Abstract
Owing to the high water content, porous structure, biocompatibility and tissue-like viscoelasticity, hydrogels have become attractive and promising biomaterials for use in drug delivery, 3D cell culture and tissue engineering applications. Various chemical approaches have been developed for hydrogel synthesis using monomers or polymers carrying reactive functional groups. For in vivo tissue repair and in vitro cell culture purposes, it is desirable that the crosslinking reactions occur under mild conditions, do not interfere with biological processes and proceed at high yield with exceptional selectivity. Additionally, the cross-linking reaction should allow straightforward incorporation of bioactive motifs or signaling molecules, at the same time, providing tunability of the hydrogel microstructure, mechanical properties, and degradation rates. In this review, we discuss various chemical approaches applied to the synthesis of complex hydrogel networks, highlighting recent developments from our group. The discovery of new chemistries and novel materials fabrication methods will lead to the development of the next generation biomimetic hydrogels with complex structures and diverse functionalities. These materials will likely facilitate the construction of engineered tissue models that may bridge the gap between 2D experiments and animal studies, providing preliminary insight prior to in vivo assessments.
Keywords: Synthetic Polymer, Hyaluronic Acid, Peptide, Hydrogel, Extracellular Matrix, Covalent Cross-Linking
1. Introduction
Hydrogels are water-swollen materials with a three-dimensional structure and viscoelastic properties.1–3 Owing to the flexible synthesis, biomimetic hydrogels have become attractive and promising materials that find diverse applications ranging from drug delivery, and tissue engineering to medical diagnosis.4–7 Central to the engineering of functional tissues is the fabrication of a synthetic matrix with excellent biocompatibility, proper nutrient and waste diffusion, robust mechanical properties, and spatial and temporal presentation of biological moieties.8–10 In vivo, tissue-specific extracellular matrices (ECM), with defined biochemical and biophysical features, exert profound influence on the proliferation, migration, force sensing and differentiation of the resident stem cells.11–13 Therefore, it is crucial to reconstitute features of the native ECM in synthetic hydrogels.14 When appropriately designed, biomimetic hydrogels provide new opportunities to gain fundamental understandings of tissue development and to address challenges in the engineering of functional tissues.
Hydrogels can be fabricated via physical or chemical cross-linking from soluble monomers, multifunctional polymers, or insoluble nano- or micro-particles. Physical cross-linking relies on molecular interactions for the maintenance of network integrity, thus can be made reversible or responsive to changes in temperature, pH, or ionic strength. Due to the high stability of covalent bonds, chemically cross-linked hydrogels are in general permanent, irreversible, and stable under physiological conditions. These networks also exhibit tunable structures and adjustable mechanical properties. Natural or synthetic polymers can be modified to fulfill most of the requirements of tissue scaffolds by manipulating chemical properties or functionalizing biomolecules, therefore, both types of materials have been used for the preparation of covalent hydrogel networks that simulate the multifunctional nature of native biological environments to regulate cell behavior, and to promote tissue growth.15–17
2. Common hydrogel building blocks.
Various polymers, including polyethylene glycol (PEG), poly(2-hydroxethly methacrylate) (PHEMA), and poly(vinyl alcohol) (PVA), polypeptides and hyaluronic acid (HA), have been used for the preparation of covalent hydrogel networks. PEG has a long history as biomedical material. Owing to the favorable biocompatibility and non-fouling properties, PEG-based hydrogels are one of the most widely investigated biomaterials.18–20 PEGylation of natural polymers and proteins using bioconjugation techniques21 improves their solubility, reduces the toxicity, and prolongs the systemic circulation time.22 PEG is commercially available with different molecular architecture, molecular weight and end-group functionality. Specifically, PEG diacrylate (PEGDA) has been widely used for 3D cell encapsulation purposes. Additionally, multi-armed PEGs and hyperbranched polyglycerols have enabled the development of more sophisticated hydrogel networks.23 To render PEG gels biodegradable, hydrolytically degradable poly(lactic acid-co-glycolic acid) (PLGA) segments24–26 or enzymatically degradable peptide sequences27–29 have been introduced between cross-linking points.
As a stoichiometric isomer of PEG, PVA is prepared by hydrolysis of poly(vinyl acetate) (PVAc).30, 31 The physical and chemical properties of PVA can be adjusted by changing the degree of hydrolysis of PVAc. The abundant hydroxyl groups on PVA are amenable to chemical derivation and functionalization. PVA hydrogels are biocompatible, chemically stable, and non-adhesive to proteins and cells, thus are widely used in drug delivery and tissue engineering.32–36 PVA-based hydrogels have been engineered to exhibit robust mechanical properties, low coefficients of friction, and structural similarity to cartilage.37
Equally popular is PHEMA, a neutral, water-insoluble polymer synthesized via free radical polymerization of 2-hydroxyethyl methacrylate (HEMA).38 Commonly known for their application as contact lenses, PHEMA hydrogels are biocompatible, transparent, and chemically stable.39, 40 PHEMA has also been used for coating medical devices to prevent protein adsorption and cell adhesion.41–43 Other synthetic polymers based on 2-hydroxyethyl acrylate (HEA), frequently produced by living polymerization techniques, thus exhibiting defined composition, architecture and molecular weight, have also be used for the preparation of complex hydrogel networks.44–46
HA, a naturally occurring polysaccharide consisting of repeating disaccharide of D-glucuronic acid and D-glucosamine, has emerged as an attractive natural polymer for the synthesis of hydrogel materials owing to the advanced bacterial fermentation process that enables the production of high molecular weight HA with high purity and relatively narrow molecular weight distribution. Low molecular weight HA can be produced by γ-irradiation or enzymatic degradation.47, 48 HA is abundantly distributed in the natural extracellular matrices (ECM) and is degraded by the enzyme hyaluronidase. In the native ECM, HA absorbs large amounts of water and expands to form a loose hydrated network, serving as a space filler, lubricant, and osmotic buffer.49 Additionally, HA interacts with cell surface receptors, such as CD44 and RHAMM, to activate and regulate intracellular events that mediate cell behaviors. HA derivatives carrying readily accessible functional groups have been utilized to form cross-linked hydrogels for potential use in biomedical applications.50–52
Peptides, or polypeptides, have also been used for hydrogel synthesis. For short peptides (less than 50 amino acids), solution-phase and solid-phase peptide syntheses (SPPS)53, 54 are most commonly used. Polypeptides have been produced chemically from polymerization of α-amino acid-N-carboxyanhydrides (NCAs),55 or biosynthetically via recombinant DNA technology.56 While considerable progress has been made in the self-assembly of peptides into physical gels,57–59 covalent cross-linking strategies afford biomimetic hydrogels with a wide range of properties useful for tissue engineering applications.60–65
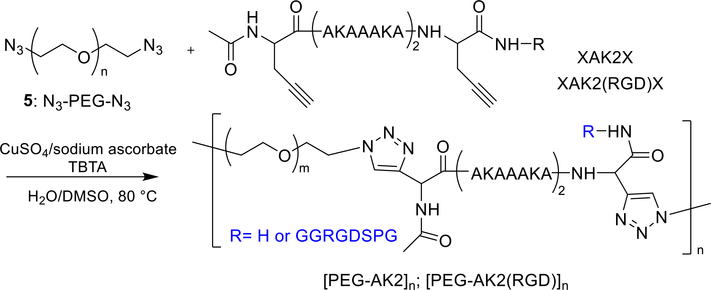
An emerging research trend is the construction of hybrid copolymers comprised of alternating peptide and synthetic polymer building blocks. The hybrid copolymers are designed to mimic the natural proteins66 in terms of their molecular architectures, dynamic responsiveness and cell-instructive properties, with the added attributes of tunability and processibility provided by the synthetic polymer constituents.67–70 Our group synthesized elastin mimetic hybrid polymers (EMHPs) by step growth, Click coupling (Figure 1) using telechelic, azide-terminated PEG and an alkyne-functionalized peptide with a sequence of (XAKAAAKA)2X, AK2, X: propargyl glycine) that is abundant in the cross-linking region of the natural elastin. The resulting multiblock copolymers, [PEG-AK2]n, had an estimated molecular weight of 34 kDa and contain an estimated 5–9 repeats of PEG-AK2. Introduction of the cell-adhesive RGD peptide at the non-natural amino acid propargyl glycine resulted in the production of EMHPs with dangling RGD moieties for cell adhesive properties.71, 72 These hybrid copolymers have been covalently crosslinked via lysine to afford hydrogels with mechanical properties comparable to those of the natural elastin.
Figure 1.
