Abstract
Purpose
Radiomic texture analysis is typically performed on images acquired under specific, homogeneous imaging conditions. These controlled conditions may not be representative of the range of imaging conditions implemented clinically. We aim to develop a two‐stage method of radiomic texture analysis that incorporates the reproducibility of individual texture features across imaging conditions to guide the development of texture signatures which are robust across mammography unit vendors.
Methods
Full‐field digital mammograms were retrospectively collected for women who underwent screening mammography on both a Hologic Lorad Selenia and GE Senographe 2000D system. Radiomic features were calculated on manually placed regions of interest in each image. In stage one (robustness assessment), we identified a set of nonredundant features that were reproducible across the two different vendors. This was achieved through hierarchical clustering and application of robustness metrics. In stage two (classification evaluation), we performed stepwise feature selection and leave‐one‐out quadratic discriminant analysis (QDA) to construct radiomic signatures. We refer to this two‐state method as robustness assessment, classification evaluation (RACE). These radiomic signatures were used to classify the risk of breast cancer through receiver operator characteristic (ROC) analysis, using the area under the ROC curve as a figure of merit in the task of distinguishing between women with and without high‐risk factors present. Generalizability was investigated by comparing the classification performance of a feature set on the images from which they were selected (intravendor) to the classification performance on images from the vendor on which it was not selected (intervendor). Intervendor and intravendor performances were also compared to the performance obtained by implementing ComBat, a feature‐level harmonization method and to the performance by implementing ComBat followed by RACE.
Results
Generalizability, defined as the difference between intervendor and intravendor classification performance, was shown to monotonically decrease as the number of clusters used in stage one increased (Mann–Kendall P < 0.001). Intravendor performance was not shown to be statistically different from ComBat harmonization while intervendor performance was significantly higher than ComBat. No significant difference was observed between either of the single methods and the use of ComBat followed by RACE.
Conclusions
A two‐stage method for robust radiomic signature construction is proposed and demonstrated in the task of breast cancer risk assessment. The proposed method was used to assess generalizability of radiomic texture signatures at varying levels of feature robustness criteria. The results suggest that generalizability of feature sets monotonically decreases as reproducibility of features decreases. This trend suggests that considerations of feature robustness in feature selection methodology could improve classifier generalizability in multifarious full‐field digital mammography datasets collected on various vendor units. Additionally, harmonization methods such as ComBat may hold utility in classification schemes and should continue to be investigated.
Keywords: radiomics, breast cancer, robustness
1. Introduction
Breast cancer is one of the most commonly screened for forms of cancer, with 65.3% of women aged 40 and over reported to having had a mammogram in the past 2 yr.1 Mammographic screening has proved useful in increasing early detection of breast cancer and reducing disease mortality.2 In addition to detecting cancer, mammograms provide imaging phenotypes which may inform lifetime risk. For example, it has been well documented that mammographic density can be useful in predicting breast cancer risk.3, 4, 5 Typically, personalized risk models include characteristics such as age, family history, and certain genetic mutations such as BRCA1/BRCA2. Developments in computer‐aided diagnosis (CAD) suggest that parenchymal texture may also help inform risk.
Quantitative measures of parenchymal texture have been successfully applied to evaluate the risk of cancer in asymptomatic females.6, 7, 8, 9, 10, 11, 12, 13, 14 These studies use radiomic texture features including fractal dimension,6 power law spectral analysis,7 absolute gray level, gray‐level histogram analysis, neighborhood gray tone difference matrix (NGTDM), and gray‐level co‐occurrence matrix (GLCM).15
Risk evaluation stands to be particularly impactful for patient care due to established high‐risk screening recommendations that have been enacted by agencies such as the American Cancer Society. These recommendations help translate identification of high‐risk individuals to actionable recommendations, which may lead to improved early detection of disease.16 The availability of specialized screening modalities such as MRI and clinical impact of supplemental screening on high‐risk populations has elevated the demand for strong risk evaluation metrics. The actionable screening steps available to women at an elevated risk of breast cancer have motivated continued research in risk assessment in order to best utilize the available specialized screening modalities.
One challenge faced in developing widely generalizable imaging phenotypes is the sensitivity of individual texture features to imaging conditions. Imaging conditions such as manufacturer, kVp, and processing algorithms may each affect radiomic feature values.17, 18, 19, 20, 21 Studies have been performed to evaluate repeatability (test–retest) and reproducibility of radiomic features in cancer imaging.
In a study by van Velden et al., the repeatability of radiomic features in nonsmall‐cell lung cancer (NSCLC) using positron emission tomography/computed tomography (PET/CT) images was investigated. The study reported high repeatability of radiomic features relative to standardized uptake value measures, and found that more features were sensitive to delineation than to reconstruction changes.
Hunter et al. studied reproducibility and redundancy of radiomic features of NSCLC patients and on a texture phantom from CT images.22 The study reported that feature redundancy and reproducibility was highly machine sensitive.
Zhao et al. used same‐day repeat CT scans of lung cancer patients to evaluate the impact of reconstruction settings, slice thickness, and reconstruction algorithms on feature repeatability.23 The study concluded that most texture features are repeatable, although they were significantly impacted by reconstruction parameters.
However, incorporation of repeatability and reproducibility into feature selection and classification construction procedures is relatively unexplored. Therefore, this study proposes methods by which to implement the findings of robustness studies to the improvement of CAD systems.
Many feature signatures for risk are developed on homogeneous databases, and reproducibility over imaging conditions is not always evaluated in imaging phenotype studies. To ensure generalizability of findings to heterogeneous imaging conditions, this study identifies a parenchymal texture signature descriptive of risk of breast cancer by emphasizing both (a) robustness across imaging manufacturer and (b) classification accuracy in feature selection methodology. This is important because in clinical practice, images are acquired on a number of different models from many manufacturers, used with a range of settings. Our study seeks to present a method of identifying features that are repeatable over full‐field digital mammography (FFDM) manufacturers and incorporate the subset of these that are descriptive and nonredundant into the construction of a classification model. We use the clinical task of classifying collectively the presence of breast cancer risk factors. For brevity, we refer to this two‐stage method as robustness assessment, classification evaluation (RACE).
2. Materials and methods
2.A. Image acquisition and database
All images included in this study were retrospectively collected from full‐field digital mammograms (FFDM) acquired under standard clinical protocols. All images were acquired at the University of Chicago Medical Center. All images used in this study were collected under an institutional review board (IRB)‐approved, Health Insurance Portability and Accountability Act (HIPAA)‐compliant protocol. All subjects were classified as either having or not having a risk factor of breast cancer. This classification was based on each subject's family history of breast cancer, family history of ovarian cancer, personal history of atypical ductal hyperplasia (ADH), and personal BRCA1/BRCA2 status. Each subject underwent screening mammography on a General Electric (GE) system and a Hologic system. The GE images were acquired on a GE Senographe 2000D at 12‐bit quantization with a pixel size of 100 × 100 μm. The Hologic images were acquired on a Hologic Lorad Selenia at 12‐bit quantization with a pixel size of 70 × 70 μm. Sets of images were separated in time by about 1 yr. The mean age of women without high‐risk factors present was 54.3 yr (range = 39–86), and the mean age of women with high‐risk factors present was 49.7 yr (range = 24–88). No breast procedures were performed on subjects between the two studies, and all images were assigned BIRADS 1 (negative) or 2 (benign) when reviewed by a clinical breast radiologist. Characteristics of the study population are summarized in Table 1.
