Abstract
Distributed word representations have become an essential foundation for biomedical natural language processing (BioNLP), text mining and information retrieval. Word embeddings are traditionally computed at the word level from a large corpus of unlabeled text, ignoring the information present in the internal structure of words or any information available in domain specific structured resources such as ontologies. However, such information holds potentials for greatly improving the quality of the word representation, as suggested in some recent studies in the general domain. Here we present BioWordVec: an open set of biomedical word vectors/embeddings that combines subword information from unlabeled biomedical text with a widely-used biomedical controlled vocabulary called Medical Subject Headings (MeSH). We assess both the validity and utility of our generated word embeddings over multiple NLP tasks in the biomedical domain. Our benchmarking results demonstrate that our word embeddings can result in significantly improved performance over the previous state of the art in those challenging tasks.
Subject terms: Machine learning, Literature mining
| Design Type(s) | data transformation objective • data integration objective • text processing and analysis objective |
| Measurement Type(s) | word representation |
| Technology Type(s) | Text_Mining |
| Factor Type(s) | |
| Sample Characteristic(s) |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Background & Summary
Distributed word representations learn dense and low-dimensional word embeddings from large unlabeled corpora and effectively capture the implicit semantics of words1–3. The low-dimensional word embedding is much more suitable for the recent neural-based deep learning models than the traditional one-hot representation. Based on word embeddings, the recent deep learning methods have been successfully applied to various natural language processing (NLP) tasks4–6. With the rapid advance in deep learning, word embeddings have become an integral part of NLP models and attracted significant attention.
In recent years, several word embedding models and pre-trained word embeddings1,7,8 have been made publicly available and successfully applied to many biomedical NLP (BioNLP) tasks. More recently, Wang et al. compared the performance of different word embeddings which were trained on four kinds of corpora including clinical notes, biomedical literature, Wikipedia and news articles9. Smalheiser et al. proposed a novel vector representation of words based on the similarity and co-occurrence frequency of words10.
However, the traditional biomedical word embeddings described above have two limitations. First, most of them were trained using the word2vec1 or GloVe model7, which uses a distinct vector to represent each word and ignores the internal structure of words. Such models are not particularly good at learning rare or out of vocabulary (OOV) words in the training data. If a word embedding model can capture the subword information and exploit the internal structure of words to augment the embedding representations in those rare or OOV words, it has the potential to greatly benefit various BioNLP applications. Bojanowski et al. recently proposed a novel embedding model11, which can effectively use the subword information to enrich the final word embedding results. In contrast to the word2vec model1, the subword embedding model makes use of the representations of character n-grams based on the unlabeled corpora, and then uses the sum of the n-gram vectors to represent the final word vector.
Moreover, existing word embedding models mainly focus on using the single source of large text corpora in PubMed and/or PubMed Central (PMC). Recently, some studies12–15 have suggested that integrating domain knowledge with the text corpora can be beneficial to improve the quality of word embeddings. In the biomedical domain, there are abundant biomedical knowledge data such as the medical subject headings (MeSH) and unified medical language system (UMLS), which could be explored to complement the textual information in the literature. Intuitively, integrating such biomedical domain knowledge should help improve the quality of word embedding such that it better captures the semantics of specialized terms and concepts.
In this work, we create BioWordVec: a new set of word vectors/embeddings using the subword embedding model on two different data sources: biomedical literature and domain knowledge in MeSH. Specifically, we construct a MeSH term graph based on the MeSH RDF data, followed by a random sampling strategy to generate a number of MeSH term sequences. Subsequently, we use the subword embedding model to learn the text sequences and MeSH term sequences in a unified n-gram embedding space. Our word embeddings are assessed for both validity and utility on multiple BioNLP tasks. As shown in our experimental results, our word embeddings outperform the current state-of-the-art word embeddings in all benchmarking tasks, suggesting that the subword information and domain knowledge is indeed able to improve the quality of biomedical word representations and better capture their semantics.
Methods
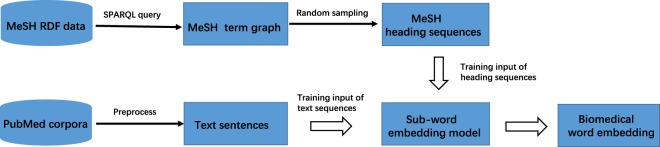
In this section, we present our method for learning biomedical word embeddings. This method consists of two steps: 1) constructing MeSH term graph based on its RDF data and sampling the MeSH term sequences and 2) employing the fastText subword embedding model to learn the distributed word embeddings based on text sequences and MeSH term sequences. A schematic overview of our method is shown in Fig. 1.
Fig. 1.
Schematic of learning word embedding based on PubMed literature and MeSH.
Sampling MeSH term sequences
Recent studies (e.g.16,17) inspired by the skip-gram model16,17 have proposed to translate network/graphs into nodes sequences to learn networks embeddings. Similarly, in this work we transfer the relations of the MeSH term graph into ordered sequences of the heading nodes. This process results in main-heading sequences from MeSH and we subsequently combine them with PubMed sentence sequences for learning word embeddings.
