Abstract
Background
Rice, which serves as a staple food for more than half of the world’s population, is very susceptible to the pathogenic fungus, Magnaporthe oryzae. However, common wild rice (Oryza rufipogon), which is the ancestor of Asian cultivated rice (O. sativa), has significant potential as a genetic source of resistance to M. oryzae. Recent studies have shown that the domestication of rice has altered its relationship to symbiotic arbuscular mycorrhizae. A comparative response of wild and domestic rice inhabited by mycorrhizae to infection by M. oryzae has not been documented.
Results
In the current study, roots of wild and cultivated rice colonized with the arbuscular mycorrhizal (AM) fungus (AMF) Rhizoglomus intraradices were used to compare the transcriptomic responses of the two species to infection by M. oryzae. Phenotypic analysis indicated that the colonization of wild and cultivated rice with R. intraradices improved the resistance of both genotypes to M. oryzae. Wild AM rice, however, was more resistant to M. oryzae than the cultivated AM rice, as well as nonmycorrhizal roots of wild rice. Transcriptome analysis indicated that the mechanisms regulating the responses of wild and cultivated AM rice to M. oryzae invasion were significantly different. The expression of a greater number of genes was changed in wild AM rice than in cultivated AM rice in response to the pathogen. Both wild and cultivated AM rice exhibited a shared response to M. oryzae which included genes related to the auxin and salicylic acid pathways; all of these play important roles in pathogenesis-related protein synthesis. In wild AM rice, secondary metabolic and biotic stress-related analyses indicated that the jasmonic acid synthesis-related α-linolenic acid pathway, the phenolic and terpenoid pathways, as well as the phenolic and terpenoid syntheses-related mevalonate (MVA) pathway were more affected by the pathogen. Genes related to these pathways were more significantly enriched in wild AM rice than in cultivated AM rice in response to M. oryzae. On the other hand, genes associated with the ‘brassinosteroid biosynthesis’ were more enriched in cultivated AM rice.
Conclusions
The AMF R. intraradices-colonized rice plants exhibited greater resistance to M. oryzae than non-AMF-colonized plants. The findings of the current study demonstrate the potential effects of crop domestication on the benefits received by the host via root colonization with AMF(s), and provide new information on the underlying molecular mechanisms. In addition, results of this study can also help develop guidelines for the applications of AMF(s) when planting rice.
Electronic supplementary material
The online version of this article (10.1186/s12284-019-0287-9) contains supplementary material, which is available to authorized users.
Keywords: Arbuscular mycorrhizal fungi, Cultivated rice, Magnaporthe oryzae, Transcriptome analysis, Wild rice
Background
Approximately two-thirds of the current human population subsist on rice (Oryza sativa) as their staple food (Talukdar et al. 2017). However, as the human population continues to increase, and land for planting rice decreases, the need to increase rice production is critical. Pathogenic fungi are one of the limiting factors for rice production. Magnaporthe oryzae, a pathogenic fungus that is the causal agent of rice blast disease, is widely distributed and causes serious reductions in rice yields worldwide (Liu et al. 2010; Wang et al. 2017). The breeding approach to create disease-resistant rice varieties is commonly used to improve rice production. Common wild rice (O. rufipogon), the source for the breeding of cultivated rice, and a relative of Asian cultivated rice, represents sources for several desirable attributes, including disease resistance (Hua et al. 2015; Mao et al. 2015; Liang et al. 2017). For example, Zhang et al. (1998) identified a new gene in common wild rice involved in bacterial blight resistance, namely the Xa21, which belongs to the R gene group and is originated from the wild rice species Oryza longistaminata (Ni et al. 2015). Another example is the R gene Pi54 that was identified in both wild and cultivated rice and shown to play a role in blast disease resistance (Zhang et al. 2018). Sources of common wild rice, however, are becoming rare due to human activities. China has protected several conservation areas to maintain the production of wild rice and preserve its genetic diversity for rice breeding efforts, as well as to provide research materials to investigate the responses of wild and cultivated varieties of rice to various abiotic and biotic stresses (Luo et al. 2017; Tian et al. 2017).
Mycorrhizae are well known for their symbiotic associations with host plants (Grove et al. 2017; Verzeaux et al. 2017; Jemo et al. 2018). More than 80% of plant species can be colonized by arbuscular mycorrhizal (AM) fungi (AMFs), which develop an endosymbiosis with their host (Feddermann et al. 2010). AMFs, among other attributes, improve the ability of host plants to capture nutrients from the soil (Grove et al. 2017; Verzeaux et al. 2017), and the fundamental aspect of the AMF symbiosis with host plants is the bidirectional exchange of nutrients (Field and Pressel 2018; Karandashov and Bucher 2005). The improvement in nutrient uptake (e.g. phosphorus) (Berdeni et al. 2018; Selvakumar et al. 2018) from soil by host plants has been reported to result from the generation of long hyphae into the soil around plant roots and the ability of AMFs to increase resistance of host plants to environmental stressors (Jones et al. 2004; Berdeni et al. 2018; Selvakumar et al. 2018; Tian et al. 2019). In turn, AMFs can obtain carbon (C) nutrients (photosynthates) from the host plants to grow and survive (Tian et al. 2010; Zhang et al. 2016). Rice domestication has been reported to have substantially changed the benefits derived from AMFs (Martín-Robles et al. 2018), suggesting that the AM mechanisms and reactions occurring in wild rice may be different than what occurs in cultivated rice. Increasing number of research has demonstrated that AMFs can improve resistance of rice plants to various pathogenic fungi, including M. oryzae (Baby 2001; Campos-Soriano et al. 2012). However, no detailed comparative studies have been conducted on the response of wild AM rice vs. cultivated AM rice during M. oryzae infection.
Although the existence of disease resistance in common wild rice has been well established (Liu et al. 2017; Stein et al. 2018), only a few studies have been conducted which demonstrate that wild rice selectively chooses and closely connects with a beneficial microbiome (Martín-Robles et al. 2018; Perez-Jaramillo et al. 2018; Shi et al. 2018a). The resistance of wild rice to M. oryzae may be partially attributable to the presence of its symbiotic AMFs (Martín-Robles et al. 2018), while the susceptibility of cultivated rice to pathogenic fungi may have arisen due to the change in the original AMF community occurred during the domestication of wild rice (Thomas et al. 2017). For instance, a recent study reported that several AMF taxa within the Glomeromycota were more enriched in wild rice in comparison with cultivated rice in the presence of M. oryzae; and thus, the enrichment of Glomeromycota was suggested to be correlated with the higher resistance of wild rice to M. oryzae (Shi et al. 2018a). These findings provided an insight into the relationship between wild and cultivated rice with regard to their associated AMFs in response to M. oryzae. The identified AMFs have been shown to stimulate metabolic pathways involved in disease resistance in wild rice (Shi et al. 2018a), suggesting that they are involved in promoting plant resistance to M. oryzae. It is then interesting to determine the mechanisms by which the identified AMFs, including the Rhizoglomus spp., benefit rice under the background of domestication when plants are infected by M. oryzae.
In the present study, experiments were conducted to assess the response of R. intraradices-colonizing wild and cultivated rice to M. oryzae to provide information on how this inoculated AMF species could contribute to the resistance of wild rice, and examine whether this R. intraradices could enhance the resistance of cultivated rice to M. oryzae. We hypothesized that 1) wild AM rice has retained some symbiotic benefits that have been lost during the domestication of cultivated AM rice which enhance resistance to M. oryzae, and unraveled that 2) the molecular mechanisms that the AM fungus R. intraradices used to enhance resistance to M. oryzae differed between wild and cultivated rice.
Methods
Plant materials and experimental design
Seedlings of cultivated rice (Oryza sativa L. ssp. Japonica) and Dongxiang wild rice (a Chinese common wild rice; Oryza rufipogon Griff.) (Li et al. 2010) were used in this study. M. oryzae was cultured on an oatmeal agar medium, and hyphae were “brushed” to induce the formation of conidia as described by Chumley and Valent (1990) and Koga et al. (2004).
The R. intraradices strain (BGC BJ09) used in this study was provided by the Institute of Plant Nutrition and Resources, Beijing Academy of Agriculture and Forestry in China (Liu et al. 2014). Spores of the AMF R. intraradices were obtained from the soil of Medicago sativa plants grown in pots that were previously inoculated with R. intraradices for 5 months. Briefly, spores were collected from the mycorrhizosphere of M. sativa using a wet-sieving method that involved placing the mycorrhizosphere in a 45-μm mesh sieve and sufficiently rinsing them with tap water at room temperature. The collected spores were then diluted to a concentration of ~ 10 spores/mL. The spores were surface-sterilized in 0.5% sodium hypochlorite for 3 min and rinsed twice in sterile deionized water prior to their use in the inoculation of the rice seedlings (Green et al. 1976).
Rice seeds were surface-sterilized with 1% sodium hypochlorite for 5 min and incubated in a dark chamber at 25 °C for 3 d. Six germinated seeds were then transplanted into each pot [18 (width) × 18 (height) cm] containing soil. The soil was collected from the experiment station of Northeast Institute of Geography and Agroecology, and was autoclaved before use. The soil characteristics were analyzed and reported previously in Tian et al. (2018) as follows: soil organic matter 61.2 g/kg, total nitrogen 651.92 mg/kg, available-nitrogen 109.20 mg/kg, available-phosphorus 7.48 mg/kg, and available-potassium 88.66 mg/kg, and pH 6.31. The pots were maintained in a growth chamber that was preset to 16-h-light (1000 μmol/m2/s)/8-h-dark photoperiod, 23–28 °C and 65% relative humidity. Seven days later, three plants with similar size were kept in each pot. For treatment with R. intraradices, the 10-day-old wild and cultivated rice plants were inoculated with R. intraradices by pouring 200 mL of its spores (~ 10 spores/mL) to each pot (W + R and C + R). Non-treated wild rice (W) and cultivated rice (C) plants were used as controls. The pots were incubated in a chamber under the light and temperature cycle described above for 45 d to guarantee the AM colonization.
The primary focus of the experiment was to examine the differences between wild and cultivated rice inoculated with the AMF R. intraradices in response to the infection with M. oryzae pathogen (p). Thus, after 45 d of growth, all four groups (W + R, C + R, W and C) were infected with the pathogenic fungus M. oryzae. Spores of M. oryzae with the concentration of 106/mL were evenly sprayed on the leaves of individual plants, and the plants were subsequently incubated in a chamber in the dark at 80% humidity and 25 °C for 2 d. Consequently, we obtained four treatment groups for this study: R. intraradices-inoculated cultivated rice infected with M. oryzae (Cp + R), R. intraradices-uninoculated cultivated rice infected with M. oryzae (Cp), R. intraradices-inoculated wild rice infected with M. oryzae (Wp + R), R. intraradices-uninoculated wild rice infected with M. oryzae (Wp) (see Fig. 1 for the scheme of experimental design). The plants of all four treatment groups (Wp + R, Cp + R, Wp and Cp) were then maintained in the chambers under the same growth conditions [8 h dark at 23 °C and 16 h light (1000 μmol/m2/s) at 28 °C] for a total of 7 d prior to sampling.
Fig. 1.
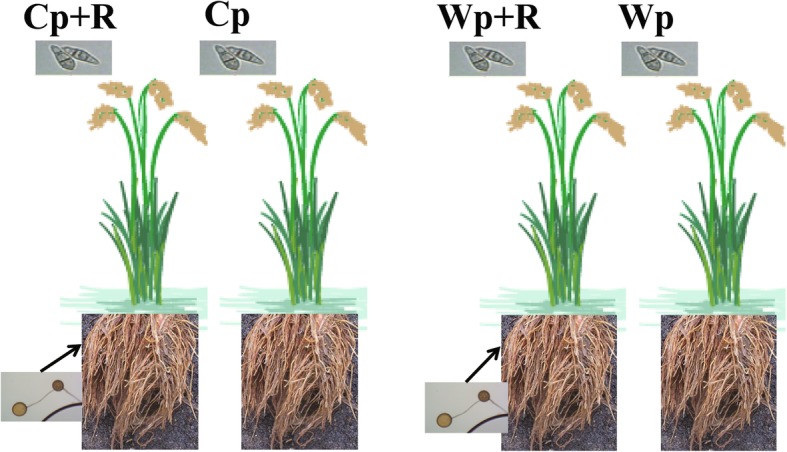
The four experimental treatment groups designed to comparatively study the responses of wild and cultivated rice plants to Magnaporthe oryzae infection with and without inoculation with the arbuscular mycorrhizal fungus Rhizoglomus intraradices. Cp + R, R. intraradices-inoculated cultivated rice infected with M. oryzae; Cp, R. intraradices-uninoculated cultivated rice infected with M. oryzae; Wp + R, R. intraradices-inoculated wild rice infected with M. oryzae; Wp, R. intraradices-uninoculated wild rice infected with M. oryzae
Sampling and phenotyping
Seven days after plants were infected with M. oryzae spores, leaves were sampled and used to analyze the leaf phenotype. The ability of the plants in resisting M. oryzae was analyzed using the typical injured leaves by using the Image J software (v.1.52i) based on lesion areas (Campos-Soriano et al. 2012). Root samples were also extracted from the soil and divided into parts. One part was cleaned with tap water and immediately placed in liquid nitrogen and stored at − 80 °C for RNA extraction. Another part of roots along with their rhizosphere was used to evaluate the presence of R. intraradices. The plant growth phenotype was recorded prior to any leaf or root sampling. Lastly, rhizosphere soils were sampled and kept at − 80 °C until DNA extraction.
