Abstract
An estimated 240 million are chronically infected with hepatitis B virus (HBV), which can lead to liver disease, cirrhosis, and hepatocellular carcinoma. Currently, HBV treatment options include only nucleoside reverse transcriptase inhibitors and the immunomodulatory agent interferon alpha, and these treatments are generally not curative. New treatments with novel mechanisms of action, therefore, are highly desired for HBV therapy. The viral core protein (Cp) has gained attention as a possible therapeutic target because of its vital roles in the HBV life cycle. Several classes of capsid assembly effectors (CAEs) have been described in detail, and these compounds all increase capsid assembly rate, but inhibit HBV replication by different mechanisms. In this study, we have developed a thermal shift-based screening method for CAE discovery and characterization, filling a much-needed gap in high-throughput screening methods for capsid-targeting molecules. Using this approach followed by cell-based screening, we identified the compound HF9C6 as a CAE with low micromolar potency against HBV replication. HF9C6 caused large multi-capsid aggregates when capsids were assembled in vitro and analyzed by transmission electron microscopy. Interestingly, when HBV-expressing cells were treated with HF9C6, Cp was excluded from cell nuclei, suggesting that this compound may inhibit nuclear entry of Cp and capsids. Furthermore, mutational scanning of Cp suggested that HF9C6 binds the known CAE binding pocket, indicating that key Cp-compound interactions within this pocket have a role in determining CAE mechanism of action.
Keywords: antiviral, hepatitis B virus, capsid
Graphical Abstract

Hepatitis B virus (HBV) is a small, enveloped DNA virus of the Hepadnaviridae family that has highly specific tropism for liver cells. Chronic infection leads to liver diseases including fibrosis, cirrhosis, hepatocellular carcinoma (HCC), and other complications. Globally, an estimated 240 million are chronically infected, and there are approximately 1 million deaths per year due to HBV- related liver conditions (1–6).
HBV has a partially double-stranded circular DNA genome, relaxed circular DNA (rcDNA), that is completed by host enzymes in the nuclei of hepatocytes to covalently closed circular DNA (cccDNA). The cccDNA is the template for viral transcripts, including a full-length pre-genomic RNA (pgRNA) that is packaged and reverse transcribed to rcDNA by the viral polymerase. The rcDNA can either enter the nucleus to amplify the cccDNA pool, or it can exit the cell in a mature virion (7,8).
Currently, HBV treatment options include only nucleoside reverse transcriptase inhibitors (NRTIs) and the immunomodulatory agent interferon alpha (IFN-α). NRTIs are able to suppress the virus to undetectable levels; however, after cessation of therapy, patients’ viral loads often rebound. This rebound is likely due to failure of NRTIs to clear the HBV cccDNA present in hepatocytes, and viral components are produced from the residual cccDNA, which can persist for decades (2,3,5,9,10). NRTI regimens, therefore, must be long-term, most likely lifelong. IFN-α treatment, on the other hand, may be able to reduce cccDNA in vitro (11), but patient cure rates are very low. IFN-α treatment also leads to adverse effects that often outweigh the potential benefits of treatment (2,3,5,9,10). New treatments, therefore, are highly desired for HBV therapy.
The viral core protein (Cp) has gained attention as a possible therapeutic target because of its vital roles in the HBV life cycle. Reverse transcription occurs exclusively inside the viral capsid, and the capsid is required for nuclear entry of rcDNA (12,13). Cp has many more functions for pathogenicity of HBV, including roles in innate immune evasion and host manipulation (14-17). Cp has also been reported to interact with cccDNA, influencing epigenetic modifications and transcriptional activity, but the effects of these cccDNA-Cp interactions are not fully understood (18-20). Several classes of capsid assembly effectors (CAEs) have been described in detail, including heteroaryldyhydropyrimidines (HAPs) (21), phenylpropenamides (PPAs) (22,23), and sulfamoylbenzamides (SBAs) (24,25). HAPs, PPAs, and SBAs all increase capsid assembly rate, but inhibit HBV replication by distinct mechanisms (24-28).
Only one high-throughput assay exists for assessment of CAE activity, and this assay requires labeling of Cp with a fluorophore and purification of the newly labeled protein (29,30). The thermal shift assay (TSA), also known as differential scanning fluorimetry, has been used extensively to study protein-ligand interactions (31), and has previously been used to evaluate the effects of Cp mutants on HBV capsid stability (32) and to evaluate binding of CAEs to wild-type preformed capsids or Cp dimer using the assembly-deficient Y132A Cp mutant (33). The assay requires only protein, ligand, and fluorescent dye, and eliminates the need for protein labeling. In this study, we describe a TSA protocol that allows monitoring of capsid assembly and use this protocol to identify a novel CAE lead compound. Through further mechanistic evaluation, we show that the identified compound can aggregate core particles in vitro and prevent nuclear entry of Cp and capsids in HepAD38 cells. Our study provides a new class of CAEs with distinct mechanism of action as well as a high-throughput method of screening specifically for CAEs and assessing the effects of compounds on capsid assembly.
RESULTS AND DISCUSSION
Development of thermal shift screening assay
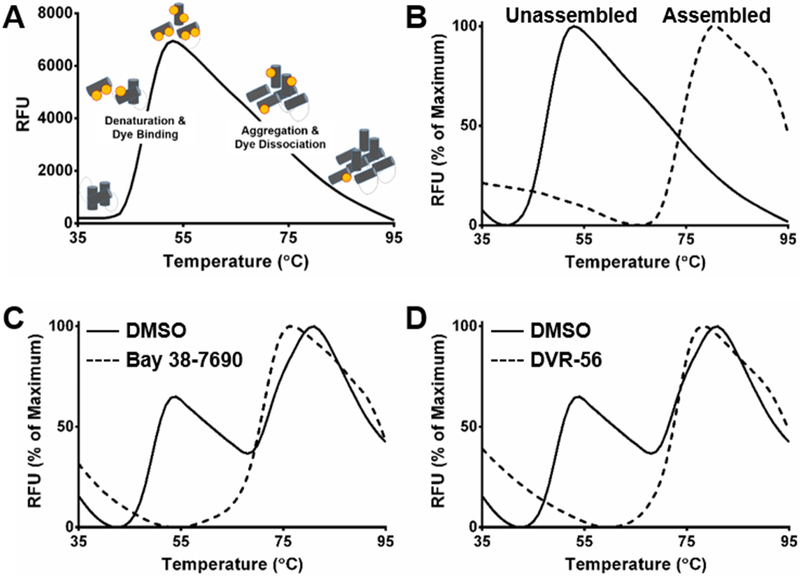
A schematic of the thermal shift assay (TSA) is shown in Figure 1A, adapted from (34). In the TSA, the fluorescent dye SYPRO Orange is initially quenched in the aqueous environment. As temperature is increased, the protein denatures, exposing hydrophobic regions to which the dye binds and becomes unquenched, resulting in fluorescence. Conditions for TSA were optimized according to signal-to-noise ratio and ease of discerning hit compounds from non-Cp-binding compounds. Ranges of Cp dimer concentrations (2.5-20 μM), pH (7.0-9.6), NaCl (0.5-2.5 M), and SYPRO Orange dye (lx-5x) were tested. Furthermore, we tested incubation conditions before thermal denaturation. Pre-incubation times of 0-30 min at 25°C and 37°C were tested. Nonassembly conditions (50 mM sodium bicarbonate, pH 9.6, no NaCl, no pre-incubation) yielded mainly one denaturation peak with a melting temperature (Tm) of 48°C, while full assembly conditions (50 mM HEPES, pH 7.5, 0.5 M NaCl, 30 min pre-incubation at 37°C) yielded one denaturation peak with Tm = 75°C (Figure 1B). To maximize potential for finding hit compounds, we chose conditions that allowed both peaks to be observed; with this method, Tm shifts can be observed on both denaturation peaks. The chosen conditions were 50 mM HEPES (pH 7.5) 0.5 M NaCl, 7.5 μM Cp dimer, and lx SYPRO Orange dye with no pre-incubation. The reaction setup and final thermal cycling parameters are shown in EXPERIMENTAL SECTION.