There are two common sampling strategies: breadth-first sampling (BFS) and depth-first sampling (DFS). BFS gives the priority to sample the immediate neighbors of the source node, whereas DFS first samples the nodes as far as possible along each branch before backtracking. Grover et al.18 proposed a random walk procedure called node2vec that efficiently samples diverse neighborhoods in a network. In this work, we adopted this strategy to sample the sequences of main-heading nodes from the MeSH term graph. Specifically, let G, N and E denote the MeSH term graph, the node and edge set, respectively. A random walk is simulated to sample the sequence of a source node from G, which is guided by two parameters p and q. Let the random walk c starts with the node u (), and , and denote the three continuous nodes t, v and x in the random walk. The generated distribution of ci is defined as follow:
| 1 |
where πvx is the transition probability from node v to x. The transition probability πvx is defined as follows:
| 2 |
where dtx is the shortest path between node t and x. Note that dtx must be one of {0, 1, 2} because nodes t, v and x are three continuous nodes in a walk.
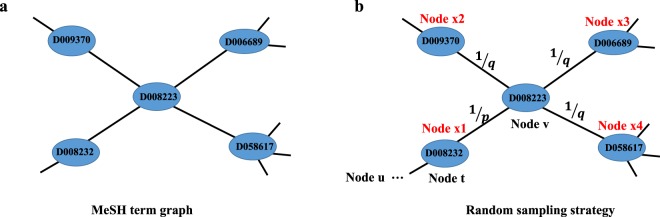
In Fig. 2, we show an example MeSH term graph that contains five MeSH term nodes. Each MeSH term node is represented by its corresponding ID. The edges between the MeSH term nodes represent the relations between MeSH terms based on the MeSH RDF data. For example, the MeSH term nodes “D008232” and “D008223” represent MeSH headings “Lymphoproliferative Disorders” and “Lymphoma”, respectively. Based on the MeSH RDF data, the relation from MeSH headings “Lymphoproliferative Disorders” to “Lymphoma” is “meshv:broaderDescriptor”, which means that “Lymphoma” is included by the higher level heading “Lymphoproliferative Disorders”. In MeSH term graph, we simply use the undirected edges to represent the relations between MeSH terms, and do not distinguish the types and direction of the relations. Figure 2b illustrates the MeSH sequences sampling strategy based on the 2nd order random walk. Suppose the random walk starting with node u just traversed from node t (“D008232”) to node v (“D008223”), and we have four choices for the next step (“D008232”, “D009370”, “D006689” and “D058617”). For the node x1 (“D008232”), the transition probability is 1/p based on the equation (1) and (2), because the shortest path distance dtx between node t (“D008232”) and node x1 (“D008232”) is 0. Similarly, the transition probability from node v (“D008223”) to node x2 (“D009370”), node x3 (“D006689”), node x4 (“D058617”) are all 1/q. Based on the equation (2), the two parameters p and q make the sampling strategy effectively combine the BFS and DFS. In our work, we applied this sampling strategy to simulate random walks starting from each main-heading node in MeSH and generated the MeSH term sequences. As a result, we transform the MeSH term graph into a set of sampling sequences.
Fig. 2.
Illustration of the MeSH sequences sampling strategy. (a) An example of MeSH term graph. (b) Random sampling strategy.
A sequence sampled from the MeSH term graph is essentially an ordered set of MeSH main-heading nodes D1, D2, …, Dl where l is the sampling length parameter. For the sampling length, we empirically set as 100. Each MeSH term node is represented by its corresponding ID such as “D008232”, “D008223” and “D058617”. For example, “…, D008232, D008223, D058617, …” may be one of the MeSH term sequences sampled from Fig. 2a. Unlike previous studies such as16–18 that aim to learn embeddings for nodes, we transform MeSH IDs sequences into word sequences so that they can be treated equally as PubMed sentences during the learning of word embeddings. In this study, we directly used the text label of each MeSH ID in the MeSH RDF data (e.g. we use “lymphoproliferative disorders” for “D008232”). As a result, the list of MeSH IDs is turned into a text sequence consisting of words only.
Subword embedding model
Bojanowski et al. proposed fastText: a subword embedding model11 based on the skip-gram model1 that learns the character n-grams distributed embeddings using unlabeled corpora where each word is represented as the sum of the vector representations of its n-grams. Compared to the word2vec model1, the subword embedding model can make effective use of the subword information and internal word structure to improve the embedding quality. In the biomedical domain, many specialized compound words, such as “deltaproteobacteria”, are rare or OOV in the training corpora, thus making them difficult to learn properly using the word2vec model. In contrast, the subword embedding model is naturally more suitable to deal with such situations. For instance, since “delta”, “proteo” and “bacteria” are common in the training corpora, the subword embedding model can learn the distributed representations of all character n-grams of “deltaproteobacteria”, and subsequently integrate the subword vectors to create the final embedding of “deltaproteobacteria”. In this study, we apply the subword embedding model to learn word embeddings from the joint text sequences of PubMed and MeSH.
