Abstract
Purpose:
Radiomics allows for powerful data-mining and feature extraction techniques to guide clinical decision making. Image segmentation is a necessary step in such pipelines and different techniques can significantly affect results. We demonstrate that a convolutional neural network (CNN) segmentation method performs comparably to expert manual segmentations in an established radiomics pipeline.
Methods:
Using the manual ROIs (regions-of-interest) of an expert radiologist (R1), a CNN was trained to segment breast lesions from DCE-MRI. Following network training, we segmented lesions for the testing set of a previously established radiomics pipeline for predicting lymph node metastases using DCE-MRI of breast cancer. Prediction accuracy of CNN segmentations relative to manual segmentations by R1 from the original study, a resident (R2) and another expert radiologist (R3) were determined. We then retrained the CNN and radiomics model using R3’s manual segmentations to determine the effects of different expert observers on end-to-end prediction.
Results:
Using R1’s ROIs, the CNN achieved a mean Dice coefficient of 0.71±0.16 in the testing set. When input to our previously published radiomics pipeline, these CNN segmentations achieved comparable prediction performance to R1’s manual ROIs, and superior performance to those of the other radiologists. Similar results were seen when training the CNN and radiomics model using R3’s ROIs.
Conclusion:
A CNN architecture is able to provide DCE-MRI breast lesion segmentations which are suitable for input to our radiomics model. Moreover, the previously established radiomics model and CNN can be accurately trained end-to-end using ground truth data provided by distinct experts.
Keywords: Radiomics, Deep learning, automated segmentation, breast cancer, DCE-MRI
Introduction
Radiomic analysis has received marked attention over the past several years, with the medical imaging community seeking to realize images as mineable data relating to a patient’s specific condition as opposed to pictorial representations of their disease or lack thereof (1). While imaging currently serves integral diagnostic and prognostic roles in oncology, high order feature extraction allows for the development of computational models of disease progression and outcome which leverage features that might be difficult or impossible for humans to perceive (2).
Whereas radiomics has been identified as an emerging tool to help advance personalized medicine without significantly impacting current routine procedures (3), these techniques require numerous processing steps which can differ in their implementation and significantly impact performance. Despite recent and encouraging endeavors to standardize quantitative imaging biomarker research (4), radiomics pipelines require a number of steps which can vary across medical centers and vendors. Images must be acquired, reconstructed, processed for viewing, segmented and finally have features automatically extracted using any of several available software packages or in house programs; finally, a predictive model comprising any number of these determined features must be developed. Given the numerous steps involved in radiomics processing, it is not surprising that achieving reproducible and reliable results—even when applied to data collected at the same center—is a primary concern within the field (5,6).
Region of interest (ROI) selection/segmentation, the explicit identification of the region to be processed, is unique among the aforementioned steps in that not only do radiomics features largely demonstrate a dependence upon segmentation method (7,8), but the requirement for segmentation is also the primary challenge for the clinical adoption of validated radiomics pipelines. Whereas simple methods based on thresholding can be used in the case of PET or MRI and CT data demonstrating high lesion contrast, the current bulk of radiomics pipelines add to physician workload by requiring manual segmentations (9). While semi-automated and interactive methods have been developed, these techniques can still be time consuming and are user-dependent, raising concern over their applicability to high throughput diseases.
Recently, convolutional neural networks (CNN) (10) have become a preeminent tool across virtually all subfields of computer vision, notably including image segmentation (11). CNNs provide a rich, neurobiologically-inspired framework for automatically learning abstract representations of training data which the network can leverage in order to make accurate inferences about prospective data after optimization. In the context of radiomics, CNNs offer the possibility of fully automated, accurate segmentation pipelines which require no human intervention after training and thus are capable of providing stable and reproducible ROIs for radiomic analysis.
Despite the demonstrated performance of CNNs in the image segmentation community, and ideality of the robust, automated segmentations provided, little work has been done to examine the possibility of using CNN-derived segmentations for input into established radiomics processing pipelines. Whereas recent publications have shown that the latent image representations extracted from the hidden layers of CNNs demonstrate predictive value for grading glioma (12) and predicting survivability in glioblastoma multiforme (13), there is a severe lack of literature assessing the viability of CNN segmentations as input for radiomics models dependent upon extraction of hand-crafted features from labeled input data.
