Abstract
Background & Aims
The epithelial response is critical for intestinal defense against Cryptosporidium, but is poorly understood. To uncover the host strategy for defense against Cryptosporidium, we examined the transcriptional response of intestinal epithelial cells (IECs) to C parvum in experimentally infected piglets by microarray. Up-regulated genes were dominated by targets of interferon (IFN) and IFN-λ3 was up-regulated significantly in infected piglet mucosa. Although IFN-λ has been described as a mediator of epithelial defense against viral pathogens, there is limited knowledge of any role against nonviral pathogens. Accordingly, the aim of the study was to determine the significance of IFN-λ3 to epithelial defense and barrier function during C parvum infection.
Methods
The significance of C parvum–induced IFN-λ3 expression was determined using an immunoneutralization approach in neonatal C57BL/6 mice. The ability of the intestinal epithelium to up-regulate IFN-λ2/3 expression in response to C parvum infection and the influence of IFN-λ2/3 on epithelial defense against C parvum invasion, intracellular development, and loss of barrier function was examined using polarized monolayers of a nontransformed porcine-derived small intestinal epithelial cell line (IPEC-J2). Specifically, changes in barrier function were quantified by measurement of transepithelial electrical resistance and transepithelial flux studies.
Results
Immunoneutralization of IFN-λ2/3 in C parvum–infected neonatal mice resulted in a significantly increased parasite burden, fecal shedding, and villus blunting with crypt hyperplasia during peak infection. In vitro, C parvum was sufficient to induce autonomous IFN-λ3 and interferon-stimulated gene 15 expression by IECs. Priming of IECs with recombinant human IFN-λ3 promoted cellular defense against C parvum infection and abrogated C parvum–induced loss of barrier function by decreasing paracellular permeability to sodium.
Conclusions
These studies identify IFN-λ3 as a key epithelial defense mechanism against C parvum infection.
Keywords: Cryptosporidiosis, Cytokine, Enterocyte, Protozoa
Abbreviations used in this paper: cDNA, complementary DNA; Ct, cycle threshold; DAPI, 4′,6-diamidino-2-phenylindole; FITC, fluorescein isothiocyanate; GBP, guanylate binding protein; IEC, intestinal epithelial cells; IFN, interferon; IPEC-J2, porcine-derived small intestinal epithelial cell line; ISG, interferon-stimulated gene; mRNA, messenger RNA; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; rHuIFN-λ3, recombinant human interferon-λ3; TEER, transepithelial electrical resistance
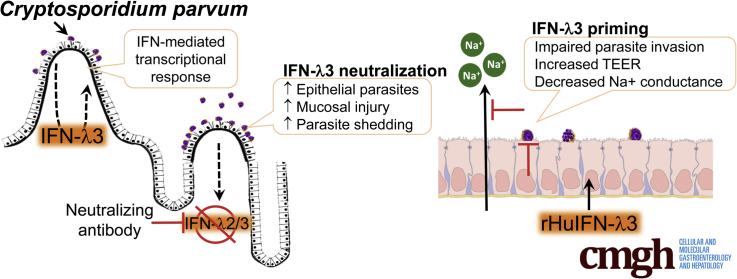
Graphical abstract
See editorial on page 149.
Summary.
We identified that interferon-λ3 promotes intestinal epithelial cell defense against Cryptosporidium parvum by inhibiting parasite invasion and mitigating loss of paracellular barrier function. These findings may lead to novel therapeutic targets by which C parvum clearance can be promoted.
Cryptosporidiosis causes unrelenting diarrhea in immunocompromised individuals worldwide and is the most common cause of infectious water and foodborne diarrheal outbreaks in the United States, resulting in an estimated 750,000 cases each year.1 In developing countries, Cryptosporidium species is the second leading cause of infectious diarrheal death in children younger than the age of 5 years and results in approximately 200,000 deaths annually.2, 3
Cryptosporidium infection occurs by fecal–oral transmission of environmentally resistant oocysts. After ingestion, oocysts release infective sporozoites that attach and become enveloped in the apical membrane of small intestinal villus intestinal epithelial cells (IECs) where they complete a complex and recurring life cycle. When Cryptosporidium infects the villus epithelial cells, they are shed rapidly, resulting in profound villus blunting. In this condition, nutrient malabsorption, intestinal secretion, and failure of barrier function result in severe diarrhea, dehydration, starvation, and, frequently, death. Despite decades of research, there are currently no consistently effective treatments for cryptosporidiosis. Clearance of the pathogen and recovery from infection are reliant on a competent host response and access to supportive care. The use of more insightful therapies to promote recovery of individuals with cryptosporidiosis is hindered by a lack of understanding of how the intestinal epithelium favorably combats the infection.
To address this gap in understanding, we began our study by investigating the transcriptional response of C parvum–infected IECs in a robust model of cryptosporidiosis, the neonatal piglet. We hypothesized that the IEC transcriptional signature would provide clues that would aid in identifying novel epithelial-derived mechanisms of innate immune activation and host defense against C parvum infection. Accordingly, we detected that IECs up-regulate numerous interferon (IFN)-stimulated genes during peak Cryptosporidium infection. By investigating the specific stimulus for this IFN response, we identified that C parvum infection induces robust mucosal expression of a type III IFN: IFN-λ3.4, 5
There is mounting evidence that IFN-λ is a critical mediator for the innate protection of epithelia against viral infection.6, 7 In addition, IFN-λ has shown extraintestinal epithelial barrier–protective effects in the face of viral infection.8 However, to date, the role of IFN-λ in defense against nonviral enteric diarrheal pathogens or in the promotion of intestinal epithelial barrier function has been limited to a single study.9 Accordingly, the aim of this study was to determine the significance of IFN-λ expression to epithelial defense and barrier function during C parvum infection.