The fastText subword embedding model11 is essentially a variant of the continuous skip-gram model1. Given a training word sequence w1, w2, …, wT, the objective function of the skip-gram model is defined as follow:
| 3 |
where Ct is the set of the surrounding words of wt. Given the current word wt, the is defined as the probability of observing its surrounding word wc.
| 4 |
where s(wt, wc) is the scoring function. The original skip-gram model defines the scoring function as scalar product, namely , where uwt and vwc are the vectors of two words wt and wc, respectively. This means the original skip-gram model can only learn a distinct vector for each word but cannot exploit subword information.
To address this issue, the subword embedding model represents a word as a bag of character n-grams. For example, the word “function” will be represented by the character 4 grams including <#fun, func, unct, ncti, ctio, tion, ion#> and the word itself <function>. The major difference between subword embedding model and the original skip-gram model is that the subword embedding model defines the s(wt, wc) as the , where (1, …, G) is the n-gram set of wt. zg is the vector of character n-gram g, and vc is the vector of word wc. Hence, the subword embedding model learns the distributed representation of character n-grams. Based on these n-grams representations, a word is represented as the sum of the vector representations of its n-grams. The advantage of the subword embedding model is that it shares the representations of n-grams across words, which is significantly helpful for learning reliable embedding for rare or OOV words.
In this work, the input of the subword model is the joint text sequences from PubMed and MeSH. For the PubMed text, the model aims to maximize the objective function , where T is the total vocabulary size. For the MeSH term sequences, the model aims to maximize the objective function , where N is the total number of MeSH main headings. We linearly combined the above two objective functions as follow:
| 5 |
The training of our model is to maximize the above objective function in (5), which will learn the joint PubMed text sequences and MeSH term sequences in word embedding. Since both the PubMed sentence sequences and MeSH term sequences are word sequences, the subword embedding model can share the n-gram representations between the PubMed text words and MeSH term terms, thus integrating the PubMed and MeSH into a unified embedding space. The JPubMed and JMeSH are trained together by the subword embedding model.
Implementation details
In our experiments, we downloaded the PubMed XML source files from https://www.nlm.nih.gov/databases/download/pubmed_medline.html. Our PubMed data contains 27,599,238 articles including the titles and abstracts. We extracted the title and abstract texts from the PubMed XML files to construct the PubMed text data. All words were converted to lowercase. The final PubMed text data contain 3,658,450,658 tokens.
For MeSH, we downloaded its RDF data (ftp://ftp.nlm.nih.gov/online/mesh/rdf/) on 3/19/2018. The MeSH terms consist of descriptor terms, qualifier terms and supplementary concept record terms. The MeSH descriptor terms are known as main headings for describing the core subjects of a PubMed article. Thus in this study, we focus on the MeSH descriptors terms. Note that it is not straightforward to handle punctuation marks such as comma in MeSH descriptor terms given their different uses. For example, comma is used differently between D001990 (“Bronchiolitis, Viral”) and D013676 (“Technology, Industry, and Agriculture”). Hence in this work, we simply removed them from the MeSH descriptor terms (this pre-processing step is to be improved in the future but is beyond the scope of this work) and converted the words to lowercase. We used SPARQL queries to retrieve the relations between descriptors terms from the MeSH RDF data, resulting in a MeSH term graph with 28,436 main heading nodes and 52,013 relations. For each main heading node, we sampled 10 MeSH term sequences, resulting in a total of 284,360 MeSH term sequences.
Our word embeddings were trained by the following hyper-parameters empirically. For the sampling strategy, the two parameters p and q were set as 2 and 1, respectively. For each node in MeSH term graph, we sampled 10 sequences of fixed length (l = 150). The dimension of the word vectors was set to be 200, and the size of the negative sample size was set to be 10. Similar to Bojanowski et al.11, all n-grams (3 ≤ n ≤ 6) were extracted by the subword model for training word representations.
Data Records
Word embeddings are commonly used and evaluated in two types of (Bio-)NLP tasks: intrinsic and extrinsic. For intrinsic tasks, word embeddings are used to calculate or predict semantic similarity between words, terms or sentences. For extrinsic tasks, word embeddings are used as the input for various downstream NLP tasks, such as relation extraction or text classification. Chiu et al.8 suggested that the extrinsic tasks benefit from smaller window size while the opposite the intrinsic tasks. In our preliminary experiments, we also observed similar results: when setting the context window size as 20 and 5, our word embedding achieved the highest performance in intrinsic and extrinsic evaluation, respectively. Hence in this work, we followed their lead and created two specialized, task-dependent sets of word embeddings via setting the context window size as 20 and 5, respectively. Our BioWordVec data are freely available on Figshare19. Both sets are in binary format and contain 2,324,849 distinct words in total where 2,309,172 words come from the PubMed and 15,677 from MeSH. All words were converted to lowercase and the number of dimensions is 200.