Root colonization rate of R. intraradices, and determination of R. intraradices levels in the rhizosphere soil by real-time quantitative PCR (qPCR)
Root colonization rate of R. intraradices was measured by the trypan blue method as described by Phillips and Hayman (1970) and Tian et al. (2019). The method described by Trouvelot et al. (1986) was used for the calculation of the arbuscule abundance in mycorrhizal parts of root fragments. For determination of the abundance of R. intraradices propagating in the rhizosphere soil during rice growth, total DNA was extracted from rhizosphere soil as previously described by Tian et al. (2018), and was then used to analyze the 18S ribosomal DNA (rDNA) gene expression by qPCR (Dai et al. 2015). Prior to the qPCR, the quality and concentrations of purified DNA samples were determined using a NanoDrop 2000 (NanoDrop Technologies, Inc., Wilmington, USA). The R. intraradices abundance was expressed as the copy number of the 18 s rDNA region of each fungus per one gram soil, which was amplified using the forward primer AMV4.5NF (5′-AAGCTCGTAGTTGAATTTCG-3′) and reverse primer AMDGR (5′-CCCAACTATCCCTATTAATCAT-3′) (Sato et al. 2005).
RNA extraction, cDNA library construction and RNA-sequencing
Roots from three biological replicates from each treatment group were collected and subjected to RNA extraction as described previously (Tian et al. 2018). The quality and concentration of extracted RNA samples were assessed spectrophotometrically using a NanoDrop 2000 (NanoDrop Technologies, Inc., Wilmington, USA). cDNA libraries were subsequently constructed as described by Chen et al. (2017) (Chen et al. 2017). For RNA-sequencing (RNA-seq), paired-end sequencing (2 × 100 bp) was conducted using an Illumina HiSeq X Ten platform (Illumina, San Diego CA, USA) at the Novogene Co., Ltd. (Beijing, China). FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Cutadapt (http://cutadapt.readthedocs.io/en/stable/) were used to determine the sequence quality. Filtered reads (~ 28 million) were mapped onto the rice reference genome using bowtie with default settings (http://bowtie-bio.sourceforge.net/index.shtml). A comparative analysis of gene expression was used to identify differentially expressed genes (DEGs). Cufflinks (http://sihua.us/Cufflinks.htm) was used to conduct a t-test (P < 0.05) and to identify DEGs in the R. intraradices-inoculated compared with the R. intraradices-non-inoculated roots of cultivated (‘Cp+R vs. Cp’) and wild rice (‘Wp+R vs. Wp’) with and without M. oryzae. A false discovery rate (FDR) of 5% (q-value < 0.05) was used to identify DEGs with at least a |log2 (fold-changes)| > 0. Gene ontology (GO) annotations were performed using Blast2GO v2.5 based on the nonredundant (Nr) protein sequences (NCBI) and Pfam (NCBI, nonredundant nucleotide sequences). A total of 20,693 annotated genes with known functions were included in the GO annotation and enrichment analyses. The KEGG Automatic Annotation Server (KAAS; http://www.genome.jp/kegg/kaas/) was used for KEGG annotations. MapMan analysis was conducted using MapMan 3.6.0 (http://mapman.gabipd.org/web/guest) software.
Validation of RNA-seq data by reverse transcription-quantitative real-time PCR (RT-qPCR)
To validate the quality of the RNA-seq analysis, RT-qPCR analysis was conducted on eight selected genes, namely Os12g0168700, Os06g0726200, Os04g0229100, Os02g0627100, Os01g0854800, Os01g0892500, Os02g0175000 and Os02g0678200, with the β-tubulin gene of O. sativa being used as a reference gene (Liu et al. 2009), as described previously (Tian et al. 2018). The gene-specific primers utilized in the RT-qPCR analysis are listed in Additional file 1: Table S1. Among the eight selected genes, Os12g0168700, Os06g0726200, Os04g0229100, Os02g0627100 and Os01g0854800 were DEGs in the ‘Wp+R vs. Wp’ comparison, while Os01g0892500, Os02g0175000 and Os02g0678200 were DEGs in the ‘Cp+R vs. Cp’ comparison.
Statistical analysis
SPSS 19.0 software was used for one-way analysis of variance to determine the statistical significance of the R. intraradices colonization rate and the abundance of R. intraradices propagating in the rhizosphere soil during rice growth, as well as for the statistical analysis of gene expression data (Student’s t-test).
Results
Colonization rate of inoculated R. intraradices and abundance of the R. intraradices propagating in the rhizosphere soil during rice growth
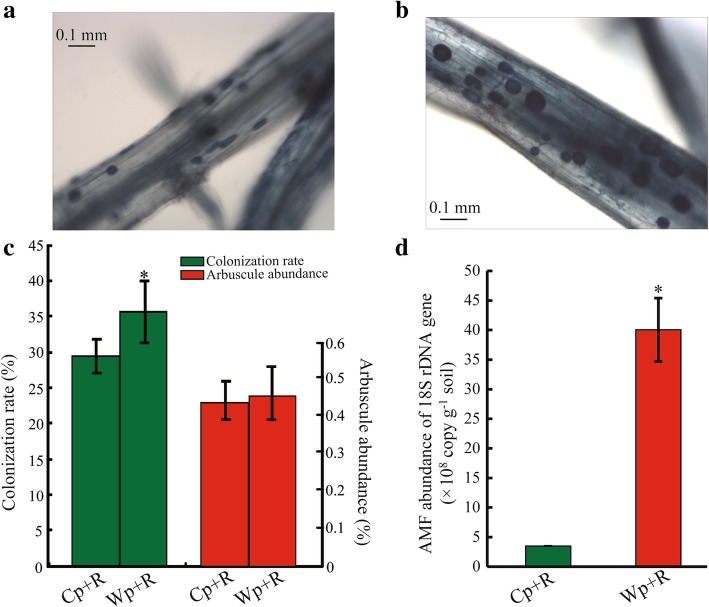
The colonization of cultivated and wild rice by R. intraradices was quantified using the trypan blue method (Phillips and Hayman 1970; Tian et al. 2019). Results indicated that R. intraradices was able to colonize both wild and cultivated rice (Fig. 2a and b). Investigation of colonization rate of roots with inoculated R. intraradices showed that Wp + R had a higher colonization rate than Cp + R after inoculation with R. intraradices and infection with M. oryzae (Fig. 2c). However, although Wp + R showed a slightly higher arbuscule abundance of R. intraradices than Cp + R, this difference is statistically insignificant, perhaps due to the very low levels of arbuscule abundance detected (Fig. 2c). The levels of R. intraradices propagating in the rhizosphere soil were also quantified using qPCR. Results indicated that the abundance of soil R. intraradices was significantly higher in the wild rice rhizosphere (roots plus surrounding soil) than in the cultivated rice rhizosphere (Fig. 2d), indicating that the wild rice rhizosphere could harbor more R. intraradices than the cultivated rice rhizosphere under our experimental conditions.
Fig. 2.
a-b Microscopic observation of root colonization of (a) wild and (b) cultivated rice inoculated with the arbuscular mycorrhizal fungus Rhizoglomus intraradices. c-d Colonization rate and arbuscule abundance of R. intraradices in roots of inoculated rice plants (c) and abundance of R. intraradices (d) in the rhizosphere soil of rice plants following soil inoculation with R. intraradices and plant infection with Magnaporthe oryzae. Data shown are the R. intraradices abundance in Cp + R and Wp + R. The error bars represent standard deviations of the means of three repeats. The asterisks above the bars indicate significant differences among the samples at P < 0.05 based on Student’s t-test. Cp + R, R. intraradices-inoculated cultivated rice infected with M. oryzae; Wp + R, R. intraradices-inoculated wild rice infected with M. oryzae
The effect of R. intraradices colonization on the response of rice plants to pathogen infection
Examination of the growth and disease symptoms of the wild and cultivated rice plants revealed that the Wp were more resistant to M. oryzae than the Cp plants (with 19.1% and 25.73% lesion areas in Wp and Cp, respectively) (Fig. 3; Additional file 2: Figure S1), and Wp + R were more resistant to M. oryzae than the Cp + R plants (with 4.77% and 7.87% lesion areas in Wp + R and Cp + R, respectively) (Fig. 3; Additional file 2: Figure S1). Notably, both the wild and cultivated rice plants inoculated with R. intraradices (Wp + R and Cp + R) exhibited a greater resistance to infection by the rice blast fungus than their corresponding Wp or Cp plants that were non-inoculated with R. intraradices (Fig. 3; Additional file 2: Figure S1). In all four treatment groups Wp + R, Wp, Cp + R and Cp, the leaf surfaces of all rice plants exhibited obvious lesion areas; however, the total lesion areas were significantly smaller in Wp + R, followed by that of Cp + R, Wp and Cp plants, suggesting the positive effect of R. intraradices in providing resistance against M. oryzae (Fig. 3b; Additional file 2: Figure S1).
Fig. 3.

Phenotypes of wild and cultivated rice plants in response to Magnaporthe oryzae infection with and without inoculation with the arbuscular mycorrhizal fungus Rhizoglomus intraradices. a plant growth and (b) infected leaves. Cp + R, R. intraradices-inoculated cultivated rice infected with M. oryzae; Cp, R. intraradices-uninoculated cultivated rice infected with M. oryzae; Wp + R, R. intraradices-inoculated wild rice infected with M. oryzae; Wp, R. intraradices-uninoculated wild rice infected with M. oryzae. Red lines are the scale bars, which represent 5.0 cm in (a) and 0.5 cm in (b)
Comparative analysis of the responses of cultivated and wild rice roots using the transcriptomic approach
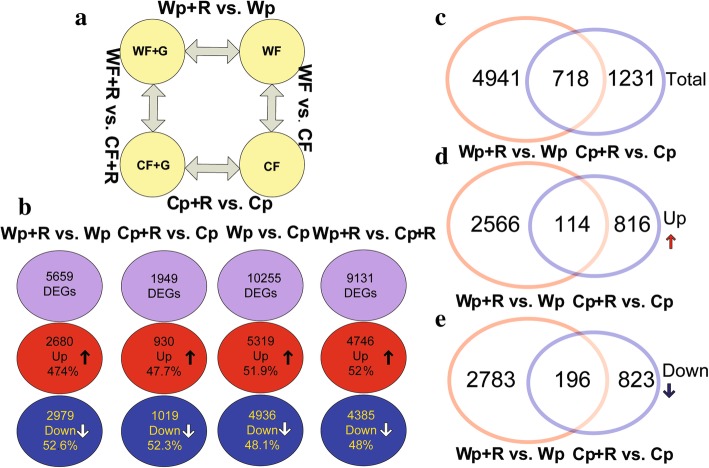
The root transcriptomes of wild and cultivated rice varieties inoculated and non-inoculated with R. intraradices were compared to elucidate their molecular response to infection by the rice blast fungus. A total of ~ 624 million raw reads were obtained from the 12 samples, with the number of reads ranging from 30.54 to 64.75 million raw reads in each sample (Table 1). After filtering out low quality reads, a total of ~ 556 million clean reads were obtained with an average of 84.18% that could be mapped to the rice reference genome Oryza_sativa.IRGSP-1.0.21 (http://asia.ensembl.org). The percentage of clean reads (144.4–146 bp in average length) from each sample that could be mapped ranged from 74.76 to 89.35% (Table 1). A total of 5659 (2680 up- and 2979 down-regulated) genes were identified as being differentially expressed genes (DEGs) in the ‘Wp+R vs. Wp’ comparison, while a total of 1949 DEGs were identified in the ‘Cp+R vs. Cp’ comparison (Fig. 4). Subsequently, 8 genes were selected and subjected to RT-qPCR analysis to confirm the results of the RNA-seq analysis. Results demonstrated that the RT-qPCR data were in general consensus with the RNA-seq data (Fig. 5; Additional file 3: Table S2).