Results
The Intestinal Epithelial Transcriptional Response to C parvum Infection In Vivo Is Dominated by Targets of Interferon Signaling
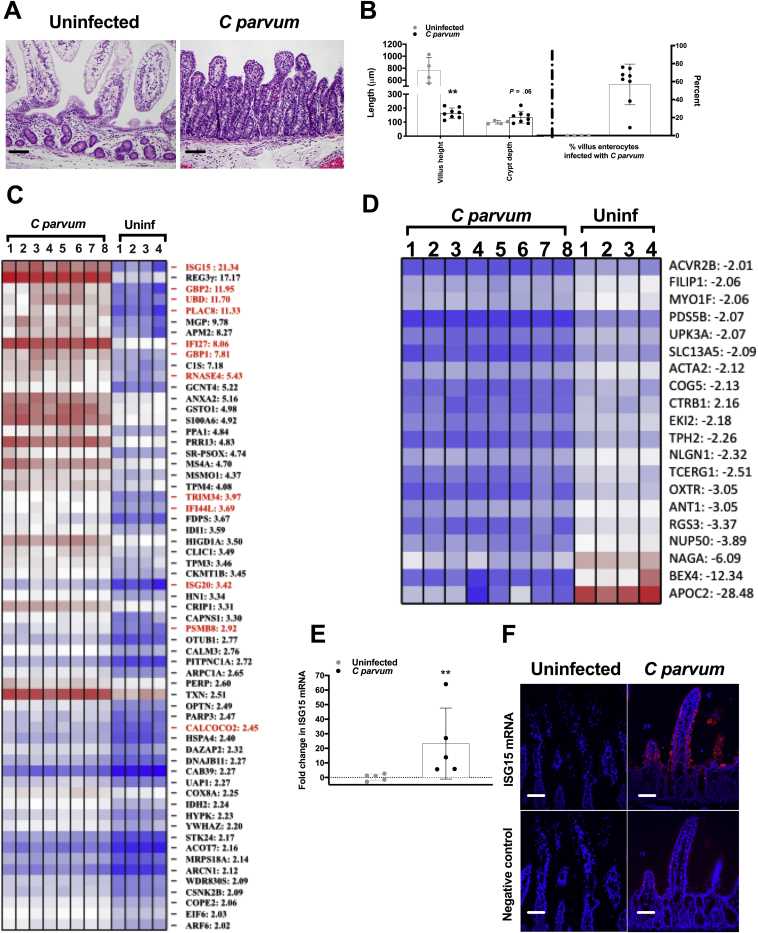
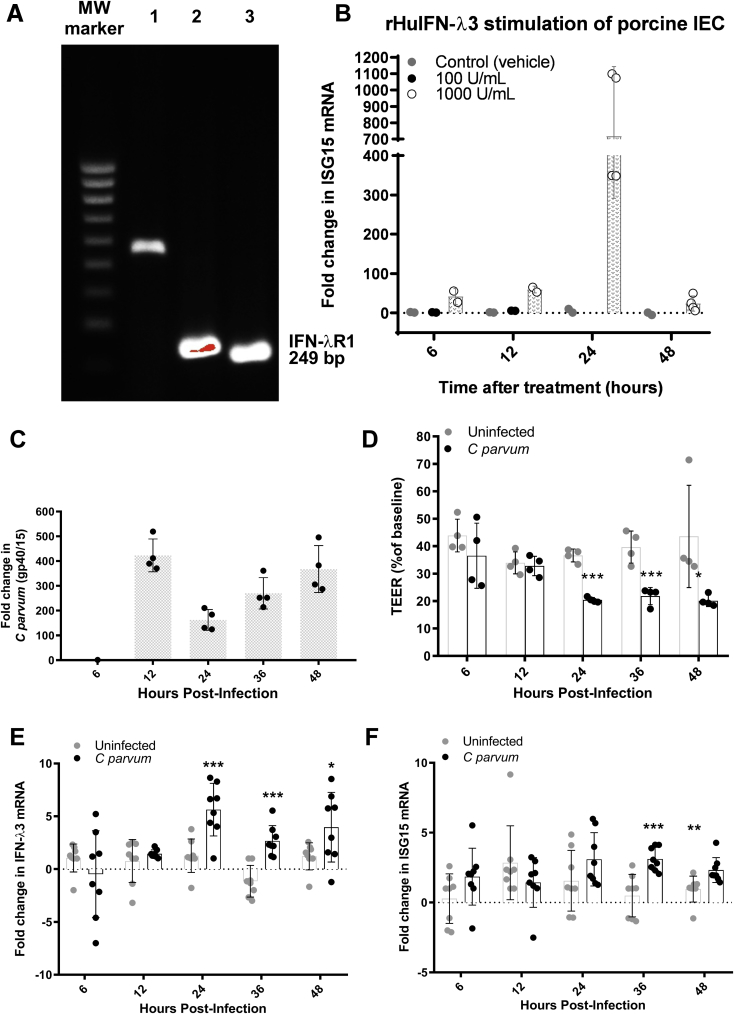
Our understanding of host strategy for epithelial defense against infection by C parvum remains poorly understood. In an effort to distinctively characterize the innate response of the intestinal epithelium to C parvum infection we used a host species that fully reproduces the diarrheal disease as observed in children with cryptosporidiosis.10 Accordingly, we infected neonatal piglets with C parvum and harvested the ileal epithelium at peak infection (days 3–5) for microarray analysis of the IEC transcriptional response. Piglets that were selected had a heavy burden of infection (57% ± 8.0% of villus enterocytes harbored C parvum) and showed marked villus blunting when compared with control piglets (P = .004) (Figure 1A and B). Based on the microarray analysis, 61 genes were up-regulated significantly and 20 genes were down-regulated significantly in IECs from C parvum–infected compared with control piglets (Figure 1C and D). Gene ontology analysis identified the IFN signaling pathway (P < .001) as the most enriched biological process in IECs from infected piglets. Twelve (20%) of the 61 significantly up-regulated genes were identified as targets of IFN signaling; specifically interferon-stimulated gene 15 (ISG15), guanylate binding protein 1 and 2 (GBP1 and GBP2), proteasome subunit-β type 8, interferon-α inducible protein 27, interferon stimulated gene-20, calcium-binding and coiled-coil domain-2, interferon-induced protein 44-like, tripartite motif containing 34, ubiquitin D, placenta-specific gene 8, and ribonuclease 4. The gene identified as most highly expressed in response to C parvum infection was ISG15 (21-fold). Significantly increased mucosal expression of ISG15 messenger RNA (mRNA) in vivo additionally was confirmed by targeted quantitative polymerase chain reaction (PCR) (Figure 1E). To further localize cellular ISG15 expression in the context of the entire ileum mucosa, fluorescence in situ hybridization was performed on uninfected control and C parvum–infected piglet ileum using custom-designed oligonucleotide probes specific for porcine ISG15. ISG15 mRNA was expressed predominately in the villus intestinal epithelial cells in infected piglets (Figure 1F).
Figure 1.