Our word embeddings can effectively integrate the MeSH term sequences to improve the representation of such terms or concepts. In Table 1, we show a randomly selected set of term pair examples from the manually-annotated UMNSRS-Sim20 and UMNSRS-Rel20 datasets and calculated the cosine similarity of the term pairs. It can be seen that all term pairs in Table 1 have relatively high scores from both UMNSRS-Sim20 and UMNSRS-Rel20. For a good word embedding method, it should yield similarly high cosine similarity scores for these pairs. Table 1 shows that the cosine similarity score calculated by our word embedding is higher than the other word embeddings1,8,11,21. For example, the cosine similarity between “mycosis” and “histoplasmosis” is 0.353, 0.544, and 0.595 by Mikolov et al.1, Pyysalo et al.21 and Chiu et al.8, respectively, but 0.706 by our word embedding. On the other hand, it is difficult to determine how high their absolute cosine similarity score should be. Hence, we further performed technical validation using the Pearson’s correlation coefficient and Spearman’s correlation coefficient in the following Section.
Table 1.
The cosine similarity of the word pair examples by different word embeddings.
| Word pair | UMNSRS-Sim20 | UMNSRS-Rel20 | Mikolov et al.1 | Pyysalo et al.21 | Chiu et al.8 | BioWordVec (win 20) |
|---|---|---|---|---|---|---|
| thalassemia, hemoglobinopathy | 1307 | 1218 | — | 0.713 | 0.754 | 0.834 |
| mycosis, histoplasmosis | 1137.25 | 1185.75 | 0.353 | 0.544 | 0.595 | 0.706 |
| thirsty, hunger | 935.75 | 1249 | 0.252 | 0.425 | 0.59 | 0.629 |
| influenza, pneumoniae | 898.5 | 1354 | 0.482 | 0.252 | 0.514 | 0.611 |
| atherosclerosis, angina | 936 | 1357.75 | 0.503 | 0.506 | 0.506 | 0.589 |
In particular, our word embeddings can make good use of the sub-word information and internal structure of words to improve the representations of the rare words, which is highly valuable for BioNLP applications. For example, the word “deltaproteobacteria” is a rare word even in the biomedical corpus. In Table 2, we gave the top 5 most similar words of “deltaproteobacteria” by our method vs. Chiu et al.8. It can be seen that our method capture the similar words of “deltaproteobacteria” better than Chiu et al.8. Due to the common sub-words “proteo” and “bacteria”, our method can easy capture the similar words such as “betaproteobacteria” and “zetaproteovacteria”.
Table 2.
The top 5 most similar words of “deltaproteobacteria”.
| BioWordVec (win20) | Chiu et al.8 | ||
|---|---|---|---|
| Top 5 similar words | Similarity score | Top 5 similar words | Similarity score |
| deltaproteobacterial | 0.985 | magnetospirilla | 0.861 |
| deltaproteobacterium | 0.963 | Thermales | 0.857 |
| betaproteobacteria | 0.952 | Acidiphilium-like | 0.854 |
| zetaproteobacteria | 0.945 | nirK1 | 0.85 |
| delta-proteobacteria | 0.939 | nostoc | 0.847 |
“win20” denotes the BioWordVec was trained by setting the context window size as 20.
Technical Validation
To validate our method, two widely-used benchmarking datasets UMNSRS-Sim20 and UMNSRS-Rel20 were employed. UMNSRS-Sim and UMNSRS-Rel respectively consist of 566 and 587 term pairs, with their corresponding relatedness/similarity scores manually judged by the domain experts from the University of Minnesota Medical School. For evaluation, we first use word embeddings to calculate a cosine similarity score for each term pair. Then, we measure the Pearson’s correlation coefficient and Spearman’s correlation coefficient between the computed scores and those provided by human experts. We compared our method with several state-of-the-art methods1,8,11,21. Mikolov et al.1 proposed the word2vec model and provided the pre-trained word embeddings on Google news. Using the same word2vec model, Chui et al.8 and Pyysalo et al.21 provide biomedical word embeddings based on PubMed and PubMed Central articles.
Our method has two variants: One was trained with only PubMed data, and the other using both PubMed and MeSH data. From Table 3, it can be seen that our method significantly outperforms the other methods. The results suggest that the subword information and MeSH data are valuable and helpful in biomedical domain. We also noticed that the biomedical corpus was more suitable in this case than the general English corpus. Both Mikolov et al.1 and Pyysalo et al.21 used the same word2vec model and default parameters, but the word embeddings trained on PubMed and PMC corpus significantly outperformed the ones trained by Google news in our results.
Table 3.
