Abstract
A TROSY-based NMR experiment is described for simultaneous measurement of the 15N longitudinal relaxation rate constant R1 and the {1H}-15N nuclear Overhauser enhancement. The experiment is based on the observation that the TROSY mixing pulse sequence element symmetrically exchanges 1H and 15N magnetizations. The accuracy of the proposed technique is validated by comparison to independent measurements of both relaxation parameters for the protein ubiquitin. The simultaneous experiment is approximately 20-33% shorter than conventional sequential measurements.
Keywords: Dynamics, Longitudinal relaxation, Nuclear Overhauser enhancement, Protein, Spin-lattice relaxation, TROSY
The 15N R1 and R2 relaxation rate constants and the steady-state {1H}-15N nuclear Overhauser enhancement (NOE) provide vital probes of the picosecond-nanosecond dynamics of backbone amide moieties in proteins 1. Measurement of these three parameters allows characterization of the spectral density function J(ω) at the frequencies 0, ωN, ωN, and ωH±ωN (the 1H single quantum and 15N single quantum frequencies frequently are approximated by a single effective frequency through reduced spectral density mapping) 2. Experimental protocols and approaches for data interpretation are well-established 1; however, particularly as measurements at multiple static magnetic fields become more commonplace 3-5, maximizing the efficiency of experimental methods remains essential.
Measurement of the steady-state {1H}-15N NOE requires two experiments: one in which the 15N steady-state magnetization is measured after irradiation of the 1H spins (saturated), and a second in which the Boltzmann equilibrium 15N magnetization is measured in the absence of 1H irradiation (unsaturated or control) 6. The ratio of the saturated to the unsaturated intensity is the steady-state {1H}-15N NOE. Although simple in the abstract, the steady-state {1H}-15N NOE experiment suffers from several factors complicating implementation. Chief among these is the inherent lower experimental sensitivity because starting magnetization originates on the low gyromagnetic 15N nucleus, rather than 1H as in the R1 and R2 experiments. In addition, the recycle delays must be set long enough (> 7-8/R1) to ensure complete recovery of equilibrium magnetization in the unsaturated control experiment. Finally, avoidance of saturation transfer through chemical exchange of labile protons requires careful manipulations of the water magnetization. Addressing these concerns to ensure accurate steady-state {1H}-15N NOE values results in long experimental times.
The TROSY-based NOE pulse sequence 7, performed without 1H irradiation, shows a unique feature: following the t1 period, a reverse-INEPT block transfers transverse 15N magnetization to 1H for subsequent detection, and simultaneously transfers longitudinal 1H magnetization to longitudinal 15N magnetization 8. In principle, this polarization-enhanced 15N magnetization could serve as the starting point for another relaxation experiment. In the present work, a second pulse sequence block utilizes this magnetization to measure the 15N R1 relaxation rate constant. Separate storage of decay curves originating with positive or negative values of the initial longitudinal 15N magnetization also allows the steady-state 15N intensity to be obtained by extrapolation during data analysis, and hence calculation of the {1H}-15N NOE. Thus, an independent measurement of the saturated spectrum is not necessary.
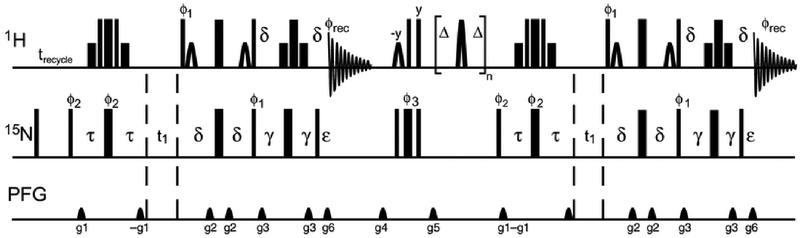
The pulse sequence for the proposed experiment is shown in Figure 1 and is based on the TROSY pulse sequences of Bax and coworkers 9. The first part of the experiment is the control measurement of equilibrium 15N magnetization. In addition, this sequence transfers longitudinal 1H magnetization to longitudinal 15N magnetization at the start of the first t2 acquisition period. Following the t2 period, two orthogonal 1H 90° hard pulses are used to purge remaining 1H magnetization and a composite 0° or 180° 15N pulse is used to generate positive or negative 15N longitudinal magnetization. The inversion-recovery relaxation period consists of repeated [Δ - 180°(1H)- Δ] blocks with Δ = 11 ms 10. The 1H 180° pulses are cosine-modulated I-BURP2 pulses to maintain water magnetization on the +z-axis 11. These 1H 180° pulses serve two purposes: (i) to remove cross-correlated relaxation effects and (ii) to obtain 15N steady-state magnetization by amide proton saturation. The partially relaxed 15N magnetization is frequency labeled and transferred to 1H for detection during the second t2 period. The experiment is performed twice for each R1 relaxation delay: the control equilibrium 15N free induction decays (FID) are co-added while the R1 FIDs with positive initial 15N longitudinal magnetization (I+(t)), or negative initial 15N longitudinal magnetization (I−(t)) are stored separately. If the inversion-recovery experiment is performed with N relaxation time points and M scans per complex point in t1 for each relaxation time point then the final data control unsaturated measurement consists of 2MN co-added FIDs per complex point in t1. The inversion-recovery curves are fit simultaneously to the equations:
| (1) |
in which ISS and R1 correspond to the steady-state 15N magnetization intensity, and the longitudinal relaxation rate constant, respectively. The initial magnetizations are treated as local variables, while ISS and R1 are treated as global parameters. The NOE is calculated as the ratio of Iss and the average intensity of the control experiment, I0.