Our group has recently developed and validated a radiomics pipeline for predicting sentinel lymph node (SLN) metastases from DCE-MRI data in women with breast cancer (14). Our model achieved a satisfying prediction performance with a high negative predictive value (NPV), providing a non-invasive method to evaluate SLN status. A high NPV could also potentially offer greater benefits for patients with negative SLN, helping them eliminate unnecessary invasive lymph node removal and associated complications. Despite the pipeline’s ability to accurately determine SLN status, its applicability to prospective clinical work is severely limited by its dependence upon manual segmentation; this limitation is shared by virtually all pipelines for radiomic modelling of breast cancer. Breast cancer is one of the most commonly encountered cases in oncology and any clinically viable radiomics pipeline must be designed to require as little physician interaction as possible in light of this.
Herein, we discuss an entirely automated CNN approach for segmenting breast lesions from the same DCE-MRI input to our previously published radiomics pipeline for predicting SLN metastases. Our approach adopts the common U-net architecture (15) and yields encouraging segmentation accuracy in a challenging testing dataset which comprises several tumor subtypes and anatomical locations within the breast. More importantly, we demonstrate that using CNN-derived segmentations for input to our radiomics pipeline not only introduces less variance than using segmentations manually drawn by additional radiologists, but also that the use of these CNN-derived segmentations does not substantially change SLN prediction accuracy. When training the CNN using manual segmentations by an expert breast radiologist, we see that radiomics prediction accuracy is higher for the automated segmentations than for manual ones drawn by either a radiology resident or a second expert radiologist.
In interesting consideration is the fact that the CNN will show some bias towards the expert whose manual segmentations were used to train it, which will affect radiomic feature calculation. Given the availability of manual segmentations from two experts, we trained both our previously established radiomics model and CNN segmentation pipeline using data from the second expert radiologist. Interesting, we saw that the end-to-end prediction accuracy for automated lesions is comparable to manual segmentations from the second radiologist and outperforms manual segmentations from the radiologists. The similar observation across both training paradigms suggests that the proposed technique is optimally conducted using CNN and radiomics training data provided by the same observer.
Methods
IRB approval was obtained for this retrospective analysis, alongside a waiver of consent.
Dataset
All MRI scans for the radiomics datasets were performed using 8-channel breast coils on 1.5T GE Signa HDxt scanners (GE Healthcare, Wauwatosa, Wisconsin). Gadolinium contrast (Magnevist; Schering, Berlin, Germany) was injected intravenously at a rate of 2ml/s for a total dose of 0.2ml/kg; Magnevist injection as followed by a 20-ml saline flush at a rate of 2ml/s.
Our retrospective dataset consisted of MRI scans from 317 women who had previously undergone DCE-MRI as part of their routine care for breast cancer at our institution. As previously described in Ref (14), out of these scans, 109 scans were used to train the radiomics model, which was tested in an independent set of 54 subjects. Acquisition for these consisted of a pre-contrast and four post-contrast frames, acquired using a sagittal VIBRANT multiband sequence (TR=4.46–7.80ms; TE=1.54–4.20ms; flip angle=10°; matrix=256×256; pixel size=0.7mm2, slice thickness=2mm). Images were acquired prior to injection of contrast, as well as 2, 4, 6 and 8 minutes after contrast administration.
In addition to the same 109 scans an additional 154 scans not used previously to train the radiomics model due to slightly different pixel sizes were also included for the training of the segmentation CNN. As such, the CNN training set contained all lesions the radiomics model was trained upon, without any exclusions, in addition to the additional 154 lesions, for a total of 263 training subjects. Acquisition parameters in these additional scans were: TR=4.19–7.80ms; TE=1.46–4.20ms; flip angle=10°; matrix=256×256; pixel size=0.63–1.0mm2, slice thickness=2–2.8mm. Temporal spacing of the frames was the same as above. Among these individuals, 32 had benign lesions which further precluded them from inclusion in the original radiomics study. The CNN model was tested using the same 54 subject datasets as the radiomics model. Figure 1 shows the schema of this study with the distribution of training and testing data.
Figure 1.