Synthesis of [PEG-AK2]n and [PEG-AK2(RGD)]n by step growth and Click coupling. TBTA: tris[(1-benzyl-1H−1,2,3-triazol-4-yl)methyl]amine
3. Chemical approaches to hydrogel synthesis
Depending on the desired structure and the targeted application, hydrogels have been synthesized employing versatile chemistries, such as radical polymerization, the formation of carbon-nitrogen double bond,73, 74 Michael-type addition reaction,75, 76 thiol-ene photochemistry,77, 78 and several types of orthogonal chemistries,79–81 and enzyme-catalyzed reaction.82, 83
3.1. Radical polymerization
Radical polymerization is frequently used for the fabrication of covalently crosslinked hydrogel networks. Monomers, such as HEMA, acrylamide (AAm), acrylic acid, and macromers, such as PEG,84 PVA, HA carrying reactive double bonds, have been used to produce hydrogels via photo- or redox-initiated polymerization. Upon exposure to visible or ultraviolet (UV) light, photoinitiators undergo homolytic cleavage or hydrogen abstraction to generate radicals to initiate the radical polymerization of monomers or macromers. Light-initiated radical polymerization enables in situ, rapid solidification of the precursor liquid under physiological conditions, and are thereby uniquely suitable for the use as injectable formulations for biomedical applications.77, 85 However, UV irradiation and free radicals are DNA damaging, thus potentially harmful to living cells.86 Adjusting the intensity of UV radiation and using initiators that decompose under visible light can alleviate this problem.87, 88
In addition to soluble macromers, micro- or nanoparticles carrying reactive acrylates have been used as microscopic cross-linkers for the preparation of macroscopic hydrogels. For example, HA-based, photo-cross-linkable hydrogel particles (HGPs) were synthesized via an inverse emulsion cross-linking process followed by chemical modification with glycidyl methacrylate (GMA). Doubly crosslinked hydrogels (DXNs) containing covalently integrated hydrogel particles were prepared by radical polymerization of GMA modified HA (HAGMA)89 in the presence of crosslinkable HGPs. The bone morphogenetic protein 2 (BMP-2), loaded into HGPs prior to photocross-linking, was released from the DXN in a controlled manner with a reduced initial burst over prolonged periods of time. The HA-based DXN not only maintained the phenotype of primary bovine chondrocytes but also fostered the production of cartilage specific extracellular matrix. These materials are promising candidates for use as bioactive matrices for cartilage tissue engineering.90
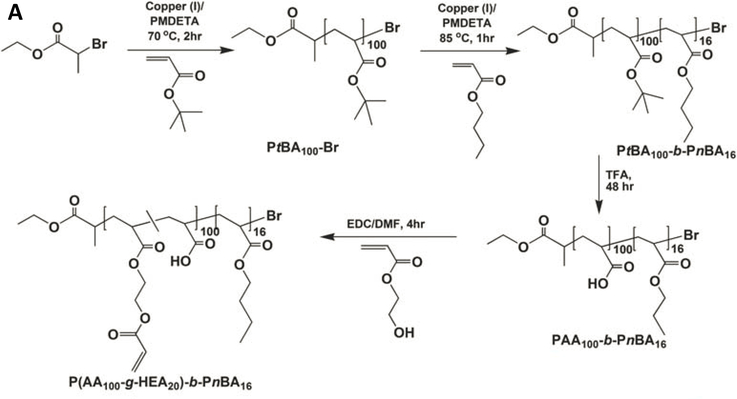
Alternatively, block copolymer micelles with a rubbery hydrophobic core have been employed for the preparation of mechano-responsive hydrogels. Specifically, poly(acrylic acid-graft-hydroxyethyl acrylate)-block-poly(n-butyl acrylate) [(PAA-g-HEA)-b-PnBA] was synthesized by atom transfer radical polymerization (ATRP) of tert-butyl acrylate (tBA) and n-butyl acrylate, followed by removal of the tert-butyl groups, and partial esterification of the carboxylic acid with HEA. (Figure 2A).91 The amphiphilic block copolymer self-assembled into radically cross-linkable block copolymer micelles (xBCMs) with a hydrophilic PAA shell and a hydrophobic PnBA core. Radical polymerization of AAm and xBCMs in the presence of a redox initiator gave rise to elastomeric hydrogels whose mechanical properties can be tuned by varying the xBCM concentration and block copolymer composition. Pyrene, loaded in the core of xBCMs, was dynamically released in response to externally applied mechanical forces. Force-induced micelle deformation was confirmed by transmission electron microscopy.92
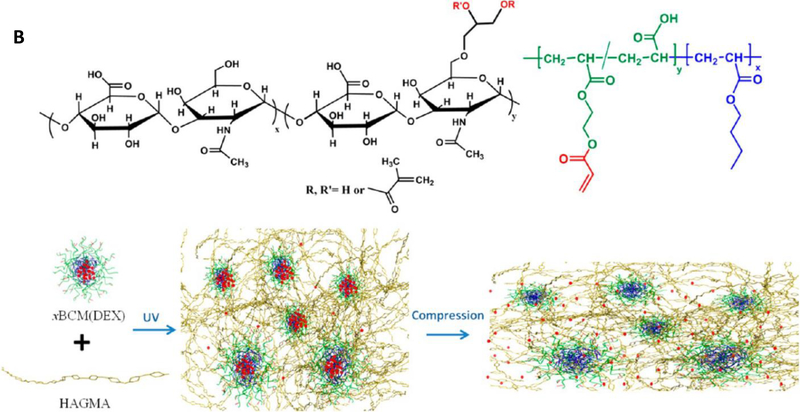
Figure 2.
Design of BCM-cross-linked mechano-responsive hydrogels. (A) Synthesis of (PAA-g-HEA)-b-PnBA. (B) Preparation of HA-based mechano-responsive hydrogels using HAGMA and DEX-loaded xBCMs. DEX was released from the PnBA core at an accelerated rate by compressive stress. Reprinted with permission from Xiao et al. 2010 91 and Xiao et al. 2013. 93
To demonstrate the utility of BCM-crosslinked hydrogels for cartilage repair, AAm was replaced with HAGMA and pyrene was replaced with an anti-inflammatory drug, dexamethasone (DEX).93 Photo-polymerization of HAGMA and DEX-loaded xBCMs produced HA-based hydrogels with covalently integrated drug depots (Figure 2B). The release kinetics of DEX can be modulated by external compressive forces that effectively deform the PnBA core. The engineered hydrogels with anti-inflammatory functions and mechano-responsiveness are attractive candidates for the repair of pathologically compromised tissue that are normally subjected to various mechanical loads.
3.2. Formation of carbon-nitrogen double bond
Reaction of aldehydes and ketones with amines occurs in absence of catalysts under physiological conditions to produce imines. N-substituted imines are often referred to as Schiff Base. Hydroxylamines and hydrazides react efficiently with aldehydes or ketones in aqueous conditions to produce the respective oxime and hydrazone linkages that are hydrolytically more stable than the corresponding imine bond. Due the mild reaction conditions, the simplicity in chemical synthesis and the reversibility of the C=N bond,94 these chemistries have been exploited for the synthesis of injectable hydrogels. While injectable hydrogels based on shear-thinning of selfassembled peptide/protein gels have been reported, 95–98 the injectability of covalently crosslinked hydrogels originates from the reversibility of the covalent bonds.99 Using synthetic and natural polymers, including PEG, polysaccharides, and (poly)peptides, injectable hydrogels have been synthesized by mixing an aldehyde-functionalized polymer with the amine-containing counterpart. 74, 100–106 For example, elastin-like polypeptides (ELP) functionalized with primary amines and aldehydes were mixed with bioglass (BG) in a high pH environment to produce ELP/BG composite hydrogels via the formation of Schiff base. The hydrogel can be further customized by fusing cell adhesive RGD peptide to ELP to promote cell spreading and to support cell growth. The injectable hydrogel may find applications in cell or drug delivery for soft tissue regeneration.106
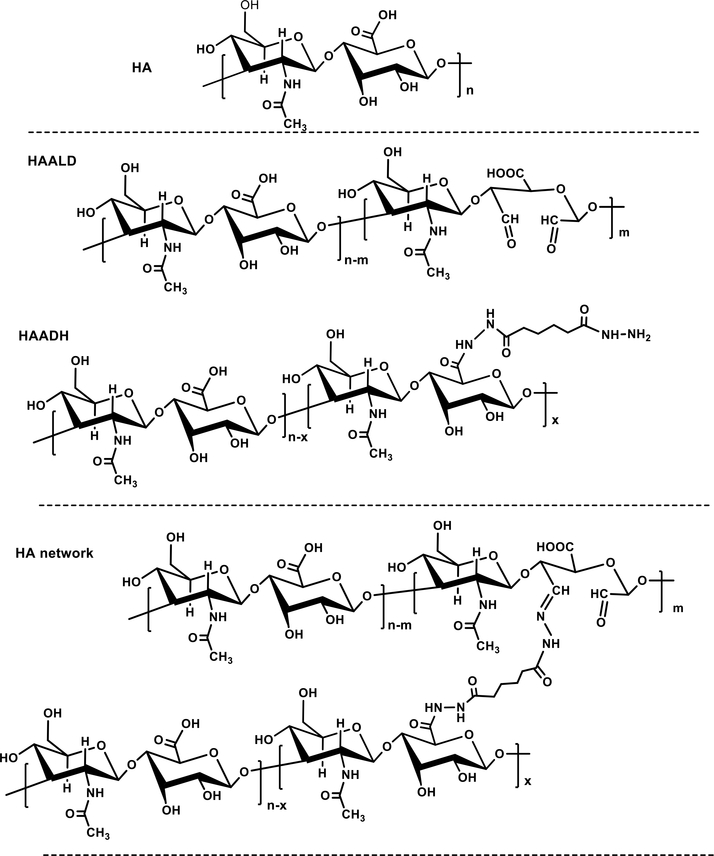
Polymers containing hydroxyamine or hydrazide groups have also been used to form hydrogels by oxime bond formation73, 107, 108 or hydrazone ligation.109–112 For example, HA derivatives carrying complementary hydrazide (HAADH) and aldehyde (HAALD) functionalities have been synthesized (Figure 3)109 via carbodiimide-mediated coupling of adipic acid dihydrazide (ADH) with the carboxyl groups of HA and sodium periodate (NaIO4) oxidation of proximal hydroxyl groups on HA, respectively. Simple mixing of HAADH and HAALD gave rise to pliable hydrogels through the formation of hydrazone linkages (Figure 3). This HA hydrogel system provides a flexible and physiologically relevant platform to study prostate cancer metastasis. As a requisite component of the tumor microenvironment, HA mediates cell migration, cancer progression and metastasis. These properties are strongly dependent on HA concentration, molecular weight, and degradation rate.113 Metastatic prostate cancer cells cultured in these hydrogels formed fingerlike structures, “invadopodia”, which is consistent with their invasive properties. The number of invadopodia, as well as cluster size, shape and convergence, can provide a quantifiable measure of invasive potential. Prostate cancer cell invasion through the HA hydrogel was dependent on HA interaction with the cell surface receptor RHAMM/CD168 and also required hyaluronidase activity. The engineered prostate cancer model is amenable to dissection of biological processes associated with cancer cell motility through HA-rich connective tissues.110, 114
Figure 3.