Table 1.
Demographics of the study population separated by risk of cancer. Data in parentheses are percentages. Radiologist‐reported breast imaging reporting and data system (BI‐RADS) density was not always consistent between the GE and Hologic imaging exam, so values in this table represent the density and age reported at the time of the GE exam. Also, summary of indication for high‐risk designation is presented. Some subjects may be designated as high‐risk for more than one factor. Also shown is a breakdown of database and inclusion. In this context, small breast is defined as breast area smaller than the size of a 512 × 512 pixel square as this limited our ability to compute features on images in this analysis
| Variable | Number of patients without risk factors present | Number of patients with risk factors present | ||
|---|---|---|---|---|
| Mean age (SD) | 54.3 (10.5) | 49.7 (11.6) | ||
| Age (yr) | ||||
| <40 | 1 | 20 | ||
| 40 to 49 | 31 | 29 | ||
| 50 to 59 | 28 | 37 | ||
| 60 to 69 | 15 | 10 | ||
| 70 to 79 | 7 | 4 | ||
| ≥80 | 1 | 2 | ||
| Breast density score | ||||
| A | 2 | 4 | ||
| B | 27 | 41 | ||
| C | 44 | 54 | ||
| D | 10 | 3 | ||
| Risk factor | ||||
| Family history of breast cancer | – | 107 | ||
| Family history of ovarian cancer | – | 9 | ||
| BRCA1/BRCA2 mutations | – | 3 | ||
| Personal history of ADH | – | 1 | ||
| Breast area exclusion | Patients | Images | Patients | Images |
| Total in database | 86 | 172 | 112 | 224 |
| # Small breast | 3 | 9 | 10 | 27 |
| # Included in study | 83 | 163 | 102 | 197 |
A small number of women were excluded from this study because the breast area in their images was smaller than that required for placement of the region of interest (ROI). Small breast area could result from small breast volume, large pixel size, or the extent of breast compression during image acquisition.
The distribution of time intervals between exams is described in Fig. 1. This histogram shows the interval of time between the GE and Hologic exam dates for each patient included in this study. The distribution of days between exams for the group with and without high‐risk factors present were not shown to be significantly different by the two‐sample t‐test (P = 0.29).24 Thus, this suggests that differences in time intervals between the two populations can be explained by random chance.
Figure 1.
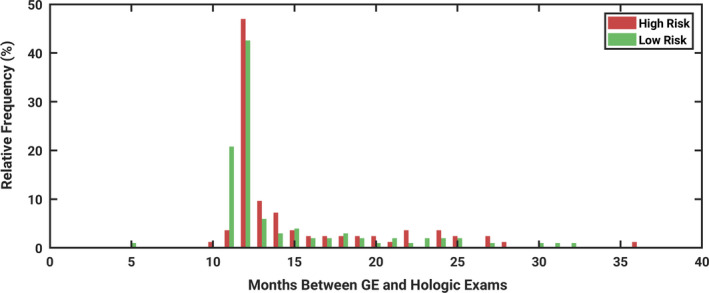
Histogram demonstrating the interval of time between the date of the GE exam and the Hologic exam, for each patient included in the study. The time between exams were not found to be significantly different between women with and without high‐risk factors present (P = 0.29). [Color figure can be viewed at wileyonlinelibrary.com]
2.B. Radiomic feature calculation
Radiomic texture features were calculated on square ROIs of size 512 × 512 pixels which were manually placed in the central breast region posterior to the nipple. Previous studies have shown that this ROI size and placement scheme performs best compared to different locations in the breast.25 Because all images had a negative or benign interpretation, ROIs were placed over normal background parenchymal tissue. Following manual ROI placement, features were automatically calculated on each ROI. The features were based on algorithmic implementations of mathematical texture descriptors which have been reported on extensively in the literature.6, 7, 15 Specifically, features were based on (a) gray‐level histogram analysis, (b) fractal dimensionality analysis including the box‐counting method and Minkowski method, (c) Fourier and power spectral analysis, (d) edge frequency analysis, and (e) GLCM. The quantity of features calculated from each group is summarized in Table 2. This set of quantitative features was evaluated because the constituent features have demonstrated utility in previous studies involving clinical classifications based on parenchyma regions in FFDM images.6, 7, 8, 25
Table 2.
Quantity of feature types included in the feature set from which features were selected for classification analysis
| Feature category | Number of features |
|---|---|
| Fourier | 148 |
| Box‐counting fractal dimension | 6 |
| Edge frequency | 4 |
| Histogram | 38 |
| Minkowski fractal dimension | 32 |
| Power law Beta | 8 |
| GLCM | 14 |
| First order | 6 |
| Total | 256 |
2.C. Robustness assessment
Hierarchical clustering was performed to identify groups of redundant features.26 Clustering was performed using the Pearson correlation coefficient as the distance metric.27 Single linkage (nearest neighbor) was used to describe the distance between objects. The number of clusters used in grouping ranged from 18 to 256 in order to evaluate the impact of varying levels of strictness in robustness considerations. The lower end of the range of the number of clusters was chosen because a total of 18 features were ultimately selected for use in classification as this is close to the optimal number of features based on our database size.28 The upper end of the range of the number of clusters was chosen because a total of 256 radiomic features were calculated in this study. Therefore, by sorting the features into 256 clusters, only a single feature persists in each cluster. This therefore would be analogous to disregarding robustness in feature selection, as nonrobust features are not removed from subsequent analyses.
A wide range of clusters was investigated in order to explore the trend in classification performance as restrictions on robustness varied, thereby permitting for an evaluation of the relevancy of robustness considerations in classification performance. However, in practical application of the proposed method, it is expected that only one number of clusters need be considered. In general, the optimal number of clusters may depend on study‐dependent factors such as the specific classification task, the redundancy between features in the full feature set explored, and the number of patients evaluated in the study.
Feature robustness across mammography vendors was evaluated using statistical metrics including (a) mean of feature ratio (MFR), (b) correlation coefficient, and (c) Kolmogorov–Smirnov test statistic.17, 29 These robustness metrics were selected to describe equivalence, correlation and sample distributions.
