Abstract
Hepatitis B virus (HBV) core protein is a small protein with 183 amino acid residues and assembles the pre-genomic (pg) RNA and viral DNA polymerase to form nucleocapsids. During the last decades, several groups have reported HBV core protein allosteric modulators (CpAMs) with distinct chemical structures. CpAMs bind to the hydrophobic HAP pocket located at the dimer-dimer interface and induce allosteric conformational changes in the core protein subunits. While type I CpAMs, heteroaryldihydropyrimidine (HAP) derivatives, misdirect core protein dimers to assemble non-capsid polymers, Type II CpAMs, represented by sulfamoylbenzamides, phenylpropenamides and several other chemotypes, induce the assembly of empty capsids with global structural alterations and faster mobility in native agarose gel electrophoresis. Through high throughput screening of an Asinex small molecule library containing 19,920 compounds, we identified 8 structurally distinct CpAMs. While 7 of those compounds are typical Type II CpAMs, a novel benzamide derivative, designated as BA-53038B, induced the formation of morphologically “normal” empty capsids with slow electrophoresis mobility. Drug resistant profile analyses indicated that BA-53038B most likely bound to the HAP pocket, but obviously modulated HBV capsid assembly in a distinct manner. BA-53038B and other CpAMs reported herein provide novel structure scaffolds for the development of core protein-targeted antiviral agents for the treatment of chronic hepatitis B.
Keywords: Hepatitis B virus, Core protein allosteric modulator, Antiviral, Capsid assembly, Nucleocapsids
Although an effective vaccine is available, there are approximately 250 million people chronically infected by hepatitis B virus (HBV) worldwide1–2. The currently available antiviral therapeutics against HBV include pegylated interferon-alpha (IFN-α) and seven nucleos(t)ide analogue viral DNA polymerase inhibitors that can efficiently reduce viral load and prevent progression to cirrhosis and hepatocellular carcinoma in a large fraction of patients treated with these drugs 3–4. However, a “functional cure”, characterized by HBV surface antigen loss or seroconversion, is rarely achieved with the current therapies 5–6. Therefore, development of novel antiviral agents that more efficiently inhibit HBV replication and accomplish a functional cure of chronic hepatitis B as monotherapeutic agents or components of combination therapy is a demanding medical need 7–9.
While the vast majority of currently available antiviral agents target virus-encoded enzymes 10, viral capsid proteins have been considered to be promising antiviral targets with high selectivity in recent years. Indeed, small molecular compounds specifically targeting the capsid proteins of human immunodeficiency virus 11–12, HBV 13–14 and dengue virus 15, have been shown to disrupt capsid assembly and/or disassembly (uncoating) and are currently under preclinical or clinical development.
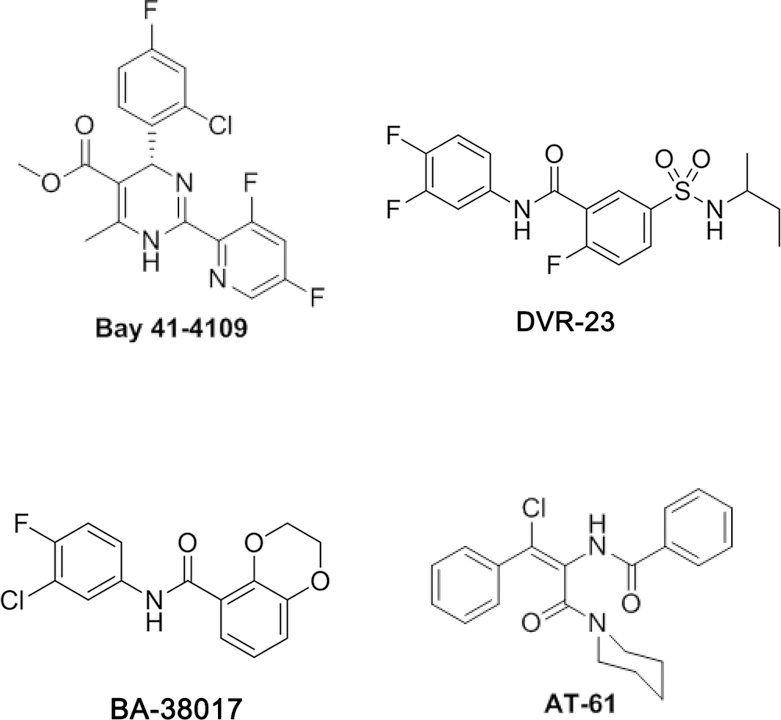
As shown in Fig. 1, several chemotypes of HBV core protein allosteric modulators (CpAMs) were reported in the last two decades. These compounds bind to a hydrophobic pocket (the HAP pocket), formed at the interface between core protein dimers and disrupt capsid assembly and packaging of the HBV pregenomic RNA (pgRNA) and DNA polymerase 16–20. Based on their pharmacological phenotypes, the CpAMs are categorized into two types. Type I CpAMs, which are heteroaryldihydropyrimidine (HAP) derivatives (represented by Bay 41–4109) 21–23, misdirect core protein dimers to form non-capsid polymers that can be either degraded intracellularly or accumulated in the nuclear PML bodies of hepatocytes 24–25. On the contrary, Type II CpAMs, including phenylpropenamides (PPAs) 26, sulfamoylbenzamides (SBAs) 27, benzamides 28 and others 29–30, induce the formation of empty capsids without packaging viral pgRNA and DNA polymerase and thus preclude viral genome replication. Moreover, both type I and type II CpAMs have been demonstrated to induce mature nucleocapsid uncoating 31 and inhibit de novo synthesis of covalently closed circular (ccc) DNA 31–32,33.
Fig. 1.
Structure of representative HBV core protein allosteric modulators (CpAMs).
In order to identify novel chemotypes of CpAMs as development leads of antiviral agents and as molecular probes to investigate the molecular mechanisms of HBV nucleocapsid assembly and disassembly, we screened 19,920 compounds from an in-house library for their ability to reduce the amount of HBV DNA in AML12HBV10 cells, an immortalized mouse hepatocyte-derived stable cell line supporting high levels of HBV DNA replication in a tetracycline inducible manner 34. Our screening effort identified six new chemotypes of compounds that, as previously reported type II CpAMs, induced the assembly of empty capsids devoid of pgRNA with faster electrophoresis mobility in a native agarose gel-based particle gel assay 35. However, a novel benzamide derivative, designated as BA-53038B, induced the formation of empty capsids with slow electrophoresis mobility. Mechanistic studies demonstrated that like other CpAMs, BA-53038B disrupted pgRNA encapsidation and associated core protein dephosphorylation 36, most likely by binding to the HAP pocket of the dimer-dimer interface. These new CpAMs reported herein provide novel structure scaffolds for the development of core protein-targeted antivirals for the treatment of chronic hepatitis B.