Figure 1.
Pulse sequence for simultaneous measurement of the steady state {1H}-15N NOE and the 15N R1 relaxation rate constant. Thin and thick solid bars represent high-power 90° and 180° pulses, respectively; short open shapes represent water-selective 90° sinc pulses; short thin solid bars represent rectangular water-selective 90° pulses; thick solid bars flanked by two thin bars indicate a 90x-210y-90x composite pulse; and the open shaped pulses during the T = n(2Δ – τ180) relaxation period are cosine-modulated I-BURP2 180° pulses of length τ180 crafted to leave the water magnetization unperturbed while maximizing inversion of amide protons. Delays: trecycle = 11 s, Δ = 11 ms, δ = τ = 2.65 ms, γ = δ – ε/2, and ε > g6. Gradients were applied as square pulses. Gradients g1 and g6 were used for coherence selection and other gradients are used for artifact suppression. Gradients: g1 (600 μs, 21.7 G/cm), g2 (300 μs, 6.8 G/cm), g3 (1000 μs, 32.9 G/cm), g4 (500 μs, 6.8 G/cm), g5 (1000 μs, 13.2 G/cm), g6 (121.60 μs, 21.7 G/cm). All pulses are x-phase unless indicated. Phase cycling: ϕ1 = y; ϕ2 = y, −y, and ϕrec = y, −y. The phase ϕ3 = y for I−(0) and −x for I+(0) in the R1 experiment. Echo-antiecho selection is obtained by inverting g1 and ϕ1. Pulse sequence and acquisition parameter files are provided as Online Resource 1 and 2, respectively
The proposed pulse sequence was used to measure 15N R1 and steady-state 15N-{1H} NOE for a 1.0 mM sample of [U-2H,15N] ubiquitin. Sample temperature was calibrated to 298 K using 98% 2H4-methanol12. Relaxation data were collected at 14.1 T on a Bruker DRX600 console equipped with a triple-resonance z-axis gradient cryogenic probe. Spectra were recorded using 4 scans per complex t1 point, and t2 × t1 of 512 × 128 complex points. The spectral width was set to 9.3 kHz × 2.4 kHz. Relaxation delays were set to T = n × 22 ms, where n = {2, 4, 10, 18, 44, 88, 240, 464}, giving T = (0.044, 0.220, 0.396, 0.968, 1.936, 5.280, 10.208} s. Spectra were processed using NMRPipe13, and in-house Python scripts were used to fit relaxation curves for calculation of R1 and Iss values. The proposed pulse sequence was validated by comparison with results from independent conventional measurements of 15N R1 and steady-state 15N-{1H} NOE using TROSY-based pulse sequences9. The conventional NOE experiment used 16 scans per complex t1 point for the control and the saturated spectra; hard 180° 1H pulses were used to saturate 1H magnetization. The conventional R1 experiment used a recycle delay of 3.5 s, 8 scans per complex t1 point, and 8 relaxation delays spaced between 0 and 1.6 s. Total acquisition times were 32, 13, and 20 hours for the proposed experiment, conventional NOE experiment, and conventional R1 experiment, respectively. Experimental uncertainties in relaxation parameters are the standard error in the mean of three replicates.