Table 1.
Summary of Illumina RNA-sequencing reads mapped to the reference genome. Cp + R, Rhizoglomus intraradices-inoculated cultivated rice infected with Magnaporthe oryzae; Cp, R. intraradices-uninoculated cultivated rice infected with M. oryzae; Wp + R, R. intraradices -inoculated wild rice infected with M. oryzae; Wp, R. intraradices-uninoculated wild rice infected with M. oryzae. M, million; G, gbase
Fig. 4.
Numbers of differentially expressed genes (DEGs). a Diagram illustrating the experimental design and comparisons. b Diagram illustrating the number of total, up-regulated, and down-regulated genes in the four comparisons. c, d, and e. A Venn analysis of the number of total, up-regulated, and down-regulated DEGs identified in the ‘Wp+R vs. Wp’ and ‘Cp+R vs. Cp’ comparisons. Cp + R, Rhizoglomus intraradices-inoculated cultivated rice infected with Magnaporthe oryzae; Cp, R. intraradices-uninoculated cultivated rice infected with M. oryzae; Wp + R, R. intraradices-inoculated wild rice infected with M. oryzae; Wp, R. intraradices-uninoculated wild rice infected with M. oryzae
Fig. 5.
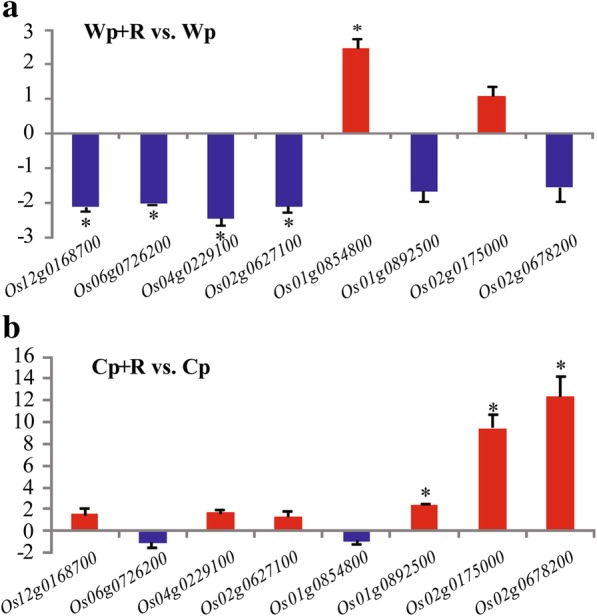
Validation of the RNA-sequencing data using reverse transcription-quantitative real-time PCR (RT-qPCR). Eight genes were selected for RT-qPCR analysis from the RNA-sequencing data. a Relative expression of the selected genes in the ‘Wp+R vs. Wp’ comparison. b Relative expression in the ‘Cp+R vs. Cp’ comparison. Cp + R, Rhizoglomus intraradices-inoculated cultivated rice infected with M. oryzae; Cp, R. intraradices-uninoculated cultivated rice infected with M. oryzae; Wp + R, R. intraradices-inoculated wild rice infected with M. oryzae; Wp, R. intraradices-uninoculated wild rice infected with M. oryzae. The error bars represent standard deviations of the means. The asterisks above the bars indicate significant change in gene expression at P < 0.05 based on Student’s t-test
Gene ontology (GO) annotation and enrichment analysis of the identified DEGs
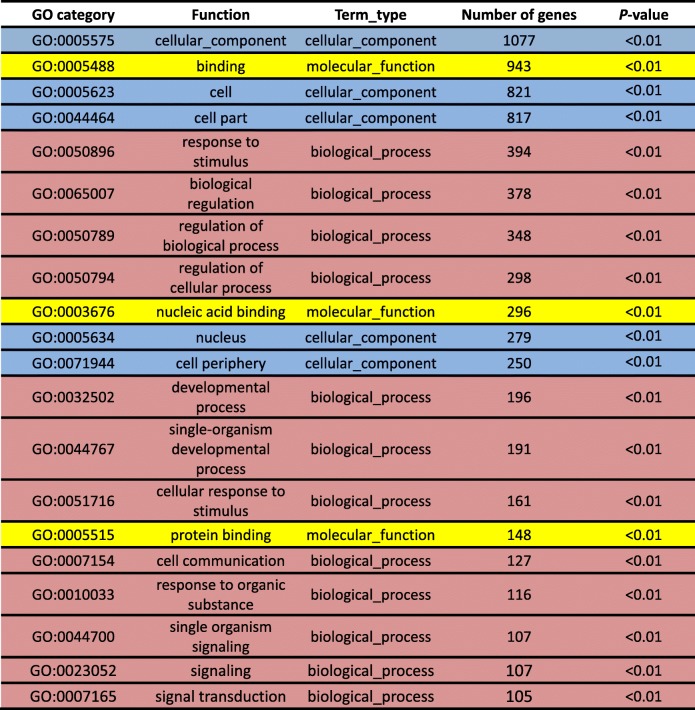
The DEGs of the ‘Wp+R vs. Wp’ and ‘Cp+R vs. Cp’ comparisons were subjected to GO analysis to assign functional terms to the identified DEGs. The top 20 enriched GO categories were then used to determine differences between the ‘Wp + R vs. Wp’ and ‘Cp+F vs. Cp’ comparisons. There were 12, 5, and 3 GO categories in the ‘Wp + R vs. Wp’ comparison labeled as ‘biological_process’, ‘cellular_component’, and ‘molecular_function’, respectively (Table 2). The GO analysis of the DEGs derived from ‘Wp + R vs. Wp’ comparison indicated that within ‘biological_process’, the terms ‘response to stimulus’, ‘biological regulation’, ‘regulation of biological process’, ‘regulation of cellular process’, ‘developmental process’, ‘single-organism developmental process’, ‘cellular response to stimulus’, ‘cell communication’, ‘response to organic substance’, ‘single organism signaling’, ‘signaling’ and ‘signal transduction’ were enriched (Table 2). As for the ‘cellular_component’ category, ‘cell’, ‘cell part’, ‘cellular_component’, ‘nucleus’ and ‘cell periphery’ were enriched in this category, whereas ‘binding’, ‘nucleic acid binding’, and ‘protein binding’ were enriched in the ‘molecular_function category’ (Table 2).
Table 2.
Gene ontology (GO) classification of differentially expressed genes (DEGs) derived from the ‘Wp+R vs. Wp’ comparison. Wp + R, Rhizoglomus intraradices-inoculated wild rice infected with Magnaporthe oryzae; Wp, R. intraradices-uninoculated wild rice infected with M. oryzae
Comparatively, the GO analysis of the DEGs obtained from ‘Cp+F vs. Cp’ comparison resulted in 18 enriched GO terms within the ‘biological_process’ category and 2 enriched GO terms within the ‘molecular_function’ category (Table 3). Specifically, the enriched terms within the ‘biological_process’ were ‘biological regulation’, ‘response to stimulus’, ‘regulation of biological process’, ‘regulation of cellular process’, ‘regulation of metabolic process’, ‘organic cyclic compound biosynthetic process’, ‘heterocycle biosynthetic process’, ‘regulation of primary metabolic process’, ‘regulation of cellular metabolic process’, ‘regulation of macromolecule metabolic process’, ‘regulation of biosynthetic process’, ‘nucleobase-containing compound biosynthetic process’, ‘response to chemical’, ‘transcription, DNA-templated’, ‘nucleic acid-templated transcription’, ‘RNA biosynthetic process’, ‘regulation of cellular biosynthetic process’ and ‘regulation of gene expression’ (Table 3), while the 2 enriched GO terms within the ‘molecular_function’ category were ‘binding’ and ‘DNA binding’ (Table 3).
Table 3.
Gene ontology (GO) classification of the differentially expressed genes (DEGs) derived from the ‘Cp+R vs. Cp’ comparison. Cp + R, Rhizoglomus intraradices-inoculated cultivated rice infected with Magnaporthe oryzae; Cp, R. intraradices-uninoculated cultivated rice infected with M. oryzae
KEGG analysis of the identified DEGs
A KEGG analysis of the ‘Wp+R vs. Wp’ comparison revealed that the pathways of ‘spliceosome valine’, ‘leucine and isoleucine degradation’, ‘fatty acid degradation’, ‘plant hormone signal transduction’, ‘phenylalanine metabolism’ and ‘α-linolenic acid metabolism’ were significantly enriched (Table 4; Additional file 4: Table S3a). It is noteworthy that α-linolenic acid metabolism is related to jasmonic acid (JA) synthesis, and JA, as one of the major biotic stress-related phytohormones, may help to induce plant induced systemic resistance (ISR) (Iwanami et al. 2017). Further analysis showed that 9 α-linolenic acid metabolism-related genes (e.g., Os11g0210300 and Os06g0346300) were up-regulated in the ‘Wp + R vs. Wp’ comparison, while no α-linolenic acid metabolism-related genes were induced in the ‘Cp+R vs. Cp’ comparison (Table 5). Comparatively, the pathways of ‘plant hormone signal transduction’, ‘brassinosteroid biosynthesis’, ‘tropane, piperidine and pyridine alkaloid biosynthesis’, ‘tyrosine metabolism’, ‘glutathione metabolism’, ‘glycine, serine and threonine metabolism’, ‘glycerolipid metabolism’ and ‘thiamine metabolism’ were significantly enriched in the ‘Cp + R vs. Cp’ comparison (Table 6; Additional file 4: Table S3b). Brassinosteroids are a group of steroid hormones with important regulatory function in plant responses to both biotic and abiotic stresses (Bari and Jones 2009; Choudhary et al. 2012), and brassinosteroids-related genes (e.g., Os03g0227700, Os05g0200400 and Os04g0469800) were up-regulated only in the ‘Cp + R vs. Cp’ comparison (Table 5; Table 6). Only the pathway ‘plant hormone signal transduction’ was shared between the ‘Wp + R vs. Wp’ (with 32 genes up-regulated) and ‘Cp + R vs. Cp’ (with 18 genes up-regulated) comparisons (Additional file 4: Table S3a and b). A more detailed analysis of the functions of the plant hormone-related genes revealed that auxin-related genes were up-regulated among the hormone-related genes common to both the ‘Wp + R vs. Wp’ and ‘Cp + R vs. Cp’ comparisons.
Table 4.
KEGG analysis of the response of Rhizoglomus intraradices-inoculated wild rice plants to infection by Magnaporthe oryzae in the ‘Wp+R vs. Wp’ comparison. Wp + R, R. intraradices-inoculated wild rice infected with M. oryzae; Wp, R. intraradices-uninoculated wild rice infected with M. oryzae
| Term | Number of genes | P-value |
|---|---|---|
| Spliceosome | 32 | < 0.01 |
| Valine, leucine and isoleucine degradation | 10 | 0.01 |
| Fatty acid degradation | 10 | 0.02 |
| Plant hormone signal transduction | 32 | 0.02 |
| Phenylalanine metabolism | 24 | 0.02 |
| α-linolenic acid metabolism | 9 | 0.04 |
Table 5.