Hydrazone ligation applied to the synthesis of HA hydrogels for tumor engineering. (A) Synthesis of HAADH and HAALD; (B) Covalent cross-linking of HAALD and HAADH in PBS via the formation of hydrazone bonds.
When hydrazone ligation was restricted to the inverse emulsion droplets, nanoporous HA HGPs with an average diameter of 10 μm were obtained. These HGPs can be readily dispersed in aqueous media, and are stable against hyaluronidase degradation. When covalently conjugated with heparin or heparan sulfate-bearing perlecan domain I (PlnDI), the HGPs effectively sequester and modulate the release of various heparin-binding growth factors. Particularly, BMP2-loaded, PlnDI-conjugated HA HGPs stimulate chondrogenic differentiation and robust production of cartilage-specific ECM both in vitro 115 and in vivo.116 The presence of residual functional groups on the particle surface also allowed for subsequent cross-linking of the HGPs with aldehyde or hydrazide-containing macromolecules to give rise to a DXN. The resultant hybrid network is hierarchically structured and mechanically robust/tunable, capable of mediating cellular activities through the spatial and temporal presentation of biological cues.117, 118
In addition to HA, oxime/hydrazone chemistry has been applied to other polysaccharides to develop injectable hydrogels for a range of biomaterials applications.119, 120 For example, hydrazide/aldehyde functionalized β-cyclodextrins and dextrans have been synthesized. 121–123 An injectable and in situ cross-linkable hydrogel was fabricated by mixing the above aldehyde and hydrazide derivatives. The presence of β-cyclodextrin in the network allows for the inclusion of hydrophobic molecules through the formation of host-guest complexes for the controlled release of therapeutic agents. This hydrogel exhibited variable degradation and mechanical properties, and can be applied to wound sites for tissue repair.119
3.3. Michael-addition reaction
The Michael addition reaction, an efficient chemistry between electron deficient olefins and nucleophiles, has been employed for the synthesis of linear, graft, hyperbranched, and dendritic polymers.124 A frequently used Michael type reaction for hydrogel synthesis is the addition of thiols to α,β-unsaturated carbonyls (such as maleimide, acrylate, acrylamide or vinylsulfone) because it occurs rapidly under mild reaction conditions with a high conversion.76, 125–127 There has been considerable research in developing synthetic ECM using thiolated HA (HA-SH) and HA or PEG carrying reactive double bonds. For instance, injectable hydrogels formed from HA-SH and 4-arm PEG vinylsulfone were prepared for potential cartilage tissue engineering.128 These hydrogels were shown to facilitate the attachment of chondrocytes and enhance the production of type II collagen, thereby supporting cartilage tissue formation.129
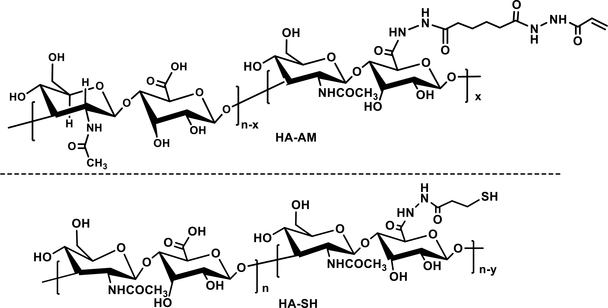
Our continued interest in the creation of physiologically relevant in vitro tumor models 130 motivated us to develop a stable and permissive HA hydrogels for the long-term 3D culture of prostate cancer cells. To this end, acrylamide-functionalized HA (HA-AM, Figure 4) was synthesized via the nucleophilic substitution of acrylic acid N-hydroxysuccinimide ester with HAADH. To obtain HA-SH (Figure 4), sulfhydryl groups were incorporated by the reaction between HA and a disulfide-containing dihydrazide reagent, followed by reduction with dithiothreitol. A HA-based hydrogel with an average elastic modulus of ~200 Pa was formed by mixing the solution of HA-AM and HA-SH under physiological conditions. To recapitulate tumorstroma crosstalk, the same HA gel containing entrapped HGPs was overlaid on the cell-laden construct.50 The HGP-containing layer served as a growth factor depot to release a heparin-binding epidermal growth factor-like growth factor (HB-EGF) in a controlled manner. LNCaP cells embedded in the bottom layer receive the growth factor signals from the top, and in response form larger tumoroids than those cultured in the absence of EGF. We discovered that prostate cancer cells cultured on HA hydrogels were more resistant to anti-cancer drug doxorubicin (DOX) treatments compared to 2D cultures.131 These tissue-engineered tumor models may bridge the gap between 2D experiments and animal studies, providing the preliminary insight into optimizing the drug formulations prior to the in vivo assessment.
Figure 4.
Chemical structures of HA-AM and HA-SH.
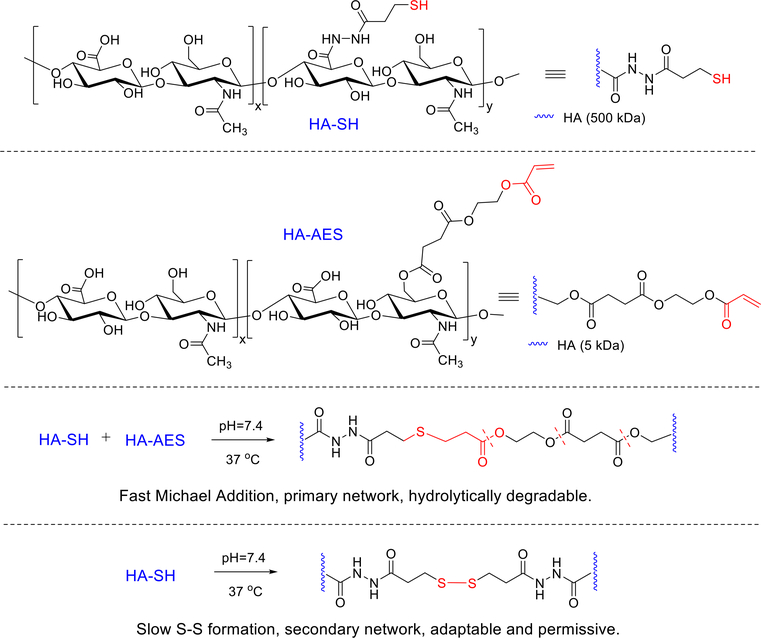
The HA hydrogel system described above facilitates the formation of tumoroids from dispersed prostate cancer cells, but does not promote the 3D organized growth of primary salivary human stem/progenitor cells (hS/PCs). We reasoned that for 3D assembly of hS/PCs, the synthetic matrix should exhibit time-dependent dynamic features to promote the cell growth and to accommodate spheroid expansion. To this end, new HA--based hydrogels (Figure 5) using HA-SH and acrylated HA (HA-AES) were developed, synthesized by reacting HA with mono-2-(acryloyloxy)ethyl succinate with a significant molecular weight and stoichiometric mismatch. Hydrogels were formulated with varying HA concentration, thiol/acrylate ratios and elastic moduli. The thiol/acrylate reaction was initiated rapidly upon mixing of HA-SH/HA-AES to establish thioether crosslinks with neighboring ester groups. Meanwhile, spontaneous sulfhydryl oxidation occurred slowly over several days to install a secondary, cell-permissive network to allow for cell expansion and aggregation. Multicellular salivary spheroids were detected in gels that are significantly softer (with a G’ < 216 Pa) and had a thiol/acrylate ratio >18. hS/PCs encapsulated in these HA gels formed organized multicellular spheroids by day 14 and reached an average diameter of 50 μm by day 28. Cells in these structures formed cell-cell junctions, secreted basement membrane proteins and maintained key stem/progenitor markers.132 Thus, incorporation of time-dependent, dynamic features into a covalently cross-linked HA network produces an adaptable hydrogel framework that promotes hS/PC assembly and supports early aspects of tissue morphogenesis.
Figure 5.
Synthesis of HA-based hydrogels via the covalent cross-linking of HA-AES and HASH. Fast thiol/acrylate Michael reaction contributes to the establishment of the primary network that is hydrolytically degradable. The disulfide crosslinks, formed gradually and spontaneously, stabilize the primary network, at the same time, introducing adaptable and dynamic features to the network to facilitate the 3D assembly of hS/PCs. Reprinted with permission from Ozdemir et al. 2016. 132
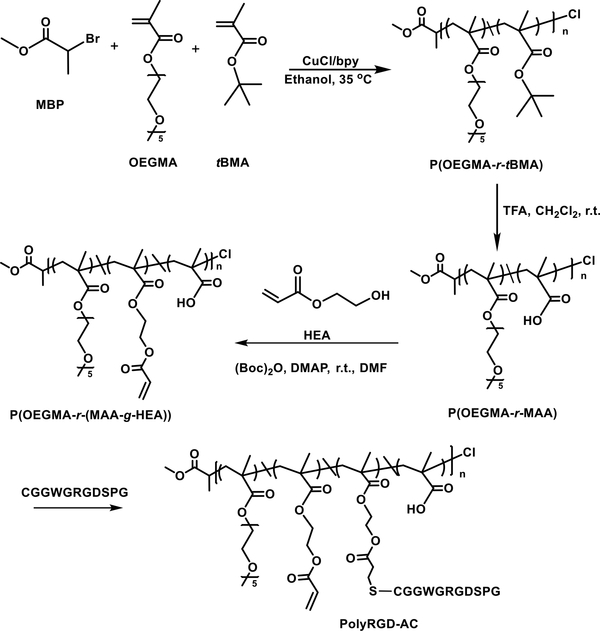
To mimic the natural ECM, biologically derived peptides have been covalently integrated in hydrogel matrices to foster cell-matrix interaction, to regulate cell signaling and to control cell differentiation.27, 133, 134 Commonly used peptides include integrin-binding peptides and matrix metalloproteinase (MMP) sensitive peptides.135–137 Recently, a biomimetic hydrogel using HA-SH and an acrylated copolymer carrying multiple copies of the cell adhesive peptide (PolyRGD-AC) was developed for the use in tumor model applications.46 Specifically, ATRP of tert-butyl methacrylate and oligomeric ethylene glycol methacrylate (OEGMA), followed by acid hydrolysis produced hydrophilic copolymers with protein-repellent OEG side chains and chemically addressable carboxylate groups. Modification of the copolymer with 2-hydroxyethyl acrylate installed reactive double bonds, through which bioactive RGD peptides were introduced. (Figure 6)
Figure 6.