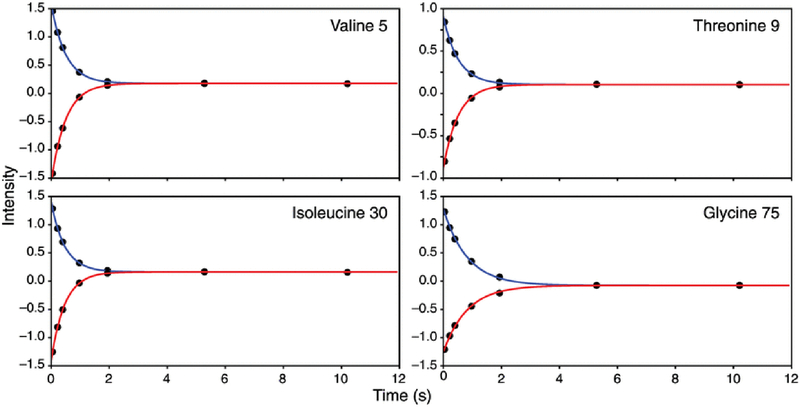
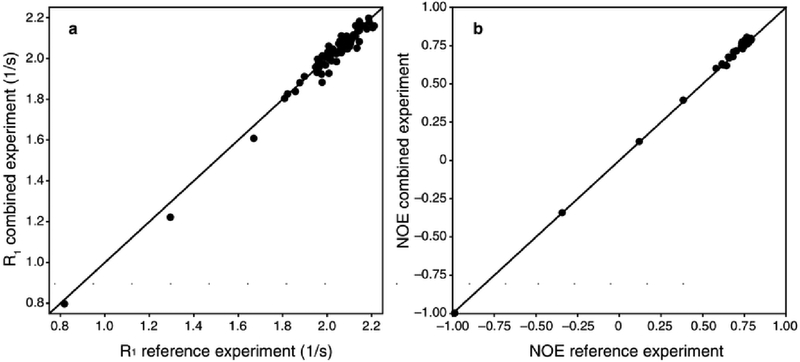
Figure 2 shows representative R1 relaxation decay curves for Val 5, Thr 9, Ile 30, and Gly 75, which are located in β-sheet, turn, α-helix, and C-terminal regions of ubiquitin, respectively. The absolute value of the initial magnetization for each set of the R1 curves differed by approximately 2% intensity, with the I+(T) curve yielding the larger intensity values. As shown in Figure 3, the R1 and NOE values from the combined experiment are in excellent agreement with reference TROSY-detected experiments for ubiquitin. Comparison of 15N R1 values between the two experiments gives a correlation coefficient of 0.974 and an RMSD of 0.032. The mean square errors in the measured R1 values are 0.012 and 0.010 for the conventional and combined experiments. Comparison of steady-state 15N-{1H} NOE values gives a correlation coefficient of 0.998 and an RMSD of 0.016. The mean square errors in the measured NOE values are 0.014 and 0.010 for the conventional and combined experiments. Corrected for differences in acquisition times, the NOE values for the conventional and combined experiments have very similar root-mean-square uncertainties of 0.00925 and 0.0102, respectively.
Figure 2.
15N R1 decay curves for Val 5, Thr 9, Ile 30, and Gly 75 in ubiquitin. Data points and fitted decay curves are shown for (blue) positive (I+(T)) and (red) negative (I−(T)) initial 15N magnetization. Data were fit using Eq. 1 using a Levenberg-Marquardt algorithm. Fitted values for R1 are (Val 5) 2.001 ± 0.001, (Thr 9) 1.927 ± 0.016, (Ile 30) 2.1484 ± 0.016, and (Gly 75) 797 ± 0.009 (uncertainties are standard error in the mean of three replicates)
Figure 3.
