site is the presence of a single chlorophyll-a molecule whose function is unrelated to light-harvesting. Crystal structure data that describe the position of the chlorophyll phytyl chain show that it partially occludes the Qp-portal of the substrate plastoquinone/ol. Documented by molecular dynamics analysis, the presence of the chlorophyll phytyl chain increases the residence time of plastoquinone in the Qp portal, implying that it exerts a plastoquinone gating function in the inter-monomer space that serves as a PQH2/PQ entry/exit portal (Hasan et al. 2014b). As described by the relatively slow (msec) reduction of cytochrome f, the rate limiting step in the linear electron transport chain is the oxidation of plastoquinol (Levine et al., 1966). Considering the structure basis for the rate limitation, it is proposed that it is a consequence of the steric restrictions on trans-membrane passage of plastoquinol imposed by the structure of the b6f complex, perhaps most obviously the narrow 11 Å long p-side channel for passage of the quinol molecule, mentioned above in the context of the TDS binding site of the TDS quinol analogue inhibitor (Fig. 7B, C). An alternative proposal for the rate-limiting step is proton transfer from the quinol to the p-side iron-sulfur protein (Wilson & Crofts, 2018).
11. Studies on the rate-limiting step in electron transport; steric considerations in quinol oxidation-reduction.
As suggested by the crystal structure (Figs. 3, 7A-C) and discussed above, the passage of the bulky lipophilic quinol to the site that allows oxidation and deprotonation by the Rieske [2Fe-2S] protein can be proposed as a rate-limiting step. As discussed above, to reach its oxidation site, the quinol must be transferred from the inter-monomer cavity between the two monomers through a narrow portal to a position within H-bond distance of the His129 imidazole ligand to the [2Fe-2S] cluster (Fe, orange, sulfur atoms yellow). Fig. 7B shows the position of the p-side quinone analogue inhibitor, tridecyl-stigmatellin, as defined in the crystal structure of the complex from M. laminosus (Yamashita et al., 2007). The portal is defined by two TMH of: the F trans-membrane helix of subunit IV (subIV) and the C-trans-membrane helix of the cyt b polypeptide. Fig. 7C shows the amino acids, almost all hydrophobic, with many valines, which line the portal.
Quinol/quinone (QH2/Q) entry/exit through the p-side portal shown in Figs. 7B, C, which would occur twice in each turnover of the Q-cycle (Mitchell, 1975; Crofts, 2004) , is hypothesized to contribute to rate limitation in the linear electron (H2O → NADP+) transport chain, depending upon the intensity of illumination and CO2 concentration. This hypothesis can be tested (Hasan and Cramer, 2012). The p-side site of plastoquinol oxidation (Qp site) lies at the end of the narrow portal formed by trans-membrane helix C of the cyt b6 subunit and the F-helix of subunit IV. Two conserved prolines are on the p-side of the subunit IV F-helix. These two residues lie on the same side of the helix, proximal to the C-helix. The two prolines cause a bend in the F-helix, away from the C-helix, thereby contributing to the size of the portal that leads into the Qp site for electron/proton donation to the Rieske iron-sulfur protein and, ultimately, to proton donation to the p-side aqueous phase.
11.1. Testing a Putative Rate-Limiting Step; the Quinone Portal.
The role of the p-side portal in the rate limitation of the linear electron transport chain can be tested by (i) mutagenesis of the F-helix in subunit IV of the b6f complex, (ii) measurement of the effect on quinol oxidation by flash kinetic spectroscopy and, (iii) if a particular mutational change is implicated by kinetic measurements, purification crystallization, and crystal structure analysis of the mutant b6f complex. It is proposed that introduction of additional proline residues via substitution mutations of other residues that lie on side of the F-helix facing the C-helix would bend the F-helix further away from the C-helix and thus enlarge the Qp site portal (Hasan and Cramer, 2012). Proline residues 105 and 112, which are conserved residues, define positions of kinks in this helix that result in a change in the aperture of the portal. Site-directed mutagenesis has been used to decrease the effective size of this quinol/quinone portal/ channel in the F-helix of subunit IV, resulting in a decreased rate of growth and oxygen evolution, and a decreased rate of cytochrome reduction (manuscript in preparation).