Synthetic pathways for the production of PolyRGD-AC. Reprinted with permission from Hao et al. 2016. 46
The resulting peptide-conjugated, chemically crosslinkable copolymer was mixed with HA-SH to form a macroscopic hydrogel with an average elastic modulus of 630 Pa. LNCaP prostate cancer cells encapsulated in HA-PolyRGD gels as dispersed single cells formed multicellular tumoroids by day 4 and reached an average diameter of ~95 μm by day 28. Cells in these structures were viable, formed cell-cell contacts through E-cadherin and displayed cortical organization of F-actin. Multivalent presentation of the RGD signal in the HA matrix increased cellular metabolism, promoted the development of larger tumoroids and enhanced E-cadherrin and integrin expression. Overall, hydrogels with multivalent immobilized RGD is a promising 3D culture platform for dissecting principles of tumorigenesis and for screening anticancer drugs.46
3.4. Thiol-ene chemistry
Thiol-ene chemistry, radical-mediated addition of a thiol to a double bond, has been extensively explored in coating, printing, adhesive, and imaging technologies.138 Considerable progress has been made using thiol-ene chemistry for hydrogel synthesis. This reaction is rapid and efficient and occurs at neutral pH under ambient conditions. The photo-initiated polymerization process allows for spatial and temporal control of gel properties.78, 139, 140 Anseth and colleagues developed a series of peptide-functionalized PEG hydrogels via thiol-ene photo-polymerization for the encapsulation, 3D culture, migration, and differentiation of human mesenchymal stem cells (hMSCs).28, 141–143 The hydrogel building blocks included the PEG-tetranorbornene (PEG4-norb) integrin binding peptide (CRGDS), and the chymotrypsin degradable peptide KCGGYRGCK). I2959 or trimethylbenzoylphosphinate (LAP) were employed as photoinitiator and radicals were generated upon UV irradiation at 365 nm. hMSCs encapsulated in this hydrogel remained viable while cell morphology and cell differentiation were shown to be dependent on culture conditions and gel degradability.144 Similar PEG/peptide gels have been reported for the 3D culture of HT-1080, a human fibrosarcoma cell line,142 and pancreatic ductal epithelial cells.143
Thiol-ene chemistry has also been applied to the synthesis of HA hydrogels. Norbornene-functionalized HA (HA-Norb) was produced through the coupling of norbornene carboxylic acid and the primary hydroxyl groups on HA in organic media in the presence of di-tert-butyl dicarbonate (Boc2O) and 4-dimethylaminopyridine (DMAP). HA-norb was combined with a bisthiol to generate a hydrogel via thiol-ene chemistry under UV light.145 By limiting the initial extent of cross-linking, the remaining pendent norbornene groups were further crosslinked with di-thiol or functionalized mono-thiol peptides. Spatial patterning of the gel stiffness and multiple peptide signals are achieved using a photomask.
3.5. Diels-Alder reaction
Diels-Alder (DA) reaction, the [4+2] cycloaddition between an electron-rich diene (e.g. furan) and an electron-deficient dienophile (e.g. maleimide),146 is selective and proceeds at a high yield without any catalyst or byproduct.147 The reaction rate can be increased by introducing electron-withdrawing substituents on the alkene and the electron-donating groups on the diene. The DA adduct is stable at ambient conditions, but can be cleaved at an elevated temperature to produce the original diene and dienophile.148, 149 The DA reaction has been exploited as an efficient strategy for hydrogel synthesis. 147, 150–157 For example, HA/PEG hydrogels with elastic moduli ranging from 275 to 680 Pa have been synthesized using furan-modified HA and PEG-bismaleimide.152 EGF was photo-patterned into the hydrogels by two-photon technology employing a photolabile coumarin-HA-furan conjugate and iodoacetamide-functionalized EGF, thereby generating spatially defined growth factor gradients to direct cell function.153 The incorporation of MMP-cleavable peptides and the RGD peptide in the HA/PEG hydrogel led to the establishment of a cancer cell invasion platform, where cells were plated on top of the hydrogel and their migration into the bulk of the hydrogel was investigated. Overall, RGD signals enhance cell proliferation whereas the MMP-cleavable sequences promote cell invasion.156 These HA-based hydrogels offer a tunable platform with varying mechanical properties and ECM environments for directing cell function and understanding the role of the microenvironment on cancer cell invasion.
3.6. CuAAC reaction
Copper(I)-catalyzed azide-alkyne cycloaddtion (CuAAC), the quintessential “click” reaction with high efficiency and outstanding orthogonal reactivity, has been gaining popularity in synthetic methodologies, bioconjugation and materials developments.158–160 Alkynes and azides are not present in nature, and the 1,2,3-triazole product is stable against oxidation, reduction and hydrolysis under acidic or basic conditions, mimicking the biological properties of the amide bond.161 The CuAAC reaction has also been exploited for gelation purposes.162–169 “Click” hydrogels have been reported using azide and alkyne functionalized PVA,162 HA,169 or PEG.167 Like gels prepared using traditional chemistries, the mechanical properties of these hydrogels can be varied by changing the concentration or functionality of the cross-linker. The advantages of the Click chemistry are highlighted in the PEG/CD system where high strength and a homogeneous network structure were observed. Azide-alkyne cycloaddition reaction occurs very slowly without Cu(I), but Cu(I)-mediated formation of reactive oxygen species can compromise the functions of native biomolecules.170, 171 Cell-compatible “Click” chemistries are being developed using novel chelating ligands,172–174 such as TBTA, tris(6-galactosyltriazomethyl)amine (TGTA), tris(hydroxypropyltriazolyl-methyl)amine (THPTA), and bathophenanthroline disul-fonate disodium salt (BPDS) to protect biomolecules from copper induced oxidation or degradation.
3.7. Copper-free Click reactions
The need for copper catalysis remains a major concern with the use of CuAAC in cell encapsulation, live cell labeling or bioconjugation purposes. The copper-free bioclick reaction developed by the Bertozzi group takes advantage of the ring strain of cyclooctyne to lower the activation barrier of the cycloaddition reaction.175 The strain-promoted azide-alkyne cycloaddition (SPAAC) reaction has enabled selective modification of biomolecules and living cells and has been applied to bioconjugation, polymer synthesis and in situ hydrogel formation in the presence of cells.176–182 “SPAAC” has been combined with other orthogonal chemistries to enable spatial and temporal modulation of matrix properties.178, 183, 184
While SPACC eliminated the need for copper catalysis, the slow kinetics (k2 10−2–10−1 M−1s−1) 185 necessitates a prolonged reaction time (months to years) to achieve 99% conversion under dilution conditions used for protein or cell labeling. Tetrazine ligation, an inverse-electron demand DielsAlder cycloaddition reaction that proceeds with exceptional kinetics and selectivity without any toxic byproducts, were reported in 2008.186–188 Among various dienenophiles reported so far, a conformationally strained trans-cyclooctene (sTCO)189 is the fastest, with a rate up to 3.3 × 106 M1s−1. With the unprecedented rate and tolerance to a broad range of biological functionalities, tetrazine ligation has become a broadly utilized bioorthogonal reaction, finding unique applications in cell labeling,190–192 cancer imaging,193, 194 and material science.80, 195–197
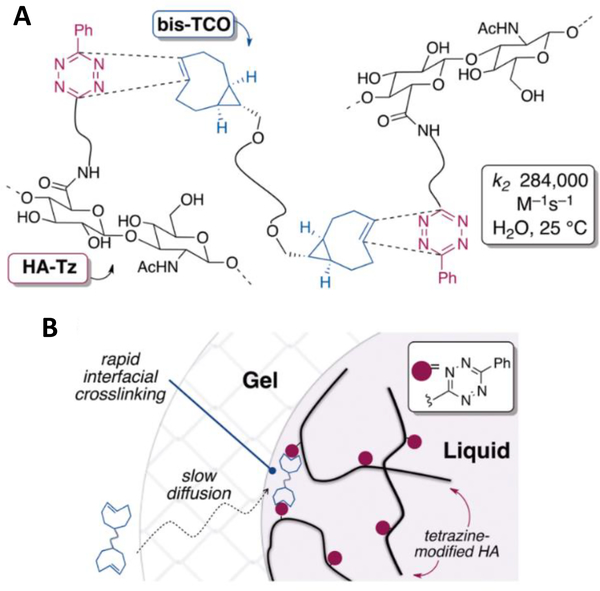
Taking advantage of the unprecedented reaction rate, a diffusion-controlled interfacial crosslinking process was established to create hydrogel matrices with well-defined spatial patterns without having to rely on free radical UV initiated chemistries and photomasks (Figure 7). Thus, s-tetrazine (Tz) and trans-cyclooctene (sTCO) were conjugated to high molecular weight HA and a low molecular weight PEG, respectively. When tetrazine-modified HA (HA-Tz, 218 kDa) is dropped into a bath of PEG-based TCO crosslinker (bis-TCO, 1253 Da), a cross-linked shell forms immediately. The smaller bis-TCO molecule diffuses readily through the gel layer to introduce additional cross-linking until HA-Tz is exhausted. The diffusion-controlled, interfacial crosslinking enables 3D patterning of the hydrogel microsphere via the alteration of the TCO bath composition as a function of time without the need for external triggers or templates. Interestingly, when bis-TCO is added to a bath of HA-Tz, water filled channels are created. Prostate cancer cells encapsulated in the microspheres have 99% viability, proliferate readily, and form aggregated clusters. The novel diffusion-controlled, interfacial process is an enabling tool for the fabrication of cell-signaling hydrogel matrices for the in vitro engineering of functional tissues. 197
Figure 7.