Comparison of simultaneous and conventional measurements of (a) 15N R1 relaxation rate constant (R2 = 0.974, RMSD = 0.032) and (b) steady-state {1H}-15N NOE (R2 = 0.998, RMSD = 0.016). Uncertainties in measured values are not shown. The mean square errors in the measured R1 values are 0.012 and 0.010 for the conventional and combined experiments. The mean square errors in the measured NOE values are 0.014 and 0.010 for the conventional and combined experiments. Data values are provided as Online Resource 3
The proposed combined experiment differs from separately acquired NOE and R1 datasets in three ways. (i) The resonance intensities for the “saturated” spectrum, needed to calculate the NOE, are not directly measured, but are extrapolated during the fitting of the positive and negative R1 relaxation curves; (ii) longitudinal 1H magnetization is inverted for a period τ + t1 prior to the TROSY transfer period; and (iii) non-equilibrium longitudinal 15N magnetization relaxes during the first acquisition t2 period prior to the start of the R1 relaxation delay. Monte Carlo simulations of these effects were simulated for a protein with tumbling time 5 ns and 15 ns, using a 600 MHz spectrometer (full protonation was assumed for simplicity, which should give an upper bound on sensitivity differences). Simulations assumed that the conventional NOE experiments were recorded with 32 scans per FID for each of the control and saturated spectra. The combined experiment used 64 scans to measure the control spectrum and simultaneously used N = 8 time points to measure R1, with 4 scans for positive initial 15N magnetization and 4 scans for negative initial magnetization. The simulations indicate that the two different NOE measurements have virtually identical sensitivity per unit time for either the 5 ns and 15 ns rotational diffusion correlation times. The R1 measurement has a lower precision by 15% and ~10% compared with a conventional experiment measured with the same number of scans and time points, for 5 ns and 15 ns tumbling times, respectively. This difference arises from the small relaxation losses noted above. The precision of the R1 experiment is ~1%, so these small differences are not a major effect on subsequent data analysis. Notably the simulations show that the values of the NOE and R1 for the combined experiment are uncorrelated, with R2 < 0.01. The theoretical calculations are in approximate agreement with the experimental results reported for ubiquitin. The combined experiment yields both the NOE and R1; thus, R1 is obtained essentially for free. The time savings afforded by the combined experiment depends on the relative lengths of the conventional NOE and R1 experiments. For example, savings in experimental time would be 20 or 33% if the conventional R1 experiment were to be recorded in 1/4 or 1/2 of the experimental time for the conventional NOE measurement.
We have described a TROSY pulse sequence for simultaneous measurement of the intensity of the Boltzmann equilibrium 15N magnetization and the 15N longitudinal relaxation rate constant R1 in a single experiment. Long-time plateau values from fitting of the R1 data serve as proxies for the usual saturated {1H}-15N intensity to allow calculation of the steady-state {1H}-15N NOE. Measurements of 15N R1 and steady-state 15N-{1H} NOE on a ubiquitin sample using both the combined experiment and conventional independent experiments are in very good agreement. The greater efficiency of the proposed pulse sequence should become even more useful for larger bio-molecular systems, for which maximizing sensitivity per unit time is critical.
Supplementary Material
Acknowledgments
Support from National Institutes of Health grants R01 GM050291 (A.G.P.) and T32 GM008281 (P.A.O.) is acknowledged gratefully. We thank Mark Rance (University of Cincinnati) for helpful discussions. Some of the work presented here was conducted at the Center on Macromolecular Dynamics by NMR Spectroscopy located at the New York Structural Biology Center, supported by a grant from the NIH National Institute of General Medical Sciences (P41 GM118302). A.G.P. is a member of the New York Structural Biology Center.
References
- 1.Palmer AG NMR characterization of the dynamics of biomacromolecules. Chem Rev 104, 3623–40 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Peng JW & Wagner G Frequency spectrum of NH bonds in eglin c from spectral density mapping at multiple fields. Biochemistry 34, 16733–52 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Abyzov A et al. Identification of dynamic modes in an intrinsically disordered protein Using temperature-dependent NMR relaxation. J Am Chem Soc 138, 6240–51 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Gill ML, Byrd RA & Palmer AG Dynamics of GCN4 facilitate DNA interaction: a model-free analysis of an intrinsically disordered region. Phys Chem Chem Phys 18, 5839–49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu A, O'Brien PA, Bhattacharya S, Rance M & Palmer AG Enhanced spectral density mapping through combined multiple-field deuterium 13CH2D methyl spin relaxation NMR spectroscopy. Methods (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavanagh J, Fairbrother WJ, Palmer Iii AG, Rance M & Skelton NJ Protein NMR Spectroscopy (Second Edition) 679–724 (Academic Press, Burlington, 2007). [Google Scholar]
- 7.Zhu G, Xia Y, Nicholson LK & Sze KH Protein dynamics measurements by TROSY-based NMR experiments. J Magn Reson 143, 423–6 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Favier A & Brutscher B Recovering lost magnetization: polarization enhancement in biomolecular NMR. J Biomol NMR 49, 9–15 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Lakomek NA, Ying J & Bax A Measurement of 15N relaxation rates in perdeuterated proteins by TROSY-based methods. J Biomol NMR 53, 209–21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrage F, Cowburn D & Ghose R Accurate sampling of high-frequency motions in proteins by steady-state 15N-{1H} nuclear Overhauser effect measurements in the presence of cross-correlated relaxation. J Am Chem Soc 131, 6048–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gairi M et al. An optimized method for 15N R1 relaxation rate measurements in non-deuterated proteins. J Biomol NMR 62, 209–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Findeisen M, Brand T & Berger SA 1H-NMR thermometer suitable for cryoprobes. Magn Reson Chem 45, 175–8 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Delaglio F et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6, 277–93 (1995). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.