List of several important differentially expressed genes derived from W + F versus W (Wp + R vs. Wp) comparison and Cp + R versus C (Cp + R vs. C) comparison. Cp + R, Rhizoglomus intraradices-inoculated cultivated rice infected with M. oryzae; Cp, R. intraradices-uninoculated cultivated rice infected with M. oryzae; Wp + G, R. intraradices-inoculated wild rice infected with M. oryzae; Wp, R. intraradices-uninoculated wild rice infected with M. oryzae
| Gene ID | Description | RNA-sequencing data | |||
|---|---|---|---|---|---|
| Wp + G vs. Wp log2(fold-change) |
q-value | Cp + G vs. Cp log2(fold-change) |
q-value | ||
| Os11g0210300 | α-linolenic acid metabolism-related genes | 1.62 | < 0.01 | unchanged | |
| Os06g0346300 | α-linolenic acid metabolism-related genes | 2.99 | < 0.01 | unchanged | |
| Os03g0227700 | brassinosteroids-related genes | −1.30 | < 0.01 | 1.41 | < 0.01 |
| Os05g0200400 | brassinosteroids-related genes | −0.61 | < 0.01 | 0.79 | < 0.01 |
| Os04g0469800 | brassinosteroids-related genes | unchanged | 0.74 | 0.03 | |
| Os05g0418100 | Mildew Resistance Locus O (MLO)-like proteins | 1.10 | < 0.01 | unchanged | |
| Os11g0181400 | Mildew Resistance Locus O (MLO)-like proteins | 1.11 | < 0.01 | unchanged | |
| Os08g0244500 | encoding 1,3-ß-glucosidases | 4.36 | < 0.01 | unchanged | |
| OS01g0713200 | encoding 1,3-ß-glucosidases | 2.75 | < 0.01 | unchanged | |
| Os07g0600700 | endo-1,3-ß-glucosidases | unchanged | 1.68 | < 0.01 | |
| Os09g0422500 | cell wall-related genes | 5.23 | < 0.01 | unchanged | |
| Os01g0750300 | cell wall-related genes | 2.31 | < 0.01 | unchanged | |
| Os01g0746700 | cell wall-related genes | 1.76 | < 0.01 | unchanged | |
| Os08g0345500 | cellulose synthases | −2.16 | 0.04 | 1.46 | < 0.01 |
| Os08g0160500 | cellulose synthases | unchanged | 1.20 | < 0.01 | |
| Os09g0428000 | cellulose synthases | unchanged | 0.84 | 0.01 | |
| Os02g0218700 | allene oxide synthase and lipoxygenase | 0.73 | 0.01 | −3.05 | < 0.01 |
| Os03g0738600 | allene oxide synthase and lipoxygenase | 1.35 | 0.02 | unchanged | |
| Os11g0256900 | carboxyl methyltransferase | 0.68 | < 0.01 | unchanged | |
| Os04g0665200 | carboxyl methyltransferase | 2.27 | < 0.01 | unchanged | |
| Os11g0260100 | carboxyl methyltransferase | −1.04 | < 0.01 | 1.34 | < 0.01 |
| Os01g0701700 | carboxyl methyltransferase | −0.73 | 0.03 | 1.65 | < 0.01 |
| Os01g0375500 | encoding a shikimate 5-dehydrogenase | 2.27 | 0.04 | unchanged | |
| Os03g0118800 | mevalonate pathway (MVA)-related gene | −1.73 | < 0.01 | unchanged | |
| Os09g0492700 | mevalonate pathway (MVA)-related gene | 0.67 | < 0.01 | unchanged | |
| OS07G0101000 | simple phenols-related gene | 1.72 | 0.01 | unchanged | |
| OS05G0458300 | simple phenols-related gene | 2.89 | < 0.01 | unchanged | |
| OS02G0749700 | phenols-related gene | unchanged | −5.05 | < 0.01 | |
| OS12G0258700 | phenols-related gene | unchanged | −2.23 | 0.04 | |
| Os02g0570400 | terpenoid pathway-related gene | 2.82 | 0.02 | −2.77 | < 0.01 |
| Os04g0344100 | terpenoid pathway-related gene | 1.53 | < 0.01 | unchanged | |
| Os02g0568700 | terpenoid pathway-related gene | unchanged | −5.34 | < 0.01 | |
| Os02g0571100 | terpenoid pathway-related gene | 4.96 | < 0.01 | −4.11 | < 0.01 |
Table 6.
KEGG analysis of the response of Rhizoglomus intraradices-inoculated cultivated rice plants to infection by Magnaporthe oryzae in the Cp + R vs. Cp comparison. Cp + R, R. intraradices-inoculated cultivated rice infected with M. oryzae; Cp, R. intraradices-uninoculated cultivated rice infected with M. oryzae
| Term | Number of genes | P-value |
|---|---|---|
| Plant hormone signal transduction | 18 | < 0.01 |
| Brassinosteroid biosynthesis | 3 | 0.01 |
| Tropane, piperidine and pyridine alkaloid biosynthesis | 4 | 0.01 |
| Tyrosine metabolism | 5 | 0.01 |
| Glutathione metabolism | 7 | 0.02 |
| Glycine, serine and threonine metabolism | 6 | 0.02 |
| Glycerolipid metabolism | 5 | 0.03 |
| Thiamine metabolism | 2 | 0.04 |
Identification of biotic stress-related genes among the identified DEGs
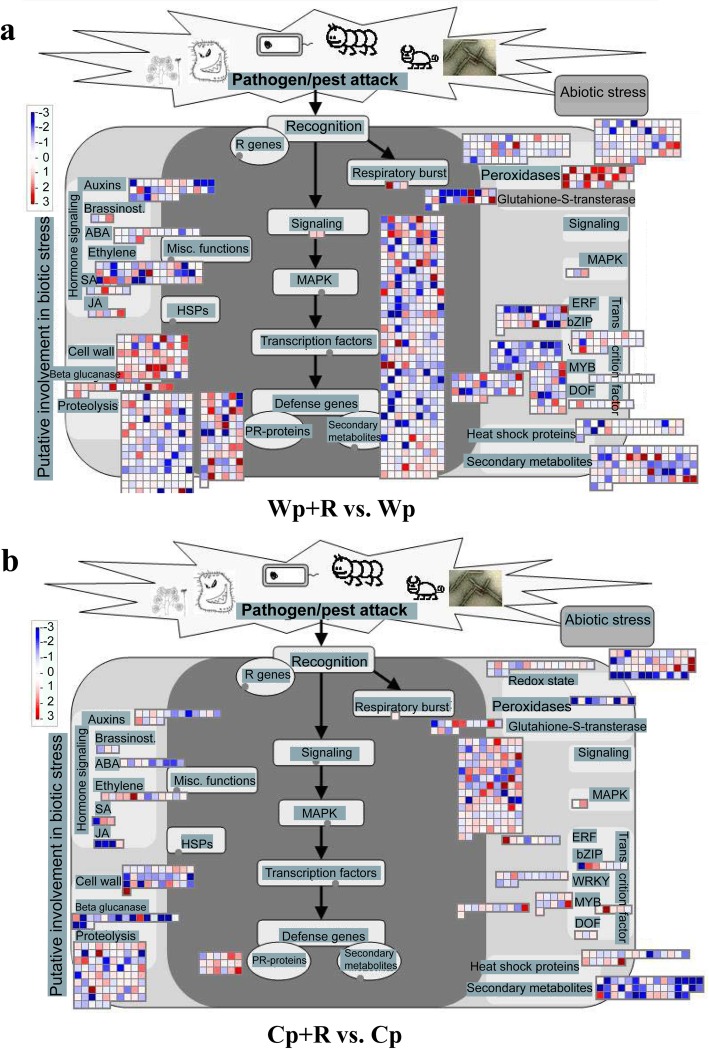
MapMan 3.6.0 software was used to identify biotic stress-related genes in the ‘Wp+R vs. Wp’ and ‘Cp+R vs. Cp’ comparisons. Results revealed that more signaling genes exhibited altered expression in the ‘Wp + R vs. Wp’ comparison than in the ‘Cp + R vs. Cp’ comparison (Fig. 6). Peroxidases encoding genes were significantly up-regulated in the ‘Wp + R vs. Wp’ comparison, while they were down-regulated in the ‘Cp + R vs. Cp’ comparison. In particular, two signaling genes (Os05g0418100 and Os11g0181400) encoding the Mildew Resistance Locus O (MLO)-like proteins were induced in the ‘Wp + R vs. Wp’ comparison. The ß-glucanase-related genes Os08g0244500 and Os01g0713200 (encoding 1,3-ß-glucosidases), were also significantly up-regulated in the ‘Wp + R vs. Wp’ comparison, while Os07g0539400 and Os07g0600700, encoding endo-1,3-ß-glucosidases, were up-regulated in the ‘Cp + R vs. Cp’ comparison (Table 5; Additional file 5: Table S4a). The cell wall-related genes Os09g0422500, Os01g0750300 and Os01g0746700 were up-regulated in the ‘Wp + R vs. Wp’ comparison, while Os08g0345500, Os08g0160500 and Os09g0428000, encoding cellulose synthases, were up-regulated in the ‘Cp + R vs. Cp’ comparison (Fig. 6; Table 5; Additional file 5: Table S4b).
Fig. 6.
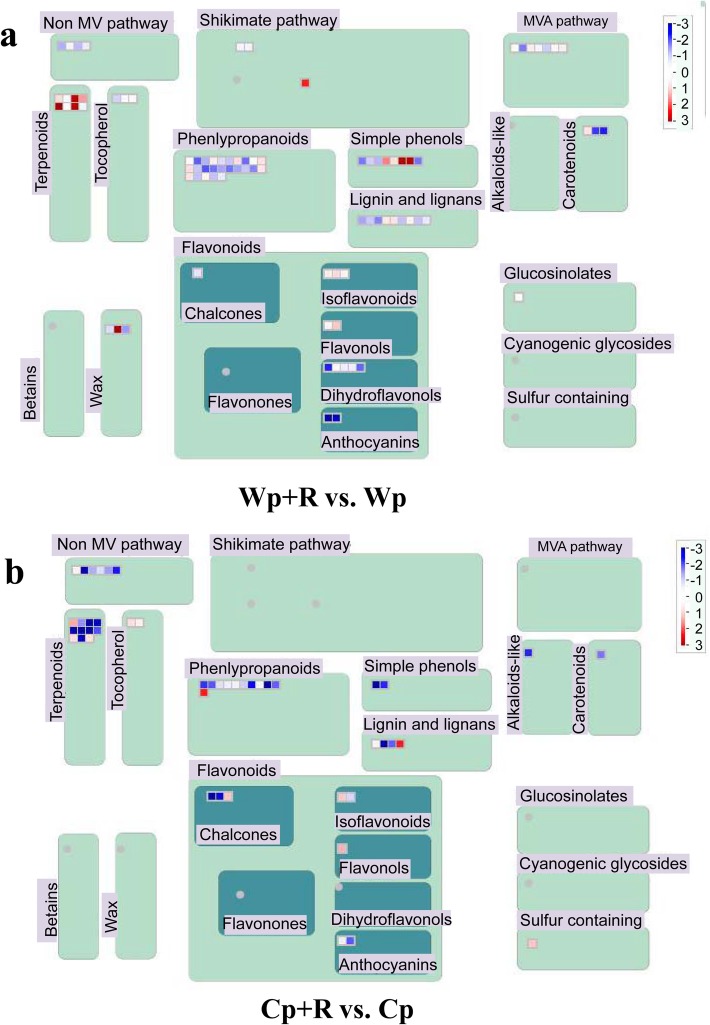
Analysis of the biotic stress response of Rhizoglomus intraradices-inoculated plants to infection by Magnaporthe oryzae using MapMan 3.6.0. ‘Wp+R vs. Wp’ (a) and ‘Cp+R vs. Cp’ (b) comparisons. Cp + R, R. intraradices-inoculated cultivated rice infected with M. oryzae; Cp, R. intraradices-uninoculated cultivated rice infected with M. oryzae; Wp + R, R. intraradices-inoculated wild rice infected with M. oryzae; Wp, R. intraradices-uninoculated wild rice infected with M. oryzae. Red color intensity indicates the levels of up-regulation by log2 (fold-change) > 0 and P value < 0.05. Blue color intensity indicates the levels of down-regulation by log2 (fold-change) < 0 and P value < 0.05
As for the genes related to JA and SA, the two hormones with well-known regulatory roles in plant responses to biotic stresses (Nasir et al. 2018; AbuQamar et al. 2017), the JA-related genes Os02g0218700 and Os03g0738600, which encode allene oxide synthase and lipoxygenase, respectively, were up-regulated in the ‘Wp+R vs. Wp’ comparison (Fig. 6; Table 5; Additional file 5: Table S4a). On the other hand, four SA-related genes encoding carboxyl methyltransferases, of which Os11g0256900 and Os04g0665200 were up-regulated in the ‘Wp + R vs. Wp’ comparison, while Os11g0260100 and Os01g0701700 were up-regulated in the ‘Cp+R vs. Cp’ comparison (Fig. 6; Table 5; Additional file 5: Table S4b).