A composite indicator (CI) was developed to merge the three robustness metrics investigated in this study so as to include multiple aspects of robustness in evaluating features. The CI was calculated by the weighted sum of metric values normalized by z‐score. Metric z‐scores were weighted by +1 when a high value indicates robustness, and by −1 when a low value indicates robustness. Therefore, the CI for feature f is defined by Eq. (1), where zcorr,f is the z‐value of the correlation coefficient for feature f, zMFR,f is the z‐value of the MFRs for feature f, and zKS,f is the z‐value of the Kolmogorov–Smirnov test statistic for feature f.
| (1) |
Relative robustness ranking of the investigated texture features was performed by ordering features based on their CI value in descending order, where more positive values of CIf suggest strong robustness, and more negative values of CIf suggest weak robustness. The features with the highest CIf in each cluster were identified and considered in the classification evaluation stage.
2.D. Classification evaluation
The classification stage involved using robust, nonredundant features to predict a woman's risk of breast cancer based on the presence of risk factors. The workflow for the proposed model is illustrated by Fig. 2.
Figure 2.
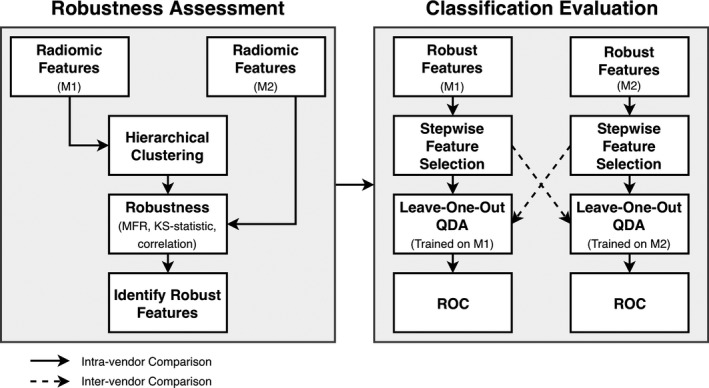
Diagrammatic illustration of steps involved in the robustness assessment, classification evaluation method. Texture features are first clustered and assessed in terms of robustness using only feature values and vendor information, remaining blinded to risk classification. The union of features identified by clustering features from M1 (machine one) and M2 (machine two) is the set considered to be robust and nonredundant. The most robust and nonredundant features are identified, and only these features are used as feature candidates in classification evaluation. Solid and dashed arrows show two different data pathways followed to evaluate the generalization of classification of the heterogeneous image datasets. The full analysis was repeated twice; once with the GE unit as M1 and the Hologic unit as M2, and then again but with the GE unit as M2 and the Hologic unit as M1.
The robust, nonredundant features identified by stage one were fed into stepwise feature selection separately for each vendor. The stepwise feature selection method employed in this study applies a stepwise regression by iteratively adding and removing features from a multilinear model.30 Features are added or removed from the model based on the statistical significance of the change in performance, with the P‐value of the f‐statistic used as the figure of merit. Feature selection was performed in a leave‐one‐out manner, and the top features were identified as those features selected the greatest number of times. The top 18 features were used in analysis, as this is near the optimal number of features for our classifier given our database size.28 Therefore, in each classification performed in this study, regardless of the number of clusters used in the robustness assessment step, exactly 18 features were ultimately selected. Note that as the number of clusters is altered, this will alter which 18 features are ultimately selected for use in the classifier, as the robustness constriction is tuned by the number of clusters.
It is standard in typical radiomics studies to perform feature selection, such as stepwise feature selection, on the full set of candidate features with no consideration for feature robustness. In our study, this standard approach is equivalent to having the number of clusters equal to the number of candidate features, thus causing all features to pass the first stage of robustness assessment. Specifically, for intravendor analyses in which the number of clusters is equal to the number of candidate features, no information from the second vendor system was used in feature selection. Therefore, in this study, intravendor classification with 256 clusters used shows the performance of a standard approach in which differences in FFDM systems is disregarded from analysis. Likewise, the intervendor classification with 256 clusters used shows the performance when the effect of vendor differences is maximized, as no robustness criteria is used in limiting candidate features considered in stepwise feature selection.
Following feature selection, selected features are used in leave‐one‐out QDA to build a model for classification. Models were built separately for GE and Hologic images. To evaluate the classification performance, the full classification evaluation analysis was performed in a leave‐one‐out manner (single fully nested loop). Receiver operating characteristic (ROC) analysis was used to calculate the area under the curve (AUC). The AUC was used as the figure of merit in this analysis.
As illustrated by Fig. 2, stepwise feature selection was performed on images from a single vendor. However, QDA was used to construct texture signatures merging the selected features on each of the two vendor image sets. Performance was evaluated, and agreement in performance was used to characterize generalizability of the model across vendors. We will refer to the vendor on whose images features were selected as machine one (M1), and the other vendor used to assess generalizability as machine two (M2). RACE was repeated with each GE and Hologic data as the primary dataset. The entire feature selection process (clustering, robustness ranking, stepwise feature selection) was performed once based on clustering features from the GE unit, and once using features from the Hologic unit. The full classification analysis (QDA, leave‐one‐out classification) was performed on each the GE unit and the Hologic unit for each the set of features identified using clustering from GE features and using clustering from Hologic features. When clusters were based on images from GE, then the GE unit was considered M1 in the analysis scheme. Likewise, when clusters were based on images from Hologic, then the Hologic unit was considered M1 in the analysis scheme.
2.E. Comparison against harmonization methods
While the approach to handling heterogeneous feature data in this paper focuses on limiting feature selection to robust features through a two‐stage analysis (RACE), other groups have approached the same issue by harmonizing (or standardizing) feature data across different imaging conditions. One such example is the ComBat harmonization method, originally developed to correct for the “batch effect” in the genomics field, and later applied to Positron Emission Tomography (PET) radiomics studies.31, 32 In a study by Orlhac et al., the ComBat harmonization method was applied to standardize radiomic features extracted from PET images of breast cancer patients acquired in two different institutions in order to identify triple negative (TN) lesions.31
As first suggested by Johnson et al. and implemented for PET radiomics by Orlhac et al., the ComBat harmonization method functions by estimating the additive scanner effect, γ, and the multiplicative scanner effect, δ, using Empirical Bayes estimates. Thus, the normalized value of features, y, are described by Eq. (2), where yij is the standardized feature for ROI j and scanner i, α is the average value for feature y, γ is the additive effect of scanner i, δ is the multiplicative scanner effect, and ε is the error term.
| (2) |
In our evaluation of the ComBat method on our data, we normalized each of the 256 examined features according to Eq. (2), and then performed stepwise feature selection and QDA for leave‐one‐out ROC analysis, mimicking the classification evaluation analysis of RACE (Fig. 2 right). For evaluation of ComBat harmonization, the robustness assessment stage (Fig. 2 left) was omitted, as feature harmonization is expected to yield all features robust across imaging conditions. To match the analysis conditions from RACE, a total of 18 features were included in the final radiomic signature construction. Furthermore, robustness metrics were computed and compared on feature values before and after ComBat harmonization used a two‐tailed t‐test.
To explore the potential interplay between the ComBat and RACE methods, we also initially applied ComBat on features for harmonization, and then used these harmonized feature values in the RACE feature selection method. To match each of the individual methods, 46 clusters were used in the RACE method, and 18 features were ultimately selected for the final radiomic signature construction.
