Abstract
The self-assembly of virus capsids is a potential target for antivirals due to its importance in the virus lifecycle. Here, we investigate the effect of phenylpropenamide derivatives B-21 and AT-130 on the assembly of hepatitis B virus (HBV) core protein. Phenylpropenamides are widely believed to yield assembly of spherical particles resembling native, empty HBV capsids. Because the details of assembly can be overlooked with ensemble measurements, we performed resistive-pulse measurements on nanofluidic devices with four pores in series to characterize the size distributions of the products in real time. With its single particle sensitivity and compatibility with typical assembly buffers, resistive-pulse sensing is well suited for analyzing virus assembly in vitro. We observed that assembly with B-21 and AT-130 produced a large fraction of partially complete virus particles that may be on-path, off-path, or trapped. For both B-21 and AT-130, capsid assembly was more sensitive to disruption under conditions where the inter-protein association energy was low at lower salt concentrations. Dilution of the reaction solutions led to the rearrangement of the incomplete particles and demonstrated that these large intermediates may be on-path, but are labile, and exist in a frustrated dynamic equilibrium. During capsid assembly, phenylpropenamide molecules modestly increase the association energy of dimers, prevent intermediates from dissociating, and lead to kinetic trapping where the formation of too many capsids has been initiated leading to both empty and incomplete particles.
Keywords: resistive-pulse sensing, hepatitis B virus, phenylpropenamides, assembly intermediates, self-assembly
Table of Contents Figure
In vivo, regulation of capsid assembly is necessary to produce infectious particles; in biotechnology, where capsids are used as carriers, regulation allows selective packaging of cargo.1 Understanding the details of the interactions between small molecules and virus capsids is thus of high importance for both pharmaceutical2–3 and engineering applications.4 For hepatitis B virus (HBV) in particular, current therapies for patients with chronic infections are non-curative. End-stage liver disease and hepatocellular carcinoma caused by HBV are responsible for 0.5 – 1 million deaths annually.5 Small molecules that disrupt the virus assembly pathway have emerged as an orthogonal approach to existing strategies.6,7 Besides the pharmaceutical applications, the study of these assembly effectors enhances the basic knowledge of the self-assembly reaction and its mechanisms,8–12 which in turn impacts the fields of virology and biomaterial science.13–14
During HBV assembly, core protein dimers react to form the icosahedral virus capsid – the protective shell of the genome. HBV capsids have icosahedral symmetry and two different sizes.15 T = 4 capsids consist of 120 dimers and have a diameter of 35.7 nm, and T = 3 capsids are composed of 90 dimers and have a diameter of 31.3 nm. The self-assembly of an empty form of the HBV capsids can be initiated in vitro without the presence of the viral genome by mixing dimers of the recombinant truncated core protein (Cp149) with a suitable electrolyte, e.g., NaCl, at an appropriate concentration, e.g., >100 mM.16 Cp149 is a truncated version of the 183-residue dimeric core protein that is missing the C-terminal nucleic acid-binding domain. During assembly, because dimer-dimer interactions are tetravalent, the weak pairwise association energy of −3.7 kcal/mol between dimers is sufficient to drive assembly. These weak interactions facilitate a process of thermodynamic editing where dimers can associate and dissociate dynamically to correct mistakes on the path to a complete icosahedron. One of the primary observables in an assembly reaction is the pseudo-critical dimer concentration, above which capsids accumulate.16 Unlike a true critical concentration, virus assembly is observed below the pseudo-critical concentration, but proceeds more slowly and has a lower yield.17
A key step for understanding HBV assembly is the characterization of assembly intermediates (incomplete capsids) that are predicted to exist only transiently and at low concentrations. A few techniques, which include resistive-pulse sensing17–19 and mass spectrometry,20–22 can resolve these assembly intermediates, which were expected to be formed in non-detectable amounts.9 Molecules that cause an abnormal formation of intermediates, either through kinetic trapping or due to improper geometric arrangements, will result in particles that are incompatible with the viral lifecycle. Thus, the ability to intentionally form intermediates generates antiviral interest and characterization of these intermediates is an area of intense development.23
Molecules that target virus assembly are collectively known as core protein allosteric modulators (CpAMs). Although all CpAMs increase the rate of assembly, the morphology of assembly products formed with CpAMs varies.24 For example, heteroaryldihydropyrimidines (HAPs) cause the formation of large and pleiomorphic particles.2,25–27 The size and shape of these products necessitate a nonuniform geometry in the subunit organization.28 For the rest of CpAMs, the assembly products appear spherical, consistent with particles which are along a path towards icosahedral geometry. Phenylpropenamides,25,29 sulfamoylbenzamides,30 and several other classes of compounds24 have been reported to produce such spherical particles.
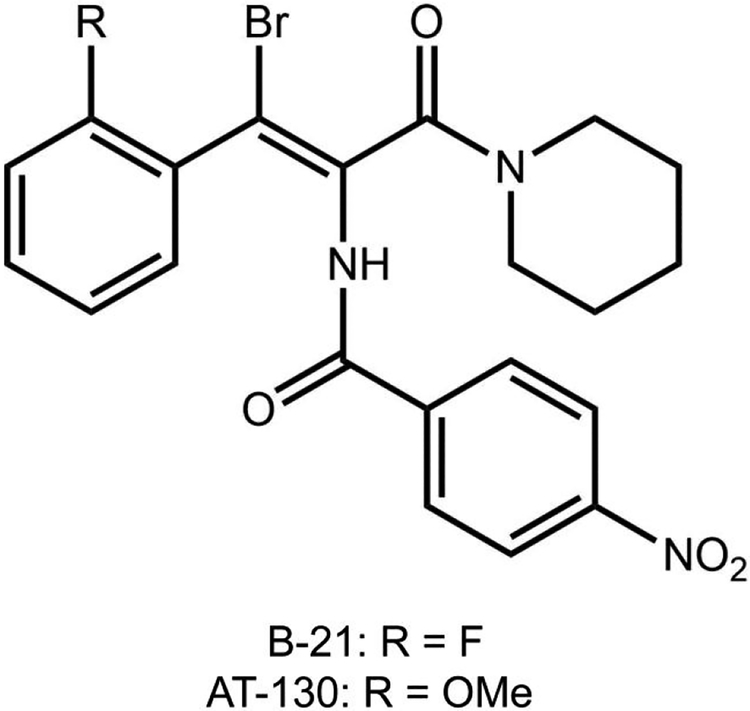
In this work, we focus on phenylpropenamides, a class of compounds effective against HBV mutants resistant to established nucleoside analog therapies.31–34 Phenylpropenamide derivatives, B-21 and AT-130 (see Figure 1 for their molecular structures) do not alter the capsid morphology when they are present in the assembly reaction.35,36 Initial studies suggested that the antiviral activity of AT-130 is a consequence of the formation of non-infectious capsids lacking the packaged RNA genome.37 In vitro studies with AT-130 and B-21 have shown that phenylpropenamides do not disrupt the geometry of inter-dimer interactions, but rather accelerate the virus assembly.35 The accelerated initiation of the assembly, perhaps premature and without the participation of viral pre-genomic RNA, was proposed to be the source of antiviral activity.35 Crystallographic studies revealed that AT-130 binds in a hydrophobic pocket in the interface between dimers in the interior of preassembled HBV capsids, which caused significant changes to the tertiary structure of the dimers.36
Figure 1. Molecular structures of B-21 and AT-130 phenylpropenamide derivatives.