Figure 1. Development and validation of thermal shift assay for Cp binders.
(A) Schematic of thermal shift assay (TSA). (B) TSA was performed under non-assembly (50 mM sodium bicarbonate, pH 9.6, no NaCl, no pre-incubation) and full assembly (50 mM HEPES, pH 7.5, 0.5 M NaCl, 30 min pre-incubation at 37°C) conditions for Cp (7.5 μM dimer). (C-D) TSA was performed for Cp (7.5 μM dimer) in the presence or absence of 20 μM (C) Bay 38-7690 or (D) DVR-56
To validate the use of our TSA conditions in screening for Cp-binding compounds, we tested the known CAEs Bay 38-7690 (a HAP) and DVR-56 (a SBA) for their ability to change the Cp melting curve. Treatment with 20 μM Bay 38–7690 or DVR-56 led to a single melting peak corresponding to assembled Cp, consistent with previous reports that HAPs and SBAs accelerate capsid assembly (Figure 1C-D). Interestingly, the single melting peak in the compound-treated reactions had a slightly lower melting temperature than the assembled capsids in the DMSO reactions, suggesting that the compound-induced assemblies may be less thermally stable than normal capsids.
TSA compound screening
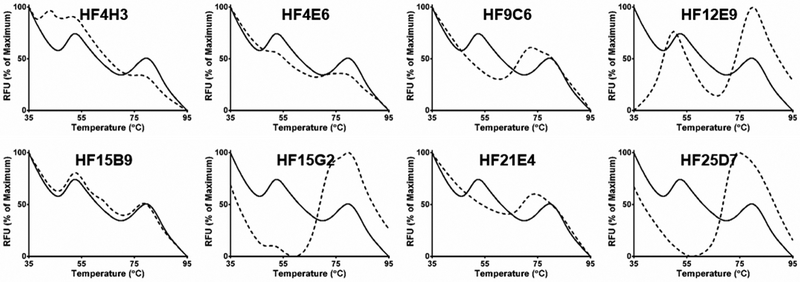
Because of the promising TSA results for Bay 38-7690 and DVR-56, we screened ~4,000 compounds from the Maybridge HitFinder chemical library. Compounds were screened at 20 μM final concentration, and melting curves were analyzed for Tm shifts or changes in the ratio of peak 1 to peak 2. While high resolution melt analysis can be used to analyze the data and discriminate hits vs. non-hits in an automated manner, we chose to inspect the curves visually for this small initial screen. ~50 hits were found in the screen, a hit rate of~1.25%. Representative graphs for assorted hit compounds are shown in Figure 2. While it is not certain that changes in Cp melting curves are due to direct Cp-compound interaction, a variety of compound-induced denaturation profiles were observed, such as appearance of additional peaks with HF4H3 or HF15B9, destabilization of the Cp dimer peak by HF12E9, or profiles resembling those of Bay 38-7690 and DVR-56
Figure 2. Screening for Cp-binding antivirals.
Cp thermal shift profiles for DMSO (solid lines) and selected hit compounds (dashed lines).
Antiviral screening of hit compounds
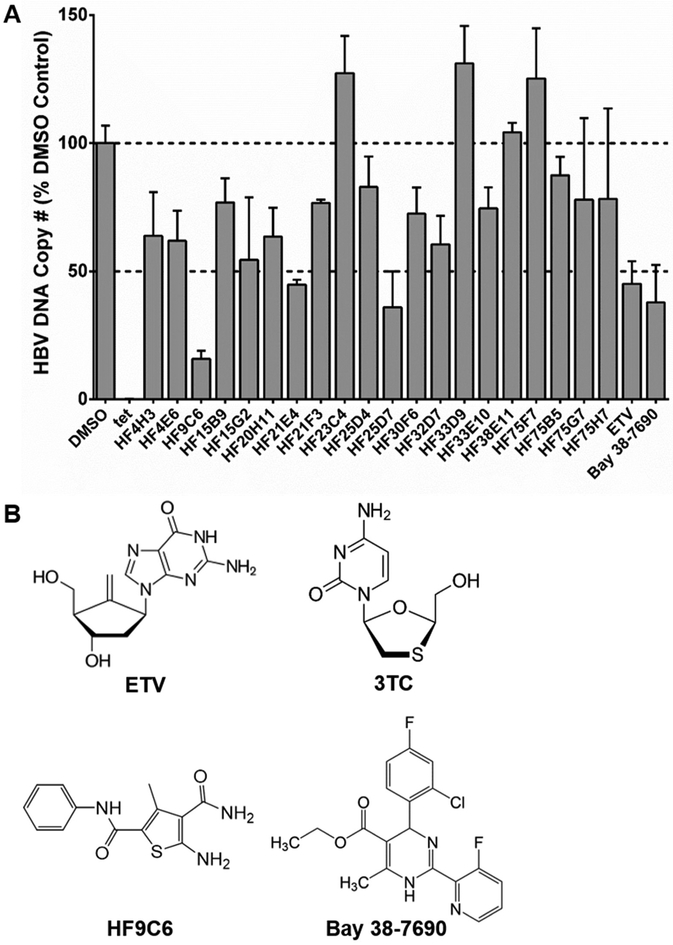
HepAD38 cells were induced and treated with 10 μM of each TSA hit compound every other day for 4 days for a total of 2 compound treatments over 4 days of induction time, and anti-HBV activity was determined by the level of viral DNA by qPCR. While many compounds significantly reduced HBV DNA, HF9C6 had the greatest effect (~80% reduction) (Figure 3A) and was chosen for further study. Dose responses for HF9C6, Bay 38-7690, 3TC, and ETV using total intracellular HBV DNA as a marker (in HepAD38 cells) and cytotoxicity with the XTT assay (in HepG2 cells) were conducted (Table 1); the chemical structures for these compounds are shown in Figure 3B. HF9C6 had a half maximal effective concentration (EC50) of 3.6 μM, compared to EC50 of 0.6 μM, 0.09 μM, and 0.004 μM for Bay 38-7690, 3TC, and ETV, respectively (Table 1). None of the compounds in Table 1 exhibited cytotoxicity at concentrations up to 100 μM (300 μM for HF9C6).