Results
Identification of novel CpAMs
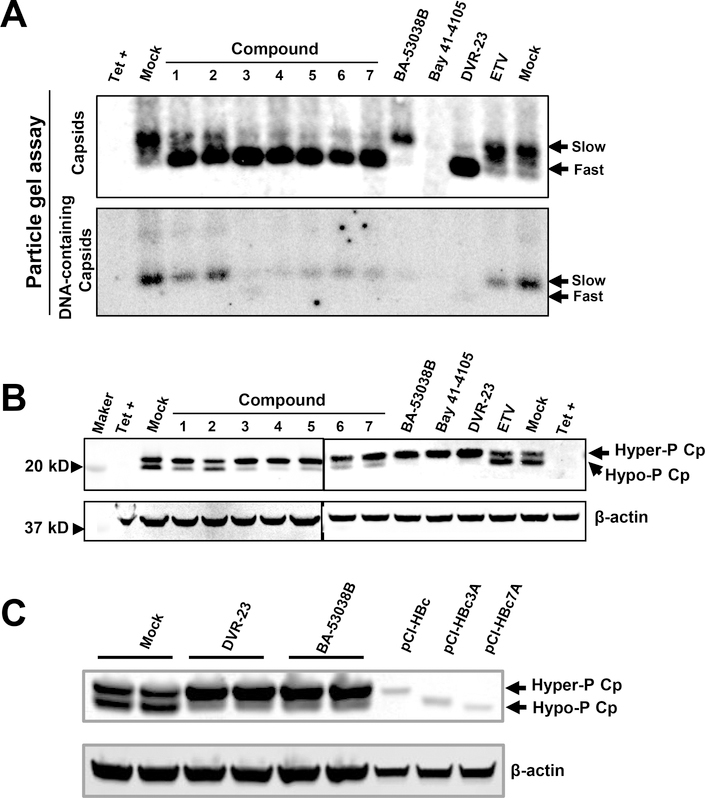
Using an AML12HBV10 cell-based assay described previously 27, 19,920 compounds from an in-house library were tested for their ability to suppress HBV DNA replication. The primary screening identified 89 compounds that reduced HBV core DNA by greater than 60% at 10 μM concentration, compared to the mock treated controls. In order to identify compounds that modulate HBV capsid assembly, taking advantage of our recent finding that CpAMs either induce the decay of mis-assembled core protein aggregates (Type I CpAMs) or assembly of capsids devoid of pgRNA with faster electrophoresis mobility (Type II CpAMs) 28, 35, all the primary “hit” compounds were tested for their effects on capsid mobility in a native agarose gel electrophoresis-based particle gel assay and identified 8 new compounds that altered the capsid electrophoresis mobility. As shown in Fig. 2A, by increasing the agarose concentration from 1% to 1.8%, HBV capsids in mock-treated cells can be separated into two species, a predominantly slow and a minor fast migrating capsid. Despite the reduction of HBV core DNA that co-migrated with the slow migrating capsids, viral DNA polymerase inhibitor entecavir (ETV) did not change the electrophoresis pattern of HBV capsids. Also as expected, Bay 41–4109 treatment induced decay of the core protein and abolished the formation of capsids. However, similar to DVR-23, a SBA chemotype of CpAM 27, treatment of cells with compounds 1 to 7 reduced the amount of capsids with slow migrating rate, but increased the amount of capsids with fast migrating rate. Also similar to DVR-23, those seven compounds reduced the amount of capsid-associated viral DNA (Fig. 2A) and core protein with hypophosphorylation (Fig. 2B). The results suggest that the seven compounds are typical type II CpAMs. However, treatment of cells with compound 8, a novel benzamide derivative and renamed as BA-53038B after re-synthesis, reduced the amount of fast migrating capsids and formed slow migrating capsids. Like other CpAMs, but distinct from viral DNA polymerase inhibitor ETV, BA-53083B treatment reduced the amount of hypophosphorylated core protein (Fig. 2B and C), suggesting the inhibition of viral pgRNA encapsidation 36. Hence, these results indicated that unlike other type II CpAMs, BA-53083B treatment promoted the assembly of empty capsids with slow electrophoresis mobility. The structure, antiviral activity and cytotoxicity of the novel CpAMs discovered in this study are presented in Table 1.
Fig. 2. The novel antiviral compounds induced the capsid/nucleocapsid mobility shift and reduced the amount of hypophosphorylated core protein.
(A and B) AML12HBV10 cells were cultured in the absence of tetracycline (tet) and treated with 5 μM of compounds 1 to 7, BA-53038B, DVR-23 or 2 μM of Bay 41–4109, 1 μM of ETV. Two days post treatment, cytoplasmic HBV capsids were resolved by a 1.8 % agarose gel electrophoresis followed by transferring on to a nitrocellulose membrane. core protein was detected by antibody Dako (A, upper panel). Capsid-associated HBV DNA was detected by using a 32P-labeled riboprobe to detect minus strand HBV DNA. (A). Intracellular core protein was detected with antibody HBc-170A in a Western blot assay. (B). (C) AML12HBV10 cells were cultured in the absence of tet and treated with 5 μM of DVR-23 or BA-53038B for 2 days. Cell lysates from HepG2 cells transfected with plasmids expressing wild-type or mutant core proteins (pCI-HBc-WT, pCI-HBc3A or pCI-HBc7A) were run in parallel to serve as size markers for core protein with different level of phosphorylation. Intracellular core protein was detected with antibody HBc-170A in a Western blot assay. β-actin served as a loading control.
Table 1.
The structure and antiviral activity of novel HBV nucleocapsid assembly inhibitors
| Chemotype | Compound Name | Structure | EC50 (μM) | CC50 (μM) |
|---|---|---|---|---|
| I | Comp. 1 | 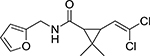 |
6.68 | >10 |
| II | Comp. 2 | 2.77 | >10 | |
| III | Comp. 3 | 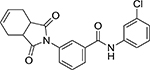 |
0.83 | >10 |
| IV | Comp. 4 | 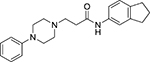 |
0.85 | >10 |
| Comp. 5 | 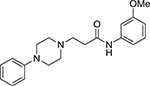 |
1.47 | >10 | |
| V | Comp. 6 | 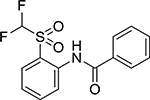 |
2.25 | >10 |
| VI | Comp. 7 | 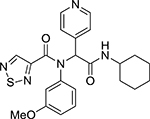 |
2.25 | >10 |
| VII | BA-53038B | 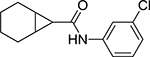 |
3.32 | >100 |
EC50 was determined by dot blot hybridization assay. CC50 was determined by MTT assay.
BA-53038B inhibits HBV nucleocapsid assembly
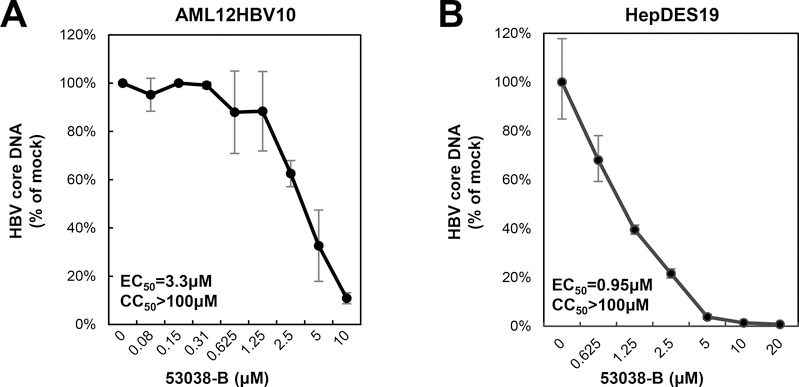
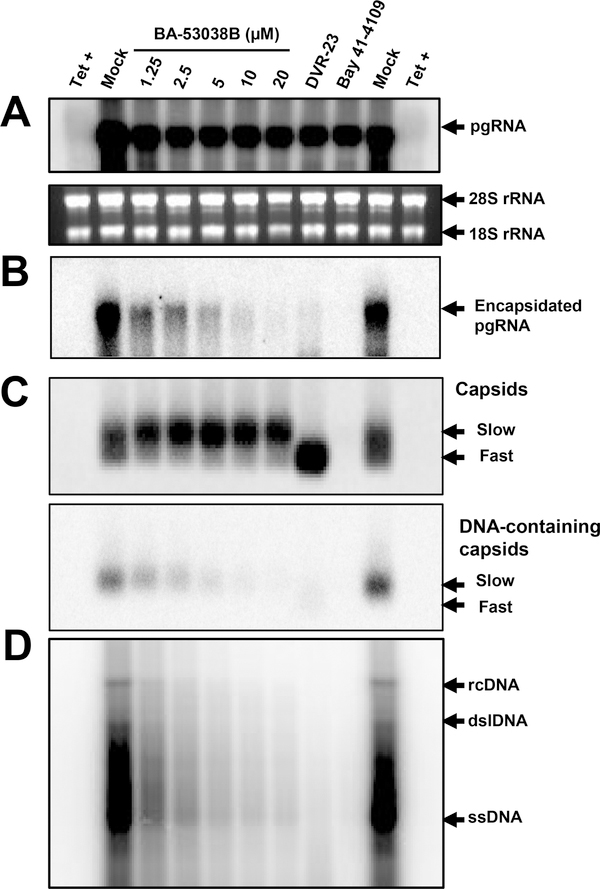
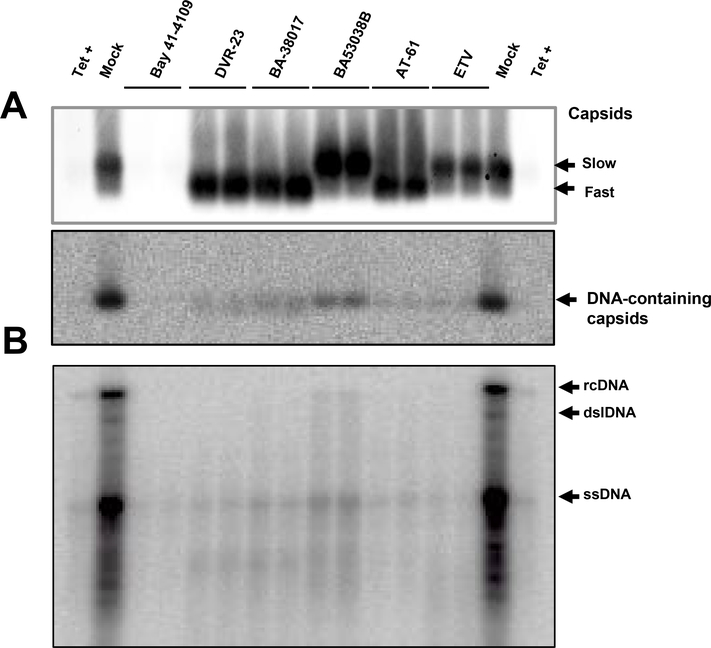
Our recent studies showed that HBV capsid electrophoresis mobility is determined by its surface charge, which is, in turn, determined by the ionization of selected charged amino acid side chains in the core protein assembly domain35. Hence, changes in capsid mobility most likely reflect capsid global structural alteration that affects the exposure and/or ionization of amino acid with charged side chains. We thus speculated that unlike other type II CpAMs, BA-53038B might uniquely interact with HBV core protein and disrupt HBV capsid assembly via distinct mechanisms. To test this hypothesis, we first showed that BA-53038B demonstrated selective antiviral activity against HBV in both AML12HBV10 and HepDES19 cells (Fig. 3). We then determined its effect on every step of the HBV replication cycle from pgRNA production to rcDNA synthesis. In this experiment, Bay 41–4109 and DVR-23, the representative type I and II CpAMs were also included. As shown in Fig. 4, the particle gel assay revealed that Bay41–4109 treatment completely abolished capsid formation (Panel C) and thus pgRNA encapsidation and DNA synthesis (panels B, C and D). On the contrary, DVR-23 treatment induced the formation of fast migrating capsids (panel C), but reduced the amount of encapsidated pgRNA (panel B) and core-associated HBV DNA replication intermediates (panels C and D). These results are in consistent with the reported antiviral mechanism of Bay 41–4109 and DVR-23. Similar to DRV-23, BA-53038B treatment did not affect the levels of pgRNA (panel A), but reduced the amount of encapsidated pgRNA (panel B) as well as core-associated HBV DNA replication intermediates (panels C and D). Interestingly, in agreement with results presented in Fig. 2, BA-53038B treatment promoted the assembly of capsids with slow electrophoresis mobility, and reduced the amount of capsids with fast electrophoresis mobility (panel C, upper). Those findings were further validated in HepDES19 cells with additional type II CpAMs (BA-38017 and AT-61) and viral DNA polymerase inhibitor ETV as positive controls (Fig. 5). Taken together, like other type II CpAMs, BA-53038B inhibited pgRNA encapsidation into nucleocapsids and consequentially suppressed viral DNA replication. However, unlike other type II CpAMs examined thus far, BA-53038B induced the assembly of structurally distinct empty capsids, as indicated by its slower electrophoresis mobility.
Fig. 3. BA-53038B inhibited HBV replication in both AML12HBV10 and HepDES19 cells.
AML12HBV10 (A) and HepDES19 (B) cells were cultured in the absence of tet and treated with BA-53038B for 2 and 6 days, respectively. (A) Cytoplasmic HBV core DNA were quantified by dot blot hybridization. (B) Cytoplasmic HBV core DNA was detected by a real-time PCR assay. Data were plotted as a fraction of that in mock-treated cells (n=3). Cytotoxicity of the compound were determined by a MTT assay.
Fig. 4. BA-53038B inhibited the assembly of pgRNA-containing nucleocapsids.
AML12HBV10 cells were cultured in the absence of tet and treated with the indicated concentrations of BA-53038B, 5 μM of DVR-23, 2 μM of Bay 41–4109 or 1 μM of ETV. Two days post treatment, total HBV RNA (A) and encapsidated pgRNA (B) were detected by Northern blot hybridization with a 32P-labeled riboprobe to detect minus-strand HBV DNA. 28S and 18S rRNA served as loading controls. (C) Cytoplasmic HBV capsids were resolved by a 1.8 % agarose gel electrophoresis followed by detection of capsid with antibody against core protein and HBV DNA, as described in Fig.2A legend. (D) HBV DNA replication intermediates were detected by Southern blot hybridization. rcDNA, dslDNA and ssDNA are relaxed circular, double-stranded linear and single-stranded forms of HBV DNA, respectively.
Fig. 5. BA-38058B induced the formation of distinct HBV capsids in HepG2 cells.
HepDES19 cells were culture in the absence of tetracycline and treated with Bay 41–4109 (2 μM), DVR-23 (5 μM), BA-38017 (5 μM), BA-53038B (5 μM), AT-61 (25 μM) or ETV (1 μM) for 6 days. (A) The total amounts of capsids and the capsid-associated HBV DNA were determined using a particle gel assay in a 1.8 % agarose gel electrophoresis as described in Fig. 2A legend. (B) HBV DNA replication intermediates were determined by Southern blot analysis. rcDNA, relaxed circular DNA. dslDNA, double-stranded linear DNA. ssDNA, single-stranded DNA.
Physical properties of capsids derived from BA-53038B-treated cells
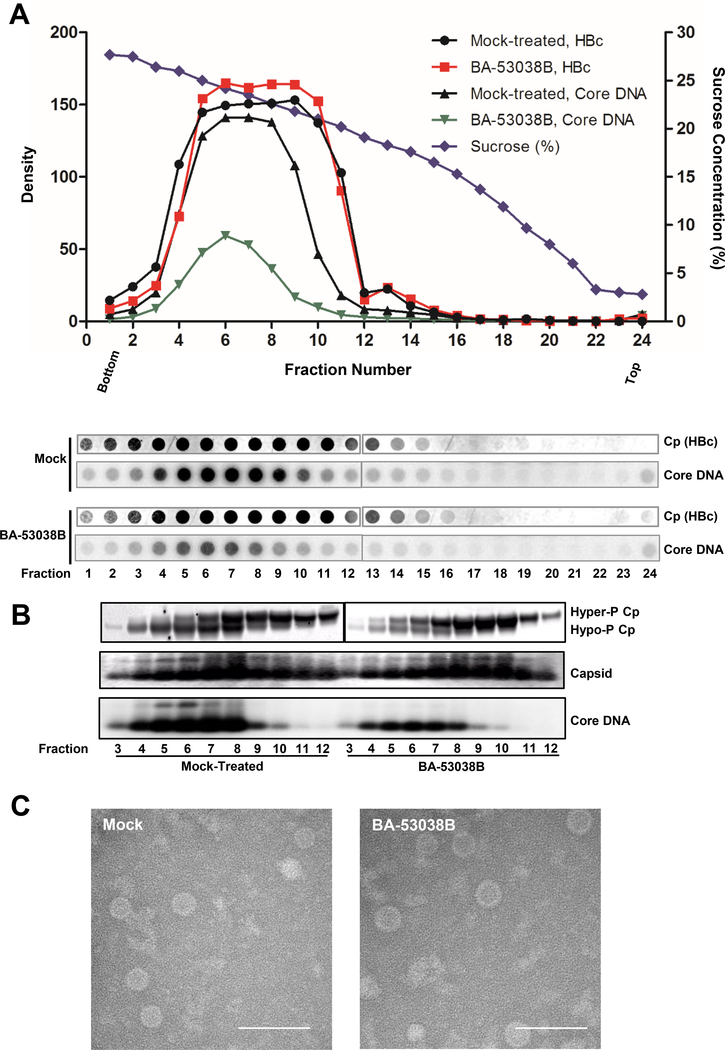
In order to determine the physical properties of HBV capsids assembled in the presence of BA-53038B, the sedimentation velocity of HBV capsids from AML12HBV10 cells with or without BA-53038B treatment were determined. As shown in Fig. 6A, similar to that from mock-treated cells, capsids assembled in the presence of BA-53038B sedimented in a sucrose gradient centrifugation with a flat peak ranging from fractions 5 to 10, and DNA-containing capsids peaking at fraction 6. Although the total amount of capsids was similar in mock-treated and BA-53038B treated cells, the amount of HBV DNA-containing capsids was significantly reduced in BA-53038 treated cells. In agreement with our recent report that core proteins are hyper-phosphorylated in empty capsids, but hypophosphorylated in pgRNA- and DNA-containing capsids 36, BA-53038B treatment inhibited pgRNA encapsidation and consequentially reduced the amount of DNA-containing capsids and hypophosphorylated core protein (Fig. 6B). Moreover, visualization of HBV capsids with electronic microscopy revealed that the capsids from mock-treated and BA-53038B treated AML12HBV10 cells are similar in morphology and size (Fig. 6C). Obviously, more advanced physics technology, such as cryo-EM structure analysis, is required to reveal the structural differences of capsids assembled in the presence of BA-38058B and other CpAMs.
Fig. 6. Physical properties of capsids derived from BA-53038B treated cells.
(A) AML12HBV10 cells were cultured in the absence of tetracycline and treated with 5 μM of BA-53038B. Two days poster treatment, the intracellular capsids were sedimented on a 15% to 30% sucrose. Total of 24fractions were collected from the bottom. 1/30 volume of each fraction was spotted on Nylon membrane for detection of HBV core protein (HBc) with Dako antibody followed by detection of minus strand HBV DNA using HBV riboprobe. (B) HBV capsids from fractions 3–12 were purified by ultracentrifugation and subjected to Western blot to detect core protein with antibody HBc-170A and analyses of capsids and their associated HBV DNA in 1.8 % native agarose gel assay. (C) Electronic microscopic graphs of the capsids purified from AML12HBV10 cells in presence and absence of 5 μM of BA-53038B treatment. Scale bar is 100 nm.
Antiviral activity of BA-53038B may be chirality and/or polarity dependent
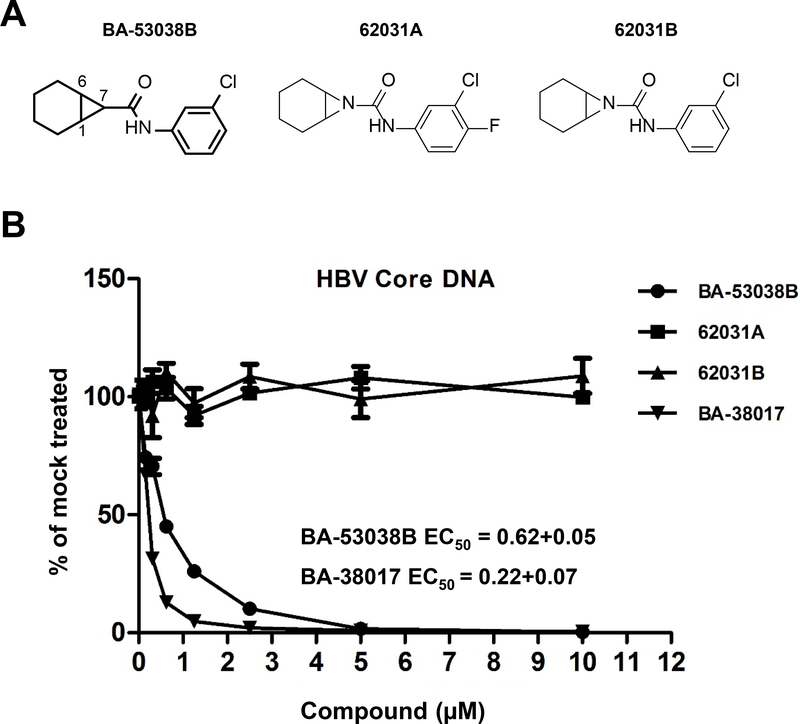
As shown in Fig. 7A, BA-53038B is a fused bicyclo[4.1.0]heptane-7-carboxamide, potentially with exo- and endo- isomers. Because many protein allosteric modulators, including CpAMs, interact with their target proteins in a chirality-dependent manner 37, we intended to separate and determine the antiviral activity of each BA-53038B enantiomer. However, the separation was not successful, we thus synthesized compounds 62031A and 62031B by substitution of the potential chiral center at C7 of BA-53038B with a nitrogen atom to eliminate the chirality at that position. Interestingly, antiviral assay performed in HepAD38 cells indicated that the two new compounds completely lost activity to inhibit HBV replication (Fig. 7B), but BA-53038B and BA-38017 demonstrated similar antiviral activities as observed in HepDES19 cells (Fig. 3) 28. These results imply that not only chirality but also polarity is important for modulators to interact with core protein dimers and disrupt nucleocapsid assembly, since 62031B (ClogP 2.62) is more polar than BA53038B (ClogP 4.15).
Fig. 7. Effects of BA-53038B derivatives on HBV replication in HepAD38 cells.
(A) Structure of BA-53038B and derivatives. (B) HepAD38 cells were cultured in the absence of tetracycline and treated with the indicated concentration of compound for 6 days. Cytoplasmic HBV core DNA was determined by a real-time PCR assay and plotted as a fraction of that in mock-treated cells (n=3).
Genetic evidence suggesting that BA-53038B may interact with core protein at HAP pocket
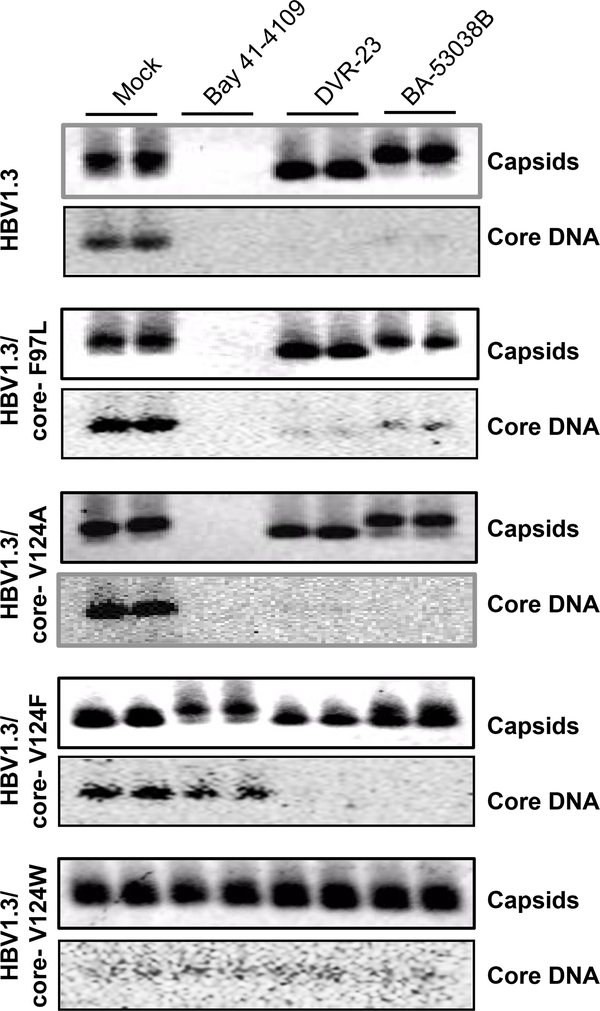
Core protein free dimer is the building block of capsids. Assembly of capsids is driven by hydrophobic interactions at the dimer-dimer interface of core protein. Recent crystal structure and cry-EM studies indicated that CpAMs bind to a hydrophobic pocket, designated as the HAP pocket, at the dimer-dimer interfaces 19. Part of the wall of the HAP pocket is formed by residue V124 of core protein. Mutation at V124, especially substitutions with amino acid residues with larger hydrophobic side chain, results in partially occupation of the HAP pocket and consequentially interferes with the assembly of capsid as well as the interaction between capsid and CpAMs38–39. To investigate whether BA-53038B disrupts capsid assembly and pgRNA encapsidation by binding to the HAP pocket, HepG2 cells transfected with pHBV1.3-derived plasmids with V124A, V124L, V124F or V124W mutations of core protein were used to examine the effect of BA-53038B on the capsid assembly and HBV replication. As a control, a natural occurring core protein mutation F97L, which is located beyond the dimer-dimer interface, was also included 40. As shown in Fig. 8, consistent with our previous report, Bay 41–4109 interfered with the accumulation of capsids and inhibited viral DNA replication in the cells transfected with pHBV1.3-derived plasmids with substitution of F97L or V124A, but not that with V124W. Interestingly, Bay 41–4109 treatment efficiently induced assembly of slower migrating capsids in cells transfected with pHBV1.3-derived plasmid with V124F mutation, without reducing the amount of HBV DNA. However, while DVR-23 and BA-53038B treatment induced the assembly of faster and slower migrating capsids in the cells transfected with pHBV1.3 or pHBV1.3 with F97L or V124A substitution, both compounds failed to alter the mobility of capsids assembled in cells transfected with plasmid with V124F or V124W substitution. Moreover, by using real-time PCR assay to quantify HBV DNA, we have demonstrated that only pHBV1.3/core-V124W supported HBV DNA replication was resistant to both DVR-23 and BA-53038B (Table 2). These experimental data thus suggest that like the SBA derivative DVR-23 and other type II CpAMs, BA-53038B modulates HBV capsid assembly by binding to the HAP pocket.
Fig. 8. Substitution of core protein valine (V) 124 with tryptophan (W) conferred resistant to BA-53038B induced capsid mobility shift and HBV DNA synthesis.
HepG2 cells were transfected with pHBV1.3 or pHBV1.3-derived plasmids expressing core protein with indicated substitution at V124. The cells were treated with 2 μM of Bay 41–4109, 5 μM of DVR-23 or BA-53038B starting at 6 h post transfection. Cytoplasmic capsids were analyzed 3 days post treatment by a particle gel assay. Capsid-associated minus-strand HBV DNA was detected with a 32P-labeled riboprobe.
Table 2.
The effect of BA-53038B on HBV capsid (HepG2 cells transiently transfected)
| Compound | EC50 (μM) | ||||
|---|---|---|---|---|---|
| HBV1.3 | HBV1.3/core-F97L | HBV1.3/core-V124A | HBV1.3/core-V124F | HBV1.3/core-V124W | |
| DVR23 | 0.37 ± 0.002 | 0.45 ± 0.06 | 1.61 ± 0.05 | 1.57 ± 0.15 | >10 |
| BA-53038B | 0.67 ± 0.17 | 2.18 ± 0.81 | 0.40 ± 0.13 | 0.32 ± 0.06 | >10 |
EC50 was determined by real-time PCR assay.
Discussion
The lack of cellular homologues of HBV core protein makes HBV capsid assembly and disassembly highly selective antiviral targets for the inhibition of HBV replication 13. Furthermore, due to targeting distinct viral proteins, CpAMs and viral DNA polymerase inhibitors have no cross resistance to each other’s drug-resistant variants and are thus ideal partners for combination therapy toward more potent or complete inhibition of viral replication, which cannot be achieved by monotherapy with either antiviral agents alone 9, 41. Currently, several type I and type II CpAMs are in clinical trials and demonstrated potent antiviral activity 42–45. It will be interesting to see if complete inhibition of HBV replication for an extended period of time will result in the exhaustion of cccDNA pool and cure of chronic hepatitis B, particularly for the patients with high basal level of antiviral immune response 9.
Extensive biophysical studies showed that although all the CpAMs bind to HAP pocket at the core protein dimer-dimer interface to disrupt capsid assembly, different CpAMs differentially alter core protein dimer assembly and disassembly 31, 46. Type I CpAMs induce core protein dimers to assemble non-capsid aggregates that are either degraded in the cytoplasm or accumulated in PML bodies in HBV infected hepatocytes 24–25. It will be interesting to investigate whether the decay of HBV core protein enhances viral antigen presentation and consequentially, the antiviral immune response. In addition, due to the important role of PML body in HBx protein regulation of cccDNA transcription 47, the possibility that accumulation of core protein aggregates in PML bodies may suppress cccDNA function should also be examined. On the contrary, binding of type II CpAMs at the HAP pocket changes the tertiary structure of the core protein dimer, which results in the assembly of empty capsids with global alteration of quaternary structure 17–18. However, how the structurally diversified type II CpAMs induce the assembly of empty capsids without encapsidation of viral pgRNA and DNA polymerase has not been fully understood. In agreement with our recent report 35, like other type II CpAMs examined thus far, compounds 1 to 7 treatment induced the assembly of capsids with increased electrophoresis mobility (Fig. 2A). The altered electrophoresis mobility is likely due to the change of surface charge of capsids, which is resulted from altered exposure and/or ionization of charged amino acid side chains in the assembly domain of core protein in capsids 35. This finding indicates that in addition to accelerating the kinetics of core protein dimers to assembly empty capsids and indirectly reducing pgRNA encapsidation 48, it is also possible that type II CpAMs induce the assembly of structurally altered nucleocapsids that do not favor the encapsidation of viral pgRNA and DNA polymerase complex 35.
Practically, type II CpAM-induced capsid electrophoresis mobility shift provides a very convenient diagnostic marker for compounds targeting core protein assembly. This assay allowed us to identify CpAMs from the “hits” of cell-based primary screening with great confidence. Moreover, the distinct electrophoresis mobility of empty capsids assembled in the presence of BA-53038B indicates that this compound induces the assembly of capsids with distinct structure feature, and thus is an important chemical probe for dissecting the capsid structure response to different CpAMs.
Conclusions
The novel chemotypes of CpAMs discovered in this screening campaign provide new leads for developing antiviral agents against HBV infection. It is particularly interesting to identify the most active chiral isomer of BA-53038B as a new chemical scaffold of next generation of CpAMs.