(A) Instantaneous cross-linking via tetrazine-TCO ligation. (B) HA-PEG Gel interface forms when a droplet of HA-Tz contacts the solution of bis-TCO. Cross-linking occurred at the gel/liquid interface is faster than the rate of diffusion through the gel interface. Reprinted with permission from Zhang et al. 2014. 197
Using a less reactive dienenophile, norbornene, other groups reported the synthesis of gelatin and PEG hydrogels via tetrazine ligation. Cross-linking occurs in bulk, upon mixing of tetrazine and norbornene modified hydrogel precursors, because of the slower kinetics compared to tetrazineTCO reaction. Subsequent modification of the network is possible when tetrazine ligation was combined with thiol-ene photochemistry.80 Alternatively, norbornene-modified gelatin was mixed with tetrazine-modified gelatin to produce an injectable hydrogel.198
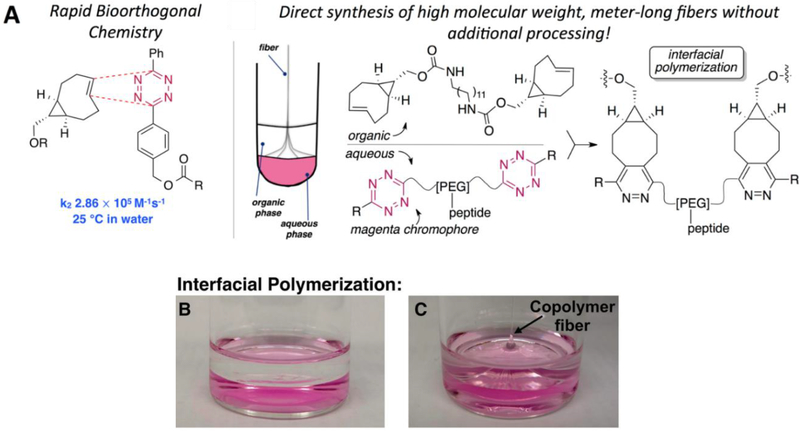
Recently, we successfully appliedtetrazine ligation to the synthesis of multiblock copolymer using homo-difunctional Tz and TCO derivatives (Figure 8). Judicious selection of the monomeric building blocks allowed for the polymerization to be carried out at an immiscible solvent interface. As the polymerization proceeded, high molecular weight polymer fibers (9–10 μm in diameter) were pulled from the interface. Compared to the corresponding solution phase polymerization, the interfacial process gave rise to a copolymer with a significantly higher fraction of high molecular weight (up to 263 kDa with 28 repeats) species. The bioorthogonal nature of the tetrazine ligation permits facile incorporation of functional peptides into the multiblock constructs. When GRGDSP was introduced to the water-soluble monomer, interfacial bioorthogonal polymerization resulted in the production of elastomeric, cell-adhesive microfibers.199 Alternatively, polymer fibers carrying latent dihydrotetrazines were catalytically activated to tetrazine and covalently modified by TCO conjugates of small molecules, peptides and proteins.200 In a related project, polymer fibers carrying latent dihydrotetrazines were catalytically activated to tetrazine and covalently modified by trans-cyclooctene conjugates of small molecules, peptides and proteins. In addition to visualization with fluorophores, fibers conjugated to a cell adhesive peptide exhibited a dramatically increased ability to mediate contact guidance of cells. Stably cross-linked tetrazine fibers have been produced using a trifunctional TCO crosslinker in place of bisTCO. Single fiber tensile testing revealed a Young’s modulus of 509 ± 141 MPa and 4.3 ± 1.7 MPa for dry and hydrated fibers, respectively. MSCs were able to attach to and elongate along the RGD-containing fibers. Fiber-guided MSC differentiation and migration are currently under investigation.
Figure 8.
(A) The tetrazine-TCO ligation involving sTCO and a diphenyl-s-tetrazine with rapid kinetics and schematic illustration of interfacial bioorthogonal to create multiblock copolymer fibers; (B) Photograph showing a phase-separated layers of bis-sTCO in ethyl acetate solution (colorless) and bis-tetrazine in an aqueous solution (pink); (C) Photograph showing colorless multiblock copolymerr fibers being pulled out of the immiscible interface. Reprinted with permission from Liu et al. 2015. 199
3.8. Enzyme-catalyzed reactions
Enzymes are known to catalyze numerous types of biochemical reactions and can be exceptionally selective for their substrates.201 Due to the mild reaction conditions, enzyme-catalyzed reactions have gained increasing popularity for hydrogel synthesis.82 Various enzymes, including horseradish peroxidase (HRP),202, 203 transglutaminase (Factor XIII),83, 204 tyrosinase (Tyr),205, 206 phosphopantetheinyl transferase (PPTase),207 lysyl oxidase208 and phosphatases209, 210 have been explored to catalyze gelation. For example, using PEG-based microgels with a uniform size and displaying transglutaminase peptide substrates K and Q, Griffin et al. produced a microporous gel via FXIIIa-catalyzed amide formation between the K and Q peptides. In vitro, FXIIIa-mediated annealing allows incorporation of living cells into the scaffolds. In vivo, the scaffold accelerated wound closure and enabled faster tissue regeneration.204 Separately, gelatin-based hydrogels catalyzed by HRP and Tyr enzymes have been developed. Hydrogels formed by HRP/Tyr exhibited 2- to 5-fold greater tissue adhesiveness as compared to those crosslinked with HRP only. Such a hydrogel formulation can be used as injectable bio-adhesives or implantable constructs to deliver cells and/or drugs for tissue-regenerative applications.211
4. Conclusion
Significant progress has been made in the development of hydrogel biomaterials for tissue engineering applications. Hydrogel composition, microstructure, mechanical properties, and biological activities can influence cell morphology, proliferation, aggregation, migration and phenotype. For in situ cell encapsulation and 3D cell culture purposes, the cross-linking reaction should be fast, cytocompatible and biorthogonal. The gelation process inevitably results in network defects that are difficult to control and quantify,212 but can impact gel properties and cell functions. Novel chemistries have enabled user-directed modulation of gel properties in a time-dependent manner, thus allowing cellular functions to evolve over time.213 Spontaneous bond formation without having to rely on external triggers leads to the reorganization of the network structure that may contribute to the desired cellular organization in 3D.132 Traditionally, covalently crosslinked hydrogels are stable unless degradable moieties are incorporated. Covalently crosslinked networks214 consisting of triggerable, reversible chemical structures have been recently highlighted. Although application of a stimulus can cause these materials to alter their shape, topography, and physical properties, their utility in 3D cell culture is not well established. With the discovery of new, orthogonal organic chemistries, novel hydrogel materials can be anticipated in the near future. Bioinspired synthetically derived hydrogels serve as a valuable tool to investigate the underlying role of physiochemical and biological factors in tissue engineering.
Acknowledgement
Work in the authors’ laboratory has been funded by grants from the National Science Foundation (DMR: 0643226, 1206310 1506613), National Institutes of Health (R01 DC008965; R01 DE022969, R01 DC011377, R01 DC014461), DuPont, Gore and the University of Delaware. We acknowledge the Delaware COBRE program (NIGMS: P20 GM103541, P30 GM110758) for instrumentation support. The authors wish to acknowledge Genzyme for generously providing HA.
References
- 1.Hoffman AS, Advanced Drug Delivery Reviews 54:3–12 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Ullah F, Othman MBH, Javed F, Ahmad Z and Akil HM, Materials Science and Engineering: C 57:414–433 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Hoffman AS, Advanced Drug Delivery Reviews 64, :18–23 (2012). [Google Scholar]
- 4.Lutolf MP, Raeber GP, Zisch AH, Tirelli N and Hubbell JA, Advanced Materials 15:888–892 (2003). [Google Scholar]
- 5.Rahmany MB and Van Dyke M, Acta Biomaterialia 9:5431–5437 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Lienemann PS, Lutolf MP and Ehrbar M, Advanced Drug Delivery Reviews 64:1078–1089 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Lee KY and Mooney DJ, Chemical Reviews 101:1869–1880 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Lutolf MP, Gilbert PM and Blau HM, Nature 462:433–441 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin H, Jo S and Mikos AG, Biomaterials 24:4353–4364 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Censi R, Di Martino P, Vermonden T and Hennink WE, Journal of Controlled Release 161:680–692 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N and Piccolo S, Nature 474:179–183 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Mouw JK, Ou G and Weaver VM, Nat Rev Mol Cell Biol 15:771–785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonnans C, Chou J and Werb Z, Nat Rev Mol Cell Biol 15:786–801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khetan S, Guvendiren M, Legant WR, Cohen DM, Chen CS and Burdick JA, Nat Mater 12:458–465 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seliktar D, Science 336:1124–1128 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Guvendiren M and Burdick JA, Current Opinion in Biotechnology 24:841–846 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pérez RA, Won J-E, Knowles JC and Kim H-W, Advanced Drug Delivery Reviews 65:471–496 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Koh W-G, Revzin A and Pishko MV, Langmuir 18:2459–2462 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Zhu J, Biomaterials 31:4639–4656 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kyburz KA and Anseth KS, Acta Biomaterialia 9:6381–6392 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hermanson GT, Bioconjugate techniques, Academic press; (2013). [Google Scholar]
- 22.Harris JM and Chess RB, Nat Rev Drug Discov 2:214–221 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Gong CY, Dong PW, Shi S, Fu SZ, Yang JL, Guo G, Zhao X, Wei YQ and Qian ZY, Journal of Pharmaceutical Sciences 98:3707–3717 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Miller RA, Brady JM and Cutright DE, Journal of Biomedical Materials Research 11:711–719 (1977). [DOI] [PubMed] [Google Scholar]
- 25.Alexander A, Ajazuddin J.Khan,Saraf Sand Saraf S, Journal of Controlled Release 172:715–729 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Wang J, Shen B, Chan CWY, Wang C, Zhao Y, Chan HN, Tian Q, Chen Y, Yao C, Hsing IM, Li RA and Wu H, Macromolecular Bioscience 15:426–436 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Patterson J and Hubbell JA, Biomaterials 31:7836–7845 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Sridhar BV, Brock JL, Silver JS, Leight JL, Randolph MA and Anseth KS, Advanced Healthcare Materials 4:702–713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Xu B, Puperi DS, Yonezawa AL, Wu Y, Tseng H, Cuchiara ML, West JL and Grande-Allen KJ, Acta Biomaterialia 14:11–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stauffer SR and Peppast NA, Polymer 33:3932–3936 (1992). [Google Scholar]
- 31.Mansur HS, Oréfice RL and Mansur AAP, Polymer 45:7193–7202 (2004). [Google Scholar]
- 32.Schmedlen RH, Masters KS and West JL, Biomaterials 23:4325–4332 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Nuttelman CR, Mortisen DJ, Henry SM and Anseth KS, Journal of Biomedical Materials Research 57:217–223 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Peppas NA and Merrill EW, Journal of Polymer Science: Polymer Chemistry Edition 14:441–457 (1976). [Google Scholar]
- 35.Asoh T-A, Takaishi K and Kikuchi A, Journal of Materials Chemistry B 3:6740–6745 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Hayes JC and Kennedy JE, Materials Science and Engineering: Part C Materials for Biological Applications 59:894–900 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Pan Y-S, Xiong D-S and Ma R-Y, Wear 262:1021–1025 (2007). [Google Scholar]
- 38.Holly FJ and Refojo MF, Journal of Biomedical Materials Research 9:315–326 (1975). [DOI] [PubMed] [Google Scholar]
- 39.Andrade-Vivero P, Fernandez-Gabriel E, Alvarez-Lorenzo C and Concheiro A, Journal of Pharmaceutical Sciences 96:802–813 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Zhao W, Lenardi C, Webb P, Liu C, Santaniello T and Gassa F, Polymer International 62:1059–1067 (2013). [Google Scholar]
- 41.Rao JK, Ramesh DV and Rao KP, Biomaterials 15:383–389 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Pertici V, Trimaille T, Laurin J, Felix M-S, Marqueste T, Pettmann B, Chauvin J-P, Gigmes D and Decherchi P, Biomaterials 35:6248–6258 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Kubinová Š, Horák D, Hejčl A, Plichta Z, Kotek J, Proks V, Forostyak S and Syková E, Journal of Tissue Engineering and Regenerative Medicine 9:1298–1309 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Khutoryanskaya OV, Mayeva ZA, Mun GA and Khutoryanskiy VV, Biomacromolecules 9:3353–3361 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Cheng X, Jin Y, Sun T, Qi R, Fan B and Li H, RSC Advances 5:4162–4170 (2015). [Google Scholar]
- 46.Hao Y, Zerdoum AB, Stuffer AJ, Rajasekaran AK and Jia X, Biomacromolecules 17:3750–3760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu X, Jha AK, Harrington DA, Farach-Carson MC and Jia X, Soft Matter 8:32803294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dicker KT, Gurski LA, Pradhan-Bhatt S, Witt RL, Farach-Carson MC and Jia X, Acta Biomaterialia 10:1558–1570 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laurent TCF, E. JR, Faseb Journal 6:2397–2404 (1992). [PubMed] [Google Scholar]
- 50.Xu X, Gurski LA, Zhang C, Harrington DA, Farach-Carson MC and Jia X, Biomaterials 33:9049–9060 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chawla K, Yu TB, Stutts L, Yen M and Guan Z, Biomaterials 33:6052–6060 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.West D, Hampson I, Arnold F and Kumar S, Science 228:1324–1326 (1985). [DOI] [PubMed] [Google Scholar]
- 53.Fields GB and Noble RL, International Journal of Peptide and Protein Research 35:161–214 (1990). [DOI] [PubMed] [Google Scholar]
- 54.El-Faham A and Albericio F, Chemical Reviews 111:6557–6602 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Deming TJ, Progress in Polymer Science 32:858–875 (2007). [Google Scholar]
- 56.Nettles DL, Chilkoti A and Setton LA, Adv Drug Deliv Rev 62:1479–1485 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kopeček J and Yang J, Acta Biomaterialia 5:805–816 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li L, Tong Z, Jia X and Kiick KL, Soft Matter 9:665–673 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li L, Mahara A, Tong Z, Levenson EA, McGann CL, Jia X, Yamaoka T and Kiick KL, Advanced Healthcare Materials 5:266–275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nowak AP, Breedveld V, Pakstis L, Ozbas B, Pine DJ, Pochan Dand Deming TJ, Nature 417:424–428 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Jing P, Rudra JS, Herr AB and Collier JH, Biomacromolecules 9:2438–2446 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Y, Pang X-H and Dong C-M, Advanced Functional Materials 20:579–586 (2010). [Google Scholar]
- 63.Deng C, Wu J, Cheng R, Meng F, Klok H-A and Zhong Z, Progress in Polymer Science 39:330–364 (2014). [Google Scholar]
- 64.Shen Y, Fu X, Fu W and Li Z, Chemical Society Reviews 44:612–622 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Ghoorchian A, Simon JR, Bharti B, Han W, Zhao X, Chilkoti A and López GP, Advanced Functional Materials 25:3122–3130 (2015). [Google Scholar]
- 66.Barker TH, Biomaterials 32:4211–4214 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Vandermeulen GWM and Klok H-A, Macromolecular Bioscience 4:383–398 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Lutolf MP and Hubbell JA, Nat Biotech 23:47–55 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Jia X and Kiick KL, Macromolecular Bioscience 9:140–156 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grieshaber SE, Paik BA, Bai S, Kiick KL and Jia X, Soft Matter 9:1589–1599 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grieshaber SEF, E. AJ; Lin–Gibson S; Kiick KL; Jia XQ, Macromolecules 42:2532–2541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grieshaber SE, Farran AJ, Bai S, Kiick KL and Jia X, Biomacromolecules 13:1774–1786 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grover GN, Lam J, Nguyen TH, Segura T and Maynard HD, Biomacromolecules 13:3013–3017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tan H, Chu CR, Payne KA and Marra KG, Biomaterials 30:2499–2506 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin R, Moreira Teixeira LS, Krouwels A, Dijkstra PJ, van Blitterswijk CA, Karperien M and Feijen J, Acta Biomaterialia 6:1968–1977 (2010). [DOI] [PubMed] [Google Scholar]
- 76.Zhang Z, Loebus A, de Vicente G, Ren F, Arafeh M, Ouyang Z and Lensen MC, Chemistry of Materials 26:3624–3630 (2014). [Google Scholar]
- 77.Nguyen KTW, J. L., Biomaterials 23:4307–4314 (2002). [DOI] [PubMed] [Google Scholar]
- 78.Kharkar PM, Rehmann MS, Skeens KM, Maverakis E and Kloxin AM, ACS Biomaterials Science & Engineering (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu SD, K. T.; Jia X, Chemical Communications 51:5218–5237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alge DL, Azagarsamy MA, Donohue DF and Anseth KS, Biomacromolecules 14:949–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iha RK, Wooley KL, Nyström AM, Burke DJ, Kade MJ and Hawker CJ, Chemical Reviews 109:5620–5686 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moreira Teixeira LS, Feijen J, van Blitterswijk CA, Dijkstra PJ and Karperien M, Biomaterials 33:1281–1290 (2012). [DOI] [PubMed] [Google Scholar]
- 83.Schense JC, Bloch J, Aebischer P and Hubbell JA, Nat Biotech 18:415–419 (2000). [DOI] [PubMed] [Google Scholar]
- 84.Kim JL, K. W.; Hefferan TE; Currier BL; Yaszemski MJ; Lu LC, Biomacromolecules 9:149–157 (2008). [DOI] [PubMed] [Google Scholar]
- 85.Pathak CPS, A. S.; Hubbell JA, Journal of the American Chemical Society 114:8311–8312 (1992). [Google Scholar]
- 86.Lobo V, Patil A, Phatak A and Chandra N, Pharmacognosy Reviews 4:118–126 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bryant SJ, Nuttelman CR and Anseth KS, Journal of Biomaterials Science, Polymer Edition 11:439–457 (2000). [DOI] [PubMed] [Google Scholar]
- 88.Shih H and Lin C-C, Macromolecular Rapid Communications 34:269–273 (2013). [DOI] [PubMed] [Google Scholar]
- 89.Jia X, Burdick JA, Kobler J, Clifton RJ,;,Rosowski RJ, Zeitels SM and Langer R, Macromolecules 37:3239–3248 (2004). [Google Scholar]
- 90.Jha AK, Malik MS, Farach-Carson MC, Duncan RL and Jia X, Soft Matter 6:5045–5055 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xiao L, Liu C, Zhu J, Pochan DJ and Jia X, Soft Matter 6:5293–5297 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiao L, Zhu J, Londono DJ, Pochan DJ and Jia X, Soft Matter 8:10233–10237 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xiao L, Tong Z, Chen Y, Pochan DJ, Sabanayagam CR and Jia X, Biomacromolecules 14:3808–3819 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xin Y and Yuan J, Polymer Chemistry 3:3045–3055 (2012). [Google Scholar]
- 95.Yan C, Altunbas A, Yucel T, Nagarkar RP, Schneider JP and Pochan DJ, Soft Matter 6:5143–5156 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Highley CB, Rodell CB and Burdick JA, Advanced Materials 27:5075–5079 (2015). [DOI] [PubMed] [Google Scholar]
- 97.Cai L, Dewi RE and Heilshorn SC, Advanced Functional Materials 25:1344–1351 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xing R, Liu K, Jiao T, Zhang N, Ma K, Zhang R, Zou Q, Ma G and Yan X Advanced Materials 28:3669–3676 (2016). [DOI] [PubMed] [Google Scholar]
- 99.Guvendiren M, Lu HD and Burdick JA, Soft Matter 8:260–272 (2012). [Google Scholar]
- 100.Nishi KK and Jayakrishnan A, Biomacromolecules 8:84–90 (2007). [DOI] [PubMed] [Google Scholar]
- 101.Zhang H, Qadeer A and Chen W, Biomacromolecules 12:1428–1437 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Balakrishnan B, Joshi N and Banerjee R, Journal of Materials Chemistry B 1:5564–5577 (2013). [DOI] [PubMed] [Google Scholar]
- 103.Ren C, Xu C, Li D, Ren H, Hao J and Yang Z, RSC Advances 4:34729–34732 (2014). [Google Scholar]
- 104.Wu X, He C, Wu Y and Chen X, Biomaterials 75:148–162 (2016). [DOI] [PubMed] [Google Scholar]
- 105.Morozowich NL, Nichol JL and Allcock HR, Journal of Polymer Science Part A: Polymer Chemistry 54:2984–2991 (2016). [Google Scholar]
- 106.Zeng Q, Desai MS, Jin H-E, Lee JH, Chang J and Lee S-W, Biomacromolecules 17:2619–2625 (2016). [DOI] [PubMed] [Google Scholar]
- 107.Schmidt P, Zhou L, Tishinov K, Zimmermann K and Gillingham D, Angewandte Chemie International Edition 53:10928–10931 (2014). [DOI] [PubMed] [Google Scholar]
- 108.Lin F, Yu J, Tang W, Zheng J, Defante A, Guo K, Wesdemiotis C and Becker ML, Biomacromolecules 14:3749–3758 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jia X, Colombo G, Padera R, Langer R and Kohane DS, Biomaterials 25:4797–4804 (2004). [DOI] [PubMed] [Google Scholar]
- 110.Gurski LA, Jha AK, Zhang C, Jia X and Farach-Carson MC, Biomaterials 30:6076–6085 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kool ET, Park DH and Crisalli P, Journal of the American Chemical Society 135:17663–17666 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Boehnke N, Cam C, Bat E, Segura T and Maynard HD, Biomacromolecules 16:2101–2108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lokeshwar VBO, C.; Soloway MS; Block N, Cancer Research 773–777 (1997). [PubMed] [Google Scholar]
- 114.Gurski LA, Xu X, Labrada LN, Nguyen NT, Xiao L, van Golen KL, Jia X and Farach-Carson MC, PLoS One 7:e50075 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jha AK, Yang W, Kirn-Safran CB, Farach-Carson MC and Jia X, Biomaterials 30:6964–6975 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Padma PS, Sarah YM, Amit KJ, Weidong Y, Xinqiao J, Mary CF-C and Catherine BKS, Biomedical Materials 7:024109 (2012).22455987 [Google Scholar]
- 117.Jia XQY, Y.; Clifton RJ; Jiao T; Kohane DS; Kobler JB; Zeitels SM; Langer R, Biomacromolecules 7:3336–3341 (2006). [DOI] [PubMed] [Google Scholar]
- 118.Jha AK, Hule RA, Jiao T, Teller SS, Clifton RJ, Duncan RL, Pochan DJ and Jia X, Macromolecules 42:537–546 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mateen R and Hoare T, Journal of Materials Chemistry B 2:5157 (2014). [DOI] [PubMed] [Google Scholar]
- 120.Luo Y, Kobler JB, Heaton JT, Jia X, Zeitels SM and Langer R, Journal of Biomedical Materials Research Part B: Applied Biomaterials 93:386–393 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hudson SP, Langer R, Fink GR and Kohane DS, Biomaterials 31:1444–1452 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sivakumaran D, Maitland D and Hoare T, Biomacromolecules 12:4112–4120 (2011). [DOI] [PubMed] [Google Scholar]
- 123.Binauld S and Stenzel MH, Chemical Communications) 49:2082–2102 (2013). [DOI] [PubMed] [Google Scholar]
- 124.Mather BD, Viswanathan K, Miller KM and Long TE, Progress in Polymer Science 31:487–531 (2006). [Google Scholar]
- 125.Zhang Y, Wang R, Hua Y, Baumgartner R and Cheng J, ACS Macro Letters 3:693–697 (2014). [DOI] [PubMed] [Google Scholar]
- 126.Bencherif SAW, N. R.; Matyjaszewski K, Biomacromolecules 10:2499–2507 (2009). [DOI] [PubMed] [Google Scholar]
- 127.Dong Y, Saeed AO, Hassan W, Keigher C, Zheng Y, Tai H, Pandit A and Wang W, Macromolecular Rapid Communications 33:120–126 (2012). [DOI] [PubMed] [Google Scholar]
- 128.Jin R, Moreira Teixeira LS, Krouwels A, Dijkstra PJ, van Blitterswijk CA, Karperien M and Feijen J, Acta Biomaterialia 6:1968–1977 (2010). [DOI] [PubMed] [Google Scholar]
- 129.Yoo HS, Lee EA, Yoon JJ and Park TG, Biomaterials 26:1925–1933 (2005). [DOI] [PubMed] [Google Scholar]
- 130.Xu X, Farach-Carson MC and Jia X, Biotechnology Advances 32:1256–1268 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xu X, Sabanayagam CR, Harrington DA, Farach-Carson MC and Jia X, Biomaterials 35:3319–3330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ozdemir T, Fowler EW, Liu S, Harrington DA, Witt RL, Farach-Carson MC, Pradhan-Bhatt S and Jia X, ACS Biomaterials Science & Engineering 2:2217–2230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nagase HW, F. J, The Journal of Biological Chemistry 274:21491–21494 (1999). [DOI] [PubMed] [Google Scholar]
- 134.Kessenbrock K, Wang CY and Werb Z, Matrix Biology 44-46:184–190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Loessner D, Stok KS, Lutolf MP, Hutmacher DW, Clements JA and Rizzi SC, Biomaterials 31:8494–8506 (2010). [DOI] [PubMed] [Google Scholar]
- 136.Bellis SL, Biomaterials 32:4205–4210 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lam J and Segura T, Biomaterials 34:3938–3947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nair DP, Podgórski M, Chatani S, Gong T, Xi W, Fenoli CR and Bowman CN, Chemistry of Materials 26:724–744 (2014). [Google Scholar]
- 139.Sawicki LA and Kloxin AM, Biomaterials Science 2:1612–1626 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shih H and Lin C-C, Biomacromolecules 16:1915–1923 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN and Anseth KS, Advanced Materials 21:5005–5010 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Singh SP, Schwartz MP, Lee JY, Fairbanks BD and Anseth KS, Biomaterials Science 2:1024–1034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tokuda EY, Leight JL and Anseth KS, Biomaterials 35:4310–4318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Anderson SB, Lin CC, Kuntzler DV and Anseth KS, Biomaterials 32:3564–3574 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gramlich WM, Kim IL and Burdick JA, Biomaterials 34:9803–9811 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nicolaou KC, Snyder SA, Montagnon T and Vassilikogiannakis G, Angewandte Chemie International Edition 41:1668–1698 (2002). [DOI] [PubMed] [Google Scholar]
- 147.Gandini A, Progress in Polymer Science 38:1–29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Inglis AJ, Nebhani L, Altintas O, Schmidt FG and Barner-Kowollik C, Macromolecules 43:5515–5520 (2010). [Google Scholar]