We performed resistive-pulse sensing on multi-pore nanofluidic devices to observe the action mechanism of B-21 and AT-130 phenylpropenamide derivatives. Resistive-pulse sensing is a single-particle analysis technique that measures changes in current caused by the translocation of individual particles through a pore.38–42 Each particle displaces a volume of electrolyte in the pore, which increases the device resistance. For the porous HBV particles, the volume of electrolyte displaced equals the protein volume, and thus, pulse amplitude measurements can be correlated with the mass of HBV particles.43 Resistive-pulse measurements allow label-free, real-time analysis of virus assembly under biologically relevant conditions.17 Multiple pores connected in series enable signal averaging and resolution enhancement.18 Compared to size exclusion chromatography (SEC)16,35 and light scattering,16,35,44 multi-pore nanofluidic devices offer superior resolution and single-particle sensitivity. In contrast to transmission electron microscopy (TEM),45 which has limited throughput, resistive-pulse sensing can measure thousands of particles individually and in real time. Mass spectrometry (MS)20–21 has single particle sensitivity, but electrospray ionization requires volatile buffers, which are not typical for assembly.
In the experiments detailed here, HBV assembly with the phenylpropenamide B-21 led to an accumulation of large assembly intermediates that may be on-path, off-path, or trapped and are consistent with a mechanism of over-initiated assembly and inefficient thermodynamic editing. The subsequent observation that kinetically trapped intermediates readily disassembled upon dilution is a confirmation of this interpretation. The second phenylpropenamide, AT-130, yielded similar effects. Thus, we establish that the mechanism described appears to be a universal property of the phenylpropenamide class of molecules.
Results and Discussion
Resistive-pulse Measurements.
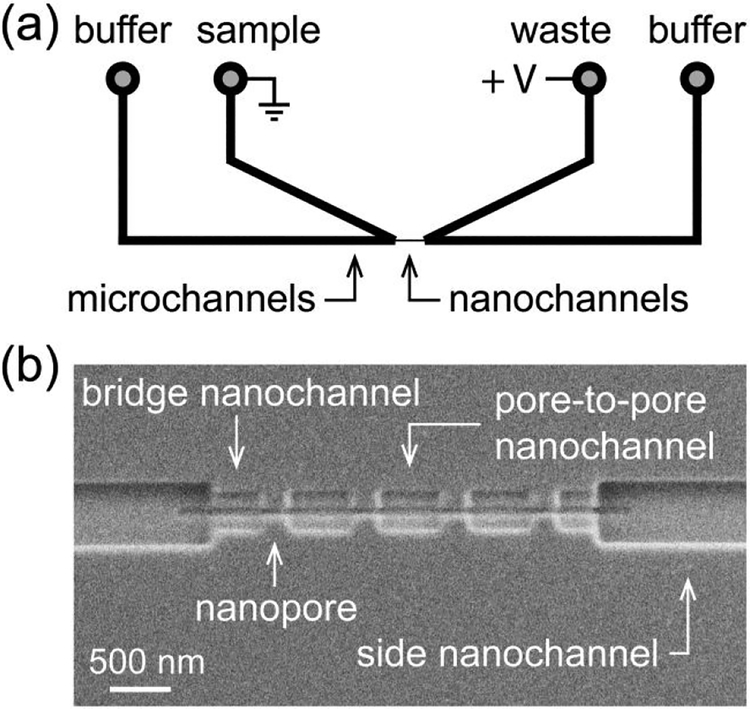
A schematic of the nanofluidic platform is illustrated in Figure 2a. A series of nanochannels and nanopores connects two V-shaped microchannels; four reservoirs on the sides facilitate filling of the microchannels with solutions. The nanochannels and nanopores (Figure 2b) were milled directly on a glass surface with a focused ion beam (FIB) instrument. The in-plane design enabled the fabrication of devices with multiple pores connected in series. Devices with four pores in series offer sufficiently high resolution to analyze assembly and have a higher signal-to-noise ratio compared to devices with a larger number of pores.18
Figure 2. Schematic of nanofluidic device and scanning electron microscope (SEM) image.
(a) Two V-shaped microchannels are connected through a series of nanopores and nanochannels. (b) SEM image of the nanochannels and nanopores milled into the glass surface. The nanopores were 60 ± 5 nm deep, 60 ± 5 nm wide, and 290 ± 6 nm long for resistive-pulse measurements of the T = 3 capsids (31.3 nm in diameter), T = 4 capsids (35.7 nm in diameter), and their intermediates.
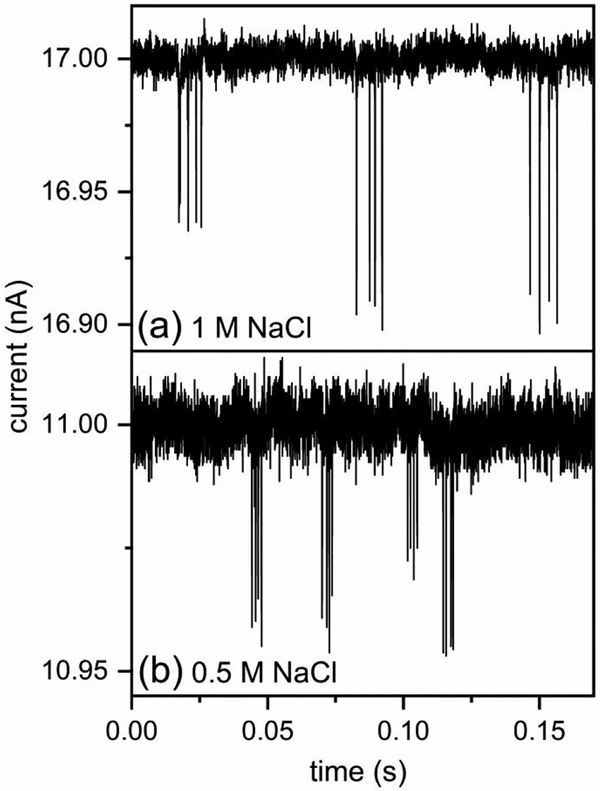
For resistive-pulse measurements, a constant voltage was applied across the side nanochannels to drive virus particles electrokinetically through the series of nanopores. The amplitude of the pulses (Δi) is proportional to the protein volume of the capsid.43 In addition, the pulse frequency is proportional to the concentration of virus particles.17 With T = 3 and T = 4 HBV capsid standards, we determined the relative standard deviation of T = 4 capsid distribution in 1.0 and 0.5 M NaCl. Current traces from measurements of 1:1 mixtures of T = 3 and T = 4 capsid standards (1 nM each) in 1 and 0.5 M NaCl are shown in Figure 3a–b, respectively. Despite the decreased resolution at the lower ionic strength (e.g., 0.5 M NaCl), T = 3 and T = 4 capsid distributions were still baseline resolved.
Figure 3. Current traces from resistive-pulse measurements on a device with 4 pores in series.
(a) Three 4-pulse sequences measured in 1 M NaCl with 900 mV applied across the nanopores and (b) four 4-pulse sequences measured in 0.5 M NaCl with 1000 mV applied across the nanopores, which were coated with a short-chain PEG-silane. Pulse sequences with smaller and larger amplitudes correspond to the translocation of T = 3 and T = 4 capsids across the 4 nanopores in series, respectively. The limits of detection are ~30 and ~70 dimers for the measurements at 1 and 0.5 M NaCl, respectively.
Assembly with Phenylpropenamides in 1 M NaCl.
HBV assembly was initiated off-chip by mixing Cp149 dimer into solutions of B-21 and NaCl. The reaction solution was then loaded onto a nanofluidic device, and after an elapsed time of ~90 s the assembly products were monitored for 20 min. In these assembly experiments, the final dimer concentration was 0.4 μM, slightly below the pseudo-critical concentration of 0.5 μM dimer in 1 M NaCl.17 Because the elongation rates of HBV particles are slower below the pseudo-critical concentration, these experiments produce unique information about reaction intermediates and reaction kinetics.
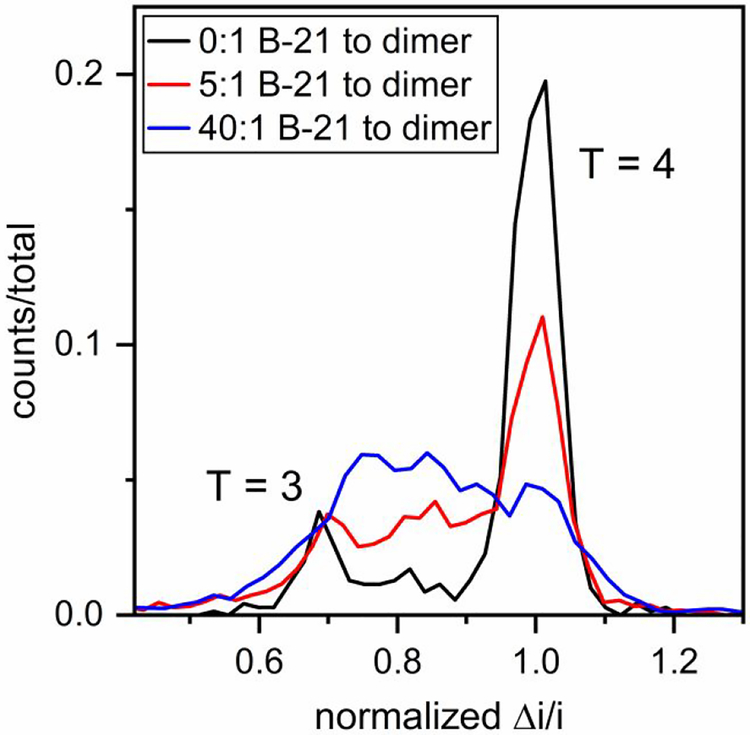
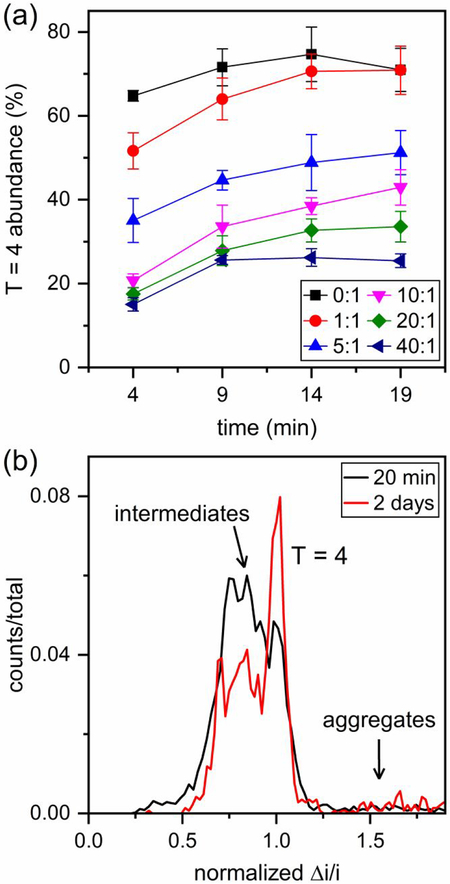
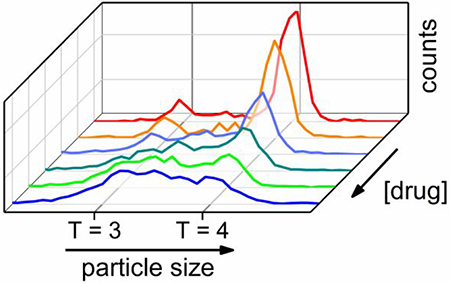
For assembly of 0.4 μM Cp149 dimer, the concentrations of B-21 tested were 0 (control), 0.4, 2, 4, 8, and 16 μM to yield B-21 to dimer molar ratios of 0:1, 1:1, 5:1, 10:1, 20:1, and 40:1. Histograms of pulse amplitudes from the 1.5 – 21.5 min of the assembly reactions are shown in Figure 4. To minimize drift in the signal, the pulse amplitudes (Δi) were divided by the baseline current (i), and to compare results from different devices, the Δi/i values were normalized by the mean pulse amplitude of T = 4 capsids. Unlike assembly of Cp149 dimer without any CpAM, where fully formed T = 3 and T = 4 capsids are the dominant products, accumulation of assembly intermediates was observed at high molar ratios of B-21 to Cp149 dimer (Figure 4). Intermediate sizes were not randomly distributed; most were larger than T = 3 capsids (e.g., 90 dimers). The limits of detection are ~30 and ~70 dimers for measurements in 1 and 0.5 M NaCl, respectively. The same experiments were conducted with AT-130, and the same effects on the HBV assembly were observed (Figure S1). Assembly products are dominated by large intermediate species, which are formed in large abundance compared to our experiments without phenylpropenamides,17 and suggest that phenylpropenamides preferentially kinetically trap intermediates during the last steps of assembly, when the capsid shell is closing.
Figure 4. Accumulation of assembly intermediates with B-21 in 1 M NaCl.
Pulse-amplitude histograms of reaction products for molar ratios of B-21 to Cp149 dimer of 0:1, 5:1, and 40:1 assembled in 1 M NaCl. As B-21 concentration increases, the abundance of large intermediates increases. Final dimer concentration was 0.4 μM, and assembly was monitored for 20 min. Pulse amplitudes are normalized to a T = 4 capsid.
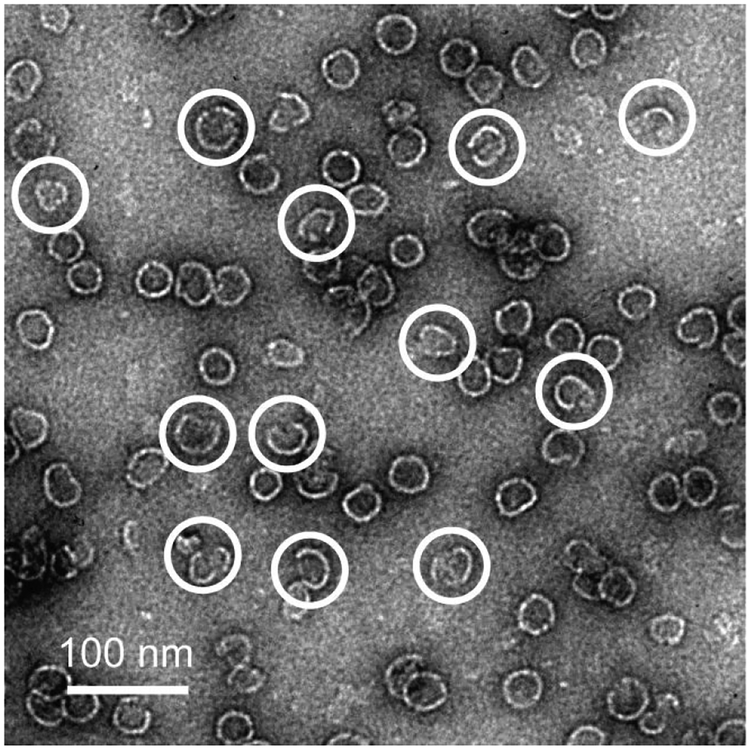
Assembly intermediates formed at a molar ratio of 40:1 B-21 to dimer were visualized with negative-stain transmission electron microscope (TEM) images. Because TEM requires relatively high particle concentrations, dimer concentrations of 2 μM were used for these experiments. Figure 5 shows the presence of large, partially formed virus capsids. However, the limited throughput, flattening of open structures, and potential distortion of fragile intermediate species during negative staining, predominantly by dehydration, impede quantifying intermediate species by TEM.
Figure 5. Transmission electron microscope (TEM) image with partially formed large intermediates.
Negative-stain TEM image of assembly products from 80 μM B-21 and 2 μM Cp149 dimer (40:1 molar ratio) in 1 M NaCl. Incomplete capsids are indicated with white circles.
For analysis of the assembly kinetics, data were binned in time increments of 5 min, and the abundance of T = 4 capsids in each increment was calculated. Figure 6a shows a gradual increase in the abundance of T = 4 capsids over the initial 1.5 – 21.5 min of the reaction. To confirm the stability of capsids and intermediate structures, the 40:1 B-21 to Cp149 dimer reaction solution was measured again after two days. Figure 6b reveals that some intermediate species rearranged over time to form T = 4 capsids or aggregates. In general, aggregation occurred more readily at high molar ratios of B-21 to Cp149 dimer. An important observation, however, is that a significant number of assembly intermediates remained trapped in the reaction on the timescale of days.
Figure 6. Time evolution of assembly reactions with B-21 in 1 M NaCl.
(a) Variation of the abundance of T = 4 capsids over the initial 1.5–21.5 min of assembly for different B-21 to Cp149 dimer ratios in 1 M NaCl. Assembly intermediates rearrange over time and form T =4 capsids. The change is more dramatic in the initial minutes of assembly. (b) Pulse-amplitude histograms of assembly products for a 40:1 ratio of B-21 to Cp149 dimer in 1 M NaCl after the initial 1.5–21.5 min and 2 days of reaction. Intermediates rearrange and form T = 4 capsids and aggregates, but a significant number of intermediate species remains remarkably stable at 1 M NaCl. Pulse amplitudes are normalized to a T = 4 capsid.
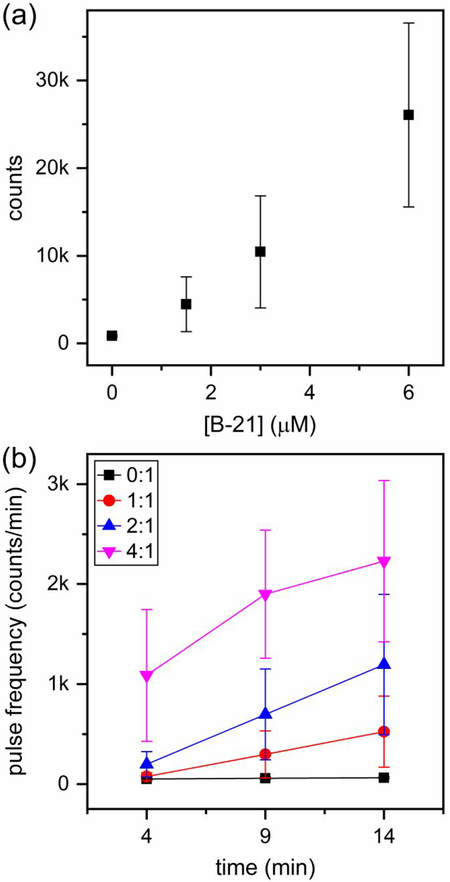
Another effect of phenylpropenamides was on the extent of HBV capsid assembly. Specifically, at a ratio of 40:1 B-21 to dimer in 1 M NaCl, the extent of capsid assembly integrated over the 20 min of the resistive-pulse measurements (initial 1.5 – 21.5 min of assembly) increased by ~6.5 times compared to the control experiment (Figure S2a). This result is consistent with the modest increase in the strength of subunit-subunit interactions observed for AT-130 and B-21.35 In addition, the rate of assembly (i.e., pulse frequency) with phenylpropenamides varied over the timescale of the measurement (Figure S2b), and our resistive-pulse devices provided sufficient temporal resolution to capture this change.
Assembly with Phenylpropenamides in 0.5 M NaCl.
The analysis of HBV assembly in lower salt concentrations offers further information about the mechanism of CpAM action. Ionic strength substantively affects the association energy between dimers and, consequently, the stability of intermediates and rate of assembly. In this series of experiments, the final NaCl concentration was 0.5 M and final Cp149 dimer concentration was 1.5 μM, which is below the pseudo-critical concentration of 1.8 μM Cp149 dimer in 0.5 M NaCl.16 This condition supports only a small amount of assembly in the absence of CpAMs. The concentrations of B-21 tested were 0 (control), 1.5, 3, and 6 μM to yield B-21 to dimer molar ratios of 0:1, 1:1, 2:1, and 4:1.
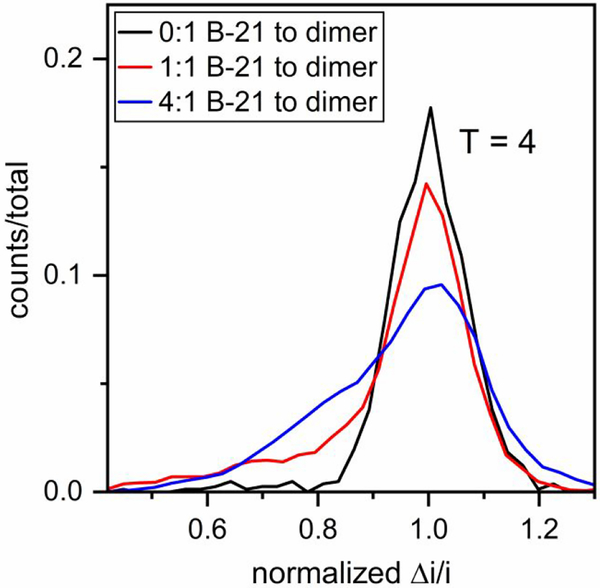
Histograms of the initial 1.5–16.5 min of the assembly reaction are shown in Figure 7. Compared to assembly in 1 M NaCl, very few T = 3 capsids were observed in 0.5 M NaCl when Cp149 dimer assembled without B-21. Similar to experiments performed in 1 M NaCl, the presence of B-21 in the reaction solution resulted in the accumulation of large assembly intermediates (Figure 7). The same experiments were repeated with AT-130, and the same effects on HBV assembly were observed (Figure S3). Thus, B-21 and AT-130 had the same effective action at both higher and lower ionic strengths.
Figure 7. Accumulation of assembly intermediates with B-21 in 0.5 M NaCl.
Pulse-amplitude histograms of reaction products for molar ratios of B-21 to Cp149 dimer of 0:1, 1:1, and 4:1 assembled in 0.5 M NaCl. Final dimer concentration was 1.5 μM, and assembly was monitored for 15 min. In contrast to experiments at 1 M NaCl, no T = 3 capsids were observed. The number of counts ranged from a few hundred for the 0:1 ratio to several thousand for the 4:1 ratio. Pulse amplitudes are normalized to a T = 4 capsid.
Analysis of the kinetics of experiments in 0.5 M NaCl showed that the abundance of T = 4 capsids increased over time (Figure S4a), which suggests movement from intermediate-sized particles to capsids. To further characterize the stability of the intermediate species, the 4:1 B-21 to Cp149 dimer reaction solution was re-measured after two days. Figure S4b shows that very few intermediates remained in the reaction solution after this two-day period. The short lifetime of intermediates at lower salt concentrations explains why their visualization and characterization is difficult with conventional analytical methods. In physiological ionic strength, where the strength of subunit-subunit interactions is even weaker, these intermediates are expected to be even more labile.
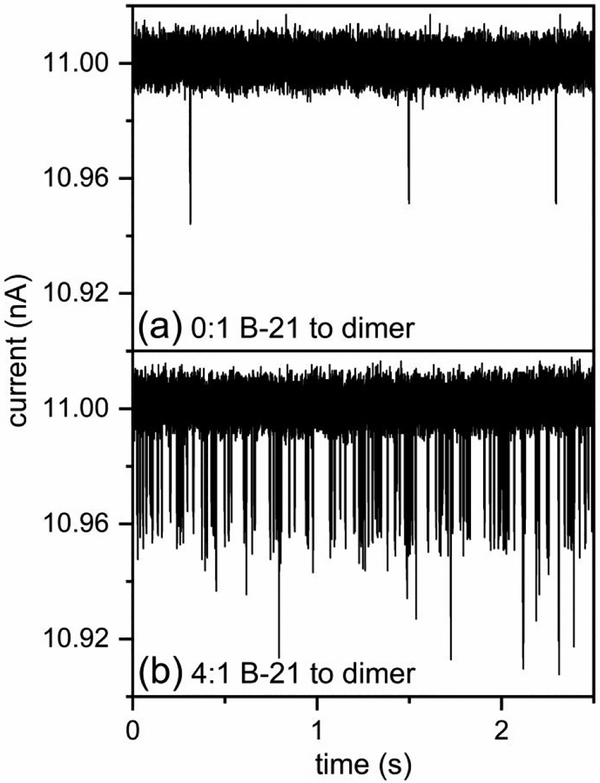
Figure 8 illustrates the effects of B-21 on the extent of Cp149 assembly in 0.5 M NaCl. A 20-fold difference in the pulse frequency is clearly visualized in the raw current traces for assembly without B-21 (Figure 8a) and assembly with a molar ratio of 4:1 B-21 to Cp149 dimer (Figure 8b). In this latter experiment, more than 15,000 counts were recorded in 15 min, which corresponds to more than 15 counts/s. In Figure 9, the total number of counts in the initial 1.5–16.5 min of assembly varied from hundreds for the ratio of 0:1 B-21 to Cp149 dimer to more than 15,000 for the ratio of 4:1 B-21 to Cp149 dimer. Of note, the increase in the extent of HBV assembly caused by B-21 was significantly greater at 0.5 M NaCl compared to 1 M NaCl. Even at a 40:1 B-21 to Cp149 dimer ratio in 1 M NaCl, the extent of HBV assembly increased by a factor of just ~6.5 times, whereas at 4:1 B-21 to Cp149 dimer ratio in 0.5 M NaCl, the extent of HBV assembly increased by ~20-fold. In higher salt concentrations, the ionic strength becomes the dominant effect driving HBV capsid assembly; consequently, the effect of B-21 on assembly is more pronounced under milder assembly conditions. In addition, the dramatic increase in the pulse frequency over the timescale of minutes was successfully captured and is illustrated in Figure 9b.
Figure 8. Dramatic increase in pulse-frequency of assembly products with B-21 in 0.5 M NaCl.
Current traces for the assembly reactions at (a) 0:1 B-21 to Cp149 dimer in 0.5 M NaCl, and at (b) 4:1 B-21 to Cp149 dimer in 0.5 M NaCl, respectively. The pulses with amplitudes > 50 pA correspond to aggregates. B-21 significantly increases the extent of HBV assembly in 0.5 M NaCl.
Figure 9. Increased rate of assembly with B-21 in 0.5 M NaCl.
(a) Change in the number of counts with B-21 concentration integrated over the 15 min of the resistive-pulse measurements (initial 1.5–16.5 min of assembly). B-21 affected the extent of HBV assembly more dramatically in 0.5 M NaCl compared to experiments in 1 M NaCl. With a molar ratio of 4:1 B-21 to Cp149 dimer, the extent of assembly increased by ~20-fold, (b) Increase in pulse frequency over the initial 1.5 – 16.5 min of the assembly reaction. Similar to the experiments at 1 M NaCl (Figure S2), the pulse frequency increased significantly over time with B-21.
Release of Trapped Assembly Products after Dilution.
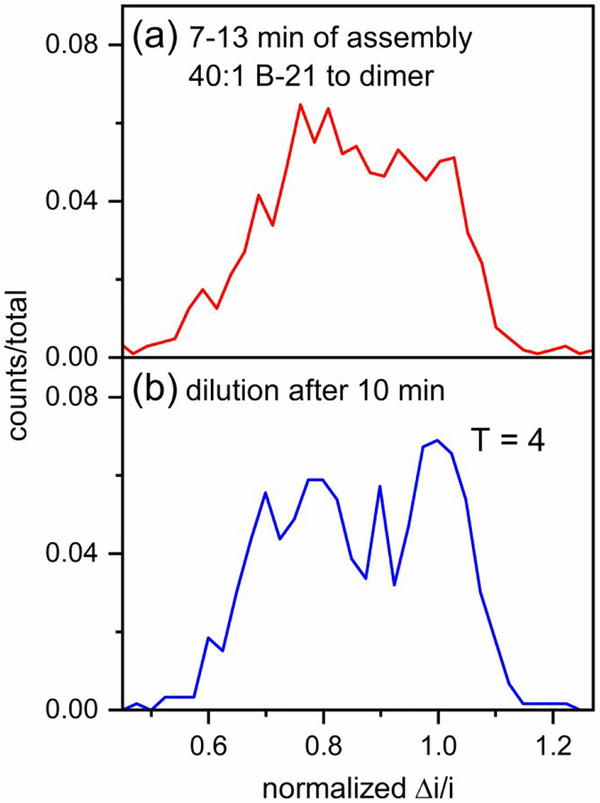
Although T = 3 and T = 4 capsids are persistent, assembly intermediates were not. A shift in the equilibrium of assembly can significantly affect the population of intermediate species in the reaction solution. Presumably, particle rearrangement is due to the ‘frayed’ edge of the intermediate that allows subunits to readily equilibrate with solution.46 To shift the equilibrium of the assembly reaction and observe changes in the population of intermediate species, dilution experiments were performed. Assembly was initiated with a 40:1 ratio of B-21 to Cp149 dimer in 1 M NaCl and 10 min after the initiation, the reaction solution was diluted 10-fold with 1 M NaCl, which brings the dimer concentration to 12-fold below the pseudo-critical concentration. The diluted sample was measured by resistive-pulse sensing until a few hundred counts were acquired, which required 1–2 h of data acquisition. After dilution, we observed a clear decrease in the population of the assembly intermediates (Figure 10). This decrease clearly indicates that intermediate species are extremely labile and able to rearrange to form free dimers, small ensembles of dimers, and fully formed capsids. On the contrary, intact capsids persist for months.47
Figure 10. Shift of large intermediates toward T = 4 capsids after dilution.
(a) Pulse-amplitude histogram from measurements of HBV assembly (7–13 min) of 16 μM B-21 and 0.4 μM Cp149 dimer (40:1 molar ratio) in 1 M NaCl. (b) Pulse-amplitude histogram of the reaction solution diluted 10-fold in 1 M NaCl 10 min after the initiation of the reaction and measured for 1–2 h. Pulse amplitudes are normalized to a T = 4 capsid.
Conclusion
We describe a mechanism of action for phenylpropenamide antivirals based on our experimental observations of single virus particles. Phenylpropenamides lead to over-initiation of assembly and induce large intermediate species to accumulate. These intermediates are extremely dynamic; a change in the equilibrium of the reaction, e.g., dilution of the reaction solution, causes rapid rearrangement. These results provide a basis for predicting the activity of phenylpropenamides and other CpAMs in the context of an infected cell. In a normal infection, assembly may be nucleated by a pgRNA-reverse transcriptase complex to yield immature RNA-filled cores, which will mature to virions. More frequently, assembly is initiated spontaneously to unproductively yield empty particles.48 CpAMs, like the phenylpropenamides, can allosterically trigger assembly, favoring the unproductive path. This behavior has been noted for phenylpropenamides.37 Here, we further show they can lead to depletion of free subunits and incomplete particles, an effect that is possible with both empty and RNA-filled defective particles. We envision a scenario where the presence of phenylpropenamide molecules modestly increases the association energy of dimers, prevents intermediates from dissociating, and leads to kinetic trapping where the formation of too many capsids has been initiated resulting in both empty and incomplete particles.
Experimental Methods
Virus Capsids.
HBV capsids were assembled from dimeric core protein Cp149 (34 kDa) expressed in E. co/iand purified.49 For the preparation of standard solutions, T = 3 capsids were purified on a 10% – 40% (w/v) continuous sucrose gradient that was centrifuged for 6 h at 150,000 g after assembly in 0.3 M NaCl with 50 mM HEPES (pH 7.5). Also, T = 4 capsids were purified after assembly in 0.3 M NaCl with 50 mM HEPES (pH 7.5) by size exclusion chromatography (SEC). For the assembly experiments, Cp149 dimer was used without further purification.
Phenylpropenamides.
B-21 and AT-130 compounds were synthesized35 and stored at −20 °C as 10 mM stock solutions diluted in dimethyl sulfoxide (DMSO). High resolution mass spectra of B-21 and AT-130 (Figure S5), high performance liquid chromatograms of B-21 and AT-130 (Figure S6), and 1H-NMR data of B-21 and AT-130 (Figure S7) confirm the identities and purities (> 90%) of the two compounds. Aliquots of the stock solutions were mixed with the assembly buffers and used for the assembly reactions. The concentration of DMSO in the final reaction solutions was kept below 2% (v/v).
Transmission Electron Microscopy (TEM).
The assembly reaction was initiated with a 40:1 ratio of B-21 to Cp149 dimer in 1 M NaCl. Ten minutes after initiation of the reaction, the sample was applied to a glow-discharged carbon-coated 300-mesh copper grid and stained with 0.75% uranyl formate. Images were acquired at 80 kV on a JEOL 1010 transmission electron microscope equipped with a Gatan 1k × 1k CCD camera.
Fabrication of Nanofluidic Devices.
On D263 glass substrates, a 120 nm thick layer of chromium was deposited by thermal evaporation (BOC Edwards Auto 306 Vacuum Coating System), and a 3 μm thick layer of S1813 photoresist was spin-coated onto the chromium layer. The microchannels were fabricated with positive UV photolithography and wet chemical etching.43 The depth of the microchannels was determined with a stylus-based profiler (P-7, KLA Tencor) before the bonding, and the channel width was determined with an optical microscope (Nikon Eclipse TE2000-E) after the bonding procedure. The microchannels were 35 ± 2 μm wide and 11.0 ± 0.5 μm deep.
To fabricate the nanochannels and nanopores, we used a focused ion beam (FIB) instrument (Auriga 60, Carl Zeiss, GmbH) controlled by the Nano-Patterning and Visualization Engine (NPVE; FIBICS, Inc.). With a 30 kV beam at 50 pA, we milled the side nanochannels with a dose of 1 nC/μm2 and the bridge and pore-to-pore nanochannels with a dose of 0.5 nC/μm2. The nanopores were milled as a single line with a 30 kV beam at 20 pA and a dose of 0.011 μC/pm. During the FIB milling, an electron flood gun (FG 15/40, SPECS, GmbH) was operated at 5 eV and 20 μA to compensate for the build-up of positive charge on the substrate surface. Dimensions of the nanochannels and nanopores were determined with an atomic force microscope (AFM; MFP-3D, Asylum Research, Inc.) and the scanning electron microscope (SEM) on the FIB instrument. In Figure 2b, the nanopores are 60 ± 5 nm deep, 60 ± 5 nm wide, and 290 ± 6 nm long; pore-to-pore nanochannels are 500 ± 7 nm long, 320 ± 10 nm wide, and 125 ± 7 nm deep; bridge nanochannels are 330 ± 10 nm long, 320 ± 10 nm wide, and 125 ± 7 nm deep; and side nanochannels are 500 ± 10 nm wide and 245 ± 10 nm deep..
For the bonding of the devices, the substrates and No.1.5 cover slips were cleaned in 1 M NaOH for 15 min, sonicated in ultrapure water for 10 min, and brought into contact with each other while still wet. The devices were dried overnight at 90 °C and annealed at 545 °C for 12 h. To hold samples and buffers and make electrical contact to them, glass reservoirs were epoxied over the access holes to the microchannels.
Operation of Nanofluidic Devices.
To rinse devices, the two reservoirs in the middle were filled with solution, and vacuum was applied to the reservoirs on the sides. Before the first use, each device was rinsed with H2O for 10 min, 0.1 M NaOH for 15 min, H2O for 5 min, and buffer solution (50 mM HEPES with 0.5 or 1 M NaCl) for 5 min. All solutions, except the samples, were filtered with 20 nm syringe filters. The sample was loaded into one of the two reservoirs in the middle, and vacuum was applied to the corresponding end reservoir for 30 s. Prior to assembly measurements, all the devices were tested and calibrated with purified standard solutions of virus capsids.
Because HBV capsids are negatively charged, their electrophoretic transport opposes the electroosmotic flow inside the nanopores. Although significantly suppressed at 1 M NaCl, electroosmotic flow is comparable in magnitude but of opposite sign to the electrophoretic mobility of HBV capsids in 0.5 M NaCl. Thus, the micro- and nanochannels in the devices used for measurements with 0.5 M NaCl were coated with a short-chain PEG-silane after the cover plate annealing step. The devices were sequentially rinsed with H2O, 0.1 M NaOH, H2O, MeOH/H2O, MeOH, toluene/MeOH, anhydrous toluene, and 0.26% PEG-silane/0.1% 12 M HCl/99.64% toluene (v/v). The PEG-silane solution was left for 4 h in the micro- and nanochannels and then sequentially rinsed with anhydrous toluene, toluene/MeOH, MeOH, MeOH/H2O, and H2O prior to filling with 50 mM HEPES buffer with NaCl.
Resistive-Pulse Measurements.
The resistive-pulse measurements were conducted inside a stainless steel Faraday cage covered with wedge foam. An Axopatch 200B (Molecular Devices, Inc.) was used to apply the potential between the two middle reservoirs through Ag/AgCl electrodes and to record the current trace. The measurements were taken at ~17 nA baseline current in 1 M NaCl and at ~11 nA baseline current in 0.5 M NaCl. The potential applied across the nanopores was in the range of 800–1000 mV. All data were collected with a sampling frequency of 40 kHz and filter frequency of 10 kHz.
For the characterization of the devices, purified T = 3 and T = 4 capsid solutions in 50 mM HEPES buffer (pH 7.5) with 0.5 or 1 M NaCl were used. For assembly reactions, the phenylpropenamide derivatives were mixed with the buffered NaCl solutions first, and Cp149 dimer was then added to the solution. A period of ~90 s elapsed between initiation of the assembly reactions and the start of the resistive-pulse measurements.
Data Analysis.
To determine the pulse amplitudes and pore-to-pore times, the raw data files were imported into MatLab R2017a (Mathworks, Inc.), and a modified version of Open Nanopore 1.2 was used for the analysis of the raw data and determination of the pulse amplitude (Δi), pulse width (w), and average baseline current adjacent to each pulse.50 The times between adjacent pulses were tabulated and plotted on a logarithmic scale to reveal two distributions: correlated times for individual capsids (pore-to-pore times) and uncorrelated times from different capsids. A Gaussian function was fitted to the pore-to-pore time distribution with Origin Pro 2018 (OriginLab Corporation), and the standard deviation of the fitted function was used as a selection criterion for correlated sets of pulses. To eliminate the drift in the signal, the pulse amplitudes (Δi) were divided by the baseline current (i), and to compare results from different devices, the Δi/i values were normalized by the mean pulse amplitude of T = 4 capsids. Events produced by the simultaneous presence of two particles inside the pores were rejected and not included in the amplitude histograms. To estimate the relative abundance of T = 4 capsids in the reaction solution, the mean of each T = 4 capsid distribution was determined by the corresponding assembly histogram and the standard deviation (σ) by measurements of purified standards. The cut-offs for quantifying T = 4 capsids were then set as the (mean) ± 2σ.
Supplementary Material
Acknowledgments.
This work was supported in part by NIH R01 GM129354, NSF CHE-0923064, and the Dorothy & Edward Bair Chair in Chemistry. The authors thank the Indiana University Nanoscale Characterization Facility and Electron Microscopy Center for use of their instruments. We also thank the NMR Facility and Mass Spectrometry Facility for analysis of B-21 and AT-130 and Fred C. Parks for his assistance with data analysis.
Footnotes
Conflict of Interest. The authors declare the following competing financial interest: A.Z. reports a financial interest in a company that develops core protein allosteric modulators (CpAMs).
Supporting Information Available. Figures of the accumulation of intermediates with AT-130 in 1 M NaCl, assembly extent with B-21 in 1 M NaCl, accumulation of intermediates with AT-130 in 0.5 M NaCl, time evolution of reactions with B-21 in 0.5 M NaCl, high resolution mass spectra of B-21 and AT-130, high performance liquid chromatograms of B-21 and AT-130, and 1H-NMR data of B-21 and AT-130. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Dragnea B Virus-Based Devices: Prospects for Allopoiesis. ACS Nano 2017, 11, 3433–3437, 10.1021/acsnano.7b01761. [DOI] [PubMed] [Google Scholar]
- (2).Deres K; Schroder CH; Paessens A; Goldmann S; Hacker HJ; Weber O; Kramer T; Niewohner U; Pleiss U; Stoltefuss J; Graef E; Koletzki D; Masantschek RNA; Reimann A; Jaeger R; Gross R; Beckermann B; Schlemmer KH; Haebich D; Rubsamen-Waigmann H Inhibition of Hepatitis B Virus Replication by Drug-Induced Depletion of Nucleocapsids. Science 2003, 299, 893–896, [DOI] [PubMed] [Google Scholar]
- (3).Prevelige PE New Approaches for Antiviral Targeting of HIV Assembly. J. Mol. Biol 2011, 410, 634–640, 10.1016/jjmb.2011.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Young M; Willits D; Uchida M; Douglas T Plant Viruses as Biotemplates for Materials and Their Use in Nanotechnology In Annu. Rev. Phytopathol, Annual Reviews: Palo Alto, 2008; Vol.46, pp 361–384. [DOI] [PubMed] [Google Scholar]
- (5).Papatheodoridis G; Buti M; Cornberg M; Janssen H; Mutimer D; Pol S; Raimondo G EASL Clinical Practice Guidelines: Management of Chronic Hepatitis B Virus Infection. J. Hepatol 2012, 57, 167–185, [DOI] [PubMed] [Google Scholar]
- (6).Stray SJ; Bourne CR; Punna S; Lewis WG; Finn MG; Zlotnick A A Heteroaryldihydropyrimidine Activates and Can Misdirect Hepatitis B Virus Capsid Assembly. Proc. Natl. Acad. Sci. U. S. A 2005, 102, 8138–8143, 10.1073/pnas.0409732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Pei YM; Wang CT; Yan SF; Liu G Past, Current, and Future Developments of Therapeutic Agents for Treatment of Chronic Hepatitis B Virus Infection. J. Med. Chem 2017, 60, 6461–6479, 10.1021/acs.jmedchem.6b01442. [DOI] [PubMed] [Google Scholar]
- (8).Zlotnick A To Build a Virus Capsid - an Equilibrium Model of the Self-Assembly of Polyhedral Protein Complexes. J. Mol. Biol 1994, 241, 59–67, [DOI] [PubMed] [Google Scholar]
- (9).Zlotnick A; Johnson JM; Wingfield PW; Stahl SJ; Endres D A Theoretical Model Successfully Identifies Features of Hepatitis B Virus Capsid Assembly. Biochemistry 1999, 38, 14644–14652, 10.1021/bi991611a. [DOI] [PubMed] [Google Scholar]
- (10).Endres D; Miyahara M; Moisant P; Zlotnick A A Reaction Landscape Identifies the Intermediates Critical for Self-Assembly of Virus Capsids and Other Polyhedral Structures. Protein Sci. 2005, 14, 1518–1525, 10.1110/ps.041314405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Hagan MF Modeling Viral Capsid Assembly In Adv. Chem. Phys: Vol. 155, John Wiley & Sons, Inc.: 2014; pp 1–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Perkett MR; Hagan MF Using Markov State Models to Study Self-Assembly. J. Chem. Phys 2014, 140, 14, 10.1063/1.4878494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Douglas T; Young M Viruses: Making Friends with Old Foes. Science 2006, 312, 873–875, 10.1126/science.1123223. [DOI] [PubMed] [Google Scholar]
- (14).DuFort CC; Dragnea B Bio-Enabled Synthesis of Metamaterials In Annu Rev. Phys. Chem, Leone SR; Cremer PS; Groves JT; Johnson MA; Richmond G, Eds. Annual Reviews: Palo Alto, 2010; Vol. 61, pp 323–344. [DOI] [PubMed] [Google Scholar]
- (15).Zlotnick A; Cheng N; Conway JF; Booy FP; Steven AC; Stahl SJ; Wingfield PT Dimorphism of Hepatitis B Virus Capsids Is Strongly Influenced by the C-Terminus of the Capsid Protein. Biochemistry 1996, 35, 7412–7421, [DOI] [PubMed] [Google Scholar]
- (16).Ceres P; Zlotnick A Weak Protein-Protein Interactions Are Sufficient to Drive Assembly of Hepatitis B Virus Capsids. Biochemistry 2002, 41, 11525–11531, 10.1021/bi0261645. [DOI] [PubMed] [Google Scholar]
- (17).Harms ZD; Selzer L; Zlotnick A; Jacobson SC Monitoring Assembly of Virus Capsids with Nanofluidic Devices. ACS Nano 2015, 9, 9087–9096, 10.1021/acsnano.5b03231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kondylis P; Zhou JS; Harms ZD; Kneller AR; Lee LS; Zlotnick A; Jacobson SC Nanofluidic Devices with 8 Pores in Series for Real-Time, Resistive-Pulse Analysis of Hepatitis B Virus Capsid Assembly. Anal. Chem 2017, 89, 4855–4862, 10.1021/acs.analchem.6b04491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhou J; Kondylis P; Haywood DG; Harms ZD; Lee LS; Zlotnick A; Jacobson SC Characterization of Virus Capsids and Their Assembly Intermediates by Multicycle Resistive-Pulse Sensing with Four Pores in Series. Anal. Chem 2018, 90, 7267–7274, 10.1021/acs.analchem.8b00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Uetrecht C; Barbu IM; Shoemaker GK; van Duijn E; Heck AJR Interrogating Viral Capsid Assembly with Ion Mobility-Mass Spectrometry. Nat. Chem 2011, 3, 126–132, 10.1038/nchem.947. [DOI] [PubMed] [Google Scholar]
- (21).Pierson EE; Keifer DZ; Selzer L; Lee LS; Contino NC; Wang JCY; Zlotnick A; Jarrold MF Detection of Late Intermediates in Virus Capsid Assembly by Charge Detection Mass Spectrometry. J. Am. Chem. Soc 2014, 136, 3536–3541, 10.1021/ja411460w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lutomski CA; Lyktey NA; Zhao ZC; Pierson EE; Zlotnick A; Jarrold MF Hepatitis B Virus Capsid Completion Occurs through Error Correction. J. Am Chem. Soc 2017, 139, 16932–16938, 10.1021/jacs.7b09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kondylis P; Schlicksup CJ; Zlotnick A; Jacobson SC Analytical Techniques to Characterize the Structure, Properties, and Assembly of Virus Capsids. Anal. Chem 2018, 10.1021/acs.analchem.8b04824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Corcuera A; Stolle K; Hillmer S; Seitz S; Lee J-Y; Bartenschlager R; Birkmann A; Urban A Novel Non-Heteroarylpyrimidine (HAP) Capsid Assembly Modifiers Have a Different Mode of Action from Haps in vitro. Antiviral Res. 2018, 158, 135–142, 10.1016/j.antiviral.2018.07.011. [DOI] [PubMed] [Google Scholar]
- (25).Li LC; Chirapu SR; Finn MG; Zlotnick A Phase Diagrams Map the Properties of Antiviral Agents Directed against Hepatitis B Virus Core Assembly. Antimicrob. Agents Chemother. 2013, 57, 1505–1508, 10.1128/aac.01766-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Schlicksup CJ; Wang JCY; Francis S; Venkatakrishnan B; Turner WW; VanNieuwenhze M; Zlotnick A Hepatitis B Virus Core Protein Allosteric Modulators Can Distort and Disrupt Intact Capsids. eLife 2018, 7, e31473, 10.7554/eLife.31473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kondylis P; Schlicksup CJ; Brunk NE; Zhou J; Zlotnick A; Jacobson SC Competition between Normative and Drug-Induced Virus Self-Assembly Observed with Single-Particle Methods. J. Am Chem. Soc 2018, 10.1021/jacs.8b10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Bourne C; Lee S; Venkataiah B; Lee A; Korba B; Finn MG; Zlotnick A Small-Molecule Effectors of Hepatitis B Virus Capsid Assembly Give Insight into Virus Life Cycle. J. Virol 2008, 82, 10262–10270, 10.1128/jvi.01360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).King RW; Ladner SK; Miller TJ; Zaifert K; Perni RB; Conway SC; Otto MJ Inhibition of Human Hepatitis B Virus Replication by AT-61, a Phenylpropenamide Derivative, Alone and in Combination with (−)Beta-L-2 ‘,3 ‘-Dideoxy-3 ‘-Thiacytidine. Antimicrob. Agents Chemother 1998, 42, 3179–3186, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Campagna MR; Liu F; Mao RC; Mills C; Cai DW; Guo F; Zhao XS; Ye H; Cuconati A; Guo HT; Chang JH; Xu XD; Block TM; Guo JT Sulfamoylbenzamide Derivatives Inhibit the Assembly of Hepatitis B Virus Nucleocapsids. J. Virol 2013, 87, 6931–6942, 10.1128/jvi.00582-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Perni RB; Conway SC; Ladner SK; Zaifert K; Otto MJ; King RW Phenylpropenamide Derivatives as Inhibitors of Hepatitis B Virus Replication. Bioorg. Med. Chem. Lett 2000, 10, 2687–2690, 10.1016/s0960-894x(00)00544-8. [DOI] [PubMed] [Google Scholar]
- (32).Wang PY; Naduthambi D; Mosley RT; Niu CR; Furman PA; Otto MJ; Sofia MJ Phenylpropenamide Derivatives: Anti-Hepatitis B Virus Activity of the Z Isomer, Sar and the Search for Novel Analogs. Bioorg. Med. Chem Lett. 2011, 21, 4642–4647, 10.1016/j.bmcl.2011.05.077. [DOI] [PubMed] [Google Scholar]
- (33).Delaney WE; Edwards R; Colledge D; Shaw T; Furman P; Painter G; Locarnini S Phenylpropenamide Derivatives AT-61 and AT-130 Inhibit Replication of Wild-Type and Lamivudine-Resistant Strains of Hepatitis B Virus in vitro. Antimicrob. Agents Chemother. 2002, 46, 3057–3060, 10.1128/aac.46.9.3057-3060.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Billioud G; Pichoud C; Puerstinger G; Neyts J; Zoulim F The Main Hepatitis B Virus (HBV) Mutants Resistant to Nucleoside Analogs Are Susceptible in vitro to Non-Nucleoside Inhibitors of HBV Replication. Antivir. Res 2011, 92, 271–276, 10.1016/j.antiviral.2011.08.012. [DOI] [PubMed] [Google Scholar]
- (35).Katen SP; Chirapu SR; Finn MG; Zlotnick A Trapping of Hepatitis B Virus Capsid Assembly Intermediates by Phenylpropenamide Assembly Accelerators. ACS Chem. Biol 2010, 5, 1125–1136, 10.1021/cb100275b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Katen SP; Tan ZN; Chirapu SR; Finn MG; Zlotnick A Assembly-Directed Antivirals Differentially Bind Quasiequivalent Pockets to Modify Hepatitis B Virus Capsid Tertiary and Quaternary Structure. Structure 2013, 21, 1406–1416, 10.1016/j.str.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Feld JJ; Colledge D; Sozzi V; Edwards R; Littlejohn M; Locarnini SA The Phenylpropenamide Derivative AT-130 Blocks HBV Replication at the Level of Viral RNA Packaging. Antiviral Res. 2007, 76, 168–177, 10.1016/j.antiviral.2007.06.014. [DOI] [PubMed] [Google Scholar]
- (38).Coulter WH Means for Counting Particles Suspended in a Fluid. 2,656,508, 1953. [Google Scholar]
- (39).DeBlois RW; Bean CP Counting and Sizing of Submicron Particles by Resistive Pulse Technique. Rev. Sci. Instrum 1970, 41, 909–916, [Google Scholar]
- (40).Luo L; German SR; Lan WJ; Holden DA; Mega TL; White HS Resistive-Pulse Analysis of Nanoparticles In Annu. Rev. Anal. Chem, Cooks RG; Pemberton JE, Eds. Annual Reviews: Palo Alto, 2014; Vol.7, pp 513–535. [DOI] [PubMed] [Google Scholar]
- (41).Howorka S; Siwy Z Nanopore Analytics: Sensing of Single Molecules. Chem. Soc. Rev 2009, 38, 2360–2384, 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- (42).Haywood DG; Saha-Shah A; Baker LA; Jacobson SC Fundamental Studies of Nanofluidics: Nanopores, Nanochannels, and Nanopipets. Anal. Chem 2015, 87, 172–187, 10.1021/ac504180h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Harms ZD; Haywood DG; Kneller AR; Selzer L; Zlotnick A; Jacobson SC Single-Particle Electrophoresis in Nanochannels. Anal Chem. 2015, 87, 699–705, 10.1021/ac503527d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Chen C; Kao CC; Dragnea B Self-Assembly of Brome Mosaic Virus Capsids: Insights from Shorter Time-Scale Experiments. J. Phys. Chem. A 2008, 112, 9405–9412, 10.1021/jp802498z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Li CL; Kneller AR; Jacobson SC; Zlotnick A Single Particle Observation of SV40 VP1 Polyanion-Induced Assembly Shows That Substrate Size and Structure Modulate Capsid Geometry. ACS Chem. Biol 2017, 12, 1327–1334, 10.1021/acschembio.6b01066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Lee LS; Brunk N; Haywood DG; Keifer D; Pierson E; Kondylis P; Wang JCY; Jacobson SC; Jarrold MF; Zlotnick A A Molecular Breadboard: Removal and Replacement of Subunits in a Hepatitis B Virus Capsid. Protein Sci. 2017, 26, 2170–2180, 10.1002/pro.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Uetrecht C; Watts NR; Stahl SJ; Wingfield PT; Steven AC; Heck AJR Subunit Exchange Rates in Hepatitis B Virus Capsids Are Geometry- and Temperature-Dependent. Phys. Chem. Chem. Phys 2010, 12, 13368–13371, 10.1039/c0cp00692k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Ning XJ; Nguyen D; Mentzer L; Adams C; Lee H; Ashley R; Hafenstein S; Hu JM Secretion of Genome-Free Hepatitis B Virus - Single Strand Blocking Model for Virion Morphogenesis of Para-Retrovirus. PLoS Pathog. 2011, 7, e1002255 10.1371/journal.ppat.1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Zlotnick A; Ceres P; Singh S; Johnson JM A Small Molecule Inhibits and Misdirects Assembly of Hepatitis B Virus Capsids. J. Virol 2002, 76, 4848–4854, 10.1128/jvi.76.10.4848-4854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Raillon C; Granjon P; Graf M; Steinbock LJ; Radenovic A Fast and Automatic Processing of Multi-Level Events in Nanopore Translocation Experiments. Nanoscale 2012, 4, 4916–4924, 10.1039/c2nr30951c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.