2.F. Statistical analysis
Data series over a number of clusters were evaluated for presence of a monotonic trend using the Mann–Kendall test for monotonic trend.33 The Mann–Kendall test evaluates the presence of a significant trend between the number of clusters used in analysis, and the direction of the trend (increasing or decreasing) was computed by the Thiel–Sen estimator.34 Therefore, the sign of the Thiel–Sen estimator indicates whether the classification performance tends to increase or decrease as the number of clusters increases.
In comparing the RACE method to ComBat harmonization, the AUC for both inter‐ and intravendor performance, when features were selected each on GE and on Hologic images, were calculated. The statistical significance of the difference between AUCs from ComBat harmonization and from the RACE was calculated using ROCKIT software.35 By applying the Holm–Bonferroni correction for multiple comparisons, P ≤ 0.17 is required to demonstrate statistical significance.
3. Results
Features found to be the most robust relative to the other features examined in this study are summarized in Table 3. Feature families that tended to have a large proportion of robust features include: box‐counting fractal dimension, power law Beta, and GLCM features. Percentage density also was robust over vendors, relative to the other features examined here.
Table 3.
List of the 20 most robust features over the two vendors examined in this study. The composite indicator is a measure of robustness, where larger values indicate a more robust feature relative to the others examined in this study. The composite indicator is computed according to Eq. (1)
| Feature name | Feature family | Composite indicator (CI) |
|---|---|---|
| Sum entropy | GLCM | 5.81 |
| Percentage density | Density | 5.52 |
| Dim 5 | Box‐counting fractal dimension | 5.33 |
| Sum variance | GLCM | 5.26 |
| Beta 3 | Power law | 5.23 |
| safmp | Fourier features | 5.18 |
| Beta 1 | Power law | 5.18 |
| Variance | GLCM | 5.14 |
| Dim 4 | Box‐counting fractal dimension | 5.11 |
| Beta 7 | Power law | 5.09 |
| IMC 2 | GLCM | 5.06 |
| Maximum correlation coefficient | GLCM | 5.01 |
| Dim 1 | Box‐counting fractal dimension | 4.99 |
| Correlation | GLCM | 4.96 |
| Sarms | Fourier features | 4.91 |
| Beta 5 | Power law | 4.81 |
| rrms | Fourier features | 4.80 |
| rfmp | Fourier features | 4.72 |
| Global Minkowski dimension | Minkowski fractal dimension | 4.71 |
| Dim | Box‐counting fractal dimension | 4.66 |
Restricting candidate features to just the most robust features was shown to have a significant impact on classification performance of the intravendor evaluation as demonstrated by a monotonic increase in AUC with increasing number of clusters (Mann–Kendall P = 0.0168 and P < 0.001 for GE and Hologic, respectively). However, the Thiel–Sen estimator of the rate of increase was still very small (0.0000586 and 0.000120 for GE and Hologic, respectively).
Restricting candidate features to just the most robust features was shown to have a significant impact on intervendor classification performance. The classification performance was observed to monotonically decrease as the number of clusters increased (Mann–Kendall P < 0.001) as shown in Fig. 2.
As more features were considered as candidate features, generalizability across vendors tended to diminish. This is demonstrated by the presence of a monotonic trend in the difference between intra‐ and intervendor classification performance as the number of clusters increased as shown in Fig. 3 (Mann–Kendall P < 0.001, Thiel–Sen Estimator = −0.000321 and −0.000191 for GE and Hologic, respectively; Fig. 4.
Figure 3.
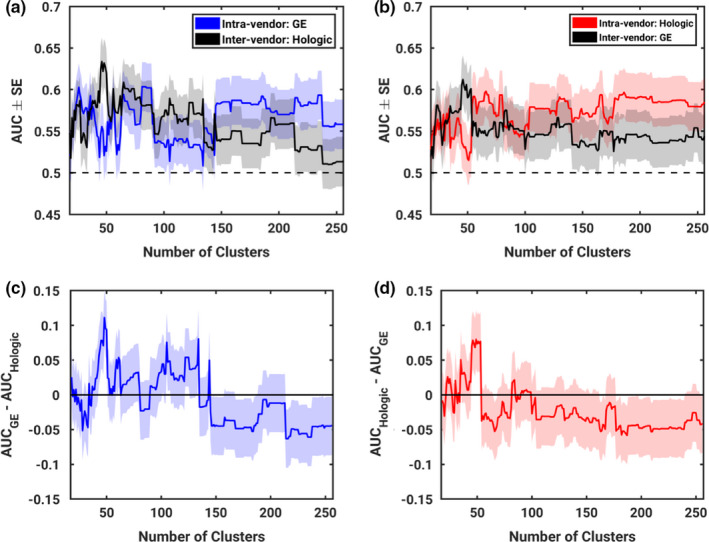
Resulting performance of classifiers trained on varying quantities of clusters and therefore varying degrees of stringency on the robustness of input features. Parts (a) and (b) show performance of intra‐ and intervendor feature selection and classifier construction as the number of clusters, and therefore stringency on robustness, is varied. Parts (c) and (d) show the difference between intra‐ and intervendor classifier performance to demonstrate generalizability. Parts (a) and (c) show results for when GE is designated M1 and Hologic is designated M2. Parts (b) and (d) show results for when Hologic is designated M1 and GE is designated M2. [Color figure can be viewed at wileyonlinelibrary.com]
Figure 4.
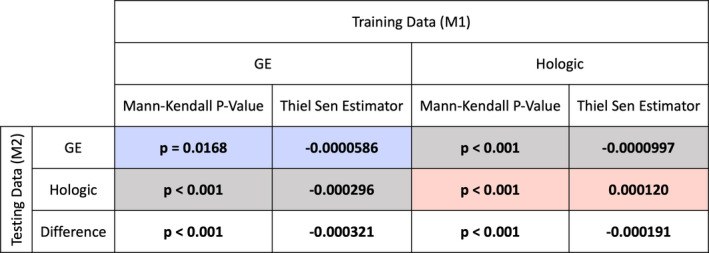
Results of the Mann–Kendall test for the presence of monotonic trends, and the Thiel–Sen Estimator of such trends for the performance as a function of the number of clusters. Statistically significant values are denoted by boldface font. Colored results (blue, red) correspond to intravendor comparisons using GE and Hologic images, respectively. Gray results correspond to intervendor comparisons. [Color figure can be viewed at wileyonlinelibrary.com]
While these trends are interesting in a research setting, a fixed number of clusters would be more useful for practical application of RACE. In this study, 46 clusters yielded peak intervendor classification performance when RACE is repeated with either GE or Hologic as M1. This is illustrated by a peak in the intervendor curves of Fig. 3 parts (a) and (b). Therefore, while the full range of numbers of clusters can help describe trends in performance, this study will also include discussion of the particular application of RACE using 46 clusters.