Comparative analysis of changes in secondary metabolism
MapMan 3.6.0 software was also used to identify changes in secondary metabolism in the ‘Wp+R vs. Wp’ and ‘Cp+R vs. Cp’ comparisons with reference to the obtained DEGs. With respect to the shikimate pathway, results indicated that the gene Os01g0375500, encoding a shikimate 5-dehydrogenase, was significantly up-regulated in the ‘Wp + R vs. Wp’ comparison, whereas no shikimate pathway-genes with altered expression was detected in ‘Cp + R vs. Cp’ comparison (Fig. 7; Table 5; Additional file 6: Table S5a). In addition, seven mevalonate pathway (MVA)-related genes (e.g., Os03g0118800 and Os09g0492700) were found to be differentially expressed (i.e. 4 up- and 3 down-regulated genes) in the ‘Wp + R vs. Wp’ comparison (Fig. 7; Table 5; Additional file 6: Table S5b), while no MVA-related DEGs were identified in ‘Cp + R vs. Cp’ comparison. A number of simple phenols-related genes (e.g., Os07g0101000 and Os05g0458300) were up-regulated in the ‘Wp + R vs. Wp’ comparison, while only down-regulated phenols-related genes (e.g., Os02g0749700 and Os12g0258700) were detected in the ‘Cp + R vs. Cp’ comparison (Fig. 7; Table 5; Additional file 6: Table S5c). As for the terpenoid pathway-related DEGs, a majority of analyzed DEGs, i.e. 7 of 8, showed up-regulated expression (e.g., Os02g0570400 and Os04g0344100) in the ‘Wp + R vs. Wp’ comparison, whereas a higher number of the terpenoid pathway-related DEGs detected in the ‘Cp + R vs. Cp’ comparison (8 of 11) were down-regulated genes (e.g., Os02g0568700 and Os02g0571100) (Fig. 7; Table 5; Additional file 6: Table S5d).
Fig. 7.
Analysis of secondary metabolic pathway of Rhizoglomus intraradices-inoculated plants to infection by Magnaporthe oryzae using MapMan 3.6.0. ‘Wp+R vs. Wp’ (a) and the ‘Cp+R vs. Cp’ (b) comparisons. Cp + R, R. intraradices-inoculated cultivated rice infected with M. oryzae; Cp, R. intraradices-uninoculated cultivated rice infected with M. oryzae; Wp + R, R. intraradices-inoculated wild rice infected with M. oryzae; Wp, R. intraradices-uninoculated wild rice infected with M. oryzae. Red color intensity indicates the DEGs with log2 (fold-change) > 0 and P value < 0.05, and blue color intensity indicates the DEGs with log2 (fold-change) < 0 and P value < 0.05 according to the scale bar
Discussion
Mycorrhizal interactions are mutually beneficial; specifically, AMFs cannot only improve nutrient absorption in their host plants (e.g., Lycopersicon esculentum and Triticum aestivum) but also increase resistance of host plants to biotic (e.g., pathogen) and abiotic (e.g., drought and water) stresses (Chitarra et al. 2016; Brito et al. 2018; Fiorilli et al. 2018; Sánchez-Romera et al. 2018; Volpe et al. 2018). Pathogenic fungi can cause serious damage to crop production (Croll and McDonald 2017; Syed Ab Rahman et al. 2018). Since rice can form a mycorrhizal relationship with AMFs, it is important to understand what mutual benefits ensue from this interaction. The present study demonstrated that both wild and cultivated rice could be effectively colonized by the AMF R. intraradices (Fig. 2a and b), which could thereby provide strong protection against M. oryzae infection (Fig. 3). In addition, our data also indicated the abundance of R. intraradices was significantly higher in the wild rice rhizosphere than it was in the rhizosphere of cultivated rice (Fig. 2d). This result may indicate that wild rice can attract, host and sustain a greater level of AMFs in their rhizosphere than cultivated rice. This finding is consistent with results of a previous study, which showed that the relative abundance of AMFs was higher in wild rice than in cultivated rice (Shi et al. 2018a). In support of these findings, a recent study reported that the domestication of different crop species, including maize (Zea mays), barley (Hordeum vulgare) and wheat (T. durum Desf.), reduced the benefits obtained from AMFs (Martín-Robles et al. 2018).
AMFs can enhance the ability of a host plant to resist pathogenic fungi by inducing plant resistance systems (Cameron et al. 2013; Pérez-de-Luque et al. 2017). The phenotypes of R. intraradices-inoculated wild and cultivated rice plants infected with M. oryzae revealed that R. intraradices could improve the resistance of both wild and cultivated rice to the pathogen (Fig. 3), clearly demonstrating that AMF(s) can help plants resist pathogenic fungi. The results of the study also demonstrated, however, that AMF-inoculated wild rice were more resistant to M. oryzae than AMF-inoculated cultivated rice (Fig. 3), indicating that R. intraradices played a more substantial role in the resistance response to M. oryzae in wild rice than it did in cultivated rice which might be associated with its higher root colonization rate and abundance observed in wild rice versus cultivated rice (Fig. 3c and d). Thus, we hypothesize that the difference in the resistant responses of wild and cultivated rice to M. oryzae (or other pathogens or abiotic stresses) may partially be due to the increased beneficial effects of the AMFs that have been retained in wild rice and lost or reduced in domesticated cultivated rice.
Transcriptome data provide information on gene expression and are commonly used to identify functional differences in plant responses to biotic and abiotic stresses (Nasir et al. 2018; Abdelrahman et al. 2018). Our data identified a greater number of DEGs in ‘Wp+R vs. Wp’ comparison (R. intraradices-inoculated wild rice infected with M. oryzae vs. R. intraradices-uninoculated wild rice infected with M. oryzae) than in ‘Cp+R vs. Cp’ comparison (R. intraradices-inoculated cultivated rice vs. R. intraradices-uninoculated cultivated rice infected with M. oryzae) (Fig. 4). This may indicate that the resistance response in R. intraradices-inoculated wild rice is more complex than that in R. intraradices-inoculated cultivated rice. The KEGG enrichment analysis revealed that in comparison with R. intraradices-uninoculated control, both the R. intraradices-inoculated wild and cultivated rice contained up-regulated genes related to auxin signal transduction or biosynthesis, with more up-regulated genes being identified in ‘Wp + R vs. Wp’ comparison than ‘Cp + R vs. Cp’ comparison (12 vs. 8) (Additional file 4: Table S3). These data suggested that the AMF R. intraradices enhanced auxin signal transduction in both the wild and cultivated rice, and perhaps higher enhancement would occur in R. intraradices-inoculated wild than R. intraradices-inoculated cultivated rice during M. oryzae infection. Auxin is one of the plant growth regulators, which supports the establishment of mycorrhizal association between R. irregularis and rice (Etemadi et al. 2014; Pozo et al. 2015). In a good agreement, we also observed higher root colonization, as well as soil R. intraradices abundance, in wild than cultivated rice inoculated with R. intraradices and infected with M. oryzae (Fig. 2a-c). Interestingly, the KEGG results showed that ‘brassinosteroid biosynthesis’ was enriched in the ‘Cp + R vs. Cp’ comparison only, with 3 up-regulated brassinosteroids-related genes being found in ‘Cp + R vs. Cp’ comparison but none in ‘Wp + R vs. Wp’ comparison (Table 6). Brassinosteroids have been shown to function as disease-resistance-related hormones by inducing the production of reactive oxygen species (ROS), which might play important roles in disease resistance in plants as signaling molecules (Xia et al. 2009), suggesting that enhancement of brassinosteroids-related pathway is essential for survival of R. intraradices-inoculated cultivated rice during the invasion of M. oryzae.
JA and SA are plant hormones that have been known to play important roles in enhancing the resistance of plants particularly to various kinds of biotic stressors (Agrawal et al. 2003; Bonnet et al. 2017; do Prado Ribeiro et al. 2018). JA synthesis requires linolenic acid as a precursor (Iwanami et al. 2017), and JA can trigger ISR in plants that helps plants survive pathogenic conditions (Mehari et al. 2015; Zebelo et al. 2016). In our study, the JA-related genes Os02g0218700 (encoding allene oxide synthase) and Os03g0738600 (encoding lipoxygenase) were up-regulated in the ‘Wp+R vs. Wp’ comparison (Fig. 6; Additional file 5: Table S4c). Allene oxide synthase and lipoxygenase are key enzymes in the biosynthesis of JA, suggesting that R. intraradices might have activated the ISR in wild rice. SA can induce the synthesis of pathogenesis-related (PR) proteins in plants that play a role in plant defense against pathogenic fungi, and are directly involved in the plant immune system (Birkenbihl et al. 2017; Mahesh et al. 2017). The SA-related genes Os11g0256900 and Os04g0665200, which encode carboxyl methyltransferases - the key enzymes in the biosynthesis of SA, were up-regulated in the ‘Wp + R vs. Wp’ comparison. On the other hand, Os11g0260100 and Os01g0701700, which were also predicted to encode carboxyl methyltransferases, were up-regulated in the ‘Cp+R vs. Cp’ comparison (Fig. 6; Additional file 5: Table S4d). These findings together suggest that SA-related pathway might be activated in both rice genotypes by R. intraradices to enhance their resistance to M. oryzae.
ß-glucanases are proteins that can degrade the cell walls of pathogenic fungi, including M. oryzae (Samalova et al. 2017). In the present study, the ß-glucanase-related genes Os08g0244500 and OS01g0713200 (encoding 1,3-ß-glucosidases) were significantly up-regulated in the ‘Wp+R vs. Wp’ comparison, while Os07g0539400 and Os07g0600700, encoding endo-1,3-ß-glucosidases, were up-regulated in the ‘Cp+R vs. Cp’ comparison (Fig. 6; Table 5; Additional file 5: Table S4a). Cellulose also plays an important role in cell wall structure and disease resistance (Kesten et al. 2017). The cellulose synthase-encoding genes Os09g0422500, Os01g0750300 and Os01g0746700 were up-regulated in the ‘Wp + R vs. Wp’ comparison, whereas other cellulose synthase-encoding genes like Os08g0345500, Os08g0160500 and Os09g0428000 were up-regulated in the ‘Cp + R vs. Cp’ comparison (Fig. 6; Table 5; Additional file 5: Table S4b). These data indicate that R. intraradices induced the plants to express more ß-glucanases and celluloses in both wild and cultivated rice in response to M. oryzae as a means to protect both varieties from the infection.
The transcriptome data indicated that the shikimate pathway-related gene Os01g0375500, encoding shikimate 5-dehydrogenase, was significantly up-regulated in the ‘Wp+R vs. Wp’ comparison but not in the ‘Cp+R vs. Cp’ comparison (Fig. 7a; Table 5; Additional file 6: Table S5a). The synthesis of the SA phytohormone proceeds through the shikimic acid pathway (Shine et al. 2016), suggesting the contribution of up-regulated shikimate pathway to enhancement of SA signaling. With respect to the MVA pathway that is the synthetic pathway for terpenoids, 4 up- and 3 down-regulated genes were identified in the ‘Wp + R vs. Wp’ but no DEGs were detected ‘Cp + R vs. Cp’ comparison (Fig. 7a; Additional file 6: Table S5b), providing evidence for the implication of the MVA pathway in response of R. intraradices-inoculated wild rice to M. oryzae. Additionally, simple phenols-related pathways might be better enhanced in R. intraradices-inoculated wild rice than in R. intraradices-inoculated cultivated rice after their infection with M. oryzae, as several up-regulated genes were recorded in the ‘Wp + R vs. Wp’ comparison, while only down-regulated genes were found in the ‘Cp + R vs. Cp’ comparison (Fig. 7). Similarly, a majority of the terpenoid pathway-related genes were up-regulated in the ‘Wp+R vs. Wp’, while down-regulated in ‘Cp + R vs. Cp’ comparison. These findings together might suggest the contribution of the enhanced shikimate, MVA, phenolic and terpenoid pathways to the higher resistance of wild rice versus that of cultivated rice to M. oryzae by the inoculation with R. intraradices (Fig. 7). In support of our results, increasing evidence has suggested that the shikimate, MVA, terpenoid and small phenols-related metabolic pathways play important roles in the ability of plants to resist fungal infection (Kuc 1995; Martinelli et al. 2015; Massalha et al. 2017; Martinez et al. 2018; Masi et al. 2018; Takemoto et al. 2018). For instance, shikimate pathway, which promotes the synthesis of phenylpropanoids and then the synthesis of SA, is involved in regulation of plant resistance to fungal pathogens (Shi et al. 2018b). Terpenoids and small phenols can serve as phytoalexins that can promote plant in resisting diseases (Meyer et al. 2017; Sun et al. 2017). AMFs can induce the MVA, which in turn may stimulate the biosynthesis of small phenols and terpenoids, thereby contributing to improving disease resistance (Fontana et al. 2009; Kapoor et al. 2017).
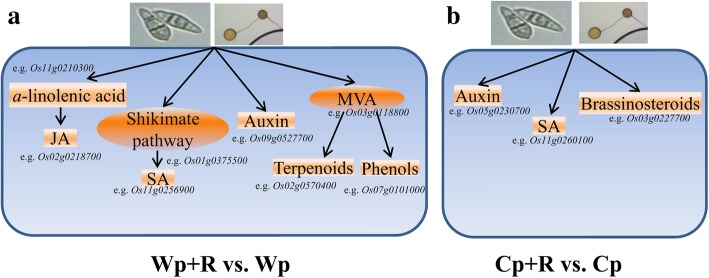
As a result of our comparative analyses, we were able to construct a model of the different responses of R. intraradices-inoculated wild and cultivated rice to M. oryzae (Fig. 8). R. intraradices contributed to the regulation of auxin and SA metabolism in both wild and cultivated rice in response to M. oryzae. Furthermore, R. intraradices might increase α-linolenic acid production, thereby improving JA synthesis in wild rice; as well as induce the shikimate pathway, leading to alterations in downstream metabolism in wild rice. R. intraradices also induced the MVA pathway, and thus increased the syntheses of terpenoids and phenols in wild rice in response to M. oryzae (Fig. 8a). On the other hand, R. intraradices enhanced the biosynthesis of brassinosteroids in cultivated rice leading to enhanced plant resistance to the pathogenic M. oryzae (Fig. 8b).
Fig. 8.
Diagrammatic model of the responses of Rhizoglomus intraradices-uninoculated and R. intraradices-inoculated wild (a) and cultivated (b) rice to Magnaporthe oryzae infection. Cp + R, R. intraradices-inoculated cultivated rice infected with M. oryzae; Cp, R. intraradices-uninoculated cultivated rice infected with M. oryzae; Wp + R, R. intraradices-inoculated wild rice infected with M. oryzae; Wp, R. intraradices-uninoculated wild rice infected with M. oryzae. SA, salicylic acid; JA, jasmonic acid; MVA, mevalonate pathway
Conclusions
In this study, we reported evidence that the benefits received by host plants from mycorrhizal associations differed between cultivated and wild rice, which might have been altered during the process of rice domestication. Our data suggest that number or level of symbiotic benefits has decreased in the cultivated rice relative to wild rice, which may result in the activation of different disease responses when cultivated rice plants are challenged by a pathogenic fungus.
Additional files
Table S1. Genes and primers used for verifying RNA-sequencing data using RT-qPCR. (DOC 46 kb)
Figure S1. Lesion areas of wild and cultivated rice plants in response to Magnaporthe oryzae infection with and without inoculation with the arbuscular mycorrhizal fungus Rhizoglomus intraradices. Cp + R, R. intraradices-inoculated cultivated rice infected with M. oryzae; Cp, R. intraradices-uninoculated cultivated rice infected with M. oryzae; Wp + R, R. intraradices-inoculated wild rice infected with M. oryzae; Wp, R. intraradices-uninoculated wild rice infected with M. oryzae. (TIF 141 kb)
Table S2. RNA-sequencing and reverse transcription-quantitative real-time PCR (RT-qPCR) data of verified genes. The log2 (fold-changes) and fold-changes shown were obtained from RNA-sequencing and RT-qPCR data derived from the ‘Cp+R vs. Cp’ comparison and ‘Wp+R vs. Wp’ comparison, respectively. Red, blue and black colors indicate the up-regulated [log2(fold-change) > 0, q-value < 0.05 in RNA-sequencing data, or fold-change ≥2 in RT-qPCR data with a P-value < 0.05], down-regulated [log2(fold-change) < 0, q-value < 0.05 in RNA-sequencing data, or fold-change ≤2 in RT-qPCR data with a P-value < 0.05] and unchanged genes, respectively. Cp + R, Rhizoglomus intraradices -inoculated cultivated rice infected with Magnaporthe oryzae; Cp, R. intraradices-uninoculated cultivated rice infected with M. oryzae; Wp + R, R. intraradices-inoculated wild rice infected with M. oryzae; Wp, R. intraradices-uninoculated wild rice infected with M. oryzae. (DOCX 32 kb)
Table S3. KEGG analysis of the response of Rhizoglomus intraradices-inoculated wild rice plants to infection by Magnaporthe oryzae in the ‘Wp+R vs. Wp’ (a) and ‘Cp+R vs. Cp’ (b) comparisons. Cp + R, AMF-inoculated cultivated rice infected with M. oryzae; Cp, AMF-uninoculated cultivated rice infected with M. oryzae; Wp + R, AMF-inoculated wild rice infected with M. oryzae; Wp, AMF-uninoculated wild rice infected with M. oryzae. (XLS 63 kb)
Table S4. MapMan 3.6.0 analysis of biotic stress response for ß-glucanase (a), cell wall (b), JA (c) and SA (d) in AMF-inoculated wild and cultivated rice in response to infection by Magnaporthe oryzae in the ‘Wp+R vs. Wp’ and ‘Cp+R vs. Cp’ comparisons. Cp + R, AMF-inoculated cultivated rice infected with M. oryzae; Cp, AMF-uninoculated cultivated rice infected with M. oryzae; Wp + R, AMF-inoculated wild rice infected with M. oryzae; Wp, AMF-uninoculated wild rice infected with M. oryzae. SA, salicylic acid; JA, jasmonic acid. (XLS 93 kb)
Table S5. MapMan 3.6.0 analysis of secondary metabolic pathway response for shikimate pathway (a), MVA (b), simple phenols (c) and terpenoids (d) in AMF-inoculated wild and cultivated rice in response to infection by Magnaporthe oryzae in the ‘Wp+R vs. Wp’ and ‘Cp+R vs. Cp’ comparisons. Cp + R, AMF-inoculated cultivated rice infected with M. oryzae; Cp, AMF-uninoculated cultivated rice infected with M. oryzae; Wp + R, AMF-inoculated wild rice infected with M. oryzae; Wp, AMF-uninoculated wild rice infected with M. oryzae. MVA, mevalonate. (XLS 46 kb)
Acknowledgements
The authors would like to thank Dr. Zhiping Song in Fudan University and Dr. Jun Rong in Nanchang University for providing the wild rice seeds. We would also like to thank the staffs in Genovogen Co., Ltd. for advice on the data analysis.
Funding
This work was financially supported by the Chinese Academic Project B (XDB15030103), National Project (2016YFC0501202), the National Natural Science Foundation of China (41571255, 31370144), and the Natural Science Foundation of Jilin Province (20140101017JC), Science and Technology Development Project of Jilin Province (20180519002JH), the Key Research Program of the Chinese Academy of Sciences (KFZD-SW-112), the Natural Science Foundation of Jilin Province (20140101017JC), and the 135 Project of Northeast Institute of Geography and Agroecology (Y6H2043001).
Availability of data and materials
All raw RNA-seq data in this article have been deposited in GeneBank with the accession number SRP150958.
Authors’ contributions
CT designed the experiment. LT, CC, LM, and FN performed the experiments; LT, CC, LM WL, and JZ analyzed the data with the input of LPT, LT, CC, LPT, and CT wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lei Tian, Email: tianlei@iga.ac.cn.
Chunling Chang, Email: changchunling0709@163.com.
Lina Ma, Email: malina@iga.ac.cn.
Fahad Nasir, Email: fa0100@nenu.edu.cn.
Jianfeng Zhang, Email: zhangjianfeng06@tsinghua.org.cn.
Weiqiang Li, Email: weiqiang.li@riken.jp.
Lam-Son Phan Tran, Email: tranplamson@duytan.edu.vn, Email: son.tran@riken.jp.
Chunjie Tian, Email: tiancj@neigae.ac.cn.
References
- Abdelrahman M, Jogaiah S, Burritt DJ, Tran L-SP. Legume genetic resources and transcriptome dynamics under abiotic stress conditions. Plant Cell Environ. 2018;41:1972–1983. doi: 10.1111/pce.13123. [DOI] [PubMed] [Google Scholar]
- AbuQamar S, Moustafa K, Tran LS. Mechanisms and strategies of plant defense against Botrytis cinerea. Crit Rev Biotechnol. 2017;37:262–274. doi: 10.1080/07388551.2016.1271767. [DOI] [PubMed] [Google Scholar]
- Agrawal GK, Tamogami S, Iwahashi H, Agrawal VP, Rakwal R. Transient regulation of jasmonic acid-inducible rice MAP kinase gene (OsBWMK1) by diverse biotic and abiotic stresses. Plant Physiol Biochem. 2003;41:355–361. doi: 10.1016/S0981-9428(03)00030-5. [DOI] [Google Scholar]
- Baby UI. Biocontrol strategies for the management of rice sheath blight disease. In: Sreenivasaprasad S, Johnson R, editors. Major fungal diseases of rice: recent advances. Dordrecht: Springer Netherlands; 2001. pp. 253–269. [Google Scholar]
- Bari R, Jones JDG. Role of plant hormones in plant defence responses. Plant Mol Biol. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- Berdeni D, Cotton TEA, Daniell TJ, Bidartondo MI, Cameron DD, Evans KL. The effects of arbuscular mycorrhizal fungal colonisation on nutrient status, growth, productivity, and canker resistance of apple (Malus pumila) Front Microbiol. 2018;9:1461. doi: 10.3389/fmicb.2018.01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Liu S, Somssich IE. Transcriptional events defining plant immune responses. Curr Opin Plant Biol. 2017;38:1–9. doi: 10.1016/j.pbi.2017.04.004. [DOI] [PubMed] [Google Scholar]
- Bonnet C, Lassueur S, Ponzio C, Gols R, Dicke M, Reymond P. Combined biotic stresses trigger similar transcriptomic responses but contrasting resistance against a chewing herbivore in Brassica nigra. BMC Plant Biol. 2017;17:127. doi: 10.1186/s12870-017-1074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito I, Goss MJ, Alho L, Brígido C, van Tuinen D, Félix MR, Carvalho M (2018) Agronomic management of AMF functional diversity to overcome biotic and abiotic stresses - the role of plant sequence and intact extraradical mycelium. Fungal Ecol. 10.1016/j.funeco.2018.06.001
- Cameron DD, Neal AL, van Wees SCM, Ton J. Mycorrhiza-induced resistance: more than the sum of its parts? Trends Plant Sci. 2013;18:539–545. doi: 10.1016/j.tplants.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Soriano L, García-Martínez J, Segundo BS. The arbuscular mycorrhizal symbiosis promotes the systemic induction of regulatory defence-related genes in rice leaves and confers resistance to pathogen infection. Mol Plant Pathol. 2012;13:579–592. doi: 10.1111/j.1364-3703.2011.00773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yang X, Huang X, Duan S, Long C, Chen J, Rong J. Leaf transcriptome analysis of a subtropical evergreen broadleaf plant, wild oil-tea camellia (Camellia oleifera), revealing candidate genes for cold acclimation. BMC Genomics. 2017;18:211. doi: 10.1186/s12864-017-3570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitarra W, Pagliarani C, Maserti B, Lumini E, Siciliano I, Cascone P, Schubert A, Gambino G, Balestrini R, Guerrieri E. Insights on the impact of arbuscular mycorrhizal symbiosis on tomato tolerance to water stress. Plant Physiol. 2016;171:1009–1023. doi: 10.1104/pp.16.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary SP, Yu J-Q, Yamaguchi-Shinozaki K, Shinozaki K, Tran L-SP. Benefits of brassinosteroid crosstalk. Trends Plant Sci. 2012;17:594–605. doi: 10.1016/j.tplants.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Chumley FG, Valent B. Genetic analysis of melanin-deficient, nonpathogenic mutants of Magnaporthe grisea. Mol Plant Microbe In. 1990;3:135–143. doi: 10.1094/MPMI-3-135. [DOI] [Google Scholar]
- Croll D, McDonald BA. The genetic basis of local adaptation for pathogenic fungi in agricultural ecosystems. Mol Ecol. 2017;26:2027–2040. doi: 10.1111/mec.13870. [DOI] [PubMed] [Google Scholar]