Materials and Methods
Cell lines and compounds.
HepG2 cell line was purchased from ATCC. HepAD38 was obtained from Dr. Christoph Seeger (Fox Chase Cancer Center, Philadelphia, USA) 49. HepDES19 and AML12HBV10 cell lines were established as we described previously 34, 50. All three cell lines support HBV pgRNA transcription and DNA replication in a tetracycline (tet)-inducible manner. DVR-23, BA-38017, BA-53038B, 62031A, 62031B and AT-61 were synthesized in-house 28. Entecavir (ETV) is a gift from Dr. William S. Mason (Fox Chase Cancer Center, Philadelphia, USA). Bay41–4109 is a gift of Dr. Lai Wei (Hepatology Institute, People’s Hospital, Peking University, China).
Plasmids.
Wild-type HBV replicon pHBV-1.3, pHBV-1.3-derived plasmids expressing core proteins with substation of F97L, V124A, V124F and V124W were described previously 28. The plasmids pCI-HBc-WT, pCI-HBc-3A and pCI-HBc-7A were reported previously 51.
Screening of compound library from Asinex
AML12HBV10 screening assay in 96-well plate was performed as previously described 27. The 19,920 small molecular compounds were purchased from Asinex and tested at 10 μM concentration. Forty-eight h post treatment, the amounts of HBV DNA in the cell lysates were determined by dot-blot hybridization with α−32P-UTP-labelled riboprobe to detect minus strand HBV DNA. The compounds that reduced HBV DNA by greater than 60% were deemed as primary “hits”.
Determination of antiviral activity of primary “hit” compounds
AML12HBV10 cells were seeded in 96-well plates in the absence of tetracycline and treated with a serial of 2-fold dilution of test compounds, ranging from 10 μM to 0.08 μM. Intracellular HBV core DNA was determined by dot blot hybridization. Quantification of dot blot was used to determine the concentration that reduces the amount of HBV DNA by 50% (EC50). Cytotoxicity of the “hit” compounds was determined by an MTT assay (Sigma) and expressed as the concentration of compound that reduced the viability of the cells by 50% (CC50).
Analysis of cytoplasmic HBV capsids by particle gel electrophoresis and ultracentrifugation.
Cells were lysed with buffer containing 10 mM Tris-HCl (pH 7.6), 100 mM NaCl, 1 mM EDTA and 0.1% NP-40. Cell debris were clarified by centrifugation at 12,000 g for 10 min. followed by electrophoresis through nondenaturing 1.8% agarose gels and transferring to a nitrocellulose filter. HBV capsids were detected with an antibody against HBV core protein (DAKO, Cat. No. B0586) 28. Capsid associated HBV DNA were detected by hybridization with an α−32P-UTP (800Ci/mmol, Perkin Elmer) labeled full-length riboprobe in positive strand polarity28.
Analyses of HBV core DNA and RNA.
Cytoplasmic core DNA from HBV replicating cell lines or replicon plasmid transfected HepG2 cells were extracted as described previously 28. HBV DNA replicative intermediates were analyzed by Southern blot hybridization with an α−32P-UTP labeled positive-stranded riboprobe 28. HBV DNA were quantified by quantitative real-time PCR assay 52. Total cellular RNA was extracted with TRIzol reagent (Invitrogen). Encapsidated HBV RNA was extracted as previously described 34. HBV mRNA and encapsidated HBV RNA were analyzed by Northern blot hybridization with an α−32P-UTP labeled full-length riboprobe in minus strand polarity.
Western blot assay.
Cells were lysed in 1 × LDS loading buffer (Invitrogen). Cell lysate was resolved in a NuPAGE™ 12% Bis-Tris Protein Gel (Invitrogen). HBV core protein was probed with a rabbit polyclonal antibody against peptide spanning the C-terminal aa 170 to 183 of HBV core protein, (GenScript, NJ, USA) 36. β-actin was detected by an antibody from Cell Signaling Technologies.
Electron microscopic analysis of capsids.
HBV capsids were purified from AML12HBV10 cells, with or without treatment with 5 μM of BA-53038B for 2 days, by sucrose gradient centrifugation 53.Electron microscopy analysis was performed after negatively stained with Uranyless (Electron Microscopy Sciences) on a FEI Tecnai 12 Spirit/Biotwin (LaB6 filament), operating at 100kV, with an AMT 2k × 2k CCD camera.
Acknowledgement
This work was supported by grants from the National Institutes of Health, USA (AI134732 and AI113267) and the Office of the Assistant Secretary of Defense for Health Affairs, through the Peer Reviewed Medical Research Program under Award No. W81XWH-17–1-0600. Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the funders.
Abbreviations
- HBV
hepatitis B virus
- HBsAg
HBV surface antigen
- pgRNA
pre-genomic RNA
- rcDNA
relaxed circular DNA
- cccDNA
covalently closed circular DNA
- CpAM
core protein allosteric modulator
- HAP
heteroaryldihydropyrimidine
- SBA
sulfamoylbenzamide
- PPA
phenylpropenamide
- ETV
entecavir
- tet
tetracycline
References
- 1.Mortalit GBD Causes of Death, C., Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015, 385 (9963), 117–71 DOI 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ott JJ; Stevens GA; Groeger J; Wiersma ST, Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine 2012, 30 (12), 2212–9 DOI 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- 3.Dienstag JL, Benefits and risks of nucleoside analog therapy for hepatitis B. Hepatology 2009, 49 (5 Suppl), S112–21 DOI 10.1002/hep.22920. [DOI] [PubMed] [Google Scholar]
- 4.Perrillo R, Benefits and risks of interferon therapy for hepatitis B. Hepatology 2009, 49 (5 Suppl), S103–11 DOI 10.1002/hep.22956 [DOI] [PubMed] [Google Scholar]
- 5.Rehermann B; Ferrari C; Pasquinelli C; Chisari FV, The hepatitis B virus persists for decades after patients’ recovery from acute viral hepatitis despite active maintenance of a cytotoxic T- lymphocyte response. Nat Med 1996, 2 (10), 1104–8. [DOI] [PubMed] [Google Scholar]
- 6.Block TM; Gish R; Guo H; Mehta A; Cuconati A; Thomas London W; Guo JT, Chronic hepatitis B: what should be the goal for new therapies? Antiviral Res 2013, 98 (1), 27–34 DOI 10.1016/j.antiviral.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu J; Cheng J; Tang L; Hu Z; Luo Y; Li Y; Zhou T; Chang J; Guo JT, Virological Basis for the Cure of Chronic Hepatitis B. ACS infectious diseases 2018. DOI 10.1021/acsinfecdis.8b00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang J; Guo F; Zhao X; Guo JT, Therapeutic strategies for a functional cure of chronic hepatitis B virus infection. Acta pharmaceutica Sinica. B 2014, 4 (4), 248–57 DOI 10.1016/j.apsb.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang L; Zhao Q; Wu S; Cheng J; Chang J; Guo JT, The current status and future directions of hepatitis B antiviral drug discovery. Expert opinion on drug discovery 2017, 12 (1), 5–15 DOI 10.1080/17460441.2017.1255195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Clercq E; Li G, Approved Antiviral Drugs over the Past 50 Years. Clin Microbiol Rev 2016, 29 (3), 695–747 DOI 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klumpp K; Crepin T, Capsid proteins of enveloped viruses as antiviral drug targets. Curr Opin Virol 2014, 5, 63–71 DOI 10.1016/j.coviro.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Carnes SK; Sheehan JH; Aiken C, Inhibitors of the HIV-1 capsid, a target of opportunity. Curr Opin HIV AIDS 2018, 13 (4), 359–365 DOI 10.1097/COH.0000000000000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zlotnick A; Venkatakrishnan B; Tan Z; Lewellyn E; Turner W; Francis S, Core protein: A pleiotropic keystone in the HBV lifecycle. Antiviral Res 2015, 121, 82–93 DOI 10.1016/j.antiviral.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole AG, Modulators of HBV capsid assembly as an approach to treating hepatitis B virus infection. Curr Opin Pharmacol 2016, 30, 131–137 DOI 10.1016/j.coph.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Byrd CM; Dai D; Grosenbach DW; Berhanu A; Jones KF; Cardwell KB; Schneider C; Wineinger KA; Page JM; Harver C; Stavale E; Tyavanagimatt S; Stone MA; Bartenschlager R; Scaturro P; Hruby DE; Jordan R, A novel inhibitor of dengue virus replication that targets the capsid protein. Antimicrob Agents Chemother 2013, 57 (1), 15–25 DOI 10.1128/AAC.01429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bourne CR; Finn MG; Zlotnick A, Global structural changes in hepatitis B virus capsids induced by the assembly effector HAP1. J Virol 2006, 80 (22), 11055–61 DOI 10.1128/JVI.00933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katen SP; Tan Z; Chirapu SR; Finn MG; Zlotnick A, Assembly-directed antivirals differentially bind quasiequivalent pockets to modify hepatitis B virus capsid tertiary and quaternary structure. Structure 2013, 21 (8), 1406–16 DOI 10.1016/j.str.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venkatakrishnan B; Katen SP; Francis S; Chirapu S; Finn MG; Zlotnick A, Hepatitis B Virus Capsids Have Diverse Structural Responses to Small-Molecule Ligands Bound to the Heteroaryldihydropyrimidine Pocket. J Virol 2016, 90 (8), 3994–4004 DOI 10.1128/JVI.03058-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klumpp K; Lam AM; Lukacs C; Vogel R; Ren S; Espiritu C; Baydo R; Atkins K; Abendroth J; Liao G; Efimov A; Hartman G; Flores OA, High-resolution crystal structure of a hepatitis B virus replication inhibitor bound to the viral core protein. Proc Natl Acad Sci U S A 2015, 112 (49), 15196–201 DOI 10.1073/pnas.1513803112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Z; Hu T; Zhou X; Wildum S; Garcia-Alcalde F; Xu Z; Wu D; Mao Y; Tian X; Zhou Y; Shen F; Zhang Z; Tang G; Najera I; Yang G; Shen HC; Young JA; Qin N, Heteroaryldihydropyrimidine (HAP) and Sulfamoylbenzamide (SBA) Inhibit Hepatitis B Virus Replication by Different Molecular Mechanisms. Sci Rep 2017, 7, 42374 DOI 10.1038/srep42374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu Z; Lin X; Zhang W; Zhou M; Guo L; Kocer B; Wu G; Zhang Z; Liu H; Shi H; Kou B; Hu T; Hu Y; Huang M; Yan SF; Xu Z; Zhou Z; Qin N; Wang YF; Ren S; Qiu H; Zhang Y; Zhang Y; Wu X; Sun K; Zhong S; Xie J; Ottaviani G; Zhou Y; Zhu L; Tian X; Shi L; Shen F; Mao Y; Zhou X; Gao L; Young JAT; Wu JZ; Yang G; Mayweg AV; Shen HC; Tang G; Zhu W, Discovery and Pre-Clinical Characterization of Third-Generation 4-H Heteroaryldihydropyrimidine (HAP) Analogues as Hepatitis B Virus (HBV) Capsid Inhibitors. J Med Chem 2017, 60 (8), 3352–3371 DOI 10.1021/acs.jmedchem. [DOI] [PubMed] [Google Scholar]
- 22.Qiu Z; Lin X; Zhou M; Liu Y; Zhu W; Chen W; Zhang W; Guo L; Liu H; Wu G; Huang M; Jiang M; Xu Z; Zhou Z; Qin N; Ren S; Qiu H; Zhong S; Zhang Y; Zhang Y; Wu X; Shi L; Shen F; Mao Y; Zhou X; Yang W; Wu JZ; Yang G; Mayweg AV; Shen HC; Tang G, Design and Synthesis of Orally Bioavailable 4-Methyl Heteroaryldihydropyrimidine Based Hepatitis B Virus (HBV) Capsid Inhibitors. J Med Chem 2016, 59 (16), 7651–66 DOI 10.1021/acs.jmedchem.6b00879. [DOI] [PubMed] [Google Scholar]
- 23.Stray SJ; Bourne CR; Punna S; Lewis WG; Finn MG; Zlotnick A, A heteroaryldihydropyrimidine activates and can misdirect hepatitis B virus capsid assembly. Proc Natl Acad Sci U S A 2005, 102 (23), 8138–43 DOI 10.1073/pnas.0409732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deres K; Schroder CH; Paessens A; Goldmann S; Hacker HJ; Weber O; Kramer T; Niewohner U; Pleiss U; Stoltefuss J; Graef E; Koletzki D; Masantschek RN; Reimann A; Jaeger R; Gross R; Beckermann B; Schlemmer KH; Haebich D; Rubsamen-Waigmann H, Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 2003, 299 (5608), 893–6 DOI 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]
- 25.Huber AD; Wolf JJ; Liu D; Gres AT; Tang J; Boschert KN; Puray-Chavez MN; Pineda DL; Laughlin TG; Coonrod EM; Yang Q; Ji J; Kirby KA; Wang Z; Sarafianos SG, The Heteroaryldihydropyrimidine Bay 38–7690 Induces Hepatitis B Virus Core Protein Aggregates Associated with Promyelocytic Leukemia Nuclear Bodies in Infected Cells. mSphere 2018, 3 (2) DOI 10.1128/mSphereDirect.00131-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King RW; Ladner SK; Miller TJ; Zaifert K; Perni RB; Conway SC; Otto MJ, Inhibition of human hepatitis B virus replication by AT-61, a phenylpropenamide derivative, alone and in combination with (−)beta-L- 2’,3’-dideoxy-3’-thiacytidine [published erratum appears in Antimicrob Agents Chemother 1999 Mar;43(3):726]. Antimicrob Agents Chemother 1998, 42 (12), 3179–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campagna MR; Liu F; Mao R; Mills C; Cai D; Guo F; Zhao X; Ye H; Cuconati A; Guo H; Chang J; Xu X; Block TM; Guo JT, Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids. J Virol 2013, 87 (12), 6931–42 DOI 10.1128/JVI.00582-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu S; Zhao Q; Zhang P; Kulp J; Hu L; Hwang N; Zhang J; Block TM; Xu X; Du Y; Chang J; Guo JT, Discovery and mechanistic study of benzamide derivatives that modulate hepatitis B virus capsid assembly. J Virol 2017, 91 (16), e0051917 DOI 10.1128/JVI.00519-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L; Wang YJ; Chen HJ; Shi LP; Tong XK; Zhang YM; Wang GF; Wang WL; Feng CL; He PL; Xu YB; Lu MJ; Tang W; Nan FJ; Zuo JP, Effect of a hepatitis B virus inhibitor, NZ-4, on capsid formation. Antiviral Res 2016, 125, 25–33 DOI 10.1016/j.antiviral.