- 149.Chen X, Dam MA, Ono K, Mal A, Shen H, Nutt SR, Sheran K and Wudl F, Science 295:1698–1702 (2002). [DOI] [PubMed] [Google Scholar]
- 150.Yu F, Cao X, Du J, Wang G and Chen X, ACS Applied Materials & Interfaces 7:24023–24031 (2015). [DOI] [PubMed] [Google Scholar]
- 151.García-Astrain C, Algar I, Gandini A, Eceiza A, Corcuera MÁ and Gabilondo N, Journal of Polymer Science Part A: Polymer Chemistry 53:699–708 (2015). [Google Scholar]
- 152.Nimmo CM, Owen SC and Shoichet MS, Biomacromolecules 12:824–830 (2011). [DOI] [PubMed] [Google Scholar]
- 153.Owen SC, Fisher SA, Tam RY, Nimmo CM and Shoichet MS, Langmuir 29:7393–7400 (2013). [DOI] [PubMed] [Google Scholar]
- 154.Hammer N, Brandl FP, Kirchhof S, Messmann V and Goepferich AM, Macromolecular Bioscience 15:405–413 (2015). [DOI] [PubMed] [Google Scholar]
- 155.Fan M, Ma Y, Zhang Z, Mao J, Tan H and Hu X, Materials Science and Engineering: Part C Materials for Biological Applications 56:311–317 (2015). [DOI] [PubMed] [Google Scholar]
- 156.Fisher SA, Anandakumaran PN, Owen SC and Shoichet MS, Advanced Functional Materials 25:7163–7172 (2015). [Google Scholar]
- 157.Stewart SA, Backholm M, Burke NA and Stover HD, Langmuir (2016). [DOI] [PubMed] [Google Scholar]
- 158.Amblard FC, h. J; Schinazi RF, Chemical Reviews 109:(2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Liang L and Astruc D, Coordination Chemistry Reviews 255:2933–2945 (2011). [Google Scholar]
- 160.Hein JE and Fokin VV, Chemical Society Reviews 39:1302–1315 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tron GC, Pirali T, Billington RA, Canonico PL, Sorba G and Genazzani AA, Medicinal Research Reviews 28:278–308 (2008). [DOI] [PubMed] [Google Scholar]
- 162.Ossipov DAH, J., Macromolecules 39:1709–1718 (2006). [Google Scholar]
- 163.Li Z, Stankevicius E, Ajami A, Raciukaitis G, Husinsky W, Ovsianikov A, Stampfl J and Liska R, Chemical Communications 49:7635–7637 (2013). [DOI] [PubMed] [Google Scholar]
- 164.Adzima BJ, Tao Y, Kloxin CJ, DeForest CA, Anseth KS and Bowman CN, Nature Chemistry 3:256–259 (2011). [DOI] [PubMed] [Google Scholar]
- 165.Truong V, Blakey I and Whittaker AK, Biomacromolecules 13:4012–4021 (2012). [DOI] [PubMed] [Google Scholar]
- 166.Assali M, Cid J-J, Fernández I and Khiar N, Chemistry of Materials 25:4250–4261 (2013). [Google Scholar]
- 167.Li Z, Zheng Z, Su S, Yu L and Wang X, Macromolecules 49:373–386 (2016). [Google Scholar]
- 168.Gandavarapu NR, Azagarsamy MA and Anseth KS, Advanced Materials 26:25212526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Crescenzi V, Cornelio L, Di Meo C, Nardecchia S and Lamanna R, Biomacromolecules 8:1844–1850 (2007). [DOI] [PubMed] [Google Scholar]
- 170.Kennedy DC, McKay CS, Legault MC, Danielson DC, Blake JA, Pegoraro AF, Stolow A, Mester Z and Pezacki JP, Journal of the American Chemical Society 133:17993–18001 (2011). [DOI] [PubMed] [Google Scholar]
- 171.Lallana E, Riguera R and Fernandez-Megia E, Angewandte Chemie International Edition 50:8794–8804 (2011). [DOI] [PubMed] [Google Scholar]
- 172.Uttamapinant C, Tangpeerachaikul A, Grecian S, Clarke S, Singh U, Slade P, Gee KR and Ting AY, Angewandte Chemie International Edition 51:5852–5856 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ekholm FS, Pynnonen H, Vilkman A, Koponen J, Helin J and Satomaa T, Organic & Biomolecular Chemistry 14:849–852 (2016). [DOI] [PubMed] [Google Scholar]
- 174.Uttamapinant C, Sanchez MI, Liu DS, Yao JZ and Ting AY, Nat Protocols 8:1620–1634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Agard NJ, Prescher JA and Bertozzi CR, Journal of the American Chemical Society 126:15046–15047 (2004). [DOI] [PubMed] [Google Scholar]
- 176.Lallana E, Fernandez-Megia E and Riguera R, Journal of the American Chemical Society 131:5748–5750 (2009). [DOI] [PubMed] [Google Scholar]
- 177.Deforest CA, Sims EAand Anseth KS, Chemistry of Materials 22:4783–4790 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.DeForest CAand Anseth KS, Nature Chemistry 3:925–931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Steinhilber D, Rossow T, Wedepohl S, Paulus F, Seiffert S and Haag R, Angewandte Chemie International Edition 52:13538–13543 (2013). [DOI] [PubMed] [Google Scholar]
- 180.Jonker AM, Bode SA, Kusters AH, van Hest JCM and Löwik DWPM, Macromolecular Bioscience 15:1338–1347 (2015). [DOI] [PubMed] [Google Scholar]
- 181.Guo J, Xie Z, Tran RT, Xie D, Jin D, Bai X and Yang J, Advanced Materials 26:1906–1911 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zheng J, Smith Callahan LA, Hao J, Guo K, Wesdemiotis C, Weiss RA and Becker ML, ACS Macro Letters 1:1071–1073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.DeForest CA and Anseth KS, Angewandte Chemie 124:1852–1855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.DeForest CA and Tirrell DA, Nat Mater 14:523–531 (2015). [DOI] [PubMed] [Google Scholar]
- 185.Sletten EM and Bertozzi CR, Accounts of Chemical Research 44:666–676 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Blackman ML, Royzen M and Fox JM, J Am Chem Soc 130:13518–13519 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sletten EM and Bertozzi CR, Angewandte Chemie International Edition 48:6974–6998 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Darko A, Wallace S, Dmitrenko O, Machovina MM, Mehl RA, Chin JW and Fox JM, Chemical Science 5:3770–3776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Taylor MT, Blackman ML, Dmitrenko O and Fox JM, Journal of the American Chemical Society 133:9646–9649 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Albu SA, Al-Karmi SA, Vito A, Dzandzi JPK, Zlitni A, Beckford-Vera D, Blacker M, Janzen N, Patel RM, Capretta A and Valliant JF, Bioconjugate Chemistry 27:207–216 (2016). [DOI] [PubMed] [Google Scholar]
- 191.Blizzard RJ, Backus DR, Brown W, Bazewicz CG, Li Y and Mehl RA, Journal of the American Chemical Society 137:10044–10047 (2015). [DOI] [PubMed] [Google Scholar]
- 192.Murrey HE, Judkins JC, am Ende CW, Ballard TE, Fang Y, Riccardi K, Di L, Guilmette ER, Schwartz JW, Fox JM and Johnson DS, Journal of the American Chemical Society 137:11461–11475 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Selvaraj R, Liu S, Hassink M, Huang C.-w., Yap L.-p., Park R, Fox JM, Li Zand Conti PS, Bioorganic & Medicinal Chemistry Letters 21:5011–5014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wu Z, Liu S, Hassink M, Nair I, Park R, Li L, Todorov I, Fox JM, Li Z, Shively JE, Conti PS and Kandeel F, Journal of Nuclear Medicine 54:244–251 (2013). [DOI] [PubMed] [Google Scholar]
- 195.Rieder U and Luedtke NW, Angewandte Chemie International Edition 53:9168–9172 (2014). [DOI] [PubMed] [Google Scholar]
- 196.Desai RM, Koshy ST, Hilderbrand SA, Mooney DJ and Joshi NS, Biomaterials 50:30–37 (2015). [DOI] [PubMed] [Google Scholar]
- 197.Zhang H, Dicker KT, Xu X, Jia X and Fox JM, ACS Macro Lett 3:727–731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Koshy ST, Desai RM, Joly P, Li J, Bagrodia RK, Lewin SA, Joshi NS and Mooney DJ, Advanced Healthcare Materials 5:541–547 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Liu S, Zhang H, Remy RA, Deng F, Mackay ME, Fox JM and Jia X, Advanced Materials 27:2783–2790 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhang H, Trout WS, Liu S, Andrade GA, Hudson DA, Scinto SL, Dicker KT, Li Y, Lazouski N, Rosenthal J, Thorpe C, Jia X and Fox JM, Journal of the American Chemical Society 138:5978–5983 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Boyer PD and Krebs EG, The enzymes, Academic Press; (1987). [Google Scholar]
- 202.Park KM, Lee Y, Son JY, Oh DH, Lee JS and Park KD, Biomacromolecules 13:604–611 (2012). [DOI] [PubMed] [Google Scholar]
- 203.Raia NR, Partlow BP, McGill M, Kimmerling EP, Ghezzi CE and Kaplan DL, Biomaterials 131:58–67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Griffin DR, Weaver WM, Scumpia PO, Di Carlo D and Segura T, Nat Mater 14:737–744 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Vazquez-Duhalt R, Tinoco R, D’Antonio P, Topoleski LDT and Payne GF, Bioconjugate Chemistry 12:301–306 (2001). [DOI] [PubMed] [Google Scholar]
- 206.Liu H-Y, Greene T, Lin T-Y, Dawes CS, Korc M and Lin C-C, Acta Biomaterialia 48:258–269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mosiewicz KA, Johnsson K and Lutolf MP, Journal of the American Chemical Society 132:5972–5974 (2010). [DOI] [PubMed] [Google Scholar]
- 208.Ohkawa K, Fujii K, Nishida A, Yamauchi T, Ishibashi H and Yamamoto H, Biomacromolecules 2:773–779 (2001). [DOI] [PubMed] [Google Scholar]
- 209.Yang Z, Liang G, Wang L and Xu B, Journal of the American Chemical Society 128:3038–3043 (2006). [DOI] [PubMed] [Google Scholar]
- 210.Wang H, Luo Z, Wang Y, He T, Yang C, Ren C, Ma L, Gong C, Li X and Yang Z, Advanced Functional Materials 26:1822–1829 (2016). [Google Scholar]
- 211.Le Thi P, Lee Y, Nguyen DH and Park KD, Journal of Materials Chemistry B 5:757–764 (2017). [DOI] [PubMed] [Google Scholar]
- 212.Zhong M, Wang R, Kawamoto K, Olsen BD and Johnson JA, Science 353:1264–1268 (2016). [DOI] [PubMed] [Google Scholar]
- 213.Tibbitt MW and Anseth KS, Science Translational Medicine 4:160ps124–160ps124 (2012). [DOI] [PubMed] [Google Scholar]
- 214.Kloxin CJ and Bowman CN, Chemical Society Reviews 42:7161–7173 (2013). [DOI] [PubMed] [Google Scholar]