12. Role of cytochrome b6f complex in trans-membrane signaling; LHCP kinase (53,54).
A trans-membrane signaling protein that transduces a redox signal on the p-side of the thylakoid membrane to function as a serine-threonine kinase (Stt7 in C. reinhardtii) on the n- or stroma side can control light energy distribution between the two photosystems (Allen et al., 1981; Barber, 1982; Millner et al., 1982; Vener et al., 1997; Lemeille et al., 2009). Oxidation of plastoquinol mediated by photosystem I and cytochrome f on the p-side of the thylakoid membrane is considered to initiate a trans-membrane activation of the kinase, leading to phosphorylation and redistribution (“state transition”) of the light-harvesting chlorophyll proteins between the two photosystems (Barber, 1982).
Stt7 has been cloned, expressed, and purified in a heterologous host (Singh et al., 2016), and was shown (a) to be active in vitro in the presence of reductant, and (b) to be purified as a tetramer, assayed by analytical ultracentrifugation, electron microscopy and electrospray-ionization mass spectrometry, to have a molecular weight of 332 kDa, consisting of four 83.4 kDa monomers (Singh et al., 2016). (c) Far-UV circular dichroism (CD) spectra show (i) Stt7 in solution to be mostly α-helical, and (ii) to interact with the b6f complex as documented by the increased thermal stability of Stt7 (function ‘d’ vs ‘a’ in Fig. 8) secondary i.e., helical, structure ( Singh et al., 2016) (Fig. 8). It can be inferred from this demonstration of perturbation of the net secondary structure of Stt7 in the presence of the b6f complex that the Stt7 can bind to/dock on the b6f complex. It has been suggested that this interaction, possibly transient, provides the structural framework for a trans-membrane configuration of the Stt7 through which plastoquinol reduction of the Stt7 at suggested redox active groups Cys 68 and 73, on the p-side of the membrane adjacent to the one hydrophobic putative span, residues 97 -123, can transmit a signal across the membrane and activate the n-side LHCP-kinase. Site-directed mutagenesis has located a specific site of interaction of the putative kinase on the n-side of subunit IV of the b6f complex (Dumas et al. 2017).Together with the far-UV CD measurements, it can be inferred that the kinase does interact with the b6f complex. However, at this the concept of a redox-activated trans-membrane signaling mechanism is questionable: (a) the binding site of the proposed p-side docking area on the Rieske protein is not defined; (b) the mechanism of redox-induced activation of the kinase has been questioned (Shapiguzov et al., 2016); (c) the putative trans-membrane structure of the kinase, particularly the C. reinhardtii Stt7 kinase, is made highly questionable by the presence of four proline residues in a 14 residue span of its putative trans-membrane hydrophobic domain.
Fig. 8. Far-UV circular dichroism spectra of Stt7.
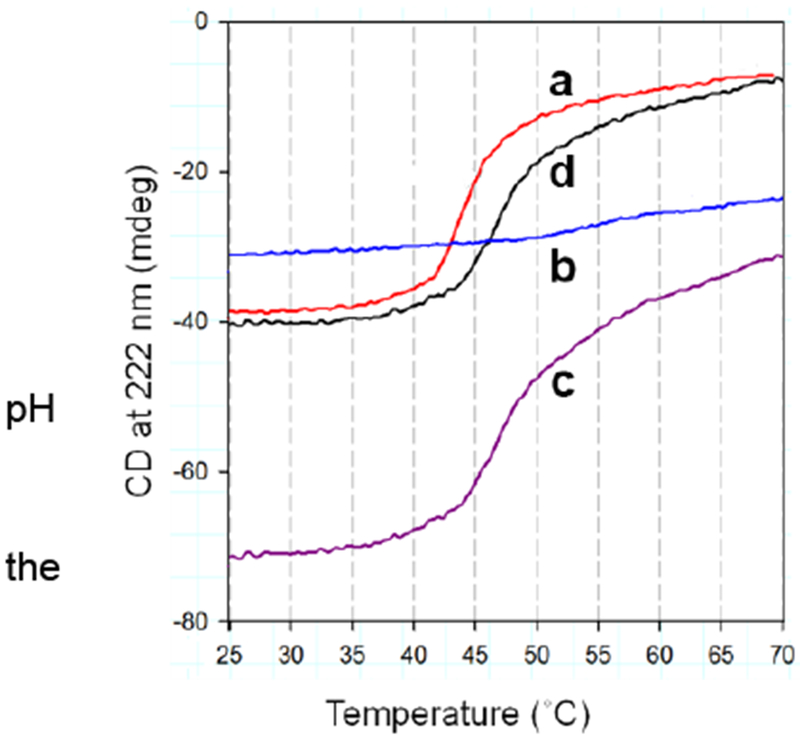
(A) Far-UV CD spectrum of purified Stt7 in 0.1 M phosphate buffer, pH 8, 12 mM βME and 0.1% UDM. The spectrum with pronounced negative peaks at 208 and 222 nm shows that the protein is primarily α-helical. (B) Temperature dependence of ellipticity of Stt7 and b6f complex, measured at 222 nm in 0.1 M phosphate buffer, 8, 0.1 % UDM of: (a) isolated Stt7 (red), (b) spinach b6f complex (blue), (c) Stt7 together with b6f complex (purple), (d) Thermally induced melting function of Stt7 in presence of b6f (black), determined by the difference between functions (c) and (b). Discussed at greater length in Singh et al. 2016.
13. PSII - b6f ‘Super-Complex’?
With respect to the sequence of electron transfer and proton translocation, the position of the b6f complex relative to the photosystem reaction center complexes I and II is shown above (Fig. 1). Although this representation is essentially conventional, it omits the recent realization that many of the individual light-harvesting and electron transport proteins in the chain function in complexes (‘super-complexes’ in the modern jargon). Thus, the b6f complex can be isolated in a complex with the photosystem I reaction center complex including FNR (Iwai et al., 2010), with an FNR-b6f sub super-complex determined separately (Zhang et al., 2001). However, no such ‘super-complex’ has yet been reported between the PSII reaction center and the b6f complex, and the question arises as to whether it exists. Of course, as shown in Fig. 1, the redox connection between PSII and b6f is unique in that the charge transfer, involving hydrogen transfer mediated by plastoquinone/ol, is of a different nature. One might then infer that this mode of intra-membrane charge transfer, the transfer involving a bulk molecule rather than an electron, cannot occur in a super-complex. In contrast, however, the entire mitochondrial electron transport chain, the ‘respirasome’, which includes the transfer of ubiquinone/ol has been isolated and visualized by cryo-electron microscopy (Kühlbrandt 2015).
A consequence of the spatial separation between the PSII and b6f complexes is the implication that plastoquinone (-ol) is transferred between the two complexes via diffusion through the lipid bilayer membrane. Historically, the competence of such a connection between the two complexes was inferred through measurement of the diffusion in membranes of plastoquinone, PQ (Blackwell and Whitmarsh 1990), or ubiquinone, UQ (Chazotte and Hackenbrock 1989). However, although this mechanism might be kinetically competent if the quinone/quinol had free access from many directions to its binding sites in the PSII reaction center and the b6f complex. However, now that structure data are available, it is clear that access to the quinone/quinol binding sites in the PSII reaction center (van Eerden et al 2017) and to the narrow niche in the inter-domain cleft, and to the quinone channel on the p-side, of the b6f complex (Fig. 7B, C) is very limited. The niche for access of PQH2 to the b6f complex is so small and restricted that the probability is very small that PQH2 produced in the PSII reaction center can find the niche in the channel that leads to the 2Fe-2S cluster. This, one inevitably thinks about the possibility of a super-complex between the PSII and b6f complexes. Although no “super-complex” of the PSII and b6f complexes has been found, EM analysis has shown fields in which clusters of PSII and b6f complexes are in close proximity (Johnson et al 2014). It is suggested that super-complexes either form transiently between PSII reaction centers and b6f complexes, or that the two complexes are arranged with little membrane lipid space between them. Investigation of this problem, proximity and interaction between the b6f and PSII complexes, is a high priority.
Acknowledgments.
Colleagues whose studies have contributed to the information and concepts obtained in our laboratory in recent years, which are reported here: R. Agarwal,7 D. Baniulis,6 S. Bhaduri,1 S. Saif Hasan,1 D. Huang (deceased), G. Kurisu,9 S.Naurin,1 S. Savikhin,2 S. K. Singh,8 J. L. Smith11, V. Stadnytskyi,2,3 A. Szczepaniak,12 J.P. Whitelegge,5 E. Yamashita,10 S.D. Zakharov,1 and Dr. H. Zhang4 who are presently associated, respectively, with the Departments of Biological Sciences1 and Physics,2 Purdue University; 3 Laboratory of Chemical Physics, NIDDK, National Institutes of Health, Bethesda, MD USA; 4SSCI, West Lafayette, IN; 5Pasarow Mass Spectrometry Laboratory, NPI-Semel Institute, David Geffen School of Medicine, UCLA; 6Institute of Horticulture, Lithuanian Research Centre for Agriculture and Forestry, Lithuania; 7Molecular Biology Division, Bhabha Atomic Research Centre, Mumbai, India; 8Department of Biological Chemistry & Molecular Pharmacology, Harvard Medical School, Boston, MA; 9Lab. of Protein Crystallography, Lab. of Protein Databases (PDB), Institute for Protein Research, Osaka University; 10Institute for Protein Research, Osaka University, Japan; 11Life Sciences Institute, University of Michigan; 12Dept. of Biotechnology, University of Wroclaw, Poland.
Colleagues whose earlier studies provided the foundation for the information on structure-function of the cytochrome b6f complex described in third report: M. T. Black, H. Böhme, R. M. Everly, P. N. Furbacher, Μ. E. Girvin, P. Horton, L. I. Krishtalik, M. Ponomarev, G. S. Tae, J. Whitmarsh, and W. R. Widger.
I thank Ms. G. Sincich for contributions to the illustrations, and express my gratitude to the NIH General Medical Sciences-038323, and Dept, of Energy, DOE DE-SC0018238), which have funded most of the research described here, and to the John Simon Guggenheim and Alexander von Humboldt Foundations for Fellowship support.
Abbreviations
- CD
circular dichroism
- Chl
chlorophyll
- cyt
cytochrome
- n, p
electrochemically negative, positive sides of membrane
- DOPC
dioleoyl-phosphatidyl choline
- EPR
electron paramagnetic resonance
- ISP
iron-sulfur protein
- PDB
protein data base
- PET
photosynthetic electron transfer
- PQ, PQH2
plastoquinone, -ol
- PSI, PSII
photosystems I, II
- TMH
trans-membrane helix
- UDM
n-undecyl-β-d-malto-pyranoside
trans-membrane proton electrochemical potential gradient
Photos of Author and Lab (2017) Colleagues

(above) moving toward a conceptual break-through; (below) after the canoe trip

References
- Agarwal R, Hasan SS, Jones LM, Stofleth JT, Ryan CM, Whitelegge JP, Kehoe DM, Cramer WA (2015) Role of domain swapping in the hetero-oligomeric cytochrome b6f lipoprotein complex. Biochemistry 54: 3151–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF, Bennett J, Steinback KE and Arntzen CJ (1981) Chloroplast protein phosphorylation couples plastoquinone redox state to distribution of excitation energy between photosystems. Nature 291,25–29.. [Google Scholar]
- Baniulis D, Hasan SS, Stofleth JT, and Cramer WA (2013) Mechanism of Enhanced Superoxide Production in the Cytochrome b6f Complex of Oxygenic Photosynthesis. Biochemistry-Us 52, 8975–8983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniulis D, Yamashita E, Whitelegge JP, Zatsman AI, Hendrich MP, Hasan SS, Ryan CM, and Cramer WA (2009) Structure-Function, Stability, and Chemical Modification of the Cyanobacterial Cytochrome b6f Complex from Nostoc sp. PCC 7120. J. Biol. Chem 284, 9861–9869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniulis D, Zhang H, Yamashita E, Zakharova T, Hasan SS, Cramer WA (2011) Purification and crystallization of the cyanobacterial cytochrome b6f complex In: Methods in Molecular Biology, Photosynthesis Research Protocols (Carpentier R , ed), Humana Press Inc, Totowa, NJ, pp 65–77 [DOI] [PubMed] [Google Scholar]
- Barber J (1982) Influence of surface charge on thylakoid structure and function. Ann. Rev. Plant Physiol , 33, 261–295 [Google Scholar]
- Barrera NP, Zhou M, Robinson CV (2013) The role of lipids in defining membrane protein interactions: insights from mass spectrometry. Trends Cell Biol 23: 1–8 [DOI] [PubMed] [Google Scholar]
- Baymann F, Giusti F, Picot D, Nitschke W.(2007) The ci/bH moiety in the b6f complex studied by EPR: a pair of strongly interacting hemes. Proc Nat Acad Sci USA 104: 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall DS (1982) Photosynthetic cytochromes of oxygenic organisms. Biochim Biophys Acta 683: 119–151 [Google Scholar]
- Bhaduri S, Stadnytski V, Zakharov SD, Hasan SS, Bujonowicz L, Sarewicz M, Savikhin S, Osyczka A, Cramer WA (2016) Pathways of Transmembrane Electron Transfer on Cytochrome bc Complexes: Dielectric Heterogeneity and Interheme Coulombic Interaxctions. J. Phys. Chem B: 121: 975–983. [DOI] [PubMed] [Google Scholar]
- Blackwell MF, and Whitmarsh J (1990) Effect of integral membrane proteins on the lateral mobility of plastoquinone in phosphatidylcholine proteoliposomes. Biophys. J 58, 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman NK, Anderson JM (1967) Fractionation of the photochemical systems of photosynthesis. II. Cytochrome and carotenoid contents of particles isolated from spinach chloroplasts. Biochim Biophys Acta 143: 187–203 [DOI] [PubMed] [Google Scholar]
- Carrell CJ, Zhang H, Cramer WA, Smith JL (1997) Biological identity and diversity in photosynthesis and respiration: structure of the lumen-side domain of the chloroplast Rieske protein. Structure 5: 1613–1625 [DOI] [PubMed] [Google Scholar]
- Carrell CJ, Schlarb BG, Bendall DS, Howe CJ, Cramer WA, Smith JL.(1999) Structure of the soluble domain of cytochrome f from the cyanobacterium, Phormidium laminosum. Biochemistry 38: 9590–9599 [DOI] [PubMed] [Google Scholar]
- Chazotte B, and Hackenbrock CR (1989) Lateral Diffusion as a Rate Limiting Step in Ubiquinone-mediated Mitochondrial Electron Transport. J. Biol. Chem 264, 4978–4985 [PubMed] [Google Scholar]
- Cramer WA, Hasan SS (2016) Structure-function of the cytochrome b6f lipoprotein complex In: Cytochrome Complexes: Evolution, Structures, Energy Transduction, and Signaling (Cramer WA, Kallas Teds.), Advances in Photosynthesis and Respiration, Vol ,41 , pp. 177–207, Springer, Dordrecht. [Google Scholar]
- Cramer WA, Martinez SE, Huang D, Tae G-S, Everly RM, Heymann JB, Cheng RH, Baker TS, Smith JL (1994) Structural aspects of the cytochrome b6f complex: Structure of the lumen-side domain of cytochrome f. J. Bioenerg Biomemb 26: 31–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer WA, Baniulis D, Yamashita E, Zhang H, Zatsman AI, Hendrich MP (2008) Structure, spectroscopy, and function of the cytochrome b6f complex: heme cn and n-side electron and proton transfer reactions In: Photosynthetic Protein Complexes: a Structural Approach (Fromme, ed), Wiley-VCH, Weinheim: pp 155–179 [Google Scholar]
- Crofts AR (2004) The cytochrome bc1 complex: Function in the context of structure. Ann. Rev. Physiol 66, 689–733 [DOI] [PubMed] [Google Scholar]
- Crofts AR, Holland JT, Victoria D, Kolling DR, Dikanov SA, Gilbreth R, Lhee S, Kuras R, Kuras MG (2008) The Q-cycle reviewed: How well does a monomeric mechanism of the bc1 complex account for the function of a dimeric complex? Biochim Biophy. Acta 1777: 1001–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashdorj N, Zhang H, Kim H, Yan J, Cramer WA, and Savikhin S (2005) The single chlorophyll a molecule in the cytochrome b6f complex: unusual optical properties protect the complex against singlet oxygen. Biophysical journal 88, 4178–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenhofer J, and Michel H (1989) Nobel lecture. The photosynthetic reaction centre from the purple bacterium Rhodopseudomonas viridis. The EMBO journal 8, 2149–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibrova DV, Cherepanov DA, Galperin MY, Skulachev VP, and Mulkidjanian AY (2013) Evolution of cytochrome bc complexes: from membrane-anchored dehydrogenases of ancient bacteria to triggers of apoptosis in vertebrates. Biochim Biophys Acta 1827, 1407–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas L, Zito F, Clangy S, Auroy P, Johnson X, Peltier G, and Alric J (2017) A stromal region of cytochrome b6f subunit IV is involved in the activation of the Stt7 kinase in Chlamydomonas Proc. Natl., Acad. Sci., U. S 114, 12063–12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duysens LNM (1955) Role of Cytochrome and Pyridine Nucleotide in Algal Photosynthesis. Science (New York., N.Y 121, 210–211 [DOI] [PubMed] [Google Scholar]
- Esser L, Zhou F, Yu C-A, and Xia D (2015) Cytochrome Complexes: Evolution, Structures, Energy Transduction, and Signaling in Cytochrome Complexes: Evolution, Structures, Energy Transduction, and Signaling (Cramer WA, and Kallas T eds.), Springer SBM NL, Dordrecht. pp. [Google Scholar]
- Ferreira KN, Iverson TM, Maghlaoui K, Barber J, and Iwata S (2004) Architecture of the photosynthetic oxygen-evolving center. Science (New York, N.Y 303, 1831–1838. [DOI] [PubMed] [Google Scholar]
- Furbacher PN, Girvin ME, and Cramer WA 1989. On the question of interheme electron transfer in the chloroplast cytochrome b6 in situ. Biochemistry, 28:8990–8998. [DOI] [PubMed] [Google Scholar]
- Hasan SS, and Cramer WA (2012) On Rate Limitations of Electron Transfer in the Photosynthetic Cytochrome b6f Complex 2012, 14, 13853 – 13860. Physical Chemistry Chemical Physics 14, 13853–13860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SS, and Cramer WA (2014) Internal lipid architecture of the hetero-oligomeric cytochrome b6f complex. Structure 22, 1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SS, Yamashita E, Ryan CM, Whitelegge JP, and Cramer WA (2011) Conservation of lipid functions in cytochrome bc complexes. J Mol Biol 414, 145–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SS, Yamashita E, Baniulis D, and Cramer WA (2013a) Quinone-dependent proton transfer pathways in the photosynthetic cytochrome b6f complex Proc. Natl. Acad. Sci., U. S. A 110, 4297–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SS, Yamashita E, and Cramer WA (2013b) Trans-membrane Signaling and Assembly of the Cytochrome b6-Lipidic Charge Transfer Complex Biochim. Biophys. Acta 1827 1295–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SS, Zakharov SD, Chauvet A, Stadnytskyi V, Savikhin S, and Cramer WA (2014a) A map of dielectric heterogeneity in a membrane protein: the hetero-oligomeric cytochrome b6f complex. J Phys Chem B 118, 6614–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SS, Proctor EA, Yamashita E, Dokholyan NV, and Cramer WA (2014b) Traffic within the Cytochrome b6f Lipoprotein Complex: Gating of the Quinone Portal. Biophysical journal 107, 1620–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimann S, Ponamarev MV, and Cramer WA. 2000. Movement of the Rieske iron-sulfur protein in the p-side bulk aqueous phase: effect of lumenal viscosity on redox reactions of the cytochrome b6f complex. Biochemistry , 39:2622–2629 [DOI] [PubMed] [Google Scholar]
- Huang D, Everly RM, Cheng RH, Heymann JB, Schagger H, Sled V, Ohnishi T, Baker TS, and Cramer WA (1994) Characterization of the chloroplast cytochrome b6f complex as a structural and functional dimer. Biochemistry-Us 33, 4401–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai M, Takizawa K, Tokutsu R, Okamuro A, Takahashi Y, and Mlnagawa J (2010) Isolation of the supercomplex that drives cyclic electron flow in photosynthesis Nature 464, 1210–1213 [DOI] [PubMed] [Google Scholar]
- Jagendorf AT, and Uribe E (1966) ATP formation caused by acid-base transition of spinach chloroplasts. Proceedings of the National Academy of Sciences of the United States of America 55, 170–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MP, Vasilev C, Olsen JD, and Hunter CN (2014) Nanodomains of cytochrome b6f and photosystem II complexes in spinach grana thylakoid membranes. The Plant Cell 26, 3051–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P, and Johnson GN (2011) Regulation of cyclic and linear electron flow in higher plants. Proceedings of the National Academy of Sciences of the United States of America 108, 13317–13322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P and Joliot A (1988) The low potential elecron transfer chain in the cytochrome bf complex. Biochim. Biophys. Acta, 933, 319–333. [Google Scholar]
- Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, and Krauss N (2001) Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411, 909–917 [DOI] [PubMed] [Google Scholar]
- Kao WC, and Hunte C (2014) The molecular evolution of the Qo motif. Genome biology and evolution 6, 1894–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Dashdorj N, Zhang H, Yan J, Cramer WA, and Savikhin S (2005) An Anomalous Distance Dependence of Intra-Protein Chlorophyll-Carotenoid Triplet Energy Transfer. Biophysical journal 89, 28–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrandt W (2015) Structure and function of mitochondrial membrane protein complexes. BMC Biol 13/89, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu G, Zhang H, Smith JL, and Cramer WA (2003) Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science (New York, N.Y 302, 1009–1014 [DOI] [PubMed] [Google Scholar]
- Lemeille S, Willig A, Depege-Fargeix N, Delessert C, Bassi R, and Rochaix JD (2009) Analysis of the Chloroplast Protein Kinase Stt7 during State Transitions. PLoS Biology 7, 664–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RP, Gorman DS, Avron M, and Butler WL (1966) Light-Induced Absorbance Changes in Wild-Type and Mutant Strains of Chlamydomonas reinhardtii. Brookhaven Symp Biol. 19, 143–148 [PubMed] [Google Scholar]
- Martinez SE, Smith JL, Huang D, Szczepaniak A, and Cramer WA (1992) Crystallographic studies of the lumen-side domain of turnip cytochrome f in Research in Photosynthesis (Murata N ed.), Kluwer Academic Publishers, Dordrecht: pp 495–498 [Google Scholar]
- Martinez SE, Huang D, Szczepaniak A, Cramer WA, and Smith JL (1994) Crystal structure of the chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation. Structure 2, 95–105 [DOI] [PubMed] [Google Scholar]
- Martinez S, Huang D, Ponamarev M, Cramer WA, and Smith JL (1996) The heme redox center of chloroplast cytochrome f is linked to a buried five-water chain. Protein Sci. 5, 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millner PA, Widger WR, Abbott MS, Cramer WA, and Dilley RA (1982) The effect of adenine nucleotides on inhibition of the thylakoid protein kinase by sulfhydryl-directed reagents. The Journal of biological chemistry 257, 1736–1742 [PubMed] [Google Scholar]
- Mitchell P (1966) Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol. Rev. 41, 445–502 [DOI] [PubMed] [Google Scholar]
- Mitchell P (1975) The protonmotive Q cycle: A general formulation. FEBS letters 59, 137–139 [DOI] [PubMed] [Google Scholar]
- Mulkidjanian A, Koonin E, Makarova K, Haselkorn R, and Galperin M (2007) The cyanobacterial genome core and the origin of photosynthesis. Photosyn. Res 91, 269–269 [Google Scholar]
- Nitschke W, van Lis R, Schoepp-Cothenet B, and Baymann F (2010) The “green” phylogenetic clade of Rieske/cyt b complexes. Photosyn. Res 104, 347–355 [DOI] [PubMed] [Google Scholar]
- Palsdottir H, and Hunte C (2004) Lipids in membrane protein structures. Biochim. Biophys. Acta 1666, 2–18 [DOI] [PubMed] [Google Scholar]
- Palmer G, and Degli-Esposti M (1994) Application of exciton coupling theory to the structure of mitochondrial cytochrome b. Biochemistry-US. 33, 176–185. [DOI] [PubMed] [Google Scholar]
- Ponamarev MV, and Cramer WA (1998) Perturbation of the internal water chain in cytochrome f of oxygenic photosynthesis: loss of the concerted reduction of cytochromes f and b6. Biochemistry-Us 37, 17199–17208 [DOI] [PubMed] [Google Scholar]
- Rieske JS, Hansen RE, and Zaugg WS (1964) Studies on the Electron Transfer System. 58. Properties of a New Oxidation-Reduction Component of the Respiratory Chain as Studied by Electron Paramagnetic Resonance Spectroscopy. J. Biol. Chem 239, 3117–3022 [PubMed] [Google Scholar]
- Rochaix JD (2014) Regulation and dynamics of the light-harvesting system. Annu Rev Plant Biol 65, 287–309 [DOI] [PubMed] [Google Scholar]
- Sainz G, Carrell CJ, Ponamarev MV, Soriano GM, Cramer WA, and Smith JL (2000) Interruption of the internal water chain of cytochrome f impairs photosynthetic function. Biochemistry-Us 39, 9164–9173 [DOI] [PubMed] [Google Scholar]
- Shapiguzov A, Chai X, Fucile G, Longoni P, Zhang L, and Rochaix JD (2016) Activation of the Stt7/STN7 Kinase through Dynamic Interactions with the Cytochrome b6f Complex. Plant physiology 171, 82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Hasan SS, Zakharov S, Naurin S, Cohn W, Ma J, Whitelegge JP, and Cramer WA (2016) Trans-membrane signaling in photosynthetic state transitions: redox and sructure-dependent interaction in vitro between Stt7 kinase and the cytochrome b6f complex. Journal of Biological Chemistry 291, 21740–21750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano GM, Ponamarev MV, Tae G-S, and Cramer WA (1996) Effect of the interdomain region of cytochrome f on its redox reactions in vivo. Biochemistry-Us 35, 14590–14598 [DOI] [PubMed] [Google Scholar]
- Soriano GM, Cramer WA, and Krishtalik LI (1997) Electrostatic effects on electron-transfer kinetics in the cytochrome f-plastocyanin complex. Biophysical journal 73, 3265–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano GM, Ponamarev ΜV, Piskorowski RA, and Cramer WA (1998) Identification of the basic residues of cytochrome f responsible for electrostatic interactions with plastocyanin in vitro: relevance to the electron transfer in vivo. Biochemistry-Us 37, 15120–15128 [DOI] [PubMed] [Google Scholar]
- Stroebel D, Choquet Y, Popot J-L, and Picot D (2003) An atypical heam in the cytochrome bef complex. Nature 426, 413–418 [DOI] [PubMed] [Google Scholar]