Evaluation results on UMNSRS datasets.
| Method | Corpus | UMNSRS-Sim | UMNSRS-Rel | ||||
|---|---|---|---|---|---|---|---|
| # | Pearson | Spearman | # | Pearson | Spearman | ||
| Mikolov et al.1 | Google news | 336 | 0.421 | 0.409 | 329 | 0.359 | 0.347 |
| Pyysalo et al.21 | PubMed + PMC | 493 | 0.549 | 0.524 | 496 | 0.495 | 0.488 |
| Chiu et al.8 | PubMed | 462 | 0.662 | 0.652 | 467 | 0.600 | 0.601 |
| BioWordVec (win20) | PubMed | 521 | 0.665 | 0.654 | 532 | 0.608 | 0.607 |
| BioWordVec (win20) | PubMed + MeSH | 521 | 0.667 | 0.657 | 532 | 0.619 | 0.617 |
“#” denotes the number of the term pairs that can be mapped by the different word embeddings. “Pearson” and “Spearman” denote the Pearson’s correlation coefficient score and Spearman’s correlation coefficient score, respectively. “win20” denotes the BioWordVec was trained by setting the context window size as 20. The highest value is shown in bold.
In Table 3, we show that our method achieves better performance on both datasets. Chui et al.8 also achieves competitive performance which significantly outperforms Mikolov et al.1 and Pyysalo et al.21. In Table 3, the correlation results are not directly comparable because the approaches were evaluated on different sets of term pairs. Hence, we show in Table 4 the results on the common set of term pairs found by both our method and Chui et al.8, which include 459 and 461 pairs in UMNSRS-Sim and UMNSRS-Rel, respectively. It can be seen that the performance improvement is greater by our method on these two common sets.
Table 4.
Comparison results on UMNSRS datasets using the common term pairs.
| Method | Corpus | UMNSRS-Sim | UMNSRS-Rel | ||||
|---|---|---|---|---|---|---|---|
| # | Pearson | Spearman | # | Pearson | Spearman | ||
| Chiu et al.8 | PubMed | 459 | 0.661 | 0.651 | 461 | 0.600 | 0.601 |
| BioWordVec (win20) | PubMed | 459 | 0.679 | 0.665 | 461 | 0.624 | 0.626 |
| BioWordVec (win20) | PubMed + MeSH | 459 | 0.681 | 0.668 | 461 | 0.633 | 0.635 |
“#” denotes the number of the term pairs that can be mapped by the different word embeddings. “Pearson” and “Spearman” denote the Pearson’s correlation coefficient score and Spearman’s correlation coefficient score, respectively. “win20” denotes the BioWordVec was trained by setting the context window size as 20. The highest value is shown in bold.
Usage Notes
We demonstrate here the application of BioWordVec in two separate use cases: finding similar sentences and extracting biomedical relations.
Use case 1: sentence pair similarity
Word embeddings are often used to calculate sentence pair similarity22. In the general domain, the SemEval Semantic Textual Similarity (SemEval STS) challenge has been organized for over five years, which calls for effective models to measure sentence similarity23. Averaged word embeddings are used as a baseline to measure sentence pair similarity in the challenges: each sentence is transformed as a vector by averaging the word vectors for each word in the sentence and sentence pair similarity is effectively measured by the similarity between the averaged vectors using common measures such as Cosine and Euclidean similarity.
Sentence similarity is also critical in biomedical and clinical domains24,25. We conducted a case study to quantify the effectiveness of the proposed embeddings in the task of computing sentence pair similarity on clinical texts. We used the BioCreative/OHNLP STS dataset, which consists of 1,068 pairs of sentences derived from clinical notes and were annotated by two medical experts on a scale of 0–5, from completely dissimilar to semantically equivalent26. The top-ranked submission model used average embeddings with different similarity functions, which was shown effective to capture sentence similarity27. We applied averaged word embedding approach and adopted Cosine, Euclidean and City Block similarity to measure the averaged vectors. The result was evaluated based on Pearson’s Correlation between the predicted similarities and gold standard labels.
Table 5 shows the evaluation results on clinical sentence pair similarity. Our proposed embeddings achieved higher correlations in all three similarity measures. This demonstrates that the proposed embeddings more effectively capture the semantic meaning. On the other hand, we also noted that “BioWordVec (win20) w/o MeSH” and “BioWordVec (win20) w/MeSH” achieved similar correlation scores in all three similarity measures, which indicates integrating MeSH was not much helpful in this task. It is likely because MeSH plays a more vital role in PubMed articles than in clinical notes.
Table 5.
Sentence pair similarity results on BioCreative/OHNLP STS dataset.
| Similarity measures | Mikolov et al.1 | Pyysalo et al.21 | Chiu et al.8 | BioWordVec (win20) w/o MeSH | BioWordVec (win20) w/MeSH |
|---|---|---|---|---|---|
| Cosine | 0.768 | 0.755 | 0.757 | 0.770 | 0.771 |
| Euclidean | 0.725 | 0.723 | 0.727 | 0.751 | 0.753 |
| Block | 0.725 | 0.722 | 0.727 | 0.750 | 0.752 |
“win20” denotes the BioWordVec was trained by setting the context window size as 20.
Use case 2: biomedical relation extraction
Given that word embeddings are often used as the input in recent deep-learning based methods for various biomedical NLP tasks4–6, below we evaluate its effect in some biomedical relation extraction tasks. In our experiments, we evaluate its effect in two biomedical relation extraction tasks: protein-protein interaction (PPI) extraction and drug-drug interaction (DDI) extraction, respectively. The former is a binary relation extraction task, whereas the latter a multi-class relation extraction task. Following previous studies4,28, we use precision, recall and F-score as evaluation metrics and choose the same baseline methods.
The public PPI corpora were used for the PPI extraction, including AIMed29, BioInfer30, IEPA31, HPRD5032 and LLL33. The detailed statistics of the PPI corpora is listed in Table 6. For this binary relation extraction task, we implemented a convolutional neural network (CNN) model and used the dropout layer with a dropout rate of 0.5 after the embedding layer and before the output layer. For the input of our CNN model, we combine the position embeddings with the word embeddings as they have been shown to be effective34. The PPI extraction experiments were evaluated with 10-fold document-level cross validation. As shown in Table 7, our method achieves the highest F-score on all datasets except one (Chiu et al.8 achieved higher F-score than our method on AIMed). Knowledge from MeSH was helpful in all datasets.
Table 6.
The statistics of the PPI corpora.
| Corpus | Sentences | Positive | Negative | Total |
|---|---|---|---|---|
| AIMed | 1955 | 1,000 | 4,834 | 5,834 |
| BioInfer | 1100 | 2,534 | 7,132 | 9,666 |
| IEPA | 145 | 335 | 482 | 817 |
| HPRD50 | 486 | 163 | 270 | 433 |
| LLL | 77 | 164 | 166 | 300 |
Table 7.
PPI extraction evaluation results on five PPI corpora.
| Data Set | Mikolov et al.1 | Pyysalo et al.21 | Chiu et al.8 | BioWordVec (win5) w/o MeSH | BioWordVec (win5) w/MeSH | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| F-Score | σ | F-Score | σ | F-Score | σ | F-Score | σ | F-Score | σ | |
| AIMed | 0.445 | 0.076 | 0.457 | 0.087 | 0.492 | 0.064 | 0.484 | 0.101 | 0.487 | 0.081 |
| BioInfer | 0.524 | 0.038 | 0.532 | 0.044 | 0.545 | 0.053 | 0.543 | 0.041 | 0.549 | 0.039 |
| IEPA | 0.603 | 0.062 | 0.597 | 0.062 | 0.615 | 0.061 | 0.617 | 0.049 | 0.623 | 0.064 |
| HPRD50 | 0.484 | 0.187 | 0.499 | 0.121 | 0.481 | 0.145 | 0.504 | 0.136 | 0.511 | 0.13 |
| LLL | 0.679 | 0.12 | 0.688 | 0.093 | 0.684 | 0.124 | 0.708 | 0.092 | 0.713 | 0.095 |
The highest value is shown in bold. “σ” denotes the standard deviation of the F-score. “win5” denotes the BioWordVec was trained by setting the context window size as 5.
For the DDI extraction, we used the DDI 2013 corpus35,36, which is manually annotated and consists of five different DDI types, including Advice, Effect, Mechanism, Int and Negative. Since the DDI extraction is a multi-class relation extraction task, we compute the micro average to evaluate the overall performances37,38. In the DDI 2013 corpus, the training set and test set contain 27,792 and 5,716 instances, respectively. We randomly split 10% of the training data as the method validation set and report the results on the test set.
In the DDI extraction experiments, we first applied the same CNN model as the PPI experiments. To further evaluate the performance of different word embeddings on more complex neural models, we also conducted a comparison experiment using a recent state-of-the-art DDI extraction model4 which is a hierarchical RNNs with a input attention layer based on the sentence sequence and shortest dependency path.
The experimental results in Table 8 show that our method achieved the highest F-score on both CNN and RNN models. We also noticed that our method achieved more significant advantage on the simple CNN model than the complex RNN model. For example, our method and Mikolov et al.1 achieved the F-score of 0.687 and 0.636 on CNN model, respectively. The advantage of F-score between our method and Mikolov et al.1 was more than 0.05. When employing the state-of-the-art RNN model, the improvement of F-score reduces to 0.033. This is likely due to the fact that the state-of-the-art DDI extraction model4 already integrates the shortest dependency path information and part-of-speech embedding, as well as using the multiple layer of bidirectional long short-term memory networks (LSTMs) to boost the performance. Taken together, these complex steps/strategies partly reduced the importance of the word embedding for the DDI extraction task.
Table 8.
DDI extraction evaluation results on DDI 2013 corpus.
| Method | Corpus | CNN model | hierarchical RNN model | ||||
|---|---|---|---|---|---|---|---|
| Precision | Recall | F-score | Precision | Recall | F-score | ||
| Mikolov et al.1 | Google news | 0.698 | 0.584 | 0.636 | 0.681 | 0.699 | 0.691 |
| Pyysalo et al.21 | PubMed + PMC | 0.689 | 0.624 | 0.655 | 0.692 | 0.727 | 0.709 |
| Chiu et al.8 | PubMed | 0.709 | 0.650 | 0.677 | 0.749 | 0.691 | 0.719 |
| BioWordVec (win5) | PubMed | 0.696 | 0.669 | 0.683 | 0.744 | 0.702 | 0.722 |
| BioWordVec (win5) | PubMed + MeSH | 0.694 | 0.679 | 0.687 | 0.757 | 0.696 | 0.724 |
The highest value is shown in bold. “win5” denotes the BioWordVec was trained by setting the context window size as 5.
ISA-Tab metadata file
Acknowledgements
This research was supported by the NIH Intramural Research Program, National Library of Medicine. We would like to thank Dr. Chih-Hsuan Wei, Dr. Robert Leaman and Dr. Kyubum Lee for their help with experiments and manuscript proofreading. We are also grateful to Dr. Neil Smalheiser for his valuable comments on this work.
Author Contributions
Y.Z. and Z.L. conceived the study. Y.Z. and Q.C. collected and processed the data. Y.Z., Q.C. and Z.L. analyzed the data. Y.Z. and Q.C. wrote the manuscript. Z.L., Z.Y. and H.L. revised the manuscript. Z.L. supervised the study.
Code Availability
The source code for generating BioWordVec is freely available at https://github.com/ncbi-nlp/BioWordVec. The PubMed data are available from https://www.nlm.nih.gov/databases/download/pubmed_medline.html. The MeSH RDF data are available from https://www.nlm.nih.gov/databases/download/mesh.html.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
ISA-Tab metadata
is available for this paper at 10.1038/s41597-019-0055-0.
References
- 1.Mikolov, T., Sutskever, I., Chen, K., Corrado, G. S. & Dean, J. Distributed representations of words and phrases and their compositionality. In Advances in Neural Information Processing systems26, 3111–3119 (NIPS, 2013).
- 2.Mnih, A. & Kavukcuoglu, K. Learning word embeddings efficiently with noise-contrastive estimation. In Advances in Neural Information Processing Systems26, 2265–2273 (2013).
- 3.Bengio Y, Ducharme R, Vincent P, Jauvin C. A neural probabilistic language model. Journal of Machine Learning Research. 2003;3:1137–1155. [Google Scholar]
- 4.Zhang Y, et al. Drug–drug interaction extraction via hierarchical RNNs on sequence and shortest dependency paths. Bioinformatics. 2018;34:828–835. doi: 10.1093/bioinformatics/btx659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang, D. et al. Learning sentiment-specific word embedding for twitter sentiment classification. In Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics. 1555–1565 (2014).
- 6.Ganguly, D., Roy, D., Mitra, M. & Jones, G. J. Word embedding based generalized language model for information retrieval. In Proceedings of the 38th International Conference on Research and Development in Information Retrieval. 795–798 (2015).
- 7.Pennington, J., Socher, R. & Manning, C. Glove: global vectors for word representation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing. 1532–1543 (2014).
- 8.Chiu, B., Crichton, G., Korhonen, A. & Pyysalo, S. How to train good word embeddings for biomedical NLP. In Proceedings of the 15th Workshop on Biomedical Natural Language Processing. 166–174 (2016).
- 9.Wang Y, et al. A comparison of word embeddings for the biomedical natural language processing. Journal of Biomedical Informatics. 2018;87:12–20. doi: 10.1016/j.jbi.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smalheiser, N. R. & Bonifield, G. Unsupervised low-dimensional vector representations for words, phrases and text that are transparent, scalable, and produce similarity metrics that are complementary to neural embeddings. Preprint at, https://arxiv.org/abs/1801.01884 (2018). [DOI] [PMC free article] [PubMed]
- 11.Bojanowski P, Grave E, Joulin A, Mikolov T. Enriching word vectors with subword information. Transactions of the Association for Computational Linguistics. 2017;5:135–146. doi: 10.1162/tacl_a_00051. [DOI] [Google Scholar]
- 12.Faruqui, M. et al. Retrofitting word vectors to semantic lexicons. In Proceedings of the 2015 Conference of the North American Chapter of the Association for Computational Linguistics. 1606–1615 (2015).
- 13.Yamada, I., Shindo, H., Takeda, H. & Takefuji, Y. Joint learning of the embedding of words and entities for named entity disambiguation. In Proceedings of the 20th SIGNLL Conference on Computational Natural Language Learning. 250–259 (2016).
- 14.Han, X., Liu, Z. & Sun, M. Joint representation learning of text and knowledge for knowledge graph completion. Preprint at, https://arxiv.org/abs/1611.04125 (2016).
- 15.Cao, Y., Huang, L., Ji, H., Chen, X. & Li, J. Bridge text and knowledge by learning multi-prototype entity mention embedding. In Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics. 1623–1633 (2017).
- 16.Perozzi, B., Al-Rfou, R. & Skiena, S. DeepWalk: online learning of social representations. In Proceedings of the 20th International Conference on Knowledge Discovery and Data Mining. 701–710 (2014).
- 17.Tang, J. et al. Line: Large-scale information network embedding. In Proceedings of the 24th International Conference on World Wide Web. 1067–1077 (2015).
- 18.Grover, A. & Leskovec, J. Node2vec: scalable feature learning for networks. In Proceedings of the 22nd International Conference on Knowledge Discovery and Data Mining. 855–864 (2016). [DOI] [PMC free article] [PubMed]
- 19.Zhang, Y., Chen, Q., Yang, Z., Lin, H. & Lu, Z. BioWordVec: improving biomedical word embeddings with subword information and MeSH ontology. Figshare, 10.6084/m9.figshare.6882647.v2 (2018).
- 20.Pakhomov, S. et al. Semantic similarity and relatedness between clinical terms: An Experimental Study. In Proceedings of the 2010 AMIA annual symposium. 572–577 (2010). [PMC free article] [PubMed]
- 21.Pyysalo Sampo, G. F., Moen, H., Salakoski, T. & Ananiadou, S. PubMed-scale event extraction for post-translational modifications, epigenetics and protein structural relations. In Proceedings of the 5th International Symposium on Languages in Biology and Medicine. 39–43 (2012).
- 22.Kenter, T. & De Rijke, M. Short text similarity with word embeddings. In Proceedings of the 24th ACM International Conference on Information and Knowledge Management. 1411–1420 (2015).
- 23.Cer, D., Diab, M., Agirre, E., Lopez-Gazpio, I. & Specia, L. SemEval-2017 task 1: semantic textual similarity-multilingual and cross-lingual focused evaluation. In Proceedings of the 11th International Workshop on Semantic Evaluation. 1–14 (2017).
- 24.Rinaldi, F. et al. Strategies towards digital and semi-automated curation in RegulonDB. Database2017 (2017). [DOI] [PMC free article] [PubMed]
- 25.Chen, Q., Kim, S., Wilbur, W. J. & Lu, Z. Sentence Similarity Measures Revisited: Ranking Sentences in PubMed Documents. In Proceedings of the 2018 International Conference on Bioinformatics, Computational Biology, and Health Informatics. 531–532 (2018).
- 26.Yanshan, W. et al. Overview of the BioCreative/OHNLP challenge 2018 task 2: clinical semantic textual similarity. In Proceedings of the BioCreative/OHNLP Challenge. 1–4 (2018).
- 27.Chen, Q., Du, J., Kim, S., Wilbur, W. J. & Lu, Z. Combining rich features and deep learning for finding similar sentences in electronic medical records. In Proceedings of the BioCreative/OHNLP Challenge. 5–8 (2018).
- 28.Peng, Y., Arighi, C., Wu, C. H. & Vijay-Shanker, K. BioC-compatible full-text passage detection for protein–protein interactions using extended dependency graph. Database2016 (2016). [DOI] [PMC free article] [PubMed]
- 29.Bunescu R, et al. Comparative experiments on learning information extractors for proteins and their interactions. Artificial Intelligence in Medicine. 2005;33:139–155. doi: 10.1016/j.artmed.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 30.Pyysalo S, et al. BioInfer: a corpus for information extraction in the biomedical domain. BMC Bioinformatics. 2007;8:50. doi: 10.1186/1471-2105-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding, J., Berleant, D., Nettleton, D. & Wurtele, E. Mining MEDLINE: abstracts, sentences, or phrases. In Proceedings of the Pacific Symposium on Biocomputing. 326–337 (2002). [DOI] [PubMed]
- 32.Fundel K, Küffner R, Zimmer R. RelEx-Relation extraction using dependency parse trees. Bioinformatics. 2006;23:365–371. doi: 10.1093/bioinformatics/btl616. [DOI] [PubMed] [Google Scholar]
- 33.Pyysalo S, et al. Comparative analysis of five protein-protein interaction corpora. BMC Bioinformatics. 2008;9:S6. doi: 10.1186/1471-2105-9-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng, D., Liu, K., Lai, S., Zhou, G. & Zhao, J. Relation classification via convolutional deep neural network. In Proceedings of the 25th International Conference on Computational Linguistics: Technical Papers. 2335–2344 (2014).
- 35.Segura-Bedmar I, Martínez P, Herrero-Zazo M. Lessons learnt from the DDIExtraction-2013 shared task. Journal of Biomedical Informatics. 2014;51:152–164. doi: 10.1016/j.jbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Herrero-Zazo M, Segura-Bedmar I, Martínez P, Declerck T. The DDI corpus: An annotated corpus with pharmacological substances and drug–drug interactions. Journal of Biomedical Informatics. 2013;46:914–920. doi: 10.1016/j.jbi.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Z, Yang Z, Luo L, Lin H, Wang J. Drug drug interaction extraction from biomedical literature using syntax convolutional neural network. Bioinformatics. 2016;32:3444–3453. doi: 10.1093/bioinformatics/btw486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng, Y., Rios, A., Kavuluru, R. & Lu, Z. Chemical-protein Relation Extraction with Ensembles of SVM, CNN, and RNN Models. In Proceedings of the BioCreative VI Workshop. 148–151 (2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source code for generating BioWordVec is freely available at https://github.com/ncbi-nlp/BioWordVec. The PubMed data are available from https://www.nlm.nih.gov/databases/download/pubmed_medline.html. The MeSH RDF data are available from https://www.nlm.nih.gov/databases/download/mesh.html.