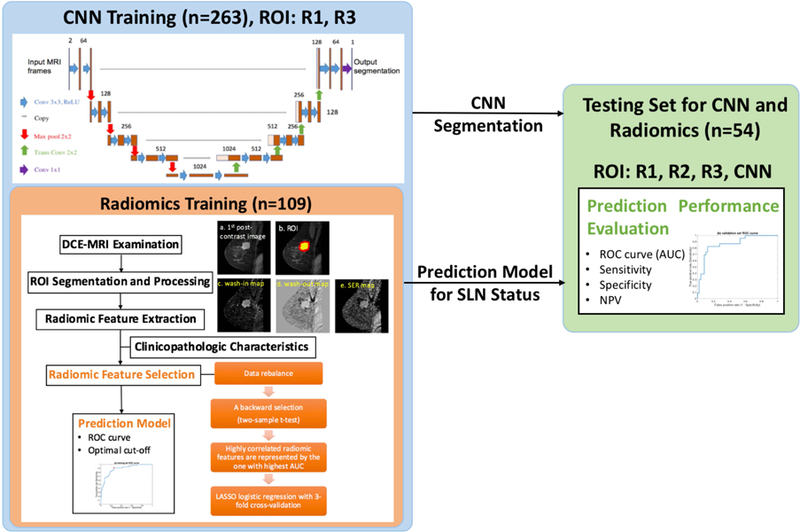
A schema of the study design, showing the distribution of training and testing data and the methods for both the CNN and radiomic analysis. Note that the radiomic training dataset is a subset of the CNN training dataset.
In all cases, only slices containing lesion volume were extracted from PACS. Lesions were subsequently segmented on the first post-contrast frame by three radiologists: an expert radiologist in breast MRI with over a decade of experience in the field (R1), a first year radiology resident (R2) who was trained by R1, and lastly another expert radiologist (R3) with similar clinical experience to R1. As R1 was the radiologist involved in the original study, they determined the slices which were extracted from PACS. R1 and R3 segmented all lesions used in this study while R2 segmented only the testing set lesions. All radiologists used the pre- and first post-contrast frame to determine lesion location, and drew their ROIs on the post-contrast frame. Figure 2 shows an example segmentation in the testing set by all three radiologists as well as the CNN.
Figure 2.
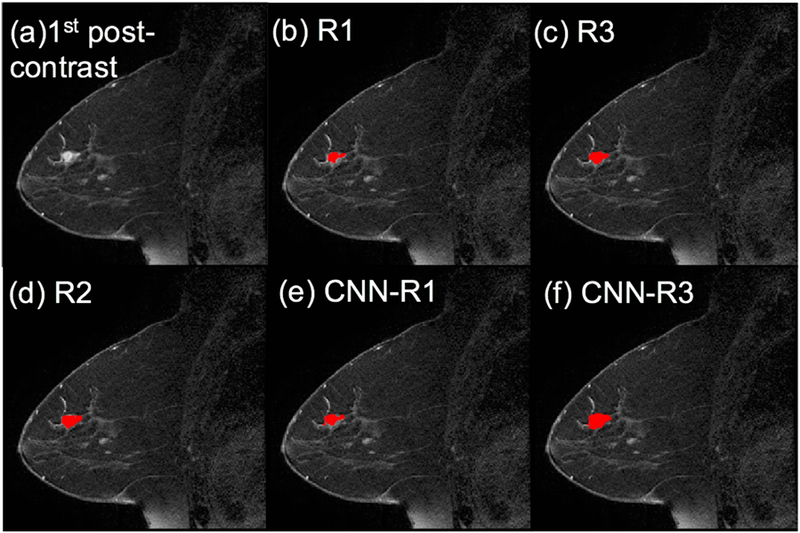
ROI comparison of an example from the testing set. (a) first post-contrast frame, (b)~(f) lesion segmentations are superimposed on the first post-contrast frame in red (the manual ROIs from the three radiologists and the CNN-based ROIs trained by R1’s and R3’s segmentations, respectively).
Convolutional Neural Network Training and Evaluation
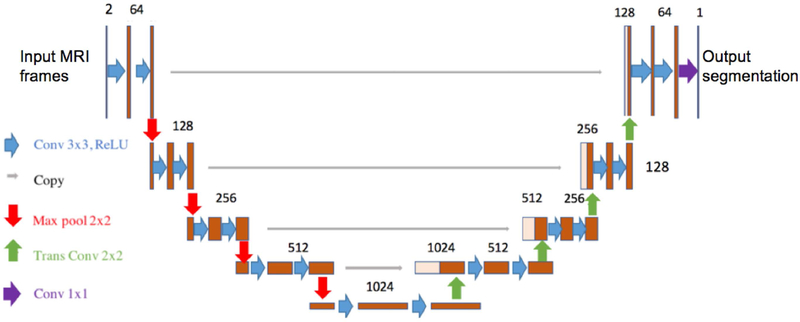
A U-net architecture was implemented in Tensorflow (16) to address the challenge of automatically segmenting breast lesions in DCE-MRI for the radiomics pipeline. Figure 3 shows the network architecture.