IECs up-regulate numerous ISGs in response to C parvum infection. Intestinal epithelium was harvested from ileum mucosa of neonatal piglets at the time of peak infection (days 3–5 after infection) (n = 8) and from age-matched uninfected controls (n = 4) for performance of gene expression analysis using microarrays. (A) Representative photomicrograph of ileum mucosa from an uninfected control and C parvum–infected piglet used for microarray analysis. H&E stain. Scale bar: 20 μm. (B) Villus height and crypt depth (μm) and the percentage of total villus IECs that were infected with C parvum in control (n = 4) and C parvum–infected (n = 8) piglets used for microarray analysis. Each data point represents the average of 5 measurements per piglet. Scale bars: means ± SD. **P < .01, Student t test comparison between uninfected and C parvum–infected piglets. Heat map of significantly (C) up-regulated and (D) down-regulated genes in infected (C parvum) and control (Uninf) exfoliated ileum villus epithelial cells. Each control and infected biological replicate is represented. Gene IDs:fold change are listed to the right of the heat map. Known ISGs are highlighted in red. (E) qRT-PCR analysis of porcine ileum mucosal total cellular mRNA for the presence of ISG15 mRNA. Samples were obtained from piglets used for microarray analysis at peak C parvum infection (days 3–5, n = 5) and age-matched controls (n = 5). For each sample, the Ct value for ISG15 was normalized to expression of the housekeeping gene cyclophilin (ΔCt). Fold-change differences between each sample were compared with a representative uninfected sample using the 2-ΔΔCt method. Scale bars: means ± SD. **P < .01, Student t test comparison between uninfected and C parvum–infected piglets. (F) Fluorescence in situ hybridization showing ISG15 mRNA (red fluorescence) in villus epithelial cells of ileum mucosa from piglets at peak C parvum infection and absent in villus epithelium of control piglets. Photomicrograph representative of results of 2 independent experiments. Scale bar: 50 μm.
Intestinal Mucosal Expression
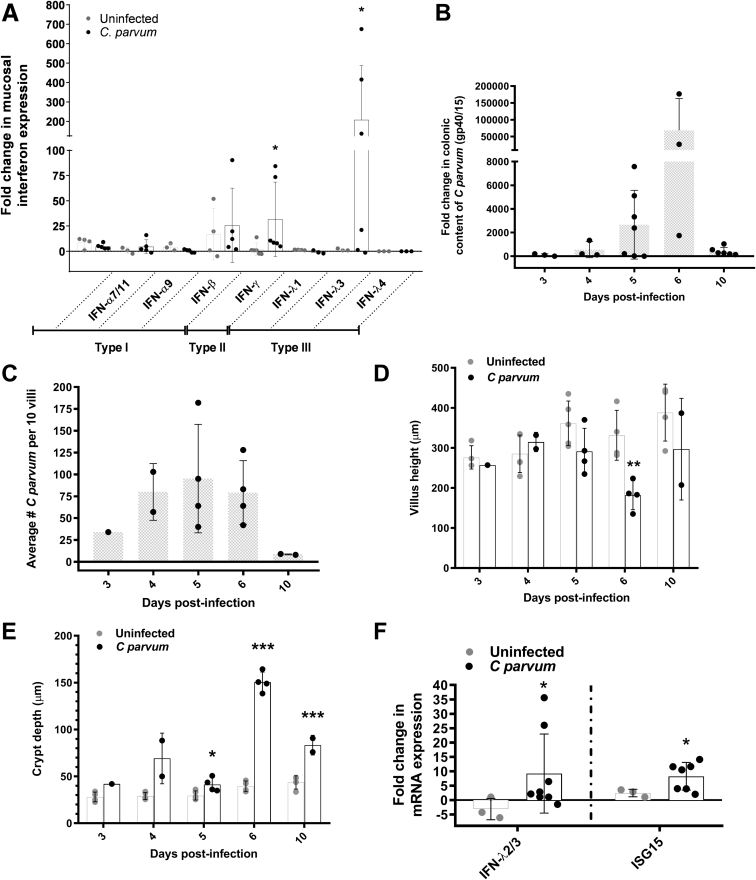
Having identified that targets of IFN signaling constitute a major transcriptional response of the intestinal epithelium to C parvum infection, we sought to determine what type(s) of IFN were likely stimulating this response in vivo. Accordingly, we used our own and published primers to quantify, by means of quantitative reverse-transcription PCR (qRT-PCR), specific members of all 3 classes of IFN (types I–III) using RNA extracted from the ileum mucosa (Figure 2A). There was no difference in expression of type I IFN (ie, IFN-α and IFN-β) observed between ileum mucosa of uninfected control and C parvum–infected piglets. Type II IFN (IFN-γ) expression was up-regulated approximately 20-fold. However, type III IFN, specifically IFN-λ3, was increased more than 200-fold in the ileum mucosa of infected piglets.
Figure 2.
C parvum stimulates ileum mucosal expression of IFN-λ3 in piglets and this response is conserved in neonatal mice. (A) qRT-PCR analysis of porcine ileum mucosal total cellular mRNA for the presence of IFN-α (all isoforms, 7/11, and 9), IFN-β, IFN-γ, and IFN-λ 1, 3, and 4. Samples were obtained from piglets at peak C parvum infection (days 3–5, n = 6–11) and age-matched controls (n = 5–7). Infected piglets showed significantly increased expression of both IFN-λ3 (∼200-fold) and IFN-γ mRNA (∼20-fold). Results obtained from 2 independent experiments were combined. *P < .05, Kruskal–Wallis analysis of variance comparison between uninfected and C parvum–infected piglets. IFN-λ4 was not amplified from any samples. For each sample, the Ct for IFN expression was normalized to expression of the housekeeping gene cyclophilin (ΔCt). For each IFN type, fold-change differences between samples are expressed as compared with a representative uninfected sample using the 2-ΔΔCt method. Bars represent means ± SD. (B–F) Characterization of a 10-day time course of C parvum infection in suckling C57BL/6 pups after orogastric gavage of 2 × 105C parvum oocysts or mock infection with PBS. Sampling took place on days 3–6, and day 10 after infection. (B) qRT-PCR analysis of Cryptosporidium glycoprotein 40/15 mRNA in total distal colon for quantification of fecal shedding of C parvum. All data points represent a fold-change comparison of individual pups with a representative day 3 infected pup. Bars represent means ± SD. (C) The average number of epithelium-associated C parvum per 10 villi per pup by light microscopy. Bars represent means ± SD. (D and E) Villus height and crypt depth as averaged for 10 villus crypt units per pup. Bars represent means ± SD. *P < .05, **P < .01, ***P < .001, Student t test compared with uninfected pups at the same time point. (F) qRT-PCR analysis of IFN-λ2/3 and ISG15 mRNA expression in samples of total ileum mucosa from control and infected pups at peak infection (days 4–6). For each sample, the Ct value for gene expression was normalized to expression of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (ΔCt). For each gene, fold-change differences between uninfected and C parvum–infected samples are expressed as compared with a representative uninfected sample using the 2-ΔΔCt method. Bars represent means ± SD. *P < .05, Student t test compared with uninfected pups. Each panel combines results obtained from 2 independent experiments.
A Type III Interferon Response Is Conserved in Suckling Mice Infected With C parvum
To further investigate the mechanistic relationship between C parvum infection, transcription of IFN-λ3, and increased expression of IFN-stimulated genes by the intestinal epithelium in vivo, we first sought to establish whether these observations were conserved in a mouse model. For these studies we used wild-type neonatal suckling mice because of their innate susceptibility to infection and its associated intestinal pathology. Over a 10-day time-course of infection, wild-type pups showed peaks in colonic content of C parvum mRNA, ileum IEC-associated C parvum organisms, and histologic indices of intestinal injury (villus blunting and crypt elongation) between 4 and 6 days after infection, with clearance typically occurring by day 10 (Figure 2B–E). At peak infection, the expression of IFN-λ2/3 and ISG15 mRNA were increased significantly in total distal jejunum/proximal ileum samples from C parvum–infected compared with uninfected mouse pups (Figure 2F).
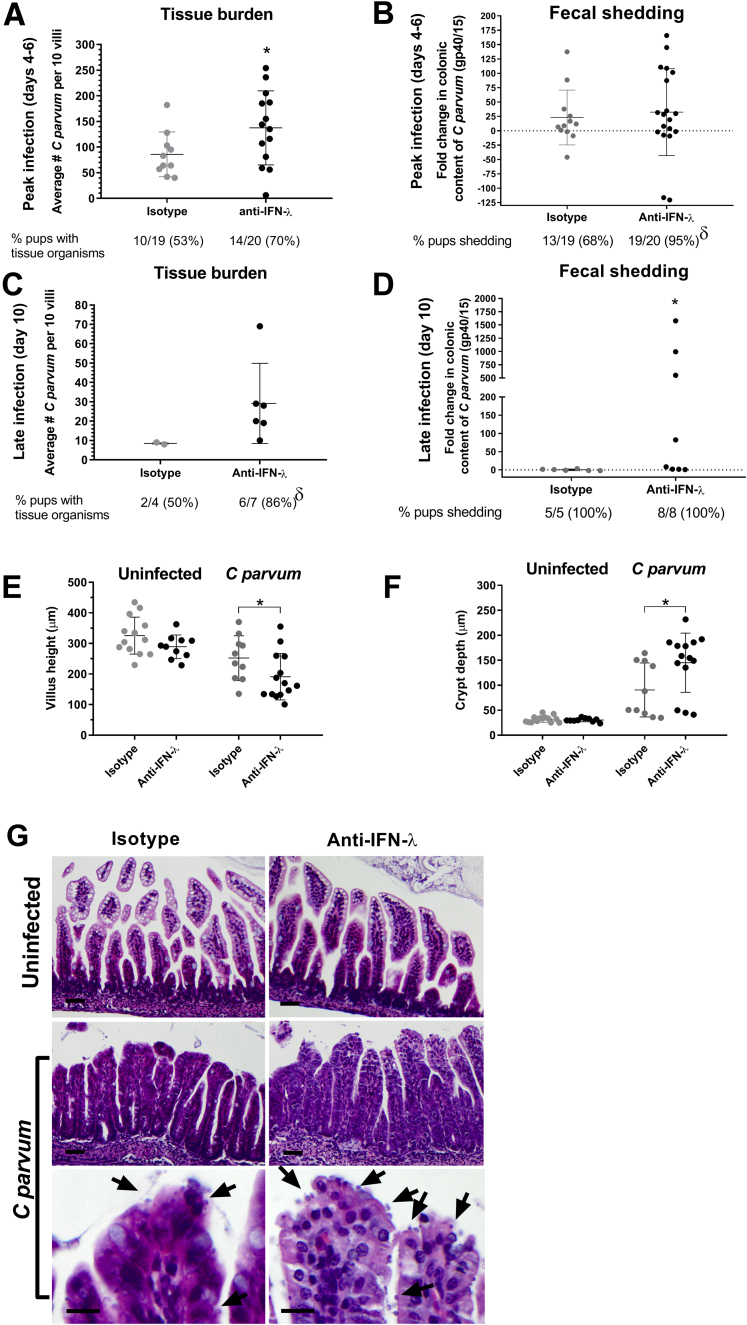
Immunoneutralization of IFN-λ2/3 Increases Severity of Epithelial Infection and Mucosal Injury in Suckling Mice Infected With C parvum
To determine the impact of IFN-λ2/3 on host defense against C parvum infection, neonatal mouse pups were treated parenterally with human anti-mouse IFN-λ2/3 immunoneutralizing vs isotype control antibodies on days –1, 0, and 3 of C parvum infection. As assessed at the time of peak infection (days 4–6), pups treated with anti–IFN-λ2/3 antibodies had a significantly greater number of organisms infecting the villus IEC (Figure 3A) and were more likely to shed C parvum in feces (19 of 20; 95%) compared with pups treated with isotype control antibodies (13 of 19; 68%), although the quantity of C parvum shed in the feces was similar between the treatment groups (Figure 3B). When examined during the recovery stage of infection (day 10), pups treated with anti–IFN-λ2/3 antibodies had retained greater numbers of organisms infecting the villous epithelium (Figure 3C) and continued to shed C parvum in feces and in significantly greater quantities compared with mice treated with isotype control antibodies (Figure 3D). In addition to a more severe epithelial burden and prolonged fecal shedding of C parvum, pups treated with anti–IFN-λ2/3 antibodies had quantitatively greater villus blunting and crypt elongation compared with infected pups treated with only the isotype control antibody (Figure 3E–G).
Figure 3.
Immunoneutralization of IFN-λ exacerbates C parvum infection. Mice were treated with either 15 μg of rat isotype or rat–anti-mouse IFN-λ2/3 neutralizing antibodies on days -1, 0, and 3 after orogastric gavage of C parvum or mock infection with PBS. (A) Quantitative assessment of C parvum epithelial burden at time of peak infection (days 4–6). Data points represent the average number of epithelium-associated C parvum per 10 villi per individual pup by light microscopy. Lines represent means ± SD. *P < .05, Student t test comparing isotype control with anti–IFN-λ–treated pups. (B) Colonic content of C parvum of individual pups at peak (days 4–6) infection as determined by qRT-PCR analysis of Cryptosporidium gp40/15 mRNA. For each sample, the Ct value for gene expression was normalized to expression of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (ΔCt). Fold-change differences between isotype and anti–IFN-λ–treated pups are expressed as compared with a representative isotype-treated pup using the 2-ΔΔCt method. Lines represent means ± SD. δP < .05, chi-square test comparing the percentage of pups shedding C parvum between isotype and anti–IFN-λ–treated groups. (C) Quantitative assessment of C parvum epithelial burden at late stage (day 10) of infection. Data points represent the average number of epithelium-associated C parvum per 10 villi per individual pup by light microscopy. Lines represent means ± SD. δP < .05, chi-square test comparing the percentage of pups with epithelial organisms between isotype and anti–IFN-λ–treated groups. (D) Colonic content of C parvum of individual pups at late stage (day 10) of infection as determined by qRT-PCR analysis of Cryptosporidium gp40/15 mRNA. Fold-change analysis as described for panel B. *P < .05, Student t test comparing isotype control with anti–IFN-λ–treated pups. (E) Average villus height and (F) crypt depth for 10 villus crypt units per individual pups at peak infection (*P < .05, Student t test comparing isotype control with anti–IFN-λ–treated pups.). Lines represent means ± SD. (A–F) Results obtained from 2 independent experiments were combined experiments. (G) Representative photomicrographs of uninfected control and C parvum–infected ileum mucosa obtained on day 6 after infection from mouse pups treated with rat isotype or rat–anti–IFN-λ2/3 neutralizing antibodies. Arrows highlight C parvum–infecting IECs. Scale bar: 20 μm for top and middle rows, and 10 μm for bottom row.
Intestinal Epithelial Cells Are Sufficient to Serve as a Source of IFN-λ and IFN-β and Autonomously Mediate an Interferon Response to C parvum Infection
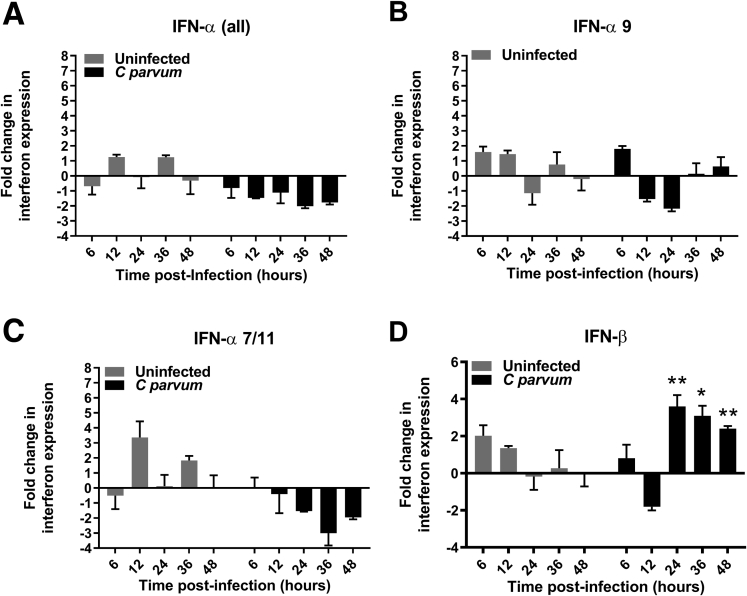
Having shown a significant and specific influence of IFN-λ in promoting intestinal defense against C parvum in immunoneutralized mouse pups, we sought to next determine the direct involvement of the intestinal epithelium in mediating the IFN response to infection. These studies were performed using a nontransformed porcine-derived small intestinal epithelial cell line (IPEC-J2) as a reductionist model for comparative studies with the neonatal piglets. We first confirmed that IPEC-J2 cells constitutively express IFN-λR1 mRNA by means of RT-PCR (Figure 4A). We subsequently examined the functionality of the receptor to stimulation using recombinant human IFN-λ3 (rHuIFN-λ3). Treatment of polarized IPEC-J2 cell monolayers with rHuIFN-λ3 resulted in a dose- and time-dependent induction of ISG15 mRNA expression that peaked at a concentration of 1000 U/mL at 24 hours after stimulation (Figure 4B). Other studies have shown that epithelial cells express IFN-λ in response to viral infection,6, 7 however, few studies have examined whether or not this response occurs with nonviral pathogens.11, 12, 13 To determine if C parvum infection is sufficient to induce a type III IFN response by the intestinal epithelium, we infected polarized monolayers of IPEC-J2 cells with C parvum and quantified the expression of IFN-λ3 and ISG15 mRNA over a 48-hour time course using qRT-PCR. The burden of C parvum infection oscillated over time within the monolayers, reflective of the phasic nature of the life cycle,14 and was accompanied by a significant decrease in transepithelial electrical resistance (TEER) beginning at 24 hours after infection (Figure 4C and D). The epithelial infection was associated with a significant increase in expression of IFN-λ3 mRNA beginning at 24 hours followed by a significant increase in expression of ISG15 mRNA beginning at 36 hours after infection (Figure 4E and F). To determine if C parvum stimulated the expression of other types of IFN that also could contribute to the observed ISG15 response, qRT-PCR also was used to quantify types I and II IFN (Figure 5). Apart from IFN-λ3, significant increases in expression also were observed for IFN-β beginning 24 hours after infection. Significant increases in expression of IFN-α were not observed at any time point after infection, despite the use of published primer pairs designed to amplify all of the known isoforms of porcine IFN-α.15 Amplification of IFN-γ was not observed in either control or C parvum–infected IPEC-J2 cells at any time point.
Figure 4.
C parvum is sufficient to stimulate IEC expression of IFN-λ3 and ISG15 mRNA. (A) Amplicons generated after PCR amplification of IFN-λR1 (249 bp) using porcine genomic DNA (lane 1), cDNA prepared from IPEC-J2 cell mRNA (lane 2), and cDNA prepared from porcine ileum mucosa mRNA (lane 3) as template. (B) qRT-PCR analysis of ISG15 mRNA expression by individual IPEC-J2 cell monolayers at time points of 6, 12, 24, or 48 hours after stimulation with rHuIFN-λ3 (100 or 1000 U/mL) or vehicle (0.1% bovine serum albumin in PBS). Bars represent means ± SD. For each monolayer, the Ct value for ISG15 gene expression was normalized to expression of the housekeeping gene cyclophilin (ΔCt). For each treatment group, fold-change differences at each time point are expressed as compared with a representative monolayer at t = 6 hours using the 2-ΔΔCt method. Figure combines results obtained from 2 independent experiments. (C) Measurement of infection burden over time on the basis of qRT-PCR analysis of Cryptosporidium glycoprotein 40/15 mRNA in infected IPEC-J2 monolayers (n = 4 unique monolayers at each time point). For each monolayer, the Ct value for gene expression was normalized to expression of the housekeeping gene cyclophilin (ΔCt). For each time point, fold-change differences between monolayers were expressed relative to a representative 6-hour infected monolayer using the 2-ΔΔCt method. Bars represent means ± SD. Data representative of 2 independent experiments. (D) TEER of control and infected IPEC-J2 monolayers (n = 4 each) expressed as a percentage of each individual monolayer’s TEER at time = 0. Bars represent means ± SD. *P < .05, ***P < .001, Student t test comparing uninfected with C parvum–infected monolayers at the same time point. Data are representative of 2 independent experiments. (E and F) qRT-PCR analysis of IFN-λ3 and ISG15 mRNA expression from control and C parvum–infected IPEC-J2 monolayers (n = 8 monolayers per treatment group per time point). For each monolayer, the Ct value for target gene expression was normalized to expression of the housekeeping gene cyclophilin (ΔCt). For each time point, fold-change differences between uninfected and infected monolayers are expressed as compared with a representative uninfected monolayer using the 2-ΔΔCt method. Bars represent means ± SD. *P < .05, **P < .01, and ***P < .001, Student t test comparing uninfected with C parvum–infected monolayers at the same time point. (E and F) Results obtained from 2 independent experiments have been combined. MW, molecular weight.
Figure 5.
IEC (IPEC-J2) monolayers express IFN-β but not IFN-α mRNA in response to C parvum infection. qRT-PCR analysis of (A) IFN-α (all) isotypes, (B) IFN-α7/11 isotypes, (C) IFN-α9 isotype, and (D) IFN-β during a 48-hour time course of C parvum infection in IPEC-J2 cells. N = 4 monolayers per treatment group and time point. For each monolayer, the Ct for target gene expression was normalized to expression of the housekeeping gene cyclophilin (ΔCt). Fold-change differences between uninfected and infected monolayers are expressed as compared with a representative uninfected monolayer at the same time point using the 2-ΔΔCt method. Bars represent means ± SD. *P < .05, **P < .01, Student t test comparison between uninfected and infected monolayers at same time point. Results were obtained from 1 independent experiment.
Intestinal Epithelial Cells Primed With IFN-λ3 Have Enhanced Resistance to C parvum Infection and Improved Barrier Function
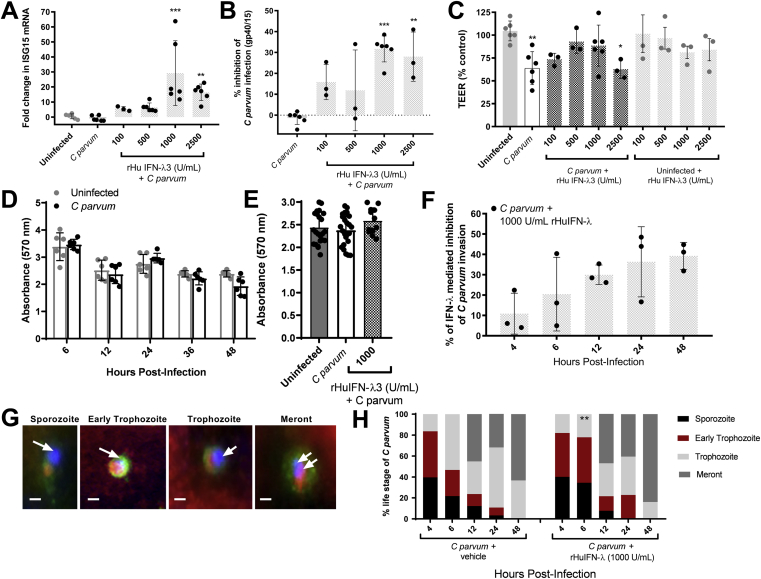
To determine if the intestinal epithelium is sufficient to mediate the anticryptosporidial and mucosal protective effects of IFN-λ, as observed to be impaired in immunoneutralized mice, we primed intestinal epithelial monolayers with rHuIFN-λ3 for 12 hours before introduction of C parvum, thereby enabling the peak IFN response (eg, ISG15 up-regulation) to coincide with establishment of cellular C parvum infection (12–24 hours after infection). A dose-response increase in ISG15 mRNA expression was observed in C parvum–infected intestinal epithelial monolayers that were primed with exogenous rHuIFN-λ3 (range, 100–2500 U/mL) (Figure 6A). Priming of monolayers with rHuIFN-λ3 was accompanied by correspondingly significant inhibitory effects on C parvum infection burden (Figure 6B). The percentage of inhibition of C parvum burden was correlated positively to up-regulation of ISG15 expression (Pearson correlation coefficient R = 0.574, P = .0082). In addition to promoting epithelial defense against infection, rHuIFN-λ3 priming significantly abrogated C parvum–associated loss of TEER at concentrations ranging from 100 to 1000 U/mL. This barrier protective effect waned, for unknown reasons, in cells treated with a higher dose of rHuIFN-λ3 (ie, 2500 U/mL) and was not observed in monolayers not infected with C parvum (Figure 6C). Importantly, neither C parvum nor rHuIFN-λ3 alone, or in combination, resulted in a loss of epithelial cells from treated monolayers (Figure 6D and E), suggesting that IFN-λ3 enhanced defense against C parvum using effector mechanisms contained within the intact monolayer.
Figure 6.
Priming of IECs with exogenous IFN-λ3 decreases the burden of C parvum infection, abrogates loss of TEER, and enhances resistance to C parvum invasion. Polarized monolayers of IPEC-J2 cells were pretreated with either vehicle, or 100, 500, 1000, or 2500 U/mL of rHuIFN-λ3 for 12 hours before C parvum infection. At the time of peak C parvum infection (12–24 hours after infection) the effect of rHuIFN-λ3 was assessed by qRT-PCR analysis of (A) ISG15 mRNA expression, (B) Cryptosporidiumgp40/15 mRNA content, and by (C) measurement of TEER. The Ct for ISG15 and Cryptosporidiumgp40/15 mRNA content of each monolayer was normalized to expression of the housekeeping gene cyclophilin (ΔCt). For each treatment group, fold-change differences in ISG15 and Cryptosporidiumgp40/15 were expressed as compared with a (A) representative uninfected monolayer or a (B) representative infected monolayer, respectively, using the 2-ΔΔCt method. **P < .01, ***P < .001, 1-way analysis of variance comparison between treatment groups and (A) uninfected or (B) infected monolayers. For TEER (C), individual data points are expressed as a percentage of a representative uninfected monolayer. *P < .05, **P < .01, 1-way analysis of variance comparison between treatment groups and uninfected monolayers. Bars represent means ± SD. (A–D) Results were obtained from 2 independent experiments and combined. (D) Spectrophotometric assay of crystal violet absorbance (570 nm) by IPEC-J2 monolayers after a 48-hour time course of C parvum infection. N = 6 monolayers per treatment and time group. Figure contains results obtained from 1 experiment. (E) Spectrophotometric assay of crystal violet absorbance in individual uninfected control and C parvum–infected monolayers 24 hours after infection and in monolayers primed with 1000 U/mL of rHuIFN-λ3 12 hours before the infection. Figure combines results obtained from 2 independent experiments. Bars represent means ± SD. (F) Intestinal epithelial (IPEC-J2) monolayers were grown on chamber slides and pretreated with either vehicle (n = 15 monolayers) or 1000 U/mL of rHuIFN-λ3 (n = 15 monolayers) for 12 hours before a 48-hour time course of C parvum infection. Three monolayers from each treatment group were examined using fluorescence microscopy at 4, 6, 12, 24, and 48 hours after infection, at which times quantification of invading C parvum parasites was determined by counting the total number of FITC–C parvum and Alexa-555 phalloidin co-labeled parasites within ten 400× fields per monolayer. At each time point, data compare the percentage reduction in C parvum invasion observed in 3 individual rHuIFN-λ3–treated monolayers compared with the average C parvum invasion observed in 3 individual vehicle-treated monolayers. Bars represent means ± SD. Figure contains results obtained from 1 experiment. (G) Representative fluorescence microscopy images of C parvum asexual intracellular life stages. Parasites shown reflect positive labeling with FITC–C parvum (green), DAPI-positive C parvum nuclei (blue and indicated by arrows), and Alexa-555–phalloidin cellular invasion sites (red). Scale bar: 1 μm. (H) The relative percentage of C parvum asexual life stages at each time point in vehicle-treated (left) and rHuIFN-λ3–pretreated infected monolayers. Parasite life stages were counted and classified within five 1000× fields for each monolayer. Data represent an average of n = 3 monolayers per time and treatment group. **P < .01, chi-square comparison of the percentage of trophozoites with vehicle-treated monolayers at the same time point.
Interferon-λ3 Decreases C parvum Invasion of IECs and Disrupts Early Intracellular Development
To gain additional insight into the cell-based mechanism(s) by which IFN-λ3 promotes epithelial defense against C parvum infection, we quantified the influence of rHuIFN-λ3 priming (1000 U/mL) on subsequent parasite invasion and cell-associated development. For these studies, infected epithelial monolayers were treated with a fluorescein isothiocyanate (FITC)-labeled anti-Cryptosporidium species antibody and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Recruitment of host–F-actin to cell-associated Cryptosporidium species16, 17 organisms was used to identify host–parasite invasion sites and the associated intracellular life stages were identified on the basis of organism size and the number of nuclei. Inhibitory effects of rHuIFN-λ3 on visualized C parvum invasion occurred within 12 hours of infection (Figure 6F). The magnitude of inhibition of C parvum invasion was similar to earlier indirect measurements of infection burden based on qRT-PCR for C parvum glycoprotein 40/15 (gp40/15) after priming of IPEC-J2 cells with rHuIFN-λ3 (Figure 6B). In unprimed cells, C parvum was observed to develop through an orderly progression of intracellular life stages over 48 hours. However, in rHuIFN-λ3–primed cells, a delay in maturation of C parvum was observed as early as 6 hours after infection with persistence of sporozoites and early trophozoites within the monolayers (Figure 6G and H). By 12 hours after infection, rHuIFN-λ3–primed monolayers resumed a distribution of life stages similar to unprimed cells but with 30% less parasites, suggesting loss of developmentally delayed sporozoites and early trophozoites from rHuIFN-λ3–treated monolayers.
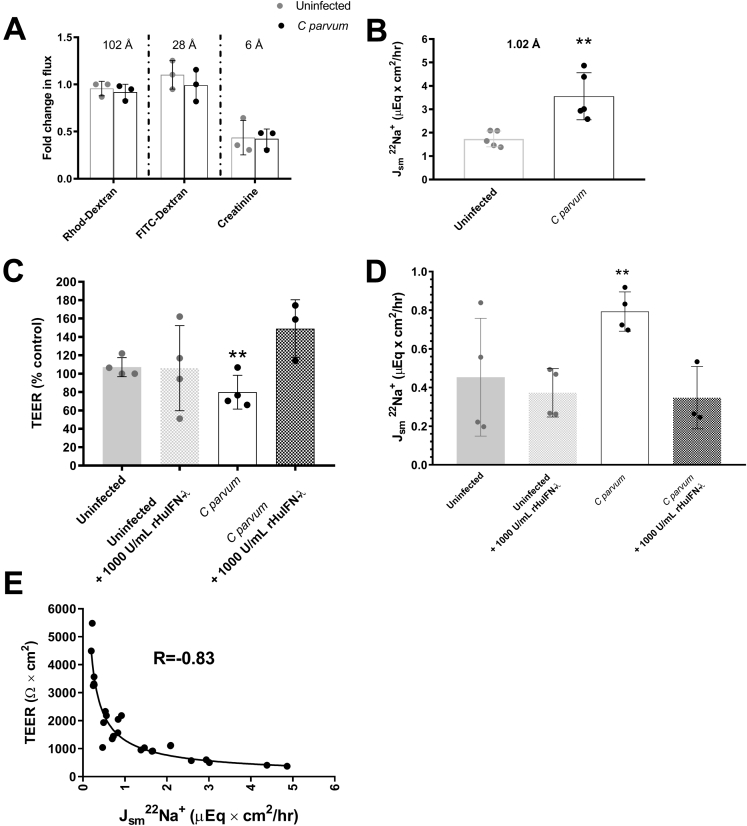
Interferon-λ3 Mitigates Loss of Barrier Function by Decreasing Paracellular Sodium Conductance of C parvum–Infected Epithelium
To further characterize the mechanism by which priming of epithelial monolayers with rHuIFN-λ3 ameliorates subsequent loss of TEER associated with C parvum infection, we conducted studies to define the nature of the barrier function defect. Having established, by means of crystal violet retention, that decreases in TEER were not associated with loss of cells from the monolayer, we focused on examination of the leak, pore, and ion/charge selective pathways, which are governed by the modification of tight junctional proteins.18, 19, 20, 21 Accordingly, we probed the pathway by which C parvum and IFN-λ3 influence barrier function by measuring the transepithelial flux of 70-kilodalton dextran, 4-kilodalton dextran, creatinine, and 22Na+. At the time of peak C parvum–induced decreases in TEER, there was no significant difference in the transepithelial flux of 70-kilodalton dextran, 4-kilodalton dextran, or creatinine across C parvum–infected compared with uninfected monolayers. The only probe identified as corresponding to changes in TEER was a significant increase in the flux of 22Na+ (Figure 7A and B). Moreover, in C parvum–infected monolayers, the mitigating effect of rHuIFN-λ3 on loss of TEER was associated similarly with a decrease in the transepithelial flux of 22Na+ (Figure 7C and D). A univariate nonlinear regression using the least-squares method was used to determine the relationship between TEER and 22Na+ conductance across all treatment groups (uninfected vs infected) and conditions (rHuIFN-λ3 vs vehicle) and identified a statistically significant, inverse relationship (correlation matrix, -0.83; P < .0001) (Figure 7E).
Figure 7.
IFN-λ3 mitigates C parvum–induced loss of TEER by decreasing sodium permeability. (A) Mucosal to serosal flux of rhodamine dextran, FITC dextran, and creatinine across uninfected control and C parvum–infected IPEC-J2 monolayers at the time of peak loss of TEER (24–48 hours after infection). Individual data points represent a mean flux of 2 technical replicates per monolayer and are expressed as a fold change difference from a representative uninfected monolayer for each probe. Figure contains results obtained from 1 experiment. (B) Serosal to mucosal flux of 22Na+ across uninfected control and C parvum–infected IPEC-J2 monolayers at time of peak loss of TEER (24–48 hours after infection). Individual data points represent a mean flux of 2 technical replicates per monolayer. **P < .01, Student t test comparison between uninfected and C parvum–infected monolayers. Data are representative of 2 independent experiments. (C) TEER at 24 hours after infection (expressed as the percentage of the mean of 24-hour uninfected controls). **P < .01, 1-way analysis of variance compared with uninfected and rHuIFN- λ3–treated C parvum–infected monolayers. Data are representative of 2 independent experiments. (D) Serosal to mucosal flux of 22Na+ across uninfected control and C parvum–infected IPEC-J2 monolayers pretreated with vehicle or 1000 U/mL of rHuIFN-λ3 for 12 hours before infection. Bars represent means ± SD. **P < .01, 1-way analysis of variance compared with rHuIFN-λ3–treated uninfected and rHuIFN-λ3–treated C parvum–infected monolayers. Data representative of 2 independent experiments. (E) Correlation between TEER (Ω × cm2) and flux of 22Na+. n = 26 observations from 3 individual experiments and includes uninfected and C parvum–infected monolayers both with and without rHuIFN-λ3 pretreatment. Correlation coefficient of -0.83, P < .0001.
Discussion
By studying IEC defense strategies against C parvum, these studies have identified that a type III IFN, IFN-λ3, is a conserved innate mediator of epithelial defense against this significant protozoal pathogen. We examined the significance of IFN-λ3 and specific mechanisms by which IFN-λ3 influences the epithelial defense against C parvum using 3 different models of infection, the neonatal piglet, suckling mouse, and nontransformed porcine intestinal epithelial cell monolayers (IPEC-J2). Results of these studies show that IFN-λ3 directly promotes epithelial resistance to parasite invasion and abrogates the loss of barrier function caused by C parvum infection. Although there is a wealth of literature supporting the role of IFN-λ in defending mucosal sites against viral pathogens,6, 7, 22 few studies have suggested a broader role for IFN-λ in mucosal defense against nonviral pathogens.9, 12, 23 This report shows the importance of IFN-λ in epithelial defense against a protozoal infection.
Cryptosporidium species is a minimally invasive epithelial pathogen that can be cleared in an immunocompetent host through an immune response largely instigated by the IECs.24 Accordingly, we hypothesized that the transcriptional responses of IEC to C parvum in vivo would provide key insights into how a competent host effectively resolves infection. By using a piglet model that uniquely recapitulates childhood cryptosporidiosis10 and microarray analysis to quantify gene expression by IECs obtained from the ileum of control and infected animals, we identified that the prevailing transcriptional response of IECs to C parvum is to up-regulate gene targets of IFN signaling. In situ localization of ISG15 mRNA to the villus IECs of C parvum–infected piglets supported that the IFN response was enriched within the epithelium. The translational relevance of our findings was recently supported by RNA sequencing data showing that targets of IFN also constitute the major transcriptional response of human small intestinal enteroids in infection by C parvum.25 Among the significantly up-regulated IFN-stimulated genes, ISG15 is most well known for its ability to promote antiviral defense by directly disrupting viral replication, budding, and release.26, 27, 28, 29, 30 Although we know that ISG15 is up-regulated during Leishmania brazilensis and Theileria annulata infections, we still have limited knowledge of its function in antiparasitic defense.31, 32 GBP1, GBP2, and tripartite motif containing 34 also were significantly up-regulated during C parvum infection; these genes have known functions in promoting host defense against nonviral pