Abstract
Both social and genetic factors contribute to cognitive impairment and decline, yet genetic factors identified through genome-wide association studies (GWAS) explain only a small portion of trait variability. This “missing heritability” may be due to rare, potentially functional, genetic variants not assessed by GWAS, as well as gene-by-social factor interactions not explicitly modeled. Gene-by-social factor interactions may also operate differently across race/ethnic groups. We selected 39 genes that had significant, replicated associations with cognition, dementia, and related traits in published GWAS. Using gene-based analysis (SKAT/iSKAT), we tested whether common and/or rare variants were associated with episodic memory performance and decline either alone or through interaction with education in >10,000 European ancestry (EA) and >2,200 African ancestry (AA) respondents from the Health and Retirement Study (HRS). Nine genes in EA and five genes in AA were associated with memory performance or decline (p<0.05), and these effects did not attenuate after adjusting for education. Interaction between education and CLPTM1 on memory performance was significant in AA (p=0.003; FDR-adjusted p=0.038) and nominally significant in EA (p=0.026). In both ethnicities, low memory performance was associated with CLPTM1 genotype (rs10416261) only for those with less than high school education, and effects persisted after adjusting for APOE ε4. For over 70% of gene-by-education interactions across the genome that were at least nominally significant in either ethnic group (p<0.05), genetic effects were only observed for those with less than high school education. These results suggest that genetic effects on memory identified in this study are not mediated by education, but there may be important gene-by-education interactions across the genome, including in the broader APOE genomic region, which operate independently of APOE ε4. This work illustrates the importance of developing theoretical frameworks and methodological approaches for integrating social and genomic data to study cognition across ethnic groups.
Keywords: Cognition, Memory, Rare Variant, Gene-environment Interaction, Education, Epidemiology, Genetics
INTRODUCTION
In 2015, 5.3 million Americans over 70 years of age (22%) were affected with cognitive impairment without dementia, 2 million (8%) with Alzheimer’s Disease, and an additional 1.4 million (5.7%) with vascular dementia (Alzheimer’s Association, 2015). Projections about the shifting demographics of the United States indicate that the number of people over 65 years of age will double between 2010 and 2040 while the number of people over 85 will almost triple (Federal Interagency Forum on Aging-Related Statistics, 2016). This population increase predicts that despite improvements in the rate of cognitive impairment and dementia, cognitive decline will be one of the major defining health concerns of the future, and identifying better predictors of cognitive decline will likely improve prevention and treatment of cognitive disorders (Langa, et al., 2017). Research in often-disparate fields has shown that cognitive impairment and decline are multifactorial and complex. In this study, we combine two areas of cognitive decline research: socioeconomic status (SES) and genetics. With few exceptions, both have remained independent, focusing on the direct effects on cognitive decline as detailed below. However, more recently, research has shown the importance of the interactive nature of genes and SES on explaining variation in cognitive decline.
Better cognitive function, reduced age-related cognitive decline, and lower rates of dementia diagnosis are persistently related to higher SES, specifically educational attainment (Albert, et al., 1995b; Cagney & Lauderdale, 2002; Evans, et al., 1997; Lee, et al., 2003; White, et al., 1994). Education protects against cognitive impairment by reducing the rate of decline and delaying the initiation of that decline (Albert, et al., 1995b; Cagney & Lauderdale, 2002; Christensen, 2001; Evans, et al., 1997; Lee, et al., 2003). Possible mechanistic pathways for the effect of higher education on cognition include brain development and function, changes in health behaviors, and general health advantages of having more wealth and opportunities to reduce risk of dementia and cognitive decline. Another explanation for the effect of education on cognitive functioning is that education may be a marker for environmental experiences that have an effect on cognition and vary with education (Albert, 1995a). In short, education has large, replicated associations with cognitive function which likely work through several mechanisms both biological and social.
The genomics of cognition and cognitive decline, while far newer compared to SES and cognition research, has changed rapidly over the last two decades. Best known are candidate genes such as APOE. Specifically, the APOE ε4 genotype has been shown to be associated with increased risks for poorer functional status (Albert, et al., 1995c) and lower cognitive performance (Bretsky, et al., 2003; Dik, et al., 2001; Fillenbaum, et al., 2001). More recently, genome-wide association studies (GWAS) have identified genotypes associated with episodic memory, memory decline, and related traits such as hippocampal volume and Alzheimer’s Disease (Davies, et al., 2015; Debette, et al., 2015; Ibrahim-Verbaas, et al., 2016; Lambert, et al., 2013). GWAS provide markers of potential nearby causal genetic effects without adjusting for environmental confounders. By utilizing GWAS “hits” (significant, replicated genotype associations), it is now possible to explore genes and genomic regions using statistical tools that jointly test large sets of genotypes, in most cases single nucleotide polymorphisms (SNPs), in order to better understand the underlying biological mechanisms (Lee, et al., 2012; Lee, et al., 2014; Wu, et al., 2011). Gene-based statistical techniques have been successfully used to identify 12 aggregate gene regions associated with cognitive decline by starting with genes identified in GWAS of Alzheimer’s Disease (Nettiksimmons, et al., 2016). The use of gene-based statistical techniques affords the examination of important genomic regions where clusters of small associations potentially indicate biological mechanisms related to cognitive decline. This may be particularly true if genetic variants are rare (i.e. found in less than 1% of the population) – which are rarely examined in GWAS studies. Finally, with the exception of APOE, it should be noted that most of the work on the genetics of cognition is cross-sectional, not population-based, and is conducted in European ancestry samples.
Despite significant achievements in genetic studies of cognition, the size of genetic effects found to date are small and explain only a small percentage (<2%) of overall cognitive variability (Davies, et al., 2015; Debette, et al., 2015). As with other complex traits, it has been hypothesized that some of the lack of explanatory power may be due to rare genetic variants having larger (but unexamined) effects, as well as gene-environment interactions that are not explicitly modeled (Eichler, et al., 2010). Both genes associated with cognition and education may share key mechanisms in influencing cognition such as brain development and function, and thus may interact. Indeed, twin studies have shown evidence for gene-environment interaction on cognitive ability from childhood to middle adulthood (Harden, et al., 2007; Kremen, et al., 2005; Tucker-Drob, et al., 2011), and education may also moderate the association between APOE ε4 and memory decline (McArdle & Prescott, 2010; Seeman, et al., 2005) or dementia-associated brain pathology (Arenaza-Urquijo, et al., 2015). However, few other genes related to cognition and cognitive decline have been tested in any systematic way to examine interactive effects with education.
Here, we utilize GWAS results to indicate genes of interest for further exploration into their effects on longitudinal cognitive decline and their moderation of the effect of education on cognitive decline. By utilizing all available genetic variation within gene regions, we seek to narrow down potential mechanisms, include rare variants, and identify clusters of genetic and gene-by-environment effects. Further, although genotype frequencies can differ by ancestry, genes have identical functions across ancestry. Thus, gene-based analyses should allow for more reasonable analyses across ancestry, even if direct comparisons are difficult. More specifically, we use a gene-based strategy to conduct association analysis of memory performance and memory decline in European ancestry (EA) and African ancestry (AA) respondents from the nationally representative Health and Retirement Study (HRS), using 39 genes known to be associated with memory phenotypes. We provide the first examination of interactions between these gene regions and educational attainment.
METHODS
Study Sample
The HRS, which began in 1992, is a nationally-representative longitudinal panel study of adults over age 50 that assesses metrics of health (including cognition), family, employment, and wealth (Sonnega, et al., 2014). Alternating face-to-face and telephone interviews are conducted biennially, and face-to-face interviews include the collection of biological and physiological measures (Ofstedal, et al., 2005; Sonnega, et al., 2014). This study includes respondents over age 50 that provided saliva samples for DNA extraction in 2006–2010. The analytic samples were comprised of those who completed at least two episodic memory assessments between 1992 and 2014 and who had genetic data (1000 Genomes Project (1000G) imputed data and/or exome chip data).
Measures
Memory Performance and Decline
To assess respondents’ memory performance, we combined measures of immediate and delayed recall. These tasks are sensitive measures of cognitive change (Small, et al., 1999) and have been shown to predict diagnosis of dementia (Crimmins, et al., 2011). After hearing a list of 10 nouns, respondents were asked to recall them. The total number of words recalled immediately and after a five-minute delay of additional test administration, ranging from 0 to 20, comprised the measure of memory performance. Previous principal components factor analysis showed that immediate and delayed recall could be combined, since they loaded onto a single factor (Ofstedal, et al., 2005). Early waves of the study (1992 and 1994) used 20 words instead of 10, so memory performance scores from these waves were normalized to a range of 0 to 20 using score distributions from respondents of similar ages in 1998. In order to minimize the effects of item-level non-response among self-respondents, we used the imputed cognition data released by HRS (Fisher, et al., 2017). For the small percentage of participants reliant on proxy at any given wave who cannot complete the cognitive assessment tasks, the composite recall score was randomly imputed between 0–4 for respondents with dementia (classified for proxy cases based on methods from Crimmins et al., 2011 and/or self-reported diagnosis of Alzheimer’s Disease or dementia in the current or previous waves) and was not imputed for respondents without evidence of dementia.
Educational Attainment
For primary analyses, respondents were characterizing as having a high school education or equivalent (high school degree) vs. having less than a high school degree. In secondary analyses, we also assessed respondents having at least a 4-year college education or equivalent (college degree) vs. having less than a college degree.
Genotype Data
HRS respondents were genotyped using the Illumina HumanOnmi2.5 array and the Illumina HumanExome-12v1 array. Genetic principal components (PCs) were calculated for each chip separately, and the first two PCs and self-reported race were used to select analytic samples of unrelated EA and AA respondents. Ethnicity-specific PCs were then calculated in the EA and AA analytic samples separately. HumanOnmi2.5 genotype data was used to impute genotypes using the 1000 Genomes Project phase I integrated variant set (v3, released March 2012).
A literature search of the NHGRI-EBI GWAS catalog (MacArthur, et al., 2017) was conducted in October, 2016, to identify genome-wide association studies (GWAS) that had at least one autosomal SNP genome-wide significantly associated (p-value < 5×10−8) with cognitive function/decline/impairment, episodic memory, memory function, hippocampal volume, Alzheimer’s Disease, vascular dementia, or closely related phenotypes. A total of 17 studies were selected, and SNPs meeting significance criteria were then selected from these studies (see Table S1 for additional details). For each SNP of interest, we next identified all those that fell within the boundaries of a gene region (including within 5kb from the start or end position of the gene). A total of 39 genes were selected (Table S1). For each gene, we then defined the gene region by selecting all SNPs between the gene start and stop sites, plus a 5kb buffer on either side. The number of SNPs included within a single gene region ranged from 30 to 8,817 for EA 1000G data, 31 to 8,627 for AA 1000G data, 1 to 43 for EA exome chip data, and 1 to 38 for AA exome chip data (Table S2). 1000G data for rs429358 was used to classify respondents as APOE ε4 allele carriers (having at least one copy of the ε4 allele) or non-carriers.
Statistical Analysis
Modeling memory trajectories
We used a series of unconditional mixed models with random effects estimated in MPLUS (Laird & Ware, 1982) to estimate the overall rate of memory change allowing random effects for individual differences from the overall pattern (Bollen & Curran, 2006; Wilson, et al., 2002). This approach accommodates the unbalanced data structure of longitudinal data and has been used successfully in previous HRS studies of cognition (McArdle, et al., 2007; Reitz & Mayeux, 2010). Age was coded as [Age at interview-65] / 10 to be approximately centered. Thus, the intercept represents the average cognitive performance at age 65 and the age coefficient represents the average change in cognitive score with each decade. Models were estimated using the full-information maximum likelihood estimation with an unstructured covariance matrix for the random effects. We compared increasingly complex models including linear, quadratic, and cubic polynomials on age as well as linear spline models in an effort to best model the pattern of memory change with age, and evaluated fit using BIC. The best fitting model included an intercept, a linear age-dependent slope, and a quadratic age slope. Better cognitive function was indicated by a higher (more positive) intercept and higher (less negative) slope. The intercept was slightly correlated with the linear slope (r=0.061) but highly correlated with the quadratic slope (r=−0.996). Thus, we adjusted the linear slope for the intercept in all analyses and did not separately analyze the quadratic slope.
Gene-based associations with memory performance and decline
We tested for gene-based association between each gene and episodic memory performance (trajectory intercept) and decline (trajectory slope) using the sequence kernel association test (SKAT) or the SKAT optimal unified test (SKAT-O) (Lee, et al., 2012; Wu, et al., 2011). SKAT is a score-based variance component test that evaluates the joint effect of multiple genetic variations in a genomic region on an outcome of interest. SKAT assumes that the effect size of each individual SNP in the region follows an arbitrary distribution with mean zero. The test statistic assesses whether the variance of this distribution deviates from zero, testing the hypothesis that at least one SNP in the region is associated with the outcome. The contribution of individual SNPs to the test statistic can be weighted by characteristics including minor allele frequency. SKAT-O performs both the SKAT test as well as a genetic burden test. The burden test is a method that evaluates whether a composite score of the number of minor alleles for the variants in the region is associated with the outcome. Burden tests are optimal when all of the rare variants in the gene have identical effect sizes and directions. Using the SKAT/SKAT-O methods, we were able to evaluate both the effects of all of the SNPs/variants within the entire gene region (including introns and regulatory regions) as well as the effects of the rare, potentially functional variants within the exome.
We performed the analyses separately for memory performance and decline, and separately by ancestry group. In each model, we included sex and the top 4 ancestry-specific genetic PCs to control for population stratification (Model 1). We also adjusted for memory performance (intercept) when modeling memory decline (slope). For analysis of the 1000G data, we used SKAT with an unweighted kernel [Beta(1,1)] to give equal weight to all SNPs/variants regardless of allele frequency (hereby referred to as “all SNPs/variants”). For analysis of the exome chip data, which is comprised primarily of rare, potentially functional variants, we used SKAT-O with a weighted kernel [Beta(1,25)] that dramatically up-weights variants with low minor allele frequencies (“rare variants”)..
To determine whether educational attainment and/or presence of the APOE ε4 allele attenuated the associations between genes with at least nominal significance and memory performance or decline (p<0.05), we further adjusted for education (Model 2), APOE ε4 status (Model 3), or both (Model 4).
Gene-by-educational attainment interactions with memory performance and decline
For each of the 39 genes, we evaluated whether genetic variation interacted with education to influence memory using analogous gene-based tests for gene-by-environment interaction, iSKAT (Lin, et al., 2016) or iSKAT-O. We were interested both in gene-based interactions that were nominally significant (p<0.05) as well as those that retained significance after multiple testing correction. For each set of results from the 39 genes (all SNPs/variants and rare variants, within each ethnic group, for each memory outcome), we calculated the False Discovery Rate (Benjamini & Hochberg, 1995). Results with FDR-adjusted p-value < 0.1 were considered significant after multiple testing correction.
As a follow-up analysis for iSKAT or iSKAT-O (ρ≤0.5) interactions with at least nominal significance (p<0.05), we modeled the interaction between education and each SNP/variant in the region to identify the specific SNPs that were most strongly contributing to the interaction using linear regression with the corresponding adjustment covariates. Results were also visualized using LocusZoom if FDR p<0.1 (Pruim, et al., 2010). For single SNP interactions of interest, we used the ESTIMATE statement in SAS (SAS Institute, Cary, NC) to calculate the effect sizes of SNP genotypes on memory performance/decline for those with and without a high school degree. Similarly, for nominally significant iSKAT-O interactions with ρ>0.5, we used the ESTIMATE statement to calculate the effect sizes of the burden scores separately by education group.
RESULTS
Descriptive statistics are provided in Table 1. For each ethnicity, sample sizes were similar between those with 1000G data and those with exome chip data, as most of the respondents had both. Respondents had an average age of 58 (EA) and 57 (AA) years at baseline, and 58% to 64% of the samples were female. More than half of respondents attained a high school degree (62% and 57% for EA and AA, respectively). Estimated memory performance at age 65 was higher in EA (10.8 ± 0.02) than AA (9.16 ± 0.04), but estimated memory decline was approximately 1.4 words per decade in both groups.
Table 1.
Descriptive statistics of demographics and memory trajectories for HRS respondents with 1000 Genomes Project imputed data and exome chip data
| European Ancestry | African Ancestry | |||
|---|---|---|---|---|
| Variablea | 1000G N=9,914 | Exome N=10,462 | 1000G N=2,223 | Exome N=2,249 |
| Age at baseline (years) | 58.3 (7.9) | 58.4 (7.9) | 56.6 (6.5) | 56.6 (6.5) |
| Gender (0=male, 1=female) | 58% | 58% | 63% | 64% |
| Educational attainment | ||||
| Less than high school degree | 13% | 12% | 30% | 30% |
| High school degree or equivalent | 62% | 62% | 57% | 57% |
| 4-year college degree or equivalent | 25% | 26% | 13% | 13% |
| Dependent Variableb | ||||
| Memory performance (trajectory intercept at age 65) | 10.80 (0.02) | 10.80 (0.02) | 9.16 (0.04) | 9.16 (0.04) |
| Memory decline (trajectory slope per decade) | −1.41 (0.01) | −1.40 (0.01) | −1.42 (0.01) | −1.42 (0.01) |
1000G = sample with 1000 Genomes Project imputed data. Exome = sample with exome chip data.
Mean (standard deviation) or percentage is presented.
Mean (standard error) is presented.
Gene-based associations with memory performance and decline
Of the 39 genes tested, 9 were at least nominally associated (p<0.05) with memory performance or decline in EA (Tables 2, S3, and S4). The most strongly associated genes were APOE and two proximal genes (TOMM40 and APOC1), which were associated with memory performance in analyses with all SNPs/variants, and with memory decline both in analyses using all SNPs/variants as well rare variants only. Rare variant associations were weighted toward SKAT (ρ=0) for APOE and TOMM40, indicating that effect directions and/or magnitudes for the variants in the region were not homogeneous, and toward a burden test (ρ=1) for APOC1, indicating relatively homogenous variant effects (p-values ranged from 0.039 to 1.5×10−14). In addition, using all SNPs/variants, INPP5D and CEACAM16 were associated with memory performance (p=0.0001 and p=0.019, respectively), and PCALM and PVRL2 with memory decline (p=0.002 and p=0.004, respectively). In rare variant analyses, MS4A6E was associated with memory performance (p=0.005, ρ=0.5) and the single SNP in HRK was associated with memory decline (p=0.049).
Table 2:
P-values for gene-based association between genes and memory trajectories (performance and decline) for European ancestry and African ancestry HRS respondents, before and after adjusting for high school education
| Gene | Memory Performance | Memory Decline | ||||||
| All SNPs/variants | Rare variants | All SNPs/variants | Rare variants | |||||
| Model 1 | Model 2 | Model 1 | Model 2 | Model 1 | Model 2 | Model 1 | Model 2 | |
| INPP5D | 0.0001 | 3.0×10−5 | - | - | - | - | - | - |
| MS4A6E | - | - | 0.005 | 0.007 | - | - | - | - |
| PICALM | - | - | - | - | 0.002 | 0.002 | - | - |
| HRK | - | - | - | - | 0.049 | 0.052 | ||
| CEACAM16 | 0.019 | 0.024 | - | - | - | - | - | - |
| PVRL2 | - | - | - | - | 0.004 | 0.004 | - | - |
| TOMM40 | 0.039 | 0.047 | - | - | 2.1×10−8 | 2.1×10−8 | 0.046 | 0.046 |
| APOE | 0.001 | 0.002 | - | - | 2.0×10−11 | 2.0×10−11 | 7.7×10−5 | 7.7×10−5 |
| APOC1 | 0.0002 | 0.0002 | - | - | 1.5×10−14 | 1.5×10−14 | 0.003 | 0.003 |
| Gene | Memory Performance | Memory Decline | ||||||
| All SNPs/variants | Rare variants | All SNPs/variants | Rare variants | |||||
| Model 1 | Model 2 | Model 1 | Model 2 | Model 1 | Model 2 | Model 1 | Model 2 | |
| INPP5D | - | - | 0.013 | 0.010 | - | - | - | - |
| TREM2 | - | - | - | - | - | - | 0.018 | 0.018 |
| MS4A6A | 0.016 | 0.088 | - | - | - | - | - | - |
| MS4A4E | 0.039 | 0.098 | - | - | - | - | - | - |
| PICALM | - | - | 0.024 | 0.020 | - | - | - | - |
| CEACAM16 | 0.031 | 0.031 | - | - | - | - | - | - |
SKAT/SKAT-O was used to model the effect of each gene region separately on memory performance (memory trajectory intercept at age 65) and decline (memory trajectory slope per decade), adjusting for sex and the top 4 ancestry-specific genetic principal components (Model 1). Additional adjustment for high school education was included in Model 2. All models for memory decline (slope) also adjusted for memory performance (intercept). SKAT using the Beta(1,1) weighting was used for 1000G data analyses (all SNPs/variants); SKAT-O using the Beta(1,25) weighting was used for the exome chip data analyses (rare variants).
P-values are reported only for gene-based associations that were significant (p<0.05) in Model 1.
In AA, 5 genes were nominally associated (p<0.05) with memory performance. In all SNP/variant analysis, MS4A6A (p=0.016), MS4A4E (p=0.039), and CEACAM16 (p=0.031) were associated, and in rare variant analysis INPP5D (p=0.013, ρ=0.04) and PICALM (p=0.024, ρ=1) were associated with memory performance. We note that MS4A6A and MS4A4E are within an LD block, so these may be marking a single association with memory performance. One gene, TREM2, was associated with memory decline in rare variant analysis only (p=0.018, ρ=0.5).
After adjusting for education (high school degree), the p-values remained nominally significant (p<0.05) for all of associations reported above except for MS4A64 and MS4A4E with memory performance in EA using all SNPs/variants, which were still suggestively significant (p<0.1) (Table 2). After adjusting for the APOE ε4 allele, associations between genes in the APOE region (PVRL2, TOMM40, APOE, and APOC1) attenuated (Table S5), indicating that associations between variation in these genes and memory was solely due to being in linkage disequilibrium with APOE ε4; however, all other genes retained association with memory. After adjusting for education and APOE ε4 allele simultaneously, no additional associations attenuated (Table S6). Adjusting for college degree gave substantively similar results (data not shown).
Gene-by-educational attainment interactions with memory performance and decline
Of the 39 genes tested, 8 and 9 genes had at least nominally significant interactions (p<0.05) with education (high school degree) on memory performance and/or decline in EA and AA, respectively (Tables 3 and S7). In EA, 3 genes showed an interaction effect with education on memory performance (BCAM and CLPTM1 using all SNPs/variants, and MS4A6E using rare variants weighted toward SKAT (ρ=0)). For memory decline, 2 genes (MS4A4E and MS4A4A) had an education interaction using all SNPs/variants, and 3 genes (TREM2, CLU, SORL1 with ρ=0.5, 1, and 0 respectively) had an education interaction using rare variants. In AA, 5 genes had an interaction effect with education on memory performance (CD2AP, BCL3, and CLPTM1 using all SNPs/variants, and FRMD4A and CEACAM16 using rare variants with burden score weighting (all ρ=1)). For memory decline, 3 genes (EPHA1, PICALM, SLC24A4) had an education interaction using all SNPs/variants, and ABCA7 had an education interaction using rare variants (ρ=1). After applying FDR correction, education had a significant (p=0.003; FDR-adjusted p=0.038) interaction with CLPTM1 on memory performance in AA using all SNPs/variants. The CLPTM1-by-education interaction was also nominally significant in EA (p=0.026) but was not significant after multiple testing correction. Additional adjustment for the APOE ε4 status did not attenuate the CLPTM1-by-education interaction in AA (p=0.004) or EA (p=0.028). No interactions were significant between genes and college degree after multiple testing correction.
Table 3:
P-values for gene-by-high school interactions on memory trajectories (performance and decline) for European ancestry and African ancestry HRS respondents
| Gene | European Ancestry | |||
| Memory Performance | Memory Decline | |||
| All SNPs/variants | Rare variants | All SNPs/variants | Rare variants | |
| TREM2 | - | - | - | 0.003 |
| CLU | - | - | - | 0.042 |
| MS4A4E | - | - | 0.028 | - |
| MS4A4A | - | - | 0.048 | - |
| MS4A6E | - | 0.033 | - | - |
| SORL1 | - | - | - | 0.038 |
| BCAM | 0.011 | - | - | - |
| CLPTM1 | 0.026 | - | - | - |
| Gene | African Ancestry | |||
| Memory Performance | Memory Decline | |||
| All SNPs/variants | Rare variants | All SNPs/variants | Rare variants | |
| CD2AP | 0.040 | - | - | - |
| EPHA1 | - | - | 0.043 | - |
| FRMD4A | - | 0.028 | - | - |
| PICALM | - | - | 0.046 | - |
| SLC24A4 | - | - | 0.013 | - |
| ABCA7 | - | - | - | 0.006 |
| CEACAM16 | - | 0.026 | - | - |
| BCL3 | 0.029 | - | - | - |
| CLPTM1 | 0.003* | - | - | - |
iSKAT/iSKAT-O was used to model the interaction between high school education and each gene region separately on memory performance (memory trajectory intercept at age 65) and decline (memory trajectory slope per decade), adjusting for sex and the top 4 ancestry-specific genetic principal components. Models for memory decline (slope) also adjusted for memory performance (intercept). iSKAT using the Beta(1,1) weighting was used for 1000G data analyses (all SNPs/variants); iSKAT-O using the Beta(1,25) weighting was used for the exome chip data analyses (rare variants).
P-values are reported only for significant gene-by-high school interactions (p<0.05).
Asterisk indicates interactions with FDR p-value<0.1.
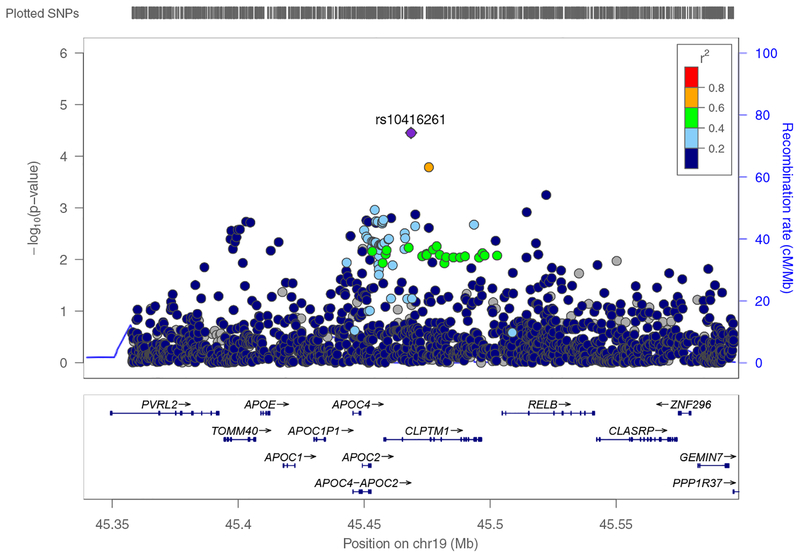
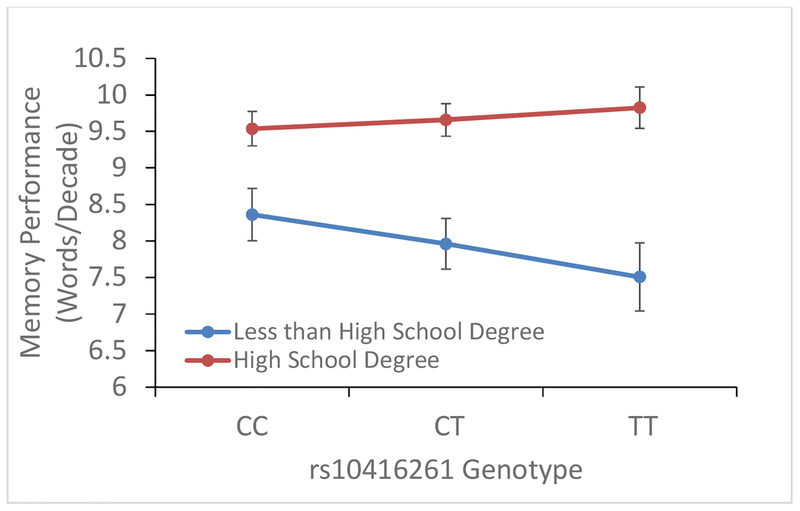
To gain a deeper understanding of the SNPs or rare variants most strongly driving the gene-by-education interactions on memory performance, we modeled each SNP-by-education interaction separately for all SNPs within the gene regions. For CLPTM1, which is in close proximity to APOE, we expanded the single SNP analysis to evaluate a larger region around CLPTM1 (±100kb). A regional plot of SNP-by-education interactions in the region for AA is shown in Figure 1. The interaction signal was restricted to CLPTM1 and the immediately proximal genes (excluding APOE). In AA, the strongest SNP-by-education interaction in CLPTM1 was with rs10416261 (p = 3.5×10−5), with the greatest risk of low memory performance for those with less than high school education who also carry the minor allele (Figure 2). rs10416261 is a common SNP (C/T) with minor allele (T) frequency of 33.3% in AA. Analysis of genetic effects separately by educational group showed that rs10416261 genotype was not associated with memory performance for those with a high school degree (p=0.10). However, for those without a high school degree, each additional copy of T allele was associated with a decrease of 0.43 words recalled at age 65, and this trend was significant (p=1.1×10−4). We also assessed interactions between the top SNP in AA, rs10416261, and education in EA. Interaction analyses in EA were consistent in that there was a non-significant effect of rs10416261 genotype (minor allele frequency = 40.5% in EA) for those with high school education (p=0.51), and a significant effect for those without high school education (p=0.004). However, the effect was in the opposite direction from AA, with each additional copy of the T allele associated with an increase of 0.22 words recalled at age 65 for those without high school education.
Figure 1: LocusZoom plot of p-values for single SNP analysis of CLPTM1-by-high school education interaction on memory performance African ancestry HRS respondents.
Left Y-axis: −log10(p-value) from interactions between high school education and SNPs in CLPTM1 and the surrounding region on memory performance (memory trajectory intercept at age 65), adjusting for sex and the top 4 ancestry-specific genetic principal components. SNPs include those within the gene plus a 100kb buffer on either side. Right Y-axis: SNP recombination rate based on 1000 Genomes Nov2014 AA panel (hg19). X-axis: chromosomal location of genes. r2: degree of linkage disequilibrium between each SNP and rs10416261 (the SNP with the strongest interaction with education in African ancestry respondents; purple diamond).
Figure 2: Interaction between rs10416261 and high school education on memory performance in African ancestry HRS respondents.
Average predicted memory performance (memory trajectory intercept at age 65) for African ancestry respondents by rs10416261 genotype, adjusting for sex and the top 4 ancestry-specific genetic principal components. Black bars indicate 95% confidence intervals for the average predicted memory performance estimates.
The genetic effects of the most significant SNP or the burden score for all interactions that showed at least nominal significance (p<0.05), separately by educational group, are reported in Table S8. Of the 17 interactions evaluated, 12 (71%) showed that genetic effects were significant (p<0.05) for those without high school education, but not for those with high school education. For two of the interactions, genetic effects within educational strata were significant and in opposite directions.
DISCUSSION
To our knowledge, this was the first study to evaluate both common and rare variants in GWAS-identified genes for cognition and related traits in EA and AA, using longitudinal measures of cognition. Using a gene-based strategy, we were able to examine the effects of all of the common variants within the entire gene region (including introns and regulatory regions) as well as the effects of the rare, potentially functional variants within the exome. Although the effects of education on cognitive function likely work through biological as well as social mechanisms, little is known about the interplay between these important social determinants and genetic variation beyond APOE. Here, we provide the first systematic assessment of the interactions between gene regions known to influence cognitive function in adulthood and educational attainment, an extraordinarily strong predictor of cognition across the lifespan. A deeper understanding of the context-dependent genetic effects on cognition in older adulthood may yield insight into the biological etiology underlying cognitive decline and dementia, provide opportunities for more effectively identifying at-risk population subgroups in order to help alleviate disparities in cognitive health, and help formulate more effective biological as well as social interventions.
This study has several other notable strengths. First, by using gene-based approaches (SKAT/iSKAT), we were able to test all genetic polymorphisms within the gene simultaneously, substantially reducing multiple testing burden. SKAT and iSKAT also allow for testing of rare variants by simultaneously assessing clusters of SNPs, which greatly enhances power (Lee, et al., 2014). These methods may also be better suited for cross-ethnic comparisons than single SNP analyses because differences in SNP correlations (linkage disequilibrium) across ethnicities do not hamper the ability to detect associations within ethnic group (Ware, et al., 2016). Second, examining memory function as well as decline allows for the identification of differential genetic relationships and interactions for each trait. Third, the genes selection for examination had significant and replicated evidence of association with cognition-related traits in large meta-analyses. This is in contrast to candidate gene approaches that rely on a priori knowledge about biological mechanisms underlying the disease. A fourth strength is that we examined genes that are associated with memory as well as a broader set of traits related to cognition including Alzheimer’s Disease, hippocampal volume, and vascular dementia. This is based on the hypothesis that the etiology of dementia may be difficult to differentiate given the overlap in symptom presentation (Karantzoulis & Galvin, 2011).
Briefly, we found that approximately 1/3 of the genes previously associated with cognition and related traits were at least nominally associated (p<0.05) with memory performance and/or decline in the HRS EA sample. A smaller number of genes showed significant effects in AA, and associated genes did not substantially overlap with those found in EA. Differences between findings in EA and AA may be due to power restrictions, the EA-centric design of the exome chip array which captures a larger proportion of rare variants in EA than AA, the selection of genes based primarily on EA GWAS, or because the genes have context-dependent effects (gene-by-environment interactions) such that the genetic effects are only operating in certain environmental contexts that also differ by ethnic group (Boardman, et al., 2013).
We also observed little overlap in the genes associated with memory performance and decline, with the exception of genes in the APOE region. This may be due to different genetic underpinnings of cognitive development during childhood and early adulthood, and potential disease-related cognitive decline at the end of life. Within these gene regions, there was evidence for effects using equal weighting of all SNPs/variants (1000G data) as well as upweighting of rare variants (exome chip data). In general, we found more associations using equal weighting of all variants, possibly because these genes were detected through GWAS which are optimized for detecting common variants. For significant associations using rare variants, SKAT-O weighted toward the burden test (ρ=1) more often than the SKAT test, indicating that the measured rare variants are more likely to have similar magnitudes and effect directions (i.e., multiple rare variants with detrimental effects).
To our knowledge, three studies have utilized SKAT to examine gene-based associations for cognitive traits. In a family-based cohort of 550 participants (mixed ethnicities) enriched for type 2 diabetes, SKAT was used on a genome-wide level to identify genes associated at p<4.7×10−6 with a range of cognitive functions in a cross-sectional analysis (Cox, et al., 2014). Although this study identified 7 genes, it failed to replicate 31 pre-selected cognition-associated SNPs, suggesting possible genetic heterogeneity in the etiology underlying cognitive function in this cohort compared to previous studies. In accordance with this, none of the 7 identified genes overlapped with the known cognition-associated genes explored in our study. Recently, SKAT was used to examine the association between previously identified genes for Alzheimer’s Disease and longitudinal cognitive decline in two single-sex cohorts (N=15,000 Caucasians) aged 65 years or older (Nettiksimmons, et al., 2016). They identified 12 genes associated with cognitive decline, 10 of which were also evaluated in our study. Of these, PICALM was significantly associated with memory decline in EA, with memory performance in AA, and had an interaction with education on memory decline in AA. MS4A6E was associated with memory performance and had an interaction with education on memory performance in EA. In addition, 3 genes (ABCA7, SLC24A4, SORL1) had a nominally significant interaction with education on memory performance/decline (p<0.05), and 3 genes (BIN1, CELF1, CR1) were suggestively associated with memory performance/decline (p<0.1) in at least one ethnic group in our study, suggesting that we were able to replicate some of the previous findings implicating Alzheimer’s Disease-associated genes in longitudinal assessment of cognitive decline. The final study applied SKAT to exome sequencing data collected from over 2,500 participants in a single gene (DMD) and cognition (Vojinovic, et al., 2015). This gene was not among those that we evaluated.
Genetic influences on cognition may be mediated by educational attainment, or may operate independently of education. In our study, adjusting for educational attainment did not substantively influence the associations for the majority of genes, indicating that these genes do not influence cognition primarily through educational attainment in this cohort. This is consistent with some studies (Cox, et al., 2014), though GWAS-identified genes for cognition and educational attainment do implicate some shared genetic influences (Davies, et al., 2015). We also examined whether the most well-known genetic risk factor for Alzheimer’s Disease, APOE ε4, was a driver of the genetic associations detected. Adjusting for ε4 attenuated the gene-based associations for genes that share a linkage disequilibrium (LD) block with APOE (PVRL2, TOMM40, APOC1); however, it did not substantively influence the associations in other genes. Thus, ε4 is the dominating factor associated with cognition in the proximal gene region, but does not influence associations with other genes.
We found CLPTM1 and education had a statistically significant interaction on memory performance in AA after accounting for multiple testing. Cleft lip and palate transmembrane protein 1 (CLPTM1) was identified as being disrupted in a family with cleft lip/palate (Yoshiura, et al., 1998), which may be associated with lower cognitive function (Roberts, et al., 2012). CLPTM1 may also play a role in cellular differentiation and T-cell development (Apweiler, et al., 2004; Roberts, et al., 2012), and overexpression of CLPTM1 has been associated with cancer risk (Rossi, et al., 2005). CLPTM1 is located on chromosome 19 adjacent to the APOE gene region. Analysis of chromatin conformation data hosted by Hi-C Unifying Genomic Interrogator shows a strong chromatin interaction between a 40kb region containing the most significant SNP in CLPTM1 (rs10416261) and the APOE gene region in multiple human tissues and cell lines (Martin, et al., 2017). Since adjustment for APOE ε4 did not attenuate this interaction, and since we did not detect an interaction between APOE and education, it is unlikely APOE SNPs/variants are driving the interaction. This indicates that SNPs within CLPTM1 may have regulatory effects on expression of APOE and/or surrounding genes due to chromatin level interactions. While some studies have suggested that high education may buffer the negative effect of APOE ε4 on dementia (Wang, et al., 2012), other studies have found that the negative effect of APOE ε4 on cognitive decline is stronger in those with higher education (Seeman, et al., 2005), and still other studies have shown inconsistent relationships (Ishioka, et al., 2016) or no interaction between education and APOE ε4 on dementia or cognitive decline (Hsiung, et al., 2004). The cumulative evidence suggests that interaction between education and genetic variations in the larger APOE gene region is likely to exist, but that interaction(s) may involve multiple genes and/or be inconsistent across populations.
Consistently in both AA and EA, CLPTM1 genotypes were associated with memory performance only in those without a high school education. However, the effect sizes were relatively small, with the most strongly associated SNP in AA conferring a per-decade loss of approximately 0.5 words for each copy of the minor allele. In addition, the effect of the top AA SNP was associated with increased memory performance in EA. This may be due to underlying differences in the LD structure of AA and EA. That is, the SNPs that showed interaction with education were not themselves causal, but were correlated with a set of causal (likely rare) variants that differed across ethnic groups. It may be possible that these causal variants were not captured on the exome chip, potentially explaining why this gene did not interact with education in the rare variants analyses that we performed.
We identified additional gene-by-high school education interactions that had at least nominal significance in EA and AA (p<0.05), but these interactions did not reach statistical significance after FDR correction. While it is critical to replicate these findings in independent samples, together they indicate that genetic effects on memory performance and decline may be operating more strongly in those without high school education. Inconsistency across ethnic groups may be a consequence of the same level of educational attainment having differential effects on cognition for EA compared to AA (Barnes, et al., 2011). While other studies have observed stronger effects of education on cognitive performance than decline (Wilson, et al., 2009), our study found a similar number of nominally significant gene-by-education interactions on performance and decline. However, the only interaction significant after FDR correction was for cognitive performance. We did not observe any gene-by-college education interactions that were significant after multiple testing correction. This may be because high school education is the strongest educational driver of dementia prevalence rates in the HRS (Langa, et al. 2017).
Our study is not without limitations. First, we assessed episodic memory, which is only one aspect of cognition. Associations with the examined genes may be different by cognitive domains. Further, we combined immediate and delayed recall into a single summary score in order to obtain better (less noisy) estimates of memory. Given this approach, we cannot determine whether observed genetic effects are driven by immediate or delayed recall, and future studies could separate the two measures and/or evaluate the residual of delayed recall after adjusting for immediate recall (for example, see Arpawong, et al., 2017). Separating the measures may also help to elucidate the biological mechanisms underlying normative cognitive aging versus disease etiology (Wolk, et al., 2011). Second, our use of gene-based methods required using a two-step process to first model cognitive trajectories and next assess the relationship between genetics and features of the trajectories; we were not able to directly model the genetic effects with the raw repeated measures data. Third, we evaluated interactions with education, which is only one aspect of SES and may differ in its relationship to SES across ethnic groups. Further, we evaluated only limited aspects of education – having a high school degree or a college degree – and due to the limitations of the SKAT methodology, were not able to evaluate education as a multi-level variable (for example, including both high school and college degree simultaneously). Fourth, although we detect statistical gene-by-education interactions, we do not have the means to examine the biological mechanisms underlying these interactions. We observe that for the majority of interactions, genetic effects are present only in those without high school education, corresponding to a stress diathesis model (Boardman, et al., 2013). This may be due to increased vulnerability to genetics as a consequence of exposure differences (i.e., social stressors, psychosocial factors, environmental toxicants), behavioral differences, reduced access to health care, or other factors. Future studies are needed to characterize the mechanisms of these interactions. Fifth, our sample is nationally representative; however there is selective mortality at older ages (Domingue et al., 2017). Also, since older individuals both with and without cognitive deficits are included in the analysis, it is possible that those with cognitive deficits are driving some of the identified associations, particularly since some genetic associations tend to be stronger with age (Lindenberger, et al., 2008). Sixth, while we examined genetic effects in multiple ethnic groups, the genes that have been identified to date are from GWAS consisting of primarily EA samples. Thus, using both common and rare variants, we found that the majority of genes had weak or no association with memory performance or decline in the AA sample, while a larger number of genes had detectable effects in the EA sample. This may also be attributable to more limited power in AA due to reduced sample size compared to EA. Inconsistent findings across analyses (all SNPs/variants vs. rare variants), both for gene-based associations and interactions, may also be partially attributable to reduced power. Finally, although we used standard variant weightings for SKAT (Wu, et al., 2011), our results may have changed under different weighting schemes.
CONCLUSION
Our study indicates that genetic effects on memory function primarily independently of educational attainment, but that there may be important interactions between education and genes in the broader APOE genomic region that are not limited to the APOE ε4 allele, as well as elsewhere across the genome. This work illustrates the importance of considering trajectories of behavioral data in genetic research, delving deeper into genetic regions discovered in GWAS to explain missing heritability, and developing theoretical frameworks and methodological approaches for integrating social and genomic data to study cognition in across ethnic groups.
Supplementary Material
Investigates genetic effects on longitudinal trajectories of memory performance
Evaluates whether education modifies genetic effects on memory trajectories
Genetic effects on memory were not mediated by educational attainment
Education may offset genetic influences on memory performance and decline
Acknowledgements:
This work was supported by the National Institute on Aging (R03 AG048806). The Health and Retirement Study (HRS) is supported by the National Institute on Aging (U01 AG009740). HRS genotyping was funded separately by the National Institute on Aging (RC2 AG036495, RC4 AG039029) and was conducted by the NIH Center for Inherited Disease Research (CIDR) at Johns Hopkins University. Genotyping quality control and final preparation of the genotype data were performed by the University of Michigan School of Public Health and the Genetics Coordinating Center at the University of Washington.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jennifer A. Smith, Email: smjenn@umich.edu.
Minjung Kho, Email: mjkho@umich.edu.
Wei Zhao, Email: zhaowei@umich.edu.
Miao Yu, Email: miayu@umich.edu.
Colter Mitchell, Email: cmsm@umich.edu.
Jessica D. Faul, Email: jfaul@umich.edu.
REFERENCES
- Albert MS (1995a). How does education affect cognitive function?. Ann Epidemiol, 5(1), 76–78. [DOI] [PubMed] [Google Scholar]
- Albert MS, Jones K, Savage CR, Berkman L, Seeman T, Blazer D et al. (1995b). Predictors of cognitive change in older persons: MacArthur studies of successful aging. Psychol Aging, 10(4), 578–589. [DOI] [PubMed] [Google Scholar]
- Albert SM, Gurland B, Maestre G, Jacobs DM, Stern Y, & Mayeux R (1995c). APOE genotype influences functional status among elderly without dementia. Am J Med Genet 60(6), 583–587. [DOI] [PubMed] [Google Scholar]
- Alzheimer’s Association. (2015). 2015 Alzheimer’s disease facts and figures. Alz Dement, 11(3), 332–384. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S et al. (2004). UniProt: The universal protein knowledgebase. Nucleic Acids Res, 32, D115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenaza-Urquijo EM, Gonneaud J, Fouquet M, Perrotin A, Mezenge F, Landeau B et al. (2015). Interaction between years of education and APOE epsilon4 status on frontal and temporal metabolism. Neurology, 85(16), 1392–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpawong TE, McArdle JJ, Gatz M, Pendleton N, Mekli K, Armoskus C, et al. (2017). Genetic variant specific to aging-related verbal memory: Insights from GWASs in a population-based cohort. PloS One, 12(8):e0182448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes LL, Wilson RS, Hebert LE, Scherr PA, Evans DA, & Mendes de Leon CF (2011). Racial differences in the association of education with physical and cognitive function in older blacks and whites. J Gerontol B Psychol Sci Soc Sci, 66(3), 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, & Hochberg Y (1995). Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc B Met, 57, 289–300. [Google Scholar]
- Boardman JD, Daw J, & Freese J (2013). Defining the environment in gene–environment research: lessons from social epidemiology. Am J Public Health, 103(S1), S64–S72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen KA, & Curran PJ (2006). Latent Curve Models: A Structural Equation Perspective, 467. [Google Scholar]
- Bretsky P, Guralnik JM, Launer L, Albert M, Seeman TE, & MacArthur Studies of Successful Aging. (2003). The role of APOE-epsilon4 in longitudinal cognitive decline: MacArthur studies of successful aging. Neurology, 60(7), 1077–1081. [DOI] [PubMed] [Google Scholar]
- Cagney KA, & Lauderdale DS (2002). Education, wealth, and cognitive function in later life. J Gerontol B Psychol Sci Soc Sci, 57(2), P163–72. [DOI] [PubMed] [Google Scholar]
- Christensen H (2001). What cognitive changes can be expected with normal ageing? Aust N Z J Psychiatry, 35(6), 768–775. [DOI] [PubMed] [Google Scholar]
- Cox AJ, Hugenschmidt CE, Raffield LM, Langefeld CD, Freedman BI, Williamson JD et al. (2014). Heritability and genetic association analysis of cognition in the diabetes heart study. Neurobiol Aging, 35(8), 1958.e3–1958.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimmins EM, Kim JK, Langa KM, & Weir DR (2011). Assessment of cognition using surveys and neuropsychological assessment: The health and retirement study and the aging, demographics, and memory study. J Gerontol B Psychol Sci Soc Sci, 66 Suppl 1, i162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Armstrong N, Bis JC, Bressler J, Chouraki V, Giddaluru S et al. (2015). Genetic contributions to variation in general cognitive function: A meta-analysis of genome-wide association studies in the CHARGE consortium (N=53949). Mol Psychiatry, 20(2), 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debette S, Ibrahim Verbaas CA, Bressler J, Schuur M, Smith A, Bis JC et al. (2015). Genome-wide studies of verbal declarative memory in nondemented older people: The cohorts for heart and aging research in genomic epidemiology consortium. Biol Psychiatry, 77(8), 749–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dik MG, Jonker C, Comijs HC, Bouter LM, Twisk JW, van Kamp GJ et al. (2001). Memory complaints and APOE-epsilon4 accelerate cognitive decline in cognitively normal elderly. Neurology, 57(12), 2217–2222. [DOI] [PubMed] [Google Scholar]
- Domingue BW, Belsky DW, Harrati A, Conley D, Weir DR, & Boardman JD (2017). Mortality selection in a genetic sample and implications for association studies. Int J Epidemiol, 46(4), 1285–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH et al. (2010). Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet, 11(6), 446–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DA, Hebert LE, Beckett LA, Scherr PA, Albert MS, Chown MJ et al. (1997). Education and other measures of socioeconomic status and risk of incident alzheimer disease in a defined population of older persons. Arch Neurol, 54(11), 1399–1405. [DOI] [PubMed] [Google Scholar]
- Federal Interagency Forum on Aging-Related Statistics. (2016). Older americans 2016 key indicators of well-being. Washington, DC: U.S. Government Printing Office. [Google Scholar]
- Fillenbaum GG, Landerman LR, Blazer DG, Saunders AM, Harris TB, & Launer LJ (2001). The relationship of APOE genotype to cognitive functioning in older african-american and caucasian community residents. J Am Geriatr Soc, 49(9), 1148–1155. [DOI] [PubMed] [Google Scholar]
- Fisher GG, Hassan H, Faul JD, Rodgers WL, & Weir D (2017). Health and retirement study imputation of cognitive functioning measures: 1992 – 2014. University of Michigan, Ann Arbor, MI. [Google Scholar]
- Harden KP, Turkheimer E, & Loehlin JC (2007). Genotype by environment interaction in adolescents’ cognitive aptitude. Behav Genet, 37(2), 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung GY, Sadovnick AD, & Feldman H (2004). Apolipoprotein E epsilon4 genotype as a risk factor for cognitive decline and dementia: Data from the canadian study of health and aging. CMAJ, 171(8), 863–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim-Verbaas CA, Bressler J, Debette S, Schuur M, Smith AV, Bis JC et al. (2016). GWAS for executive function and processing speed suggests involvement of the CADM2 gene. Mol Psychiatry, 21(2), 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishioka YL, Gondo Y, Fuku N, Inagaki H, Masui Y, Takayama M et al. (2016). Effects of the APOE epsilon4 allele and education on cognitive function in japanese centenarians. Age, 38(5–6), 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantzoulis S, & Galvin JE (2011). Distinguishing alzheimer’s disease from other major forms of dementia. Expert Rev Neurother, 11(11), 1579–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremen WS, Jacobson KC, Xian H, Eisen SA, Waterman B, Toomey R et al. (2005). Heritability of word recognition in middle-aged men varies as a function of parental education. Behav Genet, 35(4), 417–433. [DOI] [PubMed] [Google Scholar]
- Laird NM, & Ware JH (1982). Random-effects models for longitudinal data. Biometrics, 38(4), 963–974. [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C et al. (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for alzheimer’s disease. Nat Genet, 45(12), 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langa KM, Larson EB, Crimmins EM, Faul JD, Levine DA, Kabeto MU et al. (2017). A comparison of the prevalence of dementia in the United States in 2000 and 2012. JAMA Intern Med, 177(1), 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Abecasis GR, Boehnke M, & Lin X (2014). Rare-variant association analysis: Study designs and statistical tests. Am J Hum Genet, 95(1), 5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA et al. (2012). Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet, 91(2), 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kawachi I, Berkman LF, & Grodstein F (2003). Education, other socioeconomic indicators, and cognitive function. Am J Epidemiol, 157(8), 712–720. [DOI] [PubMed] [Google Scholar]
- Lin X, Lee S, Wu MC, Wang C, Chen H, Li Z et al. (2016). Test for rare variants by environment interactions in sequencing association studies. Biometrics, 72(1), 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenberger U, Nagel IE, Chicherio C, Li S-C, Heekeren HR, Bäckman L (2008). Age-related decline in brain resources modulates genetic effects on cognitive functioning. Front Neurosci. 2(39), 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E et al. (2017). The new NHGRI-EBI catalog of published genome-wide association studies (GWAS catalog). Nucleic Acids Res, 45(D1), D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JS, Xu Z, Reiner AP, Mohlke KL, Sullivan P, Ren B et al. (2017). HUGIn: Hi-C unifying genomic interrogator. Bioinformatics, 33(23), 3793–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle JJ, Fisher GG, & Kadlec KM (2007). Latent variable analyses of age trends of cognition in the health and retirement study, 1992–2004. Psychol Aging, 22(3), 525–545. [DOI] [PubMed] [Google Scholar]
- McArdle JJ, & Prescott CA (2010). Contemporary modeling of gene × environment effects in randomized multivariate longitudinal studies. Perspect Psychol Sci, 5(5), 606–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettiksimmons J, Tranah G, Evans DS, Yokoyama JS, & Yaffe K (2016). Gene-based aggregate SNP associations between candidate AD genes and cognitive decline. Age, 38(2), 41-016-9885-2. Epub 2016 Mar 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofstedal M, Fisher G, & Herzog A (2005). Documentation of cognitive functioning measures in the health and retirement study HRS documentation report (No. DR-006). University of Michigan, Ann Arbor, MI. [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP et al. (2010). LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics, 26(18), 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz C, & Mayeux R (2010). Use of genetic variation as biomarkers for mild cognitive impairment and progression of mild cognitive impairment to dementia. J Alzheimer’s Dis, 19(1), 229–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RM, Mathias JL, & Wheaton P (2012). Cognitive functioning in children and adults with nonsyndromal cleft lip and/or palate: A meta-analysis. J Pediatr Psychol, 37(7), 786–797. [DOI] [PubMed] [Google Scholar]
- Rossi MR, Hawthorn L, Platt J, Burkhardt T, Cowell JK, & Ionov Y (2005). Identification of inactivating mutations in the JAK1, SYNJ2, and CLPTM1 genes in prostate cancer cells using inhibition of nonsense-mediated decay and microarray analysis. Cancer Genet Cytogenet, 161(2), 97–103. [DOI] [PubMed] [Google Scholar]
- Seeman TE, Huang MH, Bretsky P, Crimmins E, Launer L, & Guralnik JM (2005). Education and APOE-e4 in longitudinal cognitive decline: MacArthur studies of successful aging. J Gerontol B Psychol Sci Soc Sci, 60(2), P74–83. [DOI] [PubMed] [Google Scholar]
- Small SA, Stern Y, Tang M, & Mayeux R (1999). Selective decline in memory function among healthy elderly. Neurology, 52(7), 1392–1396. [DOI] [PubMed] [Google Scholar]
- Sonnega A, Faul JD, Ofstedal MB, Langa KM, Phillips JW, & Weir DR (2014). Cohort profile: The health and retirement study (HRS). Int J Epidemiol, 43(2), 576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker-Drob EM, Rhemtulla M, Harden KP, Turkheimer E, & Fask D (2011). Emergence of a gene × socioeconomic status interaction on infant mental ability between 10 months and 2 years. Psychol Sci, 22(1), 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojinovic D, Adams HH, van der Lee SJ, Ibrahim-Verbaas CA, Brouwer R, van den Hout MC et al. (2015). The dystrophin gene and cognitive function in the general population. EJHG, 23(6), 837–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HX, Gustafson DR, Kivipelto M, Pedersen NL, Skoog I, Windblad B et al. (2012). Education halves the risk of dementia due to apolipoprotein epsilon4 allele: A collaborative study from the swedish brain power initiative. Neurobiol Aging, 33(5), 1007.e1–1007.e7. [DOI] [PubMed] [Google Scholar]
- Ware EB, Smith JA, Mukherjee B, Lee S, Kardia SL, & Diez-Roux AV (2016). Applying novel methods for assessing individual- and neighborhood-level social and psychosocial environment interactions with genetic factors in the prediction of depressive symptoms in the multi-ethnic study of atherosclerosis. Behav Genet, 46(1), 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White L, Katzman R, Losonczy K, Salive M, Wallace R, Berkman L et al. (1994). Association of education with incidence of cognitive impairment in three established populations for epidemiologic studies of the elderly. J Clin Epidemiol, 47(4), 363–374. [DOI] [PubMed] [Google Scholar]
- Wilson RS, Beckett LA, Barnes LL, Schneider JA, Bach J, Evans DA et al. (2002). Individual differences in rates of change in cognitive abilities of older persons. Psychology and Aging, 17(2), 179–193. [PubMed] [Google Scholar]
- Wilson RS, Hebert LE, Scherr PA, Barnes LL, Mendes de Leon CF, Evans DA (2009) Educational attainment and cognitive decline in old age. Neurology. 72(5), 460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk DA, Dickerson BC, Initiative AsDN. (2011). Fractionating verbal episodic memory in Alzheimer’s disease. Neuroimage, 54(2):1530–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, & Lin X (2011). Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet, 89(1), 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiura K, Machida J, Daack-Hirsch S, Patil SR, Ashworth LK, Hecht JT et al. (1998). Characterization of a novel gene disrupted by a balanced chromosomal translocation t(2;19)(q11.2;q13.3) in a family with cleft lip and palate. Genomics, 54(2), 231–240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.