Figure 3. Antiviral screening.
(A) HepAD38 cells were treated with hit compounds (10 μM), ETV (2.5 nM), or Bay 38-7690 (500 nM) and assessed for HBV DNA production. Dashed lines indicate no change and 50% reduction compared to the DMSO control. (B) The chemical structures for the NRTIs 3TC and ETV and the CAEs HF9C6 and Bay 38-7690 are shown.
Table 1.
Antiviral potency and cytotoxicity of selected compounds in HepAD38 cells.
| Compound | 1EC50 (μM) | 2CC50 (μM) | 3SI |
|---|---|---|---|
| HF9C6 | 3.6 ± 0.2 | >300 | >83 |
| Bay 38-7690 | 0.6 ± 0.2 | >100 | >167 |
| Lamivudine (3TC) | 0.09 ± 0.01 | >100 | >1,111 |
| Entecavir (ETV) | 0.004 ± 0.002 | >100 | >25,000 |
EC50, Half Maximal Effective Concentration;
CC50, Cytotoxic Concentration 50;
SI, Selectivity Index.
HF9C6 induces Cp aggregation in vitro
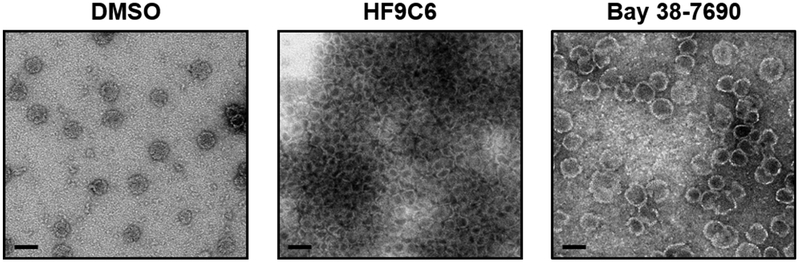
In order to evaluate the effect of HF9C6 on capsid assembly in vitro, Cp (10 μM dimer) was assembled in the presence or absence of HF9C6 (20 μM) or Bay 38-7690 (5 μM) and analyzed by transmission electron microscopy (TEM) similarly as previously described (28,35-37). As shown in Figure 4, capsids assembled in the presence of the carrier DMSO were spherical, ~40 nm in diameter, and dispersed across the TEM grid. Capsids assembled in the presence of Bay 38- 7690 were inflated and misshapen, as previously reported for other HAPs (28) and as we have previously shown (37). Capsids assembled in the presence of HF9C6, however, were heterogeneous in shape and size, and formed large inter-capsid aggregates on the grid. This effect was observed for multiple experiments, and the images shown are representative, although aggregate size did vary.
Figure 4. HF9C6 promotes aggressive aggregation of HBV capsid particles.
TEM analysis of capsids assembled in the presence and absence of 20 μM HF9C6 or 5 μM Bay 38-7690; scale bar, 50 nm. Images are representative of at least 3 independent experiments.
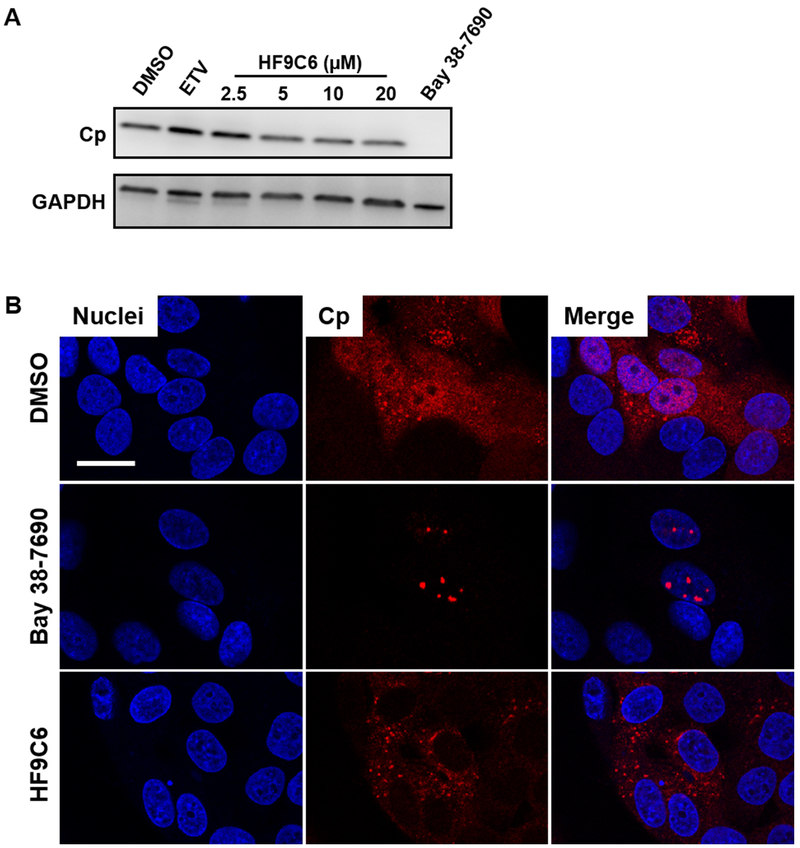
HF9C6 induces cytoplasmic localization of Cp without decreasing cellular Cp levels
HAPs have been shown to induce intracellular loss of Cp and capsids (21,37). To test if HF9C6 has the same effect, HepAD38 cells were induced and treated with HF9C6 and Bay 38-7690. While Bay 38-7690 eliminated Cp at 3 μM (~5x EC50), HF9C6 did not significantly reduce Cp levels at concentrations up to 20 μM (~5x EC50) (Figure 5A).
Figure 5. HF9C6 promotes cytoplasmic localization of Cp without significant changes in Cp levels.