Many of the same features were selected both when GE was designated as M1 and when Hologic was designated as M1. As illustrated by Fig. 5, over half of the features selected on a given vendor's data were also selected when RACE was repeated on the other vendor. Considering that 256 total features were investigated, the commonalities across the two selected feature sets suggest that features selected are descriptive on images from both vendors. However, there are some instances in which features were not selected in both analyses. For example, the feature entropy was selected when using clusters from GE but not Hologic. Energy was selected when using clusters from Hologic but not GE. While these features are calculated by different formulas, each describes image homogeneity. Furthermore, these features are each highly correlated with one another. Therefore, while features selected may have varied, the physical characteristics described by the features selected remained consistent over the two vendors.
Figure 5.

Summary of features selected for the classifier when robustness assessment, classification evaluation is performed either with GE designated as M1 or Hologic designated as M1. The results presented in this figure are specifically from selection after grouping features into 46 clusters. This number of clusters was chosen as it provides the best intervendor performance for each manufacturer. Selected features were recorded from each leave‐one‐out iteration during stepwise feature selection, and the 18 features most frequently selected for each manufacturer is recorded here. [Color figure can be viewed at wileyonlinelibrary.com]
Furthermore, not all features included in Table 3 were necessarily selected for inclusion in the classifier. Reasons for this may include redundancy, or lack of discriminatory ability of the feature of interest. Namely, robustness is not a sufficient condition for inclusion in the final set of selected features. Features that are robust and not redundant with other feature candidates are passed from the robustness assessment to classification evaluation step of RACE, but in order to be included in the final set of features, the features must also have discriminatory power in the clinical task in order to be selected by the stepwise feature selection method.
For instance, it can be observed from Table 3 and Fig. 5 that although the 20 most robust features did not include any edge frequency, first‐order, or histogram features, some features from each of these categories were ultimately chosen for inclusion in the final classifier. This happens because while the RACE method gives preference to the most robust redundant features, it does not remove features such as those with moderate robustness from the set of candidates. If a feature with moderate robustness was clustered with features that had lower robustness, that moderate feature would be considered in stepwise feature selection, and thus may be ultimately included in the final model. This can be illustrated by the selection of minima, which is a histogram feature with a CI of −0.70, suggesting that it is marginally below average in terms of its robustness. Minima was clustered with features including average, maximum cumulative distribution function (CDF), minimum CDF, seventy percent CDF, and thirty percent CDF. Each of these features had a CI between −1.05 and −1.60, suggesting even lower robustness than minima. Thus, minima would be the most robust feature of its cluster and would be considered in the next stage of classification evaluation.
Conversely, highly robust features are not guaranteed to be selected in stepwise feature selection for inclusion in a final feature set. For example, the box‐counting fractal dimension feature Dim1 was highly robust with a CI of 4.99. As the most robust feature in its cluster, it was considered in feature selection. However, during stepwise feature selection, Dim1 was not selected for inclusion in the final model.
3.A. Comparison against harmonization methods
Classification performance of the RACE method proposed in this paper was compared to the ComBat harmonization method used in previous studies.31, 32 The results, summarized in Fig. 6, suggest that while the two methods perform similarly on intravendor comparisons, RACE had significantly higher AUC than ComBat harmonization method on intervendor comparisons when training on GE images and testing on Hologic images.
Figure 6.
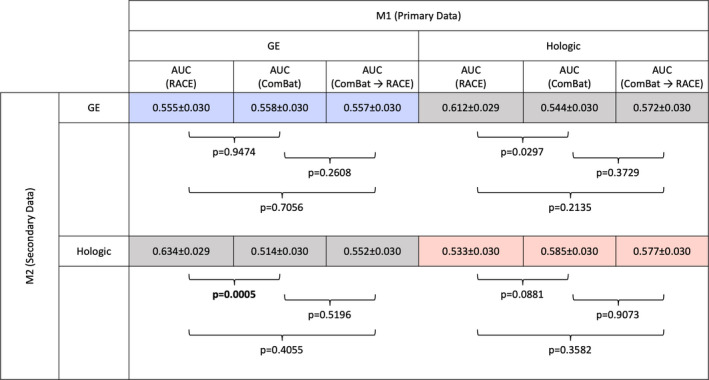
Performance in the task of classifying the presence of risk factors of breast cancer of three analysis methods: (a) robustness assessment, classification evaluation, (b) ComBat, and (c) ComBat followed by robustness assessment, classification evaluation. In each method, 18 features were included in the ultimate radiomic signature construction, and leave‐one‐out cross‐validation was performed. While intravendor comparisons were not significantly different between the three methods, intervendor comparisons were significantly different, with the two‐stage method performing better as judged by the area under the curve (AUC). Recall that M1 refers to the vendor on whose images features were selected as machine one, and M2 refers to the vendor used to assess generalizability. By the Holm–Bonferroni correction for multiple comparisons, P < 0.017 is required to demonstrate statistical significance. [Color figure can be viewed at wileyonlinelibrary.com]
When ComBat harmonization is applied and followed by RACE, the results failed to demonstrate significant differences from either ComBat alone or RACE alone. This trend held for each of the four combinations of training and testing data investigated in this study.
In comparing the robustness metrics between raw feature values and harmonized feature values, it was observed that the MFR, which characterizes the agreement in feature magnitude, was significantly changed (P < 0.001), while the correlation coefficient of the feature values between vendors was not significantly different before and after harmonization (P = 1.000) as shown in Fig. 7. This result suggests that ComBat harmonization acts to improve agreement in feature magnitude across vendors but does not impact the correlation of features across the two systems.
Figure 7.
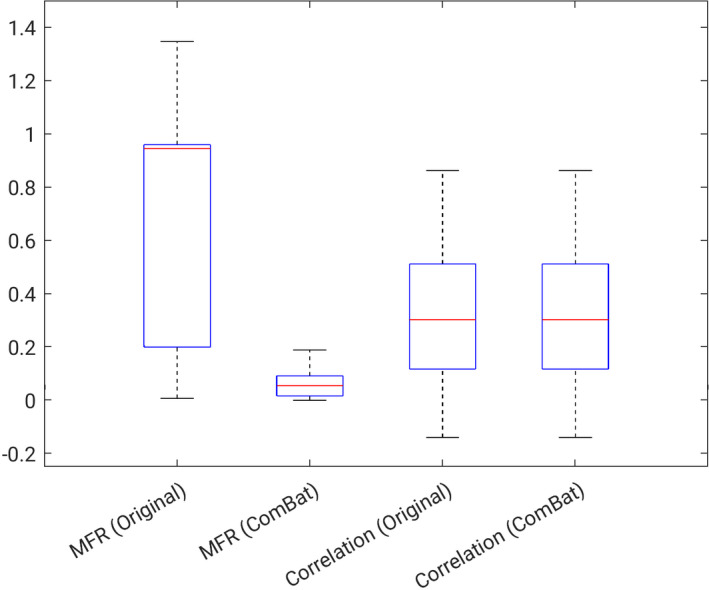
Summary of trends in robustness metrics computed on features before and after ComBat harmonization. mean of feature ratio (MFR) near zero indicates high robustness, and correlation near 1 indicates high robustness. [Color figure can be viewed at wileyonlinelibrary.com]
4. Discussion
This study found that many features describing spatial characteristics tended to be robust. Box‐counting fractal dimension features were observed to be highly robust over vendors. Fractal dimension characterizes the roughness and self‐similarity of images.36 In the case of breast parenchyma, this suggests that fractal dimension may describe the complexity of dense tissue pattern as it appears in the mammogram. Power law features were also observed to be robust over the two vendors in this study. Previous studies have suggested that power law features are related to the background parenchymal pattern of breast structure.7, 37, 38 The power law exponent, β, indirectly characterizes the frequency content of the texture pattern and can be mathematically transformed to fractal dimension.39, 40 Therefore, it would be expected that power law features would demonstrate similar trends in robustness to the fractal dimension. Derivative statistics from the GLCM matrix also demonstrated high robustness over mammographic units. GLCM features describe spatial relationships between pixels. By calculating how frequently pairs of pixels with specific values in specific spatial relationships occur throughout the ROI, descriptors such as energy, entropy, and correlation were computed.15 Studies that have investigated the robustness of GLCM features over varying region segmentations and bin sizes for discretization also found that GLCM features demonstrate high robustness.41
It is likely that the technical characteristics of the Hologic and GE unit used to acquire images influence the feature values extracted, and thus the robustness of such features. A more focused examination of specific technical parameters that differ between the two units used in this study can be found in a study by Mendel et al.24 Briefly, Mendel et al. reported that the two units differ in pixel size, anode material, detector size, detector material, and conversion method.24
The monotonic trends observed in this study suggest that considerations of feature robustness in feature selection tend to improve generalizability of models across vendors. While a trend was observed, the nature of this trend was not explored beyond the monotonic direction (increasing or decreasing). In standard practice, robustness and repeatability of radiomic features across imaging machines is typically not evaluated or included in the feature selection process. The results of this study suggest that conventional methodology may not be reproducible on data acquired on a different machine. This may also pose problems in studies which perform classification using data from multiple acquisition systems, if consideration is not given to feature robustness.
While this study observed superior intervendor results of RACE over ComBat harmonization, this could be due in part to the intended use of the two algorithms. While this study developed the two‐stage method with the goal of developing a texture signature which is robust in intervendor comparisons, the study by Orlhac et al. only performed classifications using single features, as opposed to feature signatures.31 Specifically, the calculation of a radiomic signature may accommodate for normalization differences over the two vendors, thus reducing the utility of a harmonization step prior to signature calculation. Therefore, this study suggests that RACE is useful in producing intervendor radiomic signatures, while ComBat may be useful when performing classifications using a single radiomic feature.
When applying ComBat harmonization followed by RACE, the resulting AUC values failed to show a statistical difference from those obtained when using RACE alone. The failure to observe a significant difference between ComBat harmonization alone and ComBat harmonization followed by RACE suggests that after performing feature harmonization, further application of robustness assessment does not significantly impact the classification performance. Likewise, the failure to observe a significant difference in this study between RACE alone and ComBat harmonization followed by RACE suggests that adding the standardization step of feature harmonization does not significantly improve the performance of robustness assessment for classification. A possible explanation for this could be that by following ComBat harmonization by RACE, feature reduction is being performed on the candidate feature set prior to stepwise feature selection. Thus, it is possible that the stepwise feature selection algorithm employed in this study is better suited to applications on already reduced feature sets. Another explanation could be the limited size of our dataset. Future work will investigate benefit of stepwise feature selection when applied on feature sets containing various numbers of features.
Importantly, the pixel size of images collected on the two systems was not consistent. As demonstrated separately by Mendel et al. and Mackin et al., varying the pixel size impacts the radiomic features calculated from images.17, 39 In efforts to harmonize pixel size for feature calculations, Mendel et al. found reduced feature robustness following pixel interpolation on FFDM to produce consistent pixel size, compared to no preprocessing.24 Likewise, Mackin et al. found increased feature variability following voxel resampling on computed tomography images to produce consistent pixel size, compared to no preprocessing.39 Studies have used methods such as image resizing followed by Butterworth filtering, low‐pass filters, band‐pass filters, or Gaussian filters in order to harmonize images prior to feature calculation.19, 42, 43, 44, 45 In this study, we chose not to apply these steps to force consistent pixel size as the presented method seeks to address image heterogeneities through feature selection as opposed to image processing. However, image harmonization steps may further improve feature robustness.
While this study proposes a two‐stage feature selection process for building reproducible classifiers (RACE), this study is not without limitations. Firstly, reproducibility datasets consisting of patients imaged on separate machines are relatively uncommon in medical imaging. Therefore, this initial investigation applied the proposed methods to only one task on only one dataset. In general, the proposed methods could be applied to a number of applications given that repeatability data are available.
Additionally, this study applied the methods to a challenging task, in which high performance is not necessarily expected. Several studies have used radiomic texture analysis to address risk of breast cancer based on screening images; however, even studies with well‐separated patient populations (i.e., unilateral cancer vs low risk cancer) had only moderate performance8. Previous studies have also measured reductions in classification performance when women with different types of risk factors are analyzed together (e.g., classifying BRCA2 vs low‐risk controls, compared to classifying BRCA1/2 vs low‐risk controls). In this study, the group with high‐risk factors present had a range of factors including BRCA1/2 gene mutations, family history of breast or ovarian cancer, and personal history of ADH, meaning that this group was likely heterogeneous in its true overall lifetime risk of breast cancer. Therefore, there existed greater variability within groups in this study which may have contributed to low performance values. This study did not test the classification of specific risk factors from low‐risk controls because of the limited database size. Furthermore, differences other than the presence of high‐risk factors existed between the two groups. One such measured difference was the difference in mean age (54.3 and 49.7 yr for risk factors absent and present, respectively). Parenchymal texture has been observed to change over a woman's lifetime, and therefore this confounding factor could impact the results of this study.
Another consideration for further optimization of the methods proposed in this study includes closer examination of the optimization of ROI size and location. In this study, regions with size 512 × 512 pixels were placed in the central region directly behind the nipple as this location was shown to perform well in previous risk assessment studies.25 However, because of differences in database and analysis, these parameters are not necessarily optimal in the present study. Thus, while it has not been proven that size and location used in this study are optimal, their utility in previous studies makes them logical choices. This study used radiomic features of the breast parenchyma to predict risk, and dense component, if present in the breast, is typically located in the central region immediately behind the nipple. Thus, the location used is a practical choice as it was where the tissue of interest is typically located. Furthermore, the study by Li et al. found that classification performance did not significantly decline as the ROI size changed. Instead, the study reported that there was no statistically significant difference observed as the size of the ROI decreased.25 Therefore, significant differences in the outcome of this study would not be expected if a different ROI size were used.