- Dai J, Hu J, Zhu A, Bai J, Wang J, Lin X. No tillage enhances arbuscular mycorrhizal fungal population, glomalin-related soil protein content, and organic carbon accumulation in soil macroaggregates. J Soils Sediments. 2015;15:1055–1062. doi: 10.1007/s11368-015-1091-9. [DOI] [Google Scholar]
- do Prado Ribeiro L, ALS K, JAW F, Tramontin MA, Trapp MA, Mithöfer A, Nansen C. Hyperspectral imaging to characterize plant–plant communication in response to insect herbivory. Plant Methods. 2018;14:54. doi: 10.1186/s13007-018-0322-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemadi M, Gutjahr C, Couzigou J-M, Zouine M, Lauressergues D, Timmers A, Audran C, Bouzayen M, Bécard G, Combier J-P. Auxin perception is required for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Physiol. 2014;166:281–292. doi: 10.1104/pp.114.246595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feddermann N, Finlay R, Boller T, Elfstrand M. Functional diversity in arbuscular mycorrhiza–the role of gene expression, phosphorous nutrition and symbiotic efficiency. Fungal Ecol. 2010;3:1–8. doi: 10.1016/j.funeco.2009.07.003. [DOI] [Google Scholar]
- Field Katie J., Pressel Silvia. Unity in diversity: structural and functional insights into the ancient partnerships between plants and fungi. New Phytologist. 2018;220(4):996–1011. doi: 10.1111/nph.15158. [DOI] [PubMed] [Google Scholar]
- Fiorilli V, Vannini C, Ortolani F, Garcia-Seco D, Chiapello M, Novero M, Domingo G, Terzi V, Morcia C, Bagnaresi P, Moulin L, Bracale M, Bonfante P. Omics approaches revealed how arbuscular mycorrhizal symbiosis enhances yield and resistance to leaf pathogen in wheat. Sci Rep. 2018;8:9625. doi: 10.1038/s41598-018-27622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana A, Reichelt M, Hempel S, Gershenzon J, Unsicker SB. The effects of arbuscular mycorrhizal fungi on direct and indirect defense metabolites of Plantago lanceolata L. J Chem Ecol. 2009;35:833–843. doi: 10.1007/s10886-009-9654-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green NE, Graham SO, Schenck NC. The influence of pH on the germination of vesicular-arbuscular mycorrhizal spores. Mycologia. 1976;68:929–934. doi: 10.1080/00275514.1976.12019969. [DOI] [PubMed] [Google Scholar]
- Grove S, Haubensak KA, Gehring C, Parker IM. Mycorrhizae, invasions, and the temporal dynamics of mutualism disruption. J Ecol. 2017;105:1496–1508. doi: 10.1111/1365-2745.12853. [DOI] [Google Scholar]
- Hua L, Wang DR, Tan L, Fu Y, Liu F, Xiao L, Zhu Z, Fu Q, Sun X, Gu P, Cai H, McCouch SR, Sun C. LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell. 2015;27:1875–1888. doi: 10.1105/tpc.15.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanami H, Takada N, Koda Y. Ephemerality of a spring ephemeral Gagea lutea (L.) is attributable to shoot senescence induced by free linolenic acid. Plant Cell Physiol. 2017;58:1724–1729. doi: 10.1093/pcp/pcx109. [DOI] [PubMed] [Google Scholar]
- Jemo M, Dhiba D, Hashem A, Abd Allah EF, Alqarawi AA, Tran LP. Mycorrhizal fungal community structure in tropical humid soils under fallow and cropping conditions. Sci Rep. 2018;8:17061. doi: 10.1038/s41598-018-34736-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Hodge A, Kuzyakov Y. Plant and mycorrhizal regulation of rhizodeposition. New Phytol. 2004;163:459–480. doi: 10.1111/j.1469-8137.2004.01130.x. [DOI] [PubMed] [Google Scholar]
- Kapoor R, Anand G, Gupta P, Mandal S. Insight into the mechanisms of enhanced production of valuable terpenoids by arbuscular mycorrhiza. Phytochem Rev. 2017;16:677–692. doi: 10.1007/s11101-016-9486-9. [DOI] [Google Scholar]
- Karandashov V, Bucher M. Symbiotic phosphate transport in arbuscular mycorrhizas. Trends Plant Sci. 2005;10:22–29. doi: 10.1016/j.tplants.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Kesten C, Menna A, Sánchez-Rodríguez C. Regulation of cellulose synthesis in response to stress. Curr Opin Plant Biol. 2017;40:106–113. doi: 10.1016/j.pbi.2017.08.010. [DOI] [PubMed] [Google Scholar]
- Koga H, Dohi K, Nakayachi O, Mori M. A novel inoculation method of Magnaporthe grisea for cytological observation of the infection process using intact leaf sheaths of rice plants. Physiol Mol Plant P. 2004;64:67–72. doi: 10.1016/j.pmpp.2004.07.002. [DOI] [Google Scholar]
- Kuc J. Phytoalexins, stress metabolism, and disease resistance in plants. Annu Rev Phytopathol. 1995;33:275–297. doi: 10.1146/annurev.py.33.090195.001423. [DOI] [PubMed] [Google Scholar]
- Li F, Guo S, Zhao Y, Chen D, Chong K, Xu Y. Overexpression of a homopeptide repeat-containing bHLH protein gene (OrbHLH001) from Dongxiang wild rice confers freezing and salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 2010;29:977–986. doi: 10.1007/s00299-010-0883-z. [DOI] [PubMed] [Google Scholar]
- Liang L, Wang C, Zeng L, Wang W, Feng J, Chen B, Su J, Chen S, Shang F, Zhu X, Lin F. The rice cultivar Baixiangzhan harbours a recessive gene xa42(t) determining resistance against Xanthomonas oryzae pv. Oryzae. Plant Breed. 2017;136:603–609. doi: 10.1111/pbr.12493. [DOI] [Google Scholar]
- Liu J, Wang X, Mitchell T, Hu Y, Liu X, Dai L, Wang G. Recent progress and understanding of the molecular mechanisms of the rice-Magnaporthe oryzae interaction. Mol Plant Pathol. 2010;11:419–427. doi: 10.1111/j.1364-3703.2009.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhang Q, Burton RA, Shirley NJ, Atwell BJ. Expression of vacuolar H+-pyrophosphatase (OVP3) is under control of an anoxia-inducible promoter in rice. Plant Mol Biol. 2009;72:47. doi: 10.1007/s11103-009-9549-z. [DOI] [PubMed] [Google Scholar]
- Liu W, Ghouri F, Yu H, Li X, Yu S, Shahid MQ, Liu X. Genome wide re-sequencing of newly developed Rice lines from common wild rice (Oryza rufipogon Griff.) for the identification of NBS-LRR genes. PloS One. 2017;12:e0180662. doi: 10.1371/journal.pone.0180662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Ma L, He X, Tian C. Water strategy of mycorrhizal rice at low temperature through the regulation of PIP aquaporins with the involvement of trehalose. Appl Soil Ecol. 2014;84:185–191. doi: 10.1016/j.apsoil.2014.07.010. [DOI] [Google Scholar]
- Luo W, Yang Y, Fang F, Li W, Hu F, Zhang J, Chen D, Yu L. Chronology of ancient Dongxiang wild rice (Oryza rufipogon Griff.), and the morphologies of grains, double-peaked phytoliths, and starch, in the middle Yangtze river region, China. Rev Palaeobot Palyno. 2017;244:140–147. doi: 10.1016/j.revpalbo.2017.05.008. [DOI] [Google Scholar]
- Mahesh HM, Murali M, Anup Chandra Pal M, Melvin P, Sharada MS. Salicylic acid seed priming instigates defense mechanism by inducing PR-proteins in Solanum melongena L. upon infection with Verticillium dahliae Kleb. Plant Physiol Biochem. 2017;117:12–23. doi: 10.1016/j.plaphy.2017.05.012. [DOI] [PubMed] [Google Scholar]
- Mao D, Yu L, Chen D, Li L, Zhu Y, Xiao Y, Zhang D, Chen C. Multiple cold resistance loci confer the high cold tolerance adaptation of Dongxiang wild rice (Oryza rufipogon) to its high-latitude habitat. Theor Appl Genet. 2015;128:1359–1371. doi: 10.1007/s00122-015-2511-3. [DOI] [PubMed] [Google Scholar]
- Martinelli F, Scalenghe R, Davino S, Panno S, Scuderi G, Ruisi P, Villa P, Stroppiana D, Boschetti M, Goulart LR, Davis CE, Dandekar AM. Advanced methods of plant disease detection. A review. Agron Sustain Dev. 2015;35:1–25. doi: 10.1007/s13593-014-0246-1. [DOI] [Google Scholar]
- Martinez DA, Loening UE, Graham MC. Impacts of glyphosate-based herbicides on disease resistance and health of crops: a review. Environ Sci Eur. 2018;30:2. doi: 10.1186/s12302-018-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Robles N, Lehmann A, Seco E, Aroca R, Rillig MC, Milla R. Impacts of domestication on the arbuscular mycorrhizal symbiosis of 27 crop species. New Phytol. 2018;218:322–334. doi: 10.1111/nph.14962. [DOI] [PubMed] [Google Scholar]
- Masi M, Nocera P, Boari A, Cimmino A, Zonno MC, Infantino A, Vurro M, Evidente A. Lathyroxins a and B, phytotoxic monosubstituted phenols isolated from Ascochyta lentis var. lathyri, a fungal pathogen of grass pea (Lathyrus sativus) J Nat Prod. 2018;81:1093–1097. doi: 10.1021/acs.jnatprod.7b01034. [DOI] [PubMed] [Google Scholar]
- Massalha H, Korenblum E, Tholl D, Aharoni A. Small molecules below-ground: the role of specialized metabolites in the rhizosphere. Plant J. 2017;90:788–807. doi: 10.1111/tpj.13543. [DOI] [PubMed] [Google Scholar]
- Mehari ZH, Elad Y, Rav-David D, Graber ER, Meller Harel Y. Induced systemic resistance in tomato (Solanum lycopersicum) against Botrytis cinerea by biochar amendment involves jasmonic acid signaling. Plant Soil. 2015;395:31–44. doi: 10.1007/s11104-015-2445-1. [DOI] [Google Scholar]
- Meyer J, Berger DK, Christensen SA, Murray SL. RNA-Seq analysis of resistant and susceptible sub-tropical maize lines reveals a role for kauralexins in resistance to grey leaf spot disease, caused by Cercospora zeina. BMC Plant Biol. 2017;17:197. doi: 10.1186/s12870-017-1137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir F, Tian L, Chang C, Li X, Gao Y, Tran L-SP, Tian C. Current understanding of pattern-triggered immunity and hormone-mediated defense in rice (Oryza sativa) in response to Magnaporthe oryzae infection. Semin Cell Dev Biol. 2018;83e:95–105. doi: 10.1016/j.semcdb.2017.10.020. [DOI] [PubMed] [Google Scholar]
- Ni D, Song F, Ni J, Zhang A, Wang C, Zhao K, Yang Y, Wei P, Yang J, Li L. Marker-assisted selection of two-line hybrid rice for disease resistance to rice blast and bacterial blight. Field Crop Res. 2015;184:1–8. doi: 10.1016/j.fcr.2015.07.018. [DOI] [Google Scholar]
- Pérez-de-Luque A, Tille S, Johnson I, Pascual-Pardo D, Ton J, Cameron DD. The interactive effects of arbuscular mycorrhiza and plant growth-promoting rhizobacteria synergistically enhance host plant defences against pathogens. Sci Rep. 2017;7:16409. doi: 10.1038/s41598-017-16697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Jaramillo JE, Carrion VJ, de Hollander M, Raaijmakers JM. The wild side of plant microbiomes. Microbiome. 2018;6:143. doi: 10.1186/s40168-018-0519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JM, Hayman DS. Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Tran Brit Mycol Soc. 1970;55:158–161. doi: 10.1016/S0007-1536(70)80110-3. [DOI] [Google Scholar]
- Pozo MJ, López-Ráez JA, Azcón-Aguilar C, García-Garrido JM. Phytohormones as integrators of environmental signals in the regulation of mycorrhizal symbioses. New Phytol. 2015;205:1431–1436. doi: 10.1111/nph.13252. [DOI] [PubMed] [Google Scholar]
- Samalova M, Mélida H, Vilaplana F, Bulone V, Soanes DM, Talbot NJ, Gurr SJ. The β-1,3-glucanosyltransferases (gels) affect the structure of the rice blast fungal cell wall during appressorium-mediated plant infection. Cell Microbiol. 2017;19:e12659. doi: 10.1111/cmi.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Romera B, Porcel R, Ruiz-Lozano JM, Aroca R. Arbuscular mycorrhizal symbiosis modifies the effects of a nitric oxide donor (sodium nitroprusside; SNP) and a nitric oxide synthesis inhibitor (Nω-nitro-L-arginine methyl ester;L-NAME) on lettuce plants under well watered and drought conditions. Symbiosis. 2018;74:11–20. doi: 10.1007/s13199-017-0486-3. [DOI] [Google Scholar]
- Sato K, Suyama Y, Saito M, Sugawara K. A new primer for discrimination of arbuscular mycorrhizal fungi with polymerase chain reaction-denature gradient gel electrophoresis. Grassl Sci. 2005;51:179–181. doi: 10.1111/j.1744-697X.2005.00023.x. [DOI] [Google Scholar]
- Selvakumar G, Shagol CC, Kang Y, Chung BN, Han SG, Sa TM. Arbuscular mycorrhizal fungi spore propagation using single spore as starter inoculum and a plant host. J Appl Microbiol. 2018;124:1556–1565. doi: 10.1111/jam.13714. [DOI] [PubMed] [Google Scholar]
- Shi S, Tian L, Nasir F, Li X, Li W, Phan Tran L-S, Tian C. Impact of domestication on the evolution of rhizomicrobiome of rice in response to the presence of Magnaporthe oryzae. Plant Physiol Biochem. 2018;132:156–165. doi: 10.1016/j.plaphy.2018.08.023. [DOI] [PubMed] [Google Scholar]
- Shi Y, Liu X, Fang Y, Tian Q, Jiang H, Ma H. 2, 3-Butanediol activated disease-resistance of creeping bentgrass by inducing phytohormone and antioxidant responses. Plant Physiol Biochem. 2018;129:244–250. doi: 10.1016/j.plaphy.2018.06.010. [DOI] [PubMed] [Google Scholar]
- Shine MB, Yang J-W, El-Habbak M, Nagyabhyru P, Fu D-Q, Navarre D, Ghabrial S, Kachroo P, Kachroo A. Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol. 2016;212:627–636. doi: 10.1111/nph.14078. [DOI] [PubMed] [Google Scholar]
- Stein JC, Yu Y, Copetti D, Zwickl DJ, Zhang L, Zhang C, Chougule K, Gao D, Iwata A, Goicoechea JL, Wei S, Wang J, Liao Y, Wang M, Jacquemin J, Becker C, Kudrna D, Zhang J, Londono CEM, Song X, Lee S, Sanchez P, Zuccolo A, Ammiraju JSS, Talag J, Danowitz A, Rivera LF, Gschwend AR, Noutsos C, Wu C-c, S-m K, J-w Z, F-j W, Zhao Q, Feng Q, El Baidouri M, Carpentier M-C, Lasserre E, Cooke R, Dd RF, da Maia LC, dos Santos RS, Nyberg KG, McNally KL, Mauleon R, Alexandrov N, Schmutz J, Flowers D, Fan C, Weigel D, Jena KK, Wicker T, Chen M, Han B, Henry R, Hsing Y-iC, Kurata N, de Oliveira AC, Panaud O, Jackson SA, Machado CA, Sanderson MJ, Long M, Ware D, Wing RA. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nature Genet. 2018;50:285–296. doi: 10.1038/s41588-018-0040-0. [DOI] [PubMed] [Google Scholar]
- Sun J, Janisiewicz WJ, Nichols B, Jurick Ii WM, Chen P. Composition of phenolic compounds in wild apple with multiple resistance mechanisms against postharvest blue mold decay. Postharvest Biol Tec. 2017;127:68–75. doi: 10.1016/j.postharvbio.2017.01.006. [DOI] [Google Scholar]
- Syed Ab Rahman SF, Singh E, Pieterse CMJ, Schenk PM. Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 2018;267:102–111. doi: 10.1016/j.plantsci.2017.11.012. [DOI] [PubMed] [Google Scholar]
- Takemoto D, Shibata Y, Ojika M, Mizuno Y, Imano S, Ohtsu M, Sato I, Chiba S, Kawakita K, Rin S, Camagna M. Resistance to Phytophthora infestans: exploring genes required for disease resistance in Solanaceae plants. J Gen Plant Pathol. 2018;84:312–320. doi: 10.1007/s10327-018-0801-8. [DOI] [Google Scholar]
- Talukdar PR, Rathi S, Pathak K, Chetia SK, Sarma RN. Population structure and marker-trait association in indigenous aromatic rice. Rice Sci. 2017;24:145–154. doi: 10.1016/j.rsci.2016.08.009. [DOI] [Google Scholar]
- Thomas E, Tovar E, Villafane C, Bocanegra JL, Moreno R. Distribution, genetic diversity and potential spatiotemporal scale of alien gene flow in crop wild relatives of rice (Oryza spp.) in Colombia. Rice. 2017;10:13. doi: 10.1186/s12284-017-0150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Kasiborski B, Koul R, Lammers PJ, Bucking H, Shachar-Hill Y. Regulation of the nitrogen transfer pathway in the arbuscular mycorrhizal symbiosis: gene characterization and the coordination of expression with nitrogen flux. Plant Physiol. 2010;153:1175–1187. doi: 10.1104/pp.110.156430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Shi S, Ma L, Zhou X, Luo S, Zhang J, Lu B, Tian C. The effect of Glomus intraradices on the physiological properties of Panax ginseng and on rhizospheric microbial diversity. J Ginseng Res. 2019;43:77–85. doi: 10.1016/j.jgr.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Shi S, Nasir F, Chang C, Li W, Tran L-SP, Tian C. Comparative analysis of the root transcriptomes of cultivated and wild rice varieties in response to Magnaporthe oryzae infection revealed both common and species-specific pathogen responses. Rice. 2018;11:26. doi: 10.1186/s12284-018-0211-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Zhou X, Ma L, Xu S, Nasir F, Tian C. Root-associated bacterial diversities of Oryza rufipogon and Oryza sativa and their influencing environmental factors. Arch Microbiol. 2017;199:563–571. doi: 10.1007/s00203-016-1325-2. [DOI] [PubMed] [Google Scholar]
- Trouvelot A, Kough JL, Gianinazzi-Pearson V. Physiological and Genetical Aspects of Mycorrhizae. Aspects physiologiques et genetiques des mycorhizes. Dijon: INRA; 1986. Estimation of VA mycorrhizal infection levels. Research for methods having a functional significance; pp. 217–221. [Google Scholar]
- Verzeaux J, Hirel B, Dubois F, Lea PJ, Tétu T. Agricultural practices to improve nitrogen use efficiency through the use of arbuscular mycorrhizae: basic and agronomic aspects. Plant Sci. 2017;264:48–56. doi: 10.1016/j.plantsci.2017.08.004. [DOI] [PubMed] [Google Scholar]
- Volpe V, Chitarra W, Cascone P, Volpe MG, Bartolini P, Moneti G, Pieraccini G, Di Serio C, Maserti B, Guerrieri E, Balestrini R. The association with two different arbuscular mycorrhizal fungi differently affects water stress tolerance in tomato. Front Plant Sci. 2018;9:1480–1480. doi: 10.3389/fpls.2018.01480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li L, Yin Y, Gu Z, Chai R, Wang Y, Sun G. Equol, a clinically important metabolite, inhibits the development and pathogenicity of Magnaporthe oryzae, the causal agent of rice blast disease. Molecules. 2017;22:1799. doi: 10.3390/molecules22101799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia XJ, Wang YJ, Zhou YH, Tao Y, Mao WH, Shi K, Asami T, Chen Z, Yu JQ. Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol. 2009;150:801–814. doi: 10.1104/pp.109.138230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebelo S, Song Y, Kloepper JW, Fadamiro H. Rhizobacteria activates (+)-δ-cadinene synthase genes and induces systemic resistance in cotton against beet armyworm (Spodoptera exigua) Plant Cell Environ. 2016;39:935–943. doi: 10.1111/pce.12704. [DOI] [PubMed] [Google Scholar]
- Zhang L, Nakagomi Y, Endo T, Teranishi M, Hidema J, Sato S, Higashitani A. Divergent evolution of rice blast resistance Pi54 locus in the genus Oryza. Rice. 2018;11:63. doi: 10.1186/s12284-018-0256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xu M, Liu Y, Zhang F, Hodge A, Feng G. Carbon and phosphorus exchange may enable cooperation between an arbuscular mycorrhizal fungus and a phosphate-solubilizing bacterium. New Phytol. 2016;210:1022–1032. doi: 10.1111/nph.13838. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Lin S, Zhao B, Wang C, Yang W, Zhou Y, Zhu L. Identification and tagging a new gene for resistance to bacterial blight (Xanthomonas oryzae pv. Oryzae) from O. rufipogon. Rice Genet Newsl. 1998;15:138–142. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genes and primers used for verifying RNA-sequencing data using RT-qPCR. (DOC 46 kb)
Figure S1. Lesion areas of wild and cultivated rice plants in response to Magnaporthe oryzae infection with and without inoculation with the arbuscular mycorrhizal fungus Rhizoglomus intraradices. Cp + R, R. intraradices-inoculated cultivated rice infected with M. oryzae; Cp, R. intraradices-uninoculated cultivated rice infected with M. oryzae; Wp + R, R. intraradices-inoculated wild rice infected with M. oryzae; Wp, R. intraradices-uninoculated wild rice infected with M. oryzae. (TIF 141 kb)
Table S2. RNA-sequencing and reverse transcription-quantitative real-time PCR (RT-qPCR) data of verified genes. The log2 (fold-changes) and fold-changes shown were obtained from RNA-sequencing and RT-qPCR data derived from the ‘Cp+R vs. Cp’ comparison and ‘Wp+R vs. Wp’ comparison, respectively. Red, blue and black colors indicate the up-regulated [log2(fold-change) > 0, q-value < 0.05 in RNA-sequencing data, or fold-change ≥2 in RT-qPCR data with a P-value < 0.05], down-regulated [log2(fold-change) < 0, q-value < 0.05 in RNA-sequencing data, or fold-change ≤2 in RT-qPCR data with a P-value < 0.05] and unchanged genes, respectively. Cp + R, Rhizoglomus intraradices -inoculated cultivated rice infected with Magnaporthe oryzae; Cp, R. intraradices-uninoculated cultivated rice infected with M. oryzae; Wp + R, R. intraradices-inoculated wild rice infected with M. oryzae; Wp, R. intraradices-uninoculated wild rice infected with M. oryzae. (DOCX 32 kb)
Table S3. KEGG analysis of the response of Rhizoglomus intraradices-inoculated wild rice plants to infection by Magnaporthe oryzae in the ‘Wp+R vs. Wp’ (a) and ‘Cp+R vs. Cp’ (b) comparisons. Cp + R, AMF-inoculated cultivated rice infected with M. oryzae; Cp, AMF-uninoculated cultivated rice infected with M. oryzae; Wp + R, AMF-inoculated wild rice infected with M. oryzae; Wp, AMF-uninoculated wild rice infected with M. oryzae. (XLS 63 kb)
Table S4. MapMan 3.6.0 analysis of biotic stress response for ß-glucanase (a), cell wall (b), JA (c) and SA (d) in AMF-inoculated wild and cultivated rice in response to infection by Magnaporthe oryzae in the ‘Wp+R vs. Wp’ and ‘Cp+R vs. Cp’ comparisons. Cp + R, AMF-inoculated cultivated rice infected with M. oryzae; Cp, AMF-uninoculated cultivated rice infected with M. oryzae; Wp + R, AMF-inoculated wild rice infected with M. oryzae; Wp, AMF-uninoculated wild rice infected with M. oryzae. SA, salicylic acid; JA, jasmonic acid. (XLS 93 kb)
Table S5. MapMan 3.6.0 analysis of secondary metabolic pathway response for shikimate pathway (a), MVA (b), simple phenols (c) and terpenoids (d) in AMF-inoculated wild and cultivated rice in response to infection by Magnaporthe oryzae in the ‘Wp+R vs. Wp’ and ‘Cp+R vs. Cp’ comparisons. Cp + R, AMF-inoculated cultivated rice infected with M. oryzae; Cp, AMF-uninoculated cultivated rice infected with M. oryzae; Wp + R, AMF-inoculated wild rice infected with M. oryzae; Wp, AMF-uninoculated wild rice infected with M. oryzae. MVA, mevalonate. (XLS 46 kb)
Data Availability Statement
All raw RNA-seq data in this article have been deposited in GeneBank with the accession number SRP150958.