(A) HepAD38 cells were treated with ETV (25 nM), Bay 38-7690 (3 μM), or the indicated concentrations of HF9C6 and assessed for Cp content by western blot. (B) HepAD38 cells were induced in the presence of DMSO, HF9C6 (20 μM), orBay 38-7690 (5 μM), and Cp was stained 4 days later; scale bar, 20 μm. Images are representative of at least 2 independent experiments.
Next, we studied the effect of HF9C6 treatment on Cp in HepAD38 cells, as we have previously shown that the HAP Bay 38-7690 induces nuclear Cp aggregation (37). HBV production was induced by tet withdrawal, and cells were treated with either DMSO, HF9C6 (20 μM), or Bay 38-7690 (5 μM), fixed, and stained for Cp and nuclei (Figure 5B). DMSO-treated cells had an even dispersion of Cp throughout the cell, while Cp in Bay 38-7690 -treated cells was concentrated in nuclear foci, as we previously reported (37). Surprisingly, in contrast to both DMSO- and Bay 38-7690 treated cells, Cp in HF9C6-treated cells was excluded from the nucleus. Although we cannot currently ascertain that the exclusion is due to Cp aggregation, these results suggest that HF9C6 inhibits nuclear entry of core particles, a process that is required for HBV infection and cccDNA amplification (38-41).
HF9C6 Appears to Bind Cp at the HAP Binding Pocket
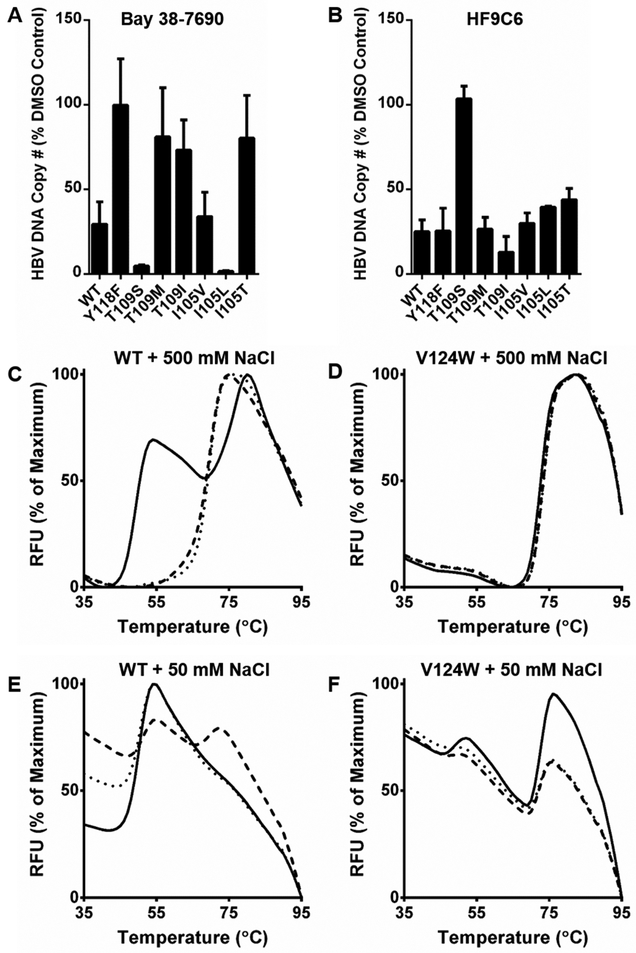
The binding site for HAPs has been found using X-ray crystallography and mutagenesis techniques (25,33,42-44). To probe this binding site, we generated HBV-expressing plasmids with mutations in Cp (Y118F, T109S, T109M, T109I, I105V, I105L, and I105T) (33). Huh7 cells were transfected with either wild-type (WT) or mutant plasmid, treated with Bay 38-7690 (500 nM) or HF9C6 (4 μM), and assessed for HBV core-associated DNA by qPCR. The compound concentrations used were sufficient to suppress viral DNA production by 75%; therefore, mutant resistance and hypersusceptibility could be easily observed at a single dose.
As shown in Figure 6A, Bay 38-7690 has the same resistance profile as previously reported for a related HAP (33); specifically, the Y118F, T109M, T109I, and I105T mutants are resistant to Bay 38-7690, while T109S and I105L are hypersusceptible. When tested against the pool of mutants, we found that the resistance profile of HF9C6 was markedly different to that of Bay 38- 7690 (Figure 6B). Specifically, while the Y118F, T109M, T109I, and I105T mutants are resistant to Bay 38-7690, there is no significant change in inhibition of these mutants by HF9C6 when compared to wild-type; in fact, T109I appears to have a slight hypersusceptibility to HF9C6. Furthermore, while T109S is hypersusceptible to Bay 38-7690 treatment, this mutation confers resistance to HF9C6.
Figure 6. HF9C6 likely binds Cp in the HAP binding pocket.
Huh7 cells were transfected with HBV-expressing plasmids harboring the indicated mutations in Cp, treated with (A) 500 nM Bay 38-7690 or (B) 4 μM HF9C6, and assessed for HBV core-associated DNA by qPCR. (C-F) Purified WT or V124W Cp was subjected to TSA in the presence of DMSO (solid lines), 20 μM Bay 38-7690 (dashed lines), or 20 μM HF9C6 (dotted lines) with 500 mM or 50 mM NaCl.
These data suggest that HF9C6 may bind Cp in the HAP binding pocket. In order to test this further, we employed a V124W Cp mutant that has been shown to be resistant to HAP treatment in vitro (45). This mutant mimics CAE-bound Cp by occupying the binding pocket with a tryptophan residue and is thus unable to support HBV genome replication. Therefore, rather than testing the mutant in cell culture, we purified V124W Cp for use in TSA. TSA with WT Cp and 500 mM NaCl yielded visible dimer and capsid peaks; in the presence of 20 μM HF9C6 or Bay 38-7690, only the capsid peak was observed (Figure 1C, Figure 2, and Figure 6C). TSA with V124W Cp and 500 mM NaCl gave a single melting peak corresponding to assembled capsids, and treatment with compounds had no effect on the profile (Figure 6D). Because of the faster assembly kinetics of the mutant, which have been reported previously (45), we performed TSA with 50 mM NaCl. With this condition, the WT Cp melting curve showed mainly dimer with a slight shoulder corresponding to capsids (Figure 6E). Interestingly, 20 μM Bay 38-7690 shifted the population to more assemblies while 20 μ HF9C6 did not. This difference may be due to the higher potency of Bay 38-7690 or to differences in mechanism. With 50 mM NaCl, V124W now had visible dimer and capsid peaks, and the melting profile was not affected by either compound (Figure 6F). This observation supports our conclusion that HF9C6 binds in the HAP binding pocket. However, the differences in mutant susceptibility to the compounds in cell culture suggests that although HAPs and HF9C6 may share the same target, they may be suitable in combination therapies to more effectively treat infections and overcome escape mutants.
CONCLUSION
Despite the availability of a vaccine and highly potent inhibitors of replication, HBV remains a highly significant global health concern with >240 million chronic infections. NRTIs such as entecavir and tenofovir greatly reduce viral replication and incidences of hepatocellular carcinoma, but do not cure; therefore, lifelong treatment is necessary. Furthermore, the HBV vaccine provides protection in an estimated 95% of cases, which leaves a significant population susceptible to infection, even if all people were to be vaccinated (1-6). Many routes are being explored for HBV treatment with ultimate goal of cure, such as viral gene expression, cccDNA formation and stability, capsid formation, and host immune modulators (46). A full cure will likely consist of a combination of approaches, and rigorous assessment of multiple targets is necessary.
In our search for new CAEs, we wished to expand the arsenal of assays available to screen for capsid-targeting agents. There is currently only one high-throughput assay for assessment of CAE activity, which provides a method for monitoring the rate and extent of capsid assembly in vitro; however, the assay requires labeling of Cp with a fluorophore and purification of the newly labeled protein (29,30). The thermal shift assay has been used extensively to study protein-ligand interactions (31), and has previously been used to evaluate the effects of Cp mutants on HBV capsid stability (32) and to evaluate binding of CAEs to wild-type preformed capsids or Cp dimer using the assembly-deficient Y132A Cp mutant (33). The assay requires only protein, ligand, and SYPRO Orange dye, and eliminates the need for protein labeling. We have modified the TSA protocol to allow simultaneous monitoring of the 2 major oligomerization states of Cp, dimer and capsid. This assay provides a high-throughput method of 1) screening for factors that affect capsid assembly and 2) assessing effects of ligands or experimental conditions on capsid assembly, and has the potential to accelerate capsid-related drug discovery and mechanistic evaluations.
Using the TSA combined with additional assays, we have identified HF9C6 as a capsid assembly effector representing a novel CAE chemical class. HF9C6 shows promising preliminary properties with an EC50 of 3.6 μM with no observed cytotoxicity up to 300 μM by XTT assay. Interestingly, HF9C6 has markedly different effects on capsid assembly than the HAP Bay 38- 7690. Whereas Bay 38-7690 induces formation of inflated and misshapen capsids, consistent with previous reports of this compound as well as related HAPs (28,37), HF9C6 induces the mass aggregation of multiple capsids in vitro when visualized by TEM (Figure 4). Furthermore, while Bay 38-7690 treatment causes nuclear aggregation of Cp in HBV-infected cells, HF9C6 promotes cytoplasmic localization of Cp (Figure 5). Until very recently, a mechanism such as this was unreported. In a characterization of several CAE chemotypes, Corcuera et al. found that compounds of the PPA and SBA class, as well as three other classes, induce Cp aggregation and nuclear Cp exclusion similar to what we have observed for HF9C6 (47).
In agreement with our observed mechanistic differences between HF9C6 and Bay 38- 7690, these compounds seem to interact differently with the binding pocket of Cp (Figure 6). The differences of mutant susceptibility to the two compound classes can not only help explain mechanistic differences, but also suggest that multiple CAEs may be suitable in combination therapies. Strikingly, the panel of mutants show complementary resistance profiles between Bay 38-7690 and HF9C6. Mutants that are resistant to Bay 38-7690 either have no change in susceptibility to HF9C6 or may be slightly hypersusceptible (T109I); likewise, the mutant that is resistant to HF9C6 (T109S) is hypersusceptible to Bay 38-7690. Future work in this area will focus on efficacy of compound combinations and how gain or loss of HAP- or HF9C6-specific interactions by compound modifications affect the mechanism of action.
EXPERIMENTAL SECTION
Compounds and Reagents
Bay 38-7690 (21) was synthesized based on patented methods (21,27,48-50), and DVR-56 was prepared according to reported procedures (51). The Maybridge Hitfinder chemical library of compounds (version 6) was purchased from Maybridge, lamivudine (3TC) was obtained through the U.S. NIH AIDS Reagent Program, and entecavir (ETV) and was purchased from Sigma- Aldrich.
HBV Cp Purification
A gBlock Gene Fragment coding for the 149 amino acid assembly domain of HBV capsid protein with an added C-terminal cysteine (C150) (29,30,42) with NdeI and BamHI restriction sites was synthesized by Integrated DNA Technologies and cloned into the pET11a expression vector (Novagen). The V124W mutant was generated by site-directed mutagenesis. HBV C150 was expressed and purified as previously described (29,30,52), with minor modifications. The C150 expression plasmid was transformed into BL21 (DE3) E. coli, grown at 37°C to an OD600 of ~0.8, and induced for 3 h with 1 mM IPTG at 37°C. Cells were pelleted and resuspended in 50 mM Tris (pH 7.5), 1 mM EDTA, 20 mM 2-mercaptoethanol (2-ME), 1 mM PMSF, 150 μg/ml lysozyme, and 0.2 mg/ml DNase I. The suspension was incubated on ice for 30 min and lysed by sonication. Polyethylenimine (PEI) was added to a final concentration of 0.15% w/v to precipitate DNA, and the lysate was centrifuged at 16,000xg for 1 h. Ammonium sulfate was added to the supernatant to 40% saturation. The solution was gently stirred for 1 h, then centrifuged at 16,000xg for 1 h. The pellet was resuspended in Buffer A [100 mM Tris (pH 7.5), 100 mM NaCl, 10 mM 2-ME] to ~10 mg/ml, centrifuged at 16,000xg for 20 min, loaded onto a Buffer A-equilibrated HiLoad 26/60 Superdex 200 prep grade (GE Healthcare) column, and eluted at 2.5 ml/min. Fractions were pooled based on the chromatogram and SDS-PAGE, concentrated to ~5 mg/ml, and dialyzed into Buffer N [50 mM sodium bicarbonate (pH 9.6), 10 mM 2-ME], Solid urea was added to 3 M and stirred for 1 h at 4°C. The solution was loaded onto a Buffer N-equilibrated HiLoad 26/60 Superdex 200 prep grade column and eluted at 2.5 ml/min. Fractions containing the C150 dimer were pooled, concentrated, and stored at −80°C. Final protein concentration was determined spectrophotometrically using an extinction coefficient of 60,900 (30).
Thermal Shift Screening Assay
The development and use of the thermal shift assay for drug discovery has been described previously (53,54). In a final reaction volume of 20 μl, 10 μl of Cp (15 μM dimer) in Buffer N was mixed with 10 μl assembly buffer [100 mM HEPES (pH 7.5), 1 M NaCl] containing 2x SYPRO Orange Protein Gel Stain (Life Technologies). Compounds were added at a final concentration of 20 μM, and reactions contained 1% DMSO. Samples were heated in a PikoReal Real-Time PCR System (Thermo Scientific) from 25°C to 95°C in steps of 1°C every 50 s. Melting curves were analyzed with PikoReal Software. For reactions containing 50 mM final [NaCl], 10 μl of Cp (15 μM dimer) in Buffer N was mixed with 10 μl assembly buffer [100 mM HEPES (pH 7.5), 100 mM NaCl] containing 2x SYPRO Orange Protein Gel Stain.
Cell Culture
HepG2 and Huh7 (ATCC) cells were maintained in complete media [Dulbecco’s Modified Eagle Medium (DMEM), 10% fetal bovine serum (FBS)]. HepAD38 cells (ATCC) (55) were maintained in tet media [complete media plus 0.4 μg/ml tetracycline (tet) and 400 μg/ml G418 (Gibco)]. All cells were incubated at 37°C with 5% CO2.
Antiviral Assays and Quantitative Polymerase Chain Reaction (qPCR) Analysis of HBV Nucleic Acids
HepAD38 cells (5 x 104) were plated in 96-well plates in tet media. The next day, cells were washed 2x with PBS, and the media was replaced with complete media containing compounds. Fresh complete media with compounds was added again after 2 days. After 2 additional days, cells were washed with PBS, trypsinized, and pelleted by centrifugation. The cell pellet was resuspended in 200 μl PBS, and total DNA was extracted using the QIAamp DNA Blood Mini Kit (Qiagen).
Forward and reverse primers for total HBV DNA quantification were 5'- CCTGGTTATCGCTGGATGTGT-3' and 5'-GGACAAACGGGCAACATACCTT-3', respectively (56). Amplification was conducted by denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min using PerfeCTa SYBR Green FastMix (Quanta Biosciences). Amplification was carried out in a PikoReal Real-Time PCR System. A standard curve was generated with dilutions of the HBV genome-containing plasmid pCMV-HBV-LE-II (a gift from Dr. John Tavis, Saint Louis University) (57). For dose responses, values were plotted in GraphPad Prism 5 and analyzed with the log (inhibitor) vs. normalized response–variable slope equation.
Cytotoxicity
HepG2 cells were plated in complete media and treated with compounds as in the antiviral screening above. At the end of treatment duration, cell viability was assessed with the Cell Proliferation Kit II (XTT) (Roche) according to the manufacturer’s instructions. Values were plotted in GraphPad Prism 5 and analyzed with the log (inhibitor) vs. normalized response – variable slope equation.
Transmission Electron Microscopy
TEM was performed as previously described (37). C150 (10 μM dimer) in buffer N was assembled in the presence of 1% DMSO or 10 μM Bay 38-7690 by addition of an equal volume of 100 mM HEPES (pH 7.5) with 1 M NaCl and incubation at room temperature (RT) for 1 h. Assemblies were absorbed to glow-discharged carbon-coated 200 mesh copper grids (Electron Microscopy Sciences), stained with 2% uranyl acetate, and imaged with a JEOL JEM-1400 transmission electron microscope.
Western Blotting
Cells were lysed by addition of RIPA buffer [50 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 0.1% SDS, 1% Triton X-100] and incubated on ice for 30 min with occasional vortexing. The lysate was centrifuged at 14,000xg for 15 min, the supernatant was collected, and protein was quantitated by Bradford assay. Protein (8 μg) in laemmli buffer was separated by SDS- PAGE and transferred to PVDF Immobilon-P membranes (Millipore). Membranes were probed with rabbit anti-HBV core (Austral Biologicals) (1:500 dilution) and mouse anti-GAPDH (Santa Cruz Biotechnology) (1:5,000) antibodies, followed by anti-mouse and anti-rabbit HRP- conjugated secondary antibodies (Sigma-Aldrich). Bound antibodies were visualized by adding Luminata Forte Western HRP substrate (Millipore) to the membrane and imaging with a Fuji camera system.
Immunofluorescence
HepAD38 cells (1 χ 104) were seeded in 96-well 2% collagen-coated image plates (BD Falcon) in tet media. The next day, the cells were washed twice with PBS, and complete media containing compounds was added to the wells with 1% final DMSO concentration. At 4 days post-induction, cells were fixed at room temperature (RT) for 15 min in 4% formaldehyde, permeabilized with 0.1% Triton X-100 in PBS, and blocked with 10% goat serum (Jackson ImmunoResearch) and 1% bovine serum albumin (BSA) in PBS. Rabbit anti-HBV core primary antibody (Dako) (1:1,000) was bound overnight at 4°C. Samples were incubated at RT for 45 min with 1:2,000 goat antirabbit Alexa Fluor 568 secondary antibody (Invitrogen) and stained for nuclei using Hoechst 33342 (Invitrogen) or DRAQ5 (Thermo Scientific). Images were taken on a confocal laser scanning Leica TCS SP8 microscope.
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Institutes of Health (NIH) (AI100890 and AI121315 to S.G.S.and Z.W.) and Trail to a Cure. A.D.H. is supported by NIH AI100890- S1, M.N.P.-C. is supported by the Fulbright Student Program, and J.J.W. is supported by a Life Sciences Fellowship from the University of Missouri. We also acknowledge partial support from Kumamoto University and the Japan Agency for Medical Research and Development. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Wang L, Zou ZQ, Liu CX, and Liu XZ (2014) Immunotherapeutic interventions in chronic hepatitis B virus infection: A review. Journal of immunological methods 407C, 1–8 [DOI] [PubMed] [Google Scholar]
- 2.Tang CM, Yau TO, and Yu J (2014) Management of chronic hepatitis B infection: Current treatment guidelines, challenges, and new developments. World journal of gastroenterology : WJG 20, 6262–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai CL, and Yuen MF (2008) Chronic hepatitis B--new goals, new treatment. The New England journal of medicine 359, 2488–2491 [DOI] [PubMed] [Google Scholar]
- 4.Dienstag JL (2008) Hepatitis B virus infection. The New England journal of medicine 359, 1486–1500 [DOI] [PubMed] [Google Scholar]
- 5.Cox N, and Tillmann H (2011) Emerging pipeline drugs for hepatitis B infection. Expert Opin Emerg Drugs 16, 713–729 [DOI] [PubMed] [Google Scholar]
- 6.Liver, E. A. F. T. S. O. T., and Cancer, E. O. F. R. A. T. O. (2012) EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. Journal of hepatology 56, 908–943 [DOI] [PubMed] [Google Scholar]
- 7.Seeger C, and Mason WS (2000) Hepatitis B virus biology. Microbiology and molecular biology reviews : MMBR 64, 51–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seeger C, and Mason WS (2015) Molecular biology of hepatitis B virus infection. Virology 479-480, 672–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.You CR, Lee SW, Jang JW, and Yoon SK (2014) Update on hepatitis B virus infection. World journal of gastroenterology : WJG 20, 13293–13305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seo Y, and Yano Y (2014) Short- and long-term outcome of interferon therapy for chronic hepatitis B infection. World journal of gastroenterology : WJG 20, 13284–13292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou WM, Thasler WE, Huser N, Durantel D, Liang TJ, Munk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, and Protzer U (2014) Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 343, 1221–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rabe B, Vlachou A, Pante N, Helenius A, and Kann M (2003) Nuclear import of hepatitis B virus capsids and release of the viral genome. Proceedings of the National Academy of Sciences of the United States of America 100, 9849–9854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beck J, and Nassal M (2007) Hepatitis B virus replication. World journal of gastroenterology : WJG 13, 48–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ait-Goughoulte M, Lucifora J, Zoulim F, and Durantel D (2010) Innate antiviral immune responses to hepatitis B virus. Viruses 2, 1394–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosmorduc O, Sirma H, Soussan P, Gordien E, Lebon P, Horisberger M, Brechot C, and Kremsdorf D (1999) Inhibition of interferon-inducible MxA protein expression by hepatitis B virus capsid protein. The Journal of general virology 80 (Pt 5), 1253–1262 [DOI] [PubMed] [Google Scholar]
- 16.Guo Y, Kang W, Lei X, Li Y, Xiang A, Liu Y, Zhao J, Zhang J, and Yan Z (2012) Hepatitis B viral core protein disrupts human host gene expression by binding to promoter regions. BMC genomics 13, 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu ZJ, Zhu Y, Huang DR, and Wang ZQ (2010) Constructing the HBV-human protein interaction network to understand the relationship between HBV and hepatocellular carcinoma. Journal of experimental & clinical cancer research : CR 29, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, and Levrero M (2006) Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 130, 823–837 [DOI] [PubMed] [Google Scholar]
- 19.Bock CT, Schwinn S, Locamini S, Fyfe J, Manns MP, Trautwein C, and Zentgraf H (2001) Structural organization of the hepatitis B virus minichromosome. Journal of molecular biology 307, 183–196 [DOI] [PubMed] [Google Scholar]
- 20.Guo YH, Li YN, Zhao JR, Zhang J, and Yan Z (2011) HBc binds to the CpG islands of HBV cccDNA and promotes an epigenetic permissive state. Epigenetics 6 720–726 [DOI] [PubMed] [Google Scholar]
- 21.Deres K, Schroder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, Kramer T, Niewohner U, Pleiss U, Stoltefuss J, Graef E, Koletzki D, Masantschek RN, Reimann A, Jaeger R, Gross R, Beckermann B, Schlemmer KH, Haebich D, and Rubsamen-Waigmann H (2003) Inhibition of hepatitis B virus replication by drug- induced depletion of nucleocapsids. Science 299, 893–896 [DOI] [PubMed] [Google Scholar]
- 22.Perni RB, Conway SC, Ladner SK, Zaifert K, Otto MJ, and King RW (2000) Phenylpropenamide derivatives as inhibitors of hepatitis B virus replication. Bioorganic & medicinal chemistry letters 10, 2687–2690 [DOI] [PubMed] [Google Scholar]
- 23.Delaney W. E. t., Edwards R, Colledge D, Shaw T, Furman P, Painter G, and Locarnini S (2002) Phenylpropenamide derivatives AT-61 and AT-130 inhibit replication of wild-type and lamivudine-resistant strains of hepatitis B virus in vitro. Antimicrobial agents and chemotherapy 46, 3057–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campagna MR, Liu F, Mao R, Mills C, Cai D, Guo F, Zhao X, Ye H, Cuconati A, Guo H, Chang J, Xu X, Block TM, and Guo JT (2013) Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids. Journal of virology 87, 6931–6942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Z, Hu T, Zhou X, Wildum S, Garcia-Alcalde F, Xu Z, Wu D, Mao Y, Tian X, Zhou Y, Shen F, Zhang Z, Tang G, Najera F, Yang G, Shen HC, Young JA, and Qin N (2017) Heteroaryldihydropyrimidine (HAP) and Sulfamoylbenzamide (SBA) Inhibit Hepatitis B Virus Replication by Different Molecular Mechanisms. Sci Rep 7, 42374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feld JJ, Colledge D, Sozzi V, Edwards R, Littlejohn M, and Locarnini SA (2007) The phenylpropenamide derivative AT-130 blocks HBV replication at the level of viral RNA packaging. Antiviral research 76, 168–177 [DOI] [PubMed] [Google Scholar]
- 27.Stray SJ, Bourne CR, Punna S, Lewis WG, Finn MG, and Zlotnick A (2005) A heteroaryldihydropyrimidine activates and can misdirect hepatitis B virus capsid assembly. Proceedings of the National Academy of Sciences of the United States of America 102, 8138–8143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stray SJ, and Zlotnick A (2006) BAY 41-4109 has multiple effects on Hepatitis B virus capsid assembly. Journal of molecular recognition : JMR 19, 542–548 [DOI] [PubMed] [Google Scholar]
- 29.Stray SJ, Johnson JM, Kopek BG, and Zlotnick A (2006) An in vitro fluorescence screen to identify antivirals that disrupt hepatitis B virus capsid assembly. Nature biotechnology 24, 358–362 [DOI] [PubMed] [Google Scholar]
- 30.Zlotnick A, Lee A, Bourne CR, Johnson JM, Domanico PL, and Stray SJ (2007) In vitro screening for molecules that affect virus capsid assembly (and other protein association reactions). Nature protocols 2, 490–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groftehauge MK, Hajizadeh NR, Swann MJ, and Pohl E (2015) Protein-ligand interactions investigated by thermal shift assays (TSA) and dual polarization interferometry (DPI). Acta Crystallogr D Biol Crystallogr 71, 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selzer L, Kant R, Wang JC, Bothner B, and Zlotnick A (2015) Hepatitis B Virus Core Protein Phosphorylation Sites Affect Capsid Stability and Transient Exposure of the C-terminal Domain. J Biol Chem 290, 28584–28593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klumpp K, Lam AM, Lukacs C, Vogel R, Ren S, Espiritu C, Baydo R, Atkins K, Abendroth J, Liao G, Efimov A, Hartman G, and Flores OA (2015) High- resolution crystal structure of a hepatitis B virus replication inhibitor bound to the viral core protein. Proceedings of the National Academy of Sciences of the United States of America 112, 15196–15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giuliani SE, Frank AM, and Collart FR (2008) Functional assignment of solutebinding proteins of ABC transporters using a fluorescence-based thermal shift assay. Biochemistry 47, 13974–13984 [DOI] [PubMed] [Google Scholar]
- 35.Bourne C, Lee S, Venkataiah B, Lee A, Korba B, Finn MG, and Zlotnick A (2008) Small-molecule effectors of hepatitis B virus capsid assembly give insight into virus life cycle. Journal of virology 82, 10262–10270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zlotnick A, Ceres P, Singh S, and Johnson JM (2002) A small molecule inhibits and misdirects assembly of hepatitis B virus capsids. Journal of virology 76, 4848–4854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huber AD, Wolf JJ, Liu D, Gres AT, Tang J, Boschert KN, Puray-Chavez MN, Pineda DL, Laughlin TG, Coonrod EM, Yang Q, Ji J, Kirby KA, Wang Z, and Sarafianos SG (2018) The Heteroaryldihydropyrimidine Bay 38-7690 Induces Hepatitis B Virus Core Protein Aggregates Associated with Promyelocytic Leukemia Nuclear Bodies in Infected Cells. mSphere 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitz A, Schwarz A, Foss M, Zhou L, Rabe B, Hoellenriegel J, Stoeber M, Pante N, and Kann M (2010) Nucleoporin 153 arrests the nuclear import of hepatitis B virus capsids in the nuclear basket. PLoS pathogens 6, e1000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabe B, Delaleau M, Bischof A, Foss M, Sominskaya I, Pumpens P, Cazenave C, Castroviejo M, and Kann M (2009) Nuclear entry of hepatitis B virus capsids involves disintegration to protein dimers followed by nuclear reassociation to capsids0. PLoS pathogens 5, e1000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuttleman JS, Pugh JC, and Summers JW (1986) In vitro experimental infection of primary duck hepatocyte cultures with duck hepatitis B virus. Journal of virology 58, 17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuttleman JS, Pourcel C, and Summers J (1986) Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 47, 451–460 [DOI] [PubMed] [Google Scholar]
- 42.Bourne CR, Finn MG, and Zlotnick A (2006) Global structural changes in hepatitis B virus capsids induced by the assembly effector HAP1. Journal of virology 80, 11055–11061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu Z, Lin X, Zhou M, Liu Y, Zhu W, Chen W, Zhang W, Guo L, Liu H, Wu G, Huang M, Jiang M, Xu Z, Zhou Z, Qin N, Ren S, Qiu H, Zhong S, Zhang Y, Zhang Y, Wu X, Shi L, Shen F, Mao Y, Zhou X, Yang W, Wu JZ, Yang G, Mayweg AV, Shen HC, and Tang G (2016) Design and Synthesis of Orally Bioavailable 4-Methyl Heteroaryldihydropyrimidine Based Hepatitis B Virus (HBV) Capsid Inhibitors. J Med Chem 59, 7651–7666 [DOI] [PubMed] [Google Scholar]
- 44.Venkatakrishnan B, Katen SP, Francis S, Chirapu S, Finn MG, and Zlotnick A (2016) Hepatitis B Virus Capsids Have Diverse Structural Responses to Small-Molecule Ligands Bound to the Heteroaryldihydropyrimidine Pocket. Journal of virology 90, 3994–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan Z, Maguire ML, Loeb DD, and Zlotnick A (2013) Genetically altering the thermodynamics and kinetics of hepatitis B virus capsid assembly has profound effects on virus replication in cell culture. Journal of virology 87, 3208–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pei Y, Wang C, Yan SF, and Liu G (2017) Past, Current, and Future Developments of Therapeutic Agents for Treatment of Chronic Hepatitis B Virus Infection. J Med Chem 60, 6461–6479 [DOI] [PubMed] [Google Scholar]
- 47.Corcuera A, Stolle K, Hillmer S, Seitz S, Lee JY, Bartenschlager R, Birkmann A, and Urban A (2018) Novel non-heteroarylpyrimidine (HAP) capsid assembly modifiers have a different mode of action from HAPs in vitro. Antiviral research [DOI] [PubMed] [Google Scholar]
- 48.Weber O, Schlemmer KH, Hartmann E, Hagelschuer I, Paessens A, Graef E, Deres K, Goldmann S, Niewoehner U, Stoltefuss J, Haebich D, Ruebsamen-Waigmann H, and Wohlfeil S (2002) Inhibition of human hepatitis B virus (HBV) by a novel non-nucleosidic compound in a transgenic mouse model. Antiviral research 54, 69–78 [DOI] [PubMed] [Google Scholar]
- 49.Stoltefuss J, Goldmann S, Paessens A, Graef E, and Lottmann S (2002) Use of dihydropyrimidines as medicaments, and novel substances. Bayer Aktiengesellschaft (Leverkusen, DE) [Google Scholar]
- 50.Stoltefuss J, Goldmann S, Paes A, Graef E, and Lottmann S (1999) New dihydropyrimidine derivatives and their corresponding mesomers useful as antiviral agents. Bayer AG [Google Scholar]
- 51.Sari O, Boucle S, Cox BD, Ozturk T, Russell OO, Bassit L, Amblard F, and Schinazi RF (2017) Synthesis of sulfamoylbenzamide derivatives as HBV capsid assembly effector. Ear J Med Chem 138 407–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zlotnick A, Cheng N, Conway JF, Booy FP, Steven AC, Stahl SJ, and Wingfield PT (1996) Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry 35, 7412–7421 [DOI] [PubMed] [Google Scholar]
- 53.Lo MC, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, and Ellestad G (2004) Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Analytical biochemistry 332, 153–159 [DOI] [PubMed] [Google Scholar]
- 54.Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, and Salemme FR (2001) High-density miniaturized thermal shift assays as a general strategy for drug discovery. Journal of biomolecular screening 6, 429–440 [DOI] [PubMed] [Google Scholar]
- 55.Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, Seeger C, and King RW (1997) Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrobial agents and chemotherapy 41, 1715–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sitnik R, Paes A, Mangueira CP, and Pinho JR (2010) A real-time quantitative assay for hepatitis B DNA virus (HBV) developed to detect all HBV genotypes. Revista do Instituto de Medicina Tropical de Sao Paulo 52, 119–124 [DOI] [PubMed] [Google Scholar]
- 57.Tavis JE, Cheng X, Hu Y, Totten M, Cao F, Michailidis E, Aurora R, Meyers MJ, Jacobsen EJ, Parniak MA, and Sarafianos SG (2013) The hepatitis B virus ribonuclease H is sensitive to inhibitors of the human immunodeficiency virus ribonuclease H and integrase enzymes. PLoS pathogens 9, e1003125. [DOI] [PMC free article] [PubMed] [Google Scholar]