As this was a retrospective study, the imaging units on which images were collected are no longer considered state‐of‐the‐art. The GE Senographe 2000D unit was first released in 2000. Compared to newer GE units, the GE Senographe 2000D has a smaller field of view and lower detective quantum efficiency (DQE) and normalized noise power spectrum (NNPS) due to improvements in electronic noise in latter models.46 The Hologic Selenia is also different from later models, as the unit used in this study had a molybdenum–molybdenum (Mo‐Mo) target‐filter. This target‐filter material has been shown to result in higher average glandular dose compared to molybdenum–rhodium (Mo‐Rh) or rhodium–rhodium (Rh‐Rh) target‐filter combinations.47 This is because Mo‐Mo target‐filter combinations results in a softer x‐ray beam. Mo‐Mo has also been shown to result in lower contrast in dense breasts compared to the other target‐filter material combinations, making it less optimal.47 Newer Hologic systems use a tungsten–silver (W‐Ag) target‐filter combination, which results in a harder x‐ray beam.48 These physical differences in image acquisition between the models used in this study and the models used clinically today may cause differences in image feature values and appearance, yet the methods proposed in this would likely remain relevant for varying image parameters or system vendor.
Additionally, the average mean glandular dose (MGD) of the two vendors’ units is different. As reported by Hendrick et al., the GE system had a MGD of 1.69 mGy per view and 4.02 mGy per exam, and the Hologic system had a MGD of 2.50 mGy per view and 5.03 mGy per exam.
Future steps include evaluation of this method on different data for different clinical tasks, such as lesion characterization or disease detection. Additionally, a wider range of radiomic features will be evaluated with this method to explore whether the proposed method may be applied to wider analytical questions. Investigation into changes in radiomic features over time will also be investigated in future studies. Studies in PET have suggested that factors such as aging and menopausal status may impact radiomic features, and a temporal set of mammograms will facilitate investigations such as this.49
5. Conclusions
This study proposed a two‐stage method (RACE) for robust radiomic signature construction. RACE was demonstrated in the task of breast cancer risk assessment. The results suggest that feature generalizability monotonically decreases as reproducibility decreases. This trend shows that considerations of feature robustness could improve classifier generalizability in multifarious datasets collected on multiple mammography units. Furthermore, the same trend was observed when either vendor was used for feature clustering thus supporting that this finding can be generalized. An investigated harmonization method (ComBat) was not shown to have strong classification performance when used on its own, but when ComBat harmonization was followed by RACE, classification results appeared similar to RACE alone. Thus, harmonization steps in conjunction with robustness assessment warrant future investigation in feature selection and classifier construction methods.
Acknowledgments
Supported, in part, by the NIBIB of the NIH under grant number T32 EB002103, the NCI of the NIH under grant number NIH QIN U01 195564. M.L.G. is a stockholder in R2 Technology/Hologic and a cofounder and shareholder in Quantitative Insights. M.L.G. receives royalties from Hologic, GE Medical Systems, MEDIAN Technologies, Riverain Medical, Mitsubishi, and Toshiba. H.L. and L.L. receive royalties from Hologic. It is the University of Chicago Conflict of Interest Policy that investigators disclose publicly actual or potential significant financial interest that would reasonably appear to be directly and significantly affected by the research activities.
References
- 1. National Center for Health Statistics (US) . Health, United States, 2016: With Chartbook on Long‐term Trends in Health [Internet]. Hyattsville (MD): National Center for Health Statistics (US); 2017 [cited 2018 May 29]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK453378/ [PubMed]
- 2. Tabár L, Vitak B, Chen TH‐H, et al. Swedish two‐county trial: impact of mammographic screening on breast cancer mortality during 3 decades. Radiology. 2011;260:658–663. [DOI] [PubMed] [Google Scholar]
- 3. Saftlas AF, Hoover RN, Brinton LA, et al. Mammographic densities and risk of breast cancer. Cancer. 1991;67:2833–2838. [DOI] [PubMed] [Google Scholar]
- 4. Boyd NF, Byng JW, Jong RA, et al. Quantitative classification of mammographic densities and breast cancer risk: results from the Canadian National Breast Screening Study. J Natl Cancer Inst. 1995;87:670–675. [DOI] [PubMed] [Google Scholar]
- 5. McCormack VA, dos Santos Silva I. Breast density and parenchymal patterns as markers of breast cancer risk: a meta‐analysis. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2006;15:1159–1169. [DOI] [PubMed] [Google Scholar]
- 6. Li H, Giger ML, Olopade OI, Lan L. Fractal analysis of mammographic parenchymal patterns in breast cancer risk assessment. Acad Radiol. 2007;14:513–521. [DOI] [PubMed] [Google Scholar]
- 7. Li H, Giger ML, Olopade OI, Chinander MR. Power spectral analysis of mammographic parenchymal patterns for breast cancer risk assessment. J Digit Imaging. 2008;21:145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li H, Giger ML, Lan L, Janardanan J, Sennett CA. Comparative analysis of image‐based phenotypes of mammographic density and parenchymal patterns in distinguishing between BRCA1/2 cases, unilateral cancer cases, and controls. J Med Imaging Bellingham Wash. 2014;1:031009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giger ML, Karssemeijer N, Schnabel JA. Breast image analysis for risk assessment, detection, diagnosis, and treatment of cancer. Annu Rev Biomed Eng. 2013;15:327–357. [DOI] [PubMed] [Google Scholar]
- 10. Oza AM, Boyd NF. Mammographic parenchymal patterns: a marker of breast cancer risk. Epidemiol Rev. 1993;15:196–208. [DOI] [PubMed] [Google Scholar]
- 11. Boyd NF, Guo H, Martin LJ, et al. Mammographic density and the risk and detection of breast cancer. N Engl J Med. 2007;356:227–236. [DOI] [PubMed] [Google Scholar]
- 12. Wang J, Kato F, Oyama‐Manabe N, et al. Identifying triple‐negative breast cancer using background parenchymal enhancement heterogeneity on dynamic contrast‐enhanced MRI: a pilot radiomics study. PLoS ONE. 2015;10:e0143308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vachon CM, Pankratz VS, Scott CG, et al. The contributions of breast density and common genetic variation to breast cancer risk. JNCI J Natl Cancer Inst. 2015;107:1–4. Available from: https://academic.oup.com/jnci/article/107/5/dju397/890191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Keller BM, Conant EF, Oh H, Kontos D. Breast cancer risk prediction via area and volumetric estimates of breast density. In: Breast Imaging. Berlin, Heidelberg: Springer; 2012:236–243.Available from: https://link.springer.com/chapter/10.1007/978-3-642-31271-7_31 [Google Scholar]