Figure 3.
A U-Net CNN architecture that was employed in this study.
Two input channels were provided, namely the pre-contrast and first post-contrast frames. This allowed the network to utilize the same information the used by the radiologist to determine ROIs. Both input channels were independently normalized using the mean value and standard deviation within that channel across the entire training set; during evaluation, normalization was again performed using mean and standard deviation values determined in the training set. During training, input MRI data were rotated by up to 5° about a random axis prior to presentation to the network for data augmentation. This data augmentation was performed on-the-fly at each iteration. The network was trained using the Adam optimizer with a learning rate of 10−5 and first and second moment exponential decay rates of 0.9 and 0.999, respectively (17). All weights were initialized using the Glorot method (18), biases were initialized to zero. The network was trained in 2D, minibatches of individual slices were randomly selected from the entire training set. The training objective was to minimize the cross entropy loss between the network’s output and radiologists’ hand drawn segmentations; this cost function was averaged over slices in each minibatch for each update step. In addition to cross entropy loss, an L2-regularization was imposed upon all network weights (regularization magnitude = 0.01). The network was trained with an Nvidia Titan XP on a workstation running Ubuntu 18.04 LTS for 50,000 iterations using a minibatch size of 32. The network was initially trained using R1 manual segmentations.
The CNN output was compressed to the range (0,1) using a sigmoid function. An optimal threshold for binarizing the CNN output was determined in the training set; 1,000 incremental values were applied as thresholds, with the optimal threshold being that which led to the highest Dice coefficient averaged at the subject-level across the training set. Binarized outputs using the optimal threshold were used as the CNN segmentation.
CNN performance was assessed in two ways: pure segmentation accuracy was assessed using the Dice coefficient between manual segmentations and the thresholded network segmentations at the subject-level; more importantly, the CNN segmentations in the testing dataset were then presented to a previously published radiomics pipeline that had been developed using R1 manual segmentations, the predictive accuracy when using CNN-derived segmentations was then assessed in comparison to evaluation using manual segmentations from R1, R2 and R3.
Following analysis using R1 manual segmentations as ground truth, both the radiomics model and segmentation network were retrained using R3 manual segmentations as ground truth. The same analyses were performed.
Radiomics
Radiomics training and testing were conducted using data from 163 patients as previously described(14). Patient characteristics are provided in Supporting Information Table S1.
Summarily, to train the radiomics model, three maps independent of original MR signal intensity were generated to allow for direct comparison across patients: wash-in maps ((S1-S0)/S0)100%, wash-out maps ((S1-S4)/S1))100% and signal enhancement ratio (SER) maps ((S1-S0)/(S4-S0))100%, where S0, S1, and S4 are the pre-contrast, first post-contrast and fourth (final) post-contrast images, respectively. The intratumoral ROIs were defined as the largest volume of the ROIs if multiple regions were segmented (for both manual and automated segmentations). Peritumoral regions were obtained by dilating the intratumoral ROI 4mm. A total of 590 radiomic features were extracted from both intratumoral and peritumoral regions on three maps for each patient, please refer to our previous publication (14) for the list of features used. Moreover, seven clinicopathologic characteristics were collected from patient medical records and combined with the radiomic features, including age, tumor location, histological type and grade of invasive carcinoma, molecular subtype, lymph-vascular invasion (LVI) and multifocality. In order to avoid overfitting, the dataset was randomly divided into two independent subsets as mentioned: a training set (~67%, 109 patients with 37 positive SLN) and a testing set (~33%, 54 patients with 18 positive SLN). The training set was used for feature selection and prediction model generation. The feature selection method, as described in our previous work (14), is provided in Supporting Information. Using the features generated from R1’s manual ROIs, six significant features were finally selected to establish the prediction model using logistic regression, these features are discussed in our previous manuscript (14). Using R3’s manual ROIs for this radiomic analysis using the same method, there were also six features selected in the final prediction model. The details of these two models derived from the ROI segmentations of different expert radiologists (R1 and R3) were provided in Supporting Information (see Supporting Information Table S2 and Supporting Information Table S3). For prediction assessment, the ROC curves were plotted with the optimal thresholds determined by maximizing the Youden index (sensitivity+specificity−1). The AUC, sensitivity, specificity and NPV were then calculated.