- Taylor RM, Sallans L, Frankel LK, and Bricker TM (2018) Natively oxidized amino acid residues in the spinach b6f complex. Photosyn Res. 137, 141–15. [DOI] [PubMed] [Google Scholar]
- Twigg AI, Baniulis D, Cramer WA, and Hendrich ΜP (2009) EPR detection of an 02 surrogate bound to heme cn of the cytochrome b6f complex. J. Am. Chem. Soc 131, 12536–12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiyaveetil FI, Zhou Y, and MacKinnon R (2002) Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry-Us 41, 10771–10777 [DOI] [PubMed] [Google Scholar]
- Van Eerden FJ, Melo MN, Frederix JM, Periole X, and Marrink SJ (2017) Exchange pathways of plastoquinone and plastoquinol in the photosystem II complex. Nature Commun, 8, 15214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vener AV, van Kan PJ, Rich PR, Ohad I, and Andersson B (1997) Plastoquinol at the quinol oxidation site of reduced cytochrome bf mediates signal transduction between light and protein phosphorylation: thylakoid protein kinase deactivation by a single-turnover flash. Proc Natl Acad Sci USA 94, 1585–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SH (2018) “Membrane Proteins of Known Structure,” www.blanco.biomol.uci.edu/mpstruc/list
- Whitelegge JP, Zhang H, Taylor R, and Cramer WA (2002) Full subunit coverage liquid chromatography electrospray-ionization mass spectrometry (LCMS+) of an oligomeric membrane protein complex: the cytochrome b6f complex from spinach and the cyanobacterium, M. laminosus. Mol. Cell Prot 1, 816–827 [DOI] [PubMed] [Google Scholar]
- Widger WR, Cramer WA, Herrmann RG, and Trebst A (1984) Sequence homology and structural similarity between the b cytochrome of mitochondrial complex III and the chloroplast b6f complex: position of the cytochrome b hemes in the membrane. Proc.Natl. Acad. Sci., U. S. A 81,674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CA, and Crofts AR (2018) Dissecting the pattern of proton release from partial process involved in ubihydroquinone oxidation in the Q-cycle Biochim Biophys Acta 1859, 531–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita E, Zhang H, and Cramer WA (2007) Structure of the cytochrome b6f complex: quinone analogue inhibitors as ligands of heme cn. J. Mol. Biol 370, 39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Dashdorj N, Baniulis D, Yamashita E, Savikhin S, and Cramer WA (2008) On the structural role of the aromatic residue environment of the chlorophyll a in the cytochrome b6f complex. Biochemistry-Us 47, 3654–3661 [DOI] [PubMed] [Google Scholar]
- Zatsman AI, Zhang H, Gunderson WA, Cramer WA, and Hendrich MP (2006) Heme-heme interactions in the cytochrome b6f complex: EPR spectroscopy and correlation with structure. J. Amer. Chem. Soc 128, 14246–14247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, and Cramer WA (2005) Problems in obtaining diffraction-quality crystals of hetero-oligomeric integral membrane proteins. J Struct Funct Genomics 6, 219–223 [DOI] [PubMed] [Google Scholar]
- Zhang H, Carrell CJ, Huang D, Sled V, Ohnishi T, Smith JL, and Cramer WA (1996) Characterization and Crystallization of the Lumen-Side Domain of the Chloroplast Rieske Iron-Sulfur Protein. J. Biol. Chem 271, 31360–31366 [DOI] [PubMed] [Google Scholar]
- Zhang H, Huang D, and Cramer WA (1999) Stoichiometrically bound beta-carotene in the cytochrome b6f complex of oxygenic photosynthesis protects against oxygen damage. J. Biol. Chem 274, 1581–1587 [DOI] [PubMed] [Google Scholar]
- Zhang H, Whitelegge JP, and Cramer WA (2001) Ferredoxin:NADP+ oxidoreductase is a subunit of the chloroplast cytochrome b6f complex. J. Biol. Chem 276, 38159–38165 [DOI] [PubMed] [Google Scholar]
- Zhang H, Kurisu G, Smith JL, and Cramer WA (2003) A defined protein-detergent-lipid complex for crystallization of integral membrane proteins: The cytochrome b6f complex of oxygenic photosynthesis. Proc. Nat. Acad. Sci. USA 100, 5160–5163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouni A, Witt H, Kern J, Fromme P, Krauss N, Saenger W, and Orth P (2001) Crystal structure of photosystem II from Synechococcus elongatus at 3.8 A resolution. Nature 409, 739–743 [DOI] [PubMed] [Google Scholar]