- 15. Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern. 1973;SMC‐3:610–621. [Google Scholar]
- 16. Saslow D, Boetes C, Burke W, et al. American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography. CA Cancer J Clin. 2007;57:75–89. [DOI] [PubMed] [Google Scholar]
- 17. Mendel KR, Li H, Lan L, et al. Quantitative texture analysis: robustness of radiomics across two digital mammography manufacturers’ systems. J Med Imaging Bellingham Wash. 2018;5:011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mackin D, Fave X, Zhang L, et al. Measuring computed tomography scanner variability of radiomics features. Invest Radiol. 2015;50:757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mackin D, Fave X, Zhang L, et al. Harmonizing the pixel size in retrospective computed tomography radiomics studies. PLoS ONE. 2017;12:e0178524. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5608195/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shafiq‐ul‐Hassan M, Zhang GG, Latifi K, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys. 2017;44:1050–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fave X, Mackin D, Yang J, et al. Can radiomics features be reproducibly measured from CBCT images for patients with non‐small cell lung cancer? Med Phys. 2015;42:6784–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hunter LA, Krafft S, Stingo F, et al. High quality machine‐robust image features: identification in nonsmall cell lung cancer computed tomography images. Med Phys. 2013;40:121916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao B, Tan Y, Tsai W‐Y, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep. 2016;6:23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ryan TH. Significance tests for multiple comparison of proportions, variances, and other statistics. Psychol Bull. 1960;57:318–328. [DOI] [PubMed] [Google Scholar]
- 25. Li H, Giger ML, Huo Z, et al. Computerized analysis of mammographic parenchymal patterns for assessing breast cancer risk: effect of ROI size and location. Med Phys. 2004;31:549–555. [DOI] [PubMed] [Google Scholar]
- 26. Ward JH Jr. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58:236–244. [Google Scholar]
- 27. Soper HE, Young AW, Cave BM, Lee A, Pearson K. On the distribution of the correlation coefficient in small samples. Appendix II to the papers of “student” and R. A. Fisher. A cooperative study. Biometrika. 1917;11:328–413. [Google Scholar]
- 28. Hua J, Xiong Z, Lowey J, Suh E, Dougherty ER. Optimal number of features as a function of sample size for various classification rules. Bioinforma Oxf Engl. 2005;21:1509–1515. [DOI] [PubMed] [Google Scholar]
- 29. Darling DA. The Kolmogorov‐Smirnov, Cramer‐von Mises tests. Ann Math. Stat. 1957;28:823–838. [Google Scholar]
- 30. Draper NR. Applied regression analysis, 3rd edn. New York: Wiley; 1998. [Google Scholar]
- 31. Orlhac F, Boughdad S, Philippe C, et al. A post‐reconstruction harmonization method for multicenter radiomic studies in PET. J Nucl Med. 2018;59:1321–1328. [DOI] [PubMed] [Google Scholar]
- 32. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. [DOI] [PubMed] [Google Scholar]
- 33. Kendall MG. A new measure of rank correlation. Biometrika. 1938;30:81–93. [Google Scholar]
- 34. Sen PK. Estimates of the regression coefficient based on Kendall's tau. J Am Stat Assoc. 1968;63:1379–1389. [Google Scholar]
- 35. Metz CE, Herman BA, Roe CA. Statistical comparison of two ROC‐curve estimates obtained from partially‐paired datasets. Med Decis Making. 1998;18:110–121. [DOI] [PubMed] [Google Scholar]
- 36. Sarkar N, Chaudhuri BB. An efficient differential box‐counting approach to compute fractal dimension of image. IEEE Trans Syst Man Cybern. 1994;24:115–120. [Google Scholar]
- 37. Burgess AE, Judy PF. Signal detection in power‐law noise: effect of spectrum exponents. J Opt Soc Am A Opt Image Sci Vis. 2007;24:B52–B60. [DOI] [PubMed] [Google Scholar]
- 38. Human observer detection experiments with mammograms and power‐law noise ‐ Burgess ‐ 2001 ‐ Medical Physics ‐ Wiley Online Library. Available from: https://aapm.onlinelibrary.wiley.com/doi/abs/10.1118/1.1355308 [DOI] [PubMed]
- 39. Burgess AE. Mammographic structure: data preparation and spatial statistics analysis. In: Medical Imaging 1999: Image Processing. International Society for Optics and Photonics; 1999:642–654. Available from: https://www.spiedigitallibrary.org/conference-proceedings-of-spie/3661/0000/Mammographic-structure-data-preparation-and-spatial-statistics-analysis/10.1117/12.348620.short
- 40. Soille P, Rivest J‐F. On the validity of fractal dimension measurements in image analysis. J Vis Commun Image Repr. 1996;7:217–229. [Google Scholar]
- 41. Lu L, Lv W, Jiang J, et al. Robustness of radiomic features in [11C]Choline and [18F]FDG PET/CT imaging of nasopharyngeal carcinoma: impact of segmentation and discretization. Mol Imaging Biol. 2016;18:935–945. [DOI] [PubMed] [Google Scholar]
- 42. Miles KA, Ganeshan B, Hayball MP. CT texture analysis using the filtration‐histogram method: what do the measurements mean? Cancer Imaging. 2013;13:400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ganeshan B, Burnand K, Young R, Chatwin C, Miles K. Dynamic contrast‐enhanced texture analysis of the liver: initial assessment in colorectal cancer. Invest Radiol. 2011;46:160–168. [DOI] [PubMed] [Google Scholar]
- 44. Coroller TP, Grossmann P, Hou Y, et al. CT‐based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol J Eur Soc Ther Radiol Oncol. 2015;114:345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Assessment of Response to Tyrosine Kinase Inhibitors in Metastatic Renal Cell Cancer: CT Texture as a Predictive Biomarker. Radiology. Available from: https://pubs.rsna.org/doi/10.1148/radiol.11110264 [DOI] [PubMed]
- 46. Ghetti C, Borrini A, Ortenzia O, Rossi R, Ordóñez PL. Physical characteristics of GE senographe essential and DS digital mammography detectors. Med Phys. 2008;35:456–463. [DOI] [PubMed] [Google Scholar]
- 47. Gingold EL, Wu X, Barnes GT. Contrast and dose with Mo‐Mo, Mo‐Rh, and Rh‐Rh target‐filter combinations in mammography. Radiology. 1995;195:639–644. [DOI] [PubMed] [Google Scholar]
- 48. (6) Comparison of Acquisition Parameters and Breast Dose in Digital Mammography and Screen‐Film Mammography in the American College of Radiology Imaging Network Digital Mammographic Imaging Screening Trial. Request PDF. ResearchGate. Available from: https://www.researchgate.net/publication/41111702_Comparison_of_Acquisition_Parameters_and_Breast_Dose_in_Digital_Mammography_and_Screen-Film_Mammography_in_the_American_College_of_Radiology_Imaging_Network_Digital_Mammographic_Imaging_Screening_Tria [DOI] [PMC free article] [PubMed]
- 49. Boughdad S, Nioche C, Orlhac F, Jehl L, Champion L, Buvat I. Influence of age on radiomic features in 18F‐FDG PET in normal breast tissue and in breast cancer tumors. Oncotarget. 2018;9:30855–30868. [DOI] [PMC free article] [PubMed] [Google Scholar]