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Wang YJ; Lu D; Xu YB; Xing WQ; Tong XK; Wang GF; Feng CL; He PL; Yang L; Tang W; Hu YH; Zuo JP, A novel pyridazinone derivative inhibits hepatitis B virus replication by inducing genome-free capsid formation. Antimicrob Agents Chemother 2015, 59 (11), 7061–72 DOI 10.1128/AAC.01558-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo F; Zhao Q; Sheraz M; Cheng J; Qi Y; Su Q; Cuconati A; Wei L; Du Y; Li W; Chang J; Guo JT, HBV core protein allosteric modulators differentially alter cccDNA biosynthesis from de novo infection and intracellular amplification pathways. PLoS Pathog 2017, 13 (9), e1006658 DOI 10.1371/journal.ppat.1006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berke JM; Dehertogh P; Vergauwen K; Van Damme E; Mostmans W; Vandyck K; Pauwels F, Capsid Assembly Modulators Have a Dual Mechanism of Action in Primary Human Hepatocytes Infected with Hepatitis B Virus. Antimicrob Agents Chemother 2017, 61 (8) DOI 10.1128/AAC.00560-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lahlali T; Berke JM; Vergauwen K; Foca A; Vandyck K; Pauwels F; Zoulim F; Durantel D, Novel potent capsid assembly modulators regulate multiple steps of the Hepatitis B virus life-cycle. Antimicrob Agents Chemother 2018. DOI 10.1128/AAC.00835-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu C; Guo H; Pan XB; Mao R; Yu W; Xu X; Wei L; Chang J; Block TM; Guo JT, Interferons accelerate decay of replication-competent nucleocapsids of hepatitis B virus. J Virol 2010, 84 (18), 9332–40 DOI 10.1128/JVI.00918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu S; Luo Y; Viswanathan U; Kulp J; Cheng J; Hu Z; Xu Q; Zhou Y; Gong GZ; Chang J; Li Y; Guo JT, CpAMs induce assembly of HBV capsids with altered electrophoresis mobility: Implications for mechanism of inhibiting pgRNA packaging. Antiviral Res 2018, 159, 1–12 DOI 10.1016/j.antiviral.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Q; Hu Z; Cheng J; Wu S; Luo Y; Chang J; Hu J; Guo JT, Hepatitis B Virus Core Protein Dephosphorylation Occurs during Pregenomic RNA Encapsidation. J Virol 2018, 92 (13) DOI 10.1128/JVI.02139-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mani N; Cole AG; Phelps JR; Ardzinski A; Cobarrubias KD; Cuconati A; Dorsey BD; Evangelista E; Fan K; Guo F; Guo H; Guo JT; Harasym TO; Kadhim S; Kultgen SG; Lee ACH; Li AHL; Long Q; Majeski SA; Mao R; McClintock KD; Reid SP; Rijnbrand R; Snead NM; Micolochick Steuer HM; Stever K; Tang S; Wang X; Zhao Q; Sofia MJ, Preclinical Profile of AB-423, an Inhibitor of Hepatitis B Virus Pregenomic RNA Encapsidation. Antimicrob Agents Chemother 2018, 62 (6) DOI 10.1128/AAC.00082-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan Z; Pionek K; Unchwaniwala N; Maguire ML; Loeb DD; Zlotnick A, The interface between hepatitis B virus capsid proteins affects self-assembly, pregenomic RNA packaging, and reverse transcription. J Virol 2015, 89 (6), 3275–84 DOI 10.1128/JVI.03545-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan Z; Maguire ML; Loeb DD; Zlotnick A, Genetically altering the thermodynamics and kinetics of hepatitis B virus capsid assembly has profound effects on virus replication in cell culture. J Virol 2013, 87 (6), 3208–16 DOI 10.1128/JVI.03014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ceres P; Stray SJ; Zlotnick A, Hepatitis B virus capsid assembly is enhanced by naturally occurring mutation F97L. J Virol 2004, 78 (17), 9538–43 DOI 10.1128/JVI.78.17.9538-9543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang P; Liu F; Guo F; Zhao Q; Chang J; Guo JT, Characterization of novel hepadnaviral RNA species accumulated in hepatoma cells treated with viral DNA polymerase inhibitors. Antiviral Res 2016, 131, 40–8 DOI 10.1016/j.antiviral.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang TJ; Block TM; McMahon BJ; Ghany MG; Urban S; Guo JT; Locarnini S; Zoulim F; Chang KM; Lok AS, Present and future therapies of hepatitis B: From discovery to cure. Hepatology 2015, 62 (6), 1893–908 DOI 10.1002/hep.28025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng S; Gao L; Han X; Hu T; Hu Y; Liu H; Thomas AW; Yan Z; Yang S; Young JAT; Yun H; Zhu W; Shen HC, Discovery of Small Molecule Therapeutics for Treatment of Chronic HBV Infection. ACS infectious diseases 2018, 4 (3), 257–277 DOI 10.1021/acsinfecdis.7b00144. [DOI] [PubMed] [Google Scholar]
- 44.Klumpp K; Shimada T; Allweiss L; Volz T; Lutgehetmann M; Hartman G; Flores OA; Lam AM; Dandri M, Efficacy of NVR 3–778, Alone and In Combination With Pegylated Interferon, vs Entecavir In uPA/SCID Mice With Humanized Livers and HBV Infection. Gastroenterology 2018, 154 (3), 652–662 e8 DOI 10.1053/j.gastro.2017. [DOI] [PubMed] [Google Scholar]
- 45.Bray M; Andrei G; Ballana E; Carter K; Durantel D; Gentry B; Janeba Z; Moffat J; Oomen CJ; Tarbet B; Riveira-Munoz E; Este JA; International Society for Antiviral, R., Meeting report: 31(st) International Conference on Antiviral Research. Antiviral Res 2018, 158, 88–102 DOI 10.1016/j.antiviral.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlicksup CJ; Wang JC; Francis S; Venkatakrishnan B; Turner WW; VanNieuwenhze M; Zlotnick A, Hepatitis B virus core protein allosteric modulators can distort and disrupt intact capsids. eLife 2018, 7 DOI 10.7554/eLife.31473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niu C; Li L; Daffis S; Lucifora J; Bonnin M; Maadadi S; Salas E; Chu R; Ramos H; Livingston CM; Beran RK; Garg AV; Balsitis S; Durantel D; Zoulim F; Delaney W. E. t.; Fletcher SP, Toll-Like Receptor 7 Agonist GS-9620 Induces Prolonged Inhibition of HBV via a Type I Interferon-Dependent Mechanism. J Hepatol 2017, 68 (5), 922–931 DOI 10.1016/j.jhep.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 48.Katen SP; Chirapu SR; Finn MG; Zlotnick A, Trapping of hepatitis B virus capsid assembly intermediates by phenylpropenamide assembly accelerators. ACS Chem Biol 2010, 5 (12), 1125–36 DOI 10.1021/cb100275b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ladner SK; Otto MJ; Barker CS; Zaifert K; Wang GH; Guo JT; Seeger C; King RW, Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother 1997, 41 (8), 1715–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo H; Jiang D; Zhou T; Cuconati A; Block TM; Guo JT, Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J Virol 2007, 81 (22), 12472–84 DOI 10.1128/JVI.01123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ludgate L; Liu K; Luckenbaugh L; Streck N; Eng S; Voitenleitner C; Delaney W. E. t.; Hu J, Cell-Free Hepatitis B Virus Capsid Assembly Dependent on the Core Protein C-Terminal Domain and Regulated by Phosphorylation. J Virol 2016, 90 (12), 5830–44 DOI 10.1128/JVI.00394-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo F; Tang L; Shu S; Sehgal M; Sheraz M; Liu B; Zhao Q; Cheng J; Zhao X; Zhou T; Chang J; Guo JT, Activation of STING in hepatocytes suppresses the replication of hepatitis B virus. Antimicrob Agents Chemother 2017, 61 (10), e00771–17 DOI 10.1128/AAC.00771-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo H; Mao R; Block TM; Guo JT, Production and function of the cytoplasmic deproteinized relaxed circular DNA of hepadnaviruses. J Virol 2010, 84 (1), 387–96 DOI 10.1128/JVI.01921-09. [DOI] [PMC free article] [PubMed] [Google Scholar]