Task-based assessment of CNN segmentation
The prediction models trained by R1 and R3 were further tested in the independent testing set using the features from the ROIs generated by R1, R2, R3 and CNN, respectively. The same thresholds determined in the training set was applied in the testing set. The corresponding ROC curves, AUC, sensitivity, specificity and NPV were calculated in MATLAB R2017b. The AUCs derived from the radiologists’ manual segmentations and CNN-based automated segmentation were compared using DeLong test(19) in MedCalc (Version 18.11.6). The level of statistically significant difference was set at p<0.05.
Results
CNN Segmentation Accuracy
For the CNN trained using R1’s manual ROIs, the optimal threshold for the CNN output was determined when the highest Dice index of 0.99±0.02 was achieved in the training set. After applying this optimal threshold to the testing set, the CNN segmentations achieved a mean Dice coefficient of 0.71±0.16 relative to R1’s manual segmentations. The CNN’s Dice coefficient relative to R2 was 0.61±0.17 and for R3 was 0.67±0.18.
Similarly, for the CNN trained using R3’s manual ROIs, the optimal threshold for the CNN output was determined when the highest Dice index of 0.98±0.02 was achieved in the training set. After applying this optimal threshold to the testing set, the CNN segmentations achieved a mean Dice coefficient of 0.67±0.20 relative to R3’s manual segmentations. When trained by R3, the CNN’s Dice coefficient relative to R2 was 0.64±0.22 and for R1 was 0.61±0.18.
To contextualize the CNN’s performances in both instances, the mean Dice coefficient between R1 and R2 is 0.61±0.17, between R1 and R3 is 0.68±0.15 and between R2 and R3 is 0.63±0.19. The CNN trained on R1 significantly outperformed the agreement between R1 and R2 (p<10−5, paired t-test), no other significant differences are observed when comparing automated lesions to the radiologist they were trained upon to the other radiologists.
Figure 2 shows an example from the testing set with the manual ROIs from three radiologists and the automated ROIs from the network when train by R1 and R3. It is noted that the CNN-based automated segmentation mimics the drawing behavior of the radiologist whose ROIs were used to train the CNN.
Figure 4 shows the frequency histograms of Dice coefficients between R1 and CNN trained by R1, and between R3 and CNN trained by R3 in the testing set.
Figure 4.
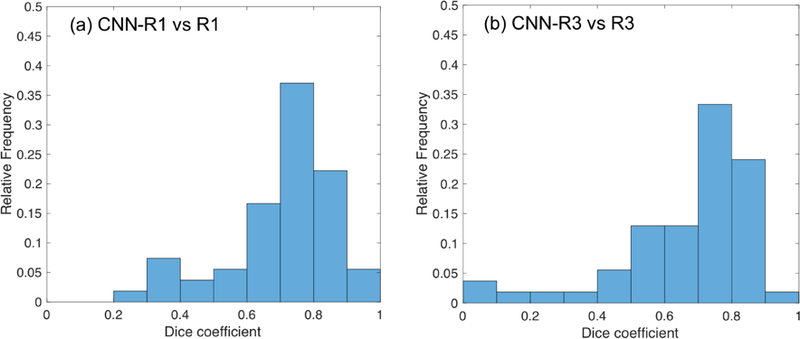
Frequency histograms of Dice coefficients in the testing dataset, (a) CNN-R1 (CNN-based ROIs trained by R1) vs R1, (b) CNN-R3 (CNN-based ROIs trained by R3) vs R3.
Radiomics Results
The original radiomics model was trained using R1’s manual segmentations. For this original model, Figure 5(a) demonstrates the ROC curves of using features from ROIs generated by R1, R2, R3 and the CNN trained by R1, respectively. The corresponding prediction results are demonstrated in Table 1, with the DeLong test p value between the AUCs derived from R1 and the CNN trained by R1. Figure 5(b) demonstrates the same ROC curves yielded by training both the radiomics model and CNN using R3’s manual segmentations. The corresponding prediction results are demonstrated in Table 2.
Figure 5.
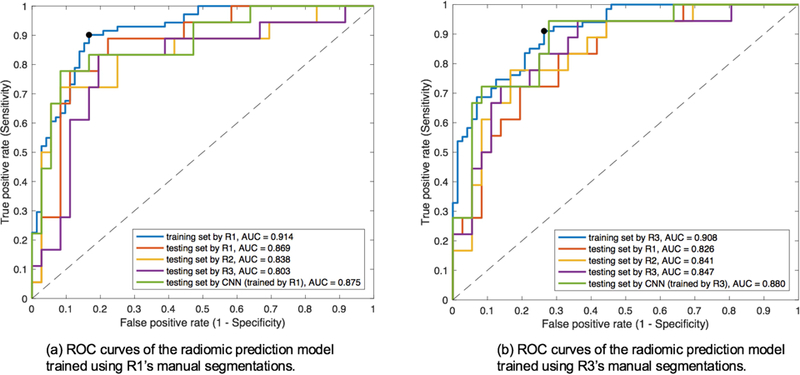
ROC curves with AUC corresponding to Table 1(a) and Table 2(b).
Table 1.
Performance metrics for SLN prediction based on the radiomic model trained by R1’s ROIs in the training dataset alongside testing set performance with R1, R2, R3 and CNN segmentations trained by R1’s ROIs.
| Training set R1 | Testing set | ||||
|---|---|---|---|---|---|
| R1 | R2 | R3 | CNN (trained by R1)* | ||
| AUC | 0.914 | 0.869 | 0.838 | 0.803 | 0.875 |
| Sensitivity | 0.901 | 0.778 | 0.722 | 0.833 | 0.778 |
| Specificity | 0.833 | 0.861 | 0.806 | 0.722 | 0.833 |
| NPV | 0.896 | 0.886 | 0.853 | 0.897 | 0.882 |
p=0.869, DeLong test between AUCs of R1 and CNN trained by R1.
Table 2.
Performance metrics for SLN prediction based on the radiomic model trained by R3’s ROIs in the training dataset alongside testing set performance with R1, R2, R3 and CNN segmentations trained by R3’s ROIs.
| Training set R3 | Testing set | ||||
|---|---|---|---|---|---|
| R1 | R2 | R3 | CNN (trained by R3)* | ||
| AUC | 0.908 | 0.826 | 0.841 | 0.847 | 0.880 |
| Sensitivity | 0.910 | 0.722 | 0.778 | 0.722 | 0.722 |
| Specificity | 0.736 | 0.778 | 0.778 | 0.861 | 0.889 |
| NPV | 0.898 | 0.848 | 0.875 | 0.861 | 0.865 |
p=0.395, DeLong test between AUCs of R3 and CNN trained by R3.
As shown in Table 1 and 2, the CNN-based ROIs exhibit very comparable task-based performance in the radiomic model when compared to manual ROIs from the radiologist whose data were used to train the model and CNN (no statistically significant difference was found in the AUCs using DeLong test). Moreover, in both instances, automated ROI prediction performance is slightly superior to the manual ROIs drawn by other radiologists.
Discussion
The primary conclusion of this task-based assessment is that CNN segmentations are sufficient for the radiomics processing task in the presented pipeline. Additional validation for separate cases would be beneficial to the radiomics community. The conclusion is supported by two primary observations. Most importantly, the U-net approach achieved predictive accuracy similar to expert radiologists’ manual segmentations for this specific task. Outside of this, comparison to other radiologists, with varying levels of experience, is one of the strongest methodological aspects of this work, and when possible should be included in future task-based assessments. It is extremely encouraging to note that our previously published model can be trained “end-to-end” using data from different expert radiologists to serve as a standalone, fully automated, prediction pipeline.
Despite a suboptimal training dataset, which was limited in size and contained some heterogeneity of MRI parameters, the U-net achieved comparable Dice coefficient performance to recently published methods (9,20). This segmentation accuracy is achieved in a realistic, clinically challenging dataset which comprises several breast cancer subtypes and presentations. A consequence of the limited dataset size is the fact that the CNN, when trained using data from either R1 or R3, generally overfit the training data, yielding a higher Dice coefficient across the training dataset than the testing set in both instances. Despite this, testing set batch statistics were logged throughout training and the maximum Dice coefficient was seen to plateau as opposed to degrade overtime, suggesting that the near perfect training set performance did not come at the expense of testing set performance. In the future, an expanded dataset will allow for a held-out validation population, enabling hyperparameter and network tuning in a manner free of bias towards testing set performance, as well as selection of segmentation thresholds outside of the training set.
A substantial strength of this approach is that the presented network is in no way dependent upon modelling of gadolinium kinetics, or any other assumption about the underlying behavior of lesions in an image, unlike many other comparably performing candidate strategies (21). As such, neural network approaches have the benefit of not only being fully automated but also being adaptable to training with multiple contrasts such as diffusion or T1/T2 weighted MRI as are commonly acquired in routine clinical cancer imaging. Furthermore, giving this flexibility, transfer learning could be used to apply our now fully automated model to other disease cases while requiring a smaller amount of data to train the segmentation pipeline. CNN models are the ideal framework for radiomics segmentation in many regards, including notably the results herein, and the community will benefit from their adoption and task-based assessment in future radiomics works.
In this study, we utilized the manual segmentations from two expert radiologists and repeated the study (both CNN segmentation and radiomic analysis) using the same method. Our results show that the CNN-based ROIs exhibit very comparable or even slightly better prediction performance in the radiomic model than manual ROIs from the same radiologist who had drawn the CNN training set, and also the prediction performance is superior than the manual ROIs drawn by other radiologists. Our segmentation accuracy approaches that of contemporary state of the art hand-crafted, semi-automated methods(22). When training with either R1 or R3 ground truth, the CNN shows testing set segmentation accuracy which is strictly noninferior to that demonstrated by separate radiologists’ hand drawn segmentations. At the same time, our method achieves this impressive result without the need for semi-automated approaches or use case-specific modelling of acquisition physics or gadolinium kinetics, a key facet of other DCE-MRI segmentation pipelines. Previous radiomics-oriented segmentation work has been conducted using semi-automated pipelines which are more heavily dependent upon specific modelling of DCE-MRI acquisition (9). On contrast, our purely CNN framework shows promise for general use in radiomics processing; this is, in part, supported by our ability to retrain the model end-to-end using ground truth data from R3.
Our study admits a handful of limitations which must be noted. The first among these is the fact that we were required to train our CNN in 2D given the relative paucity of training data. This, in combination with data augmentation, allowed the network to undergo more distinct update steps than 3D training would provide. Despite this, a 3D CNN would likely be able to leverage interslice spatial information to achieve more accurate predictions. Further improvements may be afforded by the integration of recurrent neural network models into our U-net approach, which may allow for the exploitation of gadolinium kinetics beyond the two frames used in presented segmentation, although this would come at the expense of the ability to easily adapt our network for other disease cases where temporal data might not be available. The application of our end-to-end pipeline to other radiomics tasks will be a key avenue moving forward. An additional limitation of our study is the fact that DCE-MRI data were originally extracted solely to develop the radiomics model and, as such, only slices that R1 deemed to contain lesion were included. This limits the network’s applicability slightly in the sense that it never was exposed to genuinely negative data and, moreover, slightly biases the dataset towards R1’s interpretation of lesion area. Additional fine-tuning using only lesion-free slices will likely be necessary before application to prospective datasets.
In recent years, deep learning-based techniques have been published which either synergize hand crafted features with automated feature representations achieved using deep learning (23), or simply extract features exclusively using deep CNNs (12). However, traditional radiomics support is valuable for many reasons. Primarily, a large amount of preexisting work is underpinned by more hand-crafted methods such as our original pipeline. In demonstrating the suitability of fully automatic prediction, from segmentation to clinical endpoint, such models can achieve many of the same discussed benefits of deep learning-predicated pipelines, namely robustness against interobserver segmentation variance and general convenience for physicians utilizing the pipelines. Moreover, hand crafted feature pipelines benefit from a reduced need for training data and better interpretability given their predication upon analytically derived features. In the future, we will investigate methods to synergize our current two step approach, although our primary interests moving forward will be the adaptation of this end-to-end automated pipeline to other clinical decision cases.
Supplementary Material
Supporting Information Table S1. Clinical and histopathological characteristics.
Supporting Information Table S2. Selected features in the prediction model trained by R1’s segmentations.
Supporting Information Table S3. Selected features in the prediction model trained by R3’s segmentations.
Acknowledgements
This work in part supported by NIH R03CA223052, Carol M. Baldwin Foundation for Breast Cancer, Walk for Beauty Foundation Award, NVIDIA GPU Grant Program.
References
- 1.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2015;278(2):563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chicklore S, Goh V, Siddique M, Roy A, Marsden PK, Cook GJ. Quantifying tumour heterogeneity in 18 F-FDG PET/CT imaging by texture analysis. European journal of nuclear medicine and molecular imaging 2013;40(1):133–140. [DOI] [PubMed] [Google Scholar]
- 3.Lambin P, Leijenaar RT, Deist TM, Peerlings J, de Jong EE, van Timmeren J, Sanduleanu S, Larue RT, Even AJ, Jochems A. Radiomics: the bridge between medical imaging and personalized medicine. Nature Reviews Clinical Oncology 2017;14(12):749. [DOI] [PubMed] [Google Scholar]
- 4.Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative arXiv preprint arXiv:161207003 2016.
- 5.Hatt M, Tixier F, Pierce L, Kinahan PE, Le Rest CC, Visvikis D. Characterization of PET/CT images using texture analysis: the past, the present… any future? European journal of nuclear medicine and molecular imaging 2017;44(1):151–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yip SS, Aerts HJ. Applications and limitations of radiomics. Physics in Medicine & Biology 2016;61(13):R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larue RT, Defraene G, De Ruysscher D, Lambin P, Van Elmpt W. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. The British journal of radiology 2017;90(1070):20160665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altazi BA, Zhang GG, Fernandez DC, Montejo ME, Hunt D, Werner J, Biagioli MC, Moros EG. Reproducibility of F18‐FDG PET radiomic features for different cervical tumor segmentation methods, gray‐level discretization, and reconstruction algorithms. Journal of applied clinical medical physics 2017;18(6):32–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veeraraghavan H, Dashevsky BZ, Onishi N, Sadinski M, Morris E, Deasy JO, Sutton EJ. Appearance Constrained Semi-Automatic Segmentation from DCE-MRI is Reproducible and Feasible for Breast Cancer Radiomics: A Feasibility Study. Scientific reports 2018;8(1):4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proceedings of the IEEE 1998;86(11):2278–2324. [Google Scholar]
- 11.Garcia-Garcia A, Orts-Escolano S, Oprea S, Villena-Martinez V, Garcia-Rodriguez J. A review on deep learning techniques applied to semantic segmentation arXiv preprint arXiv:170406857 2017.
- 12.Li Z, Wang Y, Yu J, Guo Y, Cao W. Deep learning based radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Scientific reports 2017;7(1):5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lao J, Chen Y, Li Z-C, Li Q, Zhang J, Liu J, Zhai G. A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Scientific reports 2017;7(1):10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, Ding J, Spuhler K, Gao Y, Serrano Sosa M, Moriarty M, Hussain S, He X, Liang C, Huang C. Preoperative prediction of sentinel lymph node metastasis in breast cancer by radiomic signatures from dynamic contrast‐enhanced MRI. Journal of Magnetic Resonance Imaging 2018. [DOI] [PMC free article] [PubMed]
- 15.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation 2015. Springer; p 234–241. [Google Scholar]
- 16.Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M. Tensorflow: Large-scale machine learning on heterogeneous distributed systems arXiv preprint arXiv:160304467 2016.
- 17.Kingma D, Ba J. Adam: A method for stochastic optimization arXiv preprint arXiv:14126980 2014.
- 18.Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks 2010. p 249–256.
- 19.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44(3):837–845. [PubMed] [Google Scholar]
- 20.Maicas G, Carneiro G, Bradley AP. Globally optimal breast mass segmentation from DCE-MRI using deep semantic segmentation as shape prior 2017. IEEE; p 305–309. [Google Scholar]
- 21.Jayender J, Chikarmane S, Jolesz FA, Gombos E. Automatic segmentation of invasive breast carcinomas from dynamic contrast‐enhanced MRI using time series analysis. Journal of Magnetic Resonance Imaging 2014;40(2):467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vesal S, Diaz-Pinto A, Ravikumar N, Ellmann S, Davari A, Maier A. Semi-automatic algorithm for breast MRI lesion segmentation using marker-controlled watershed transformation arXiv preprint arXiv:171205200 2017.
- 23.Cha KH, Hadjiiski L, Chan H-P, Weizer AZ, Alva A, Cohan RH, Caoili EM, Paramagul C, Samala RK. Bladder cancer treatment response assessment in CT using radiomics with deep-learning. Scientific reports 2017;7(1):8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Table S1. Clinical and histopathological characteristics.
Supporting Information Table S2. Selected features in the prediction model trained by R1’s segmentations.
Supporting Information Table S3. Selected features in the prediction model trained by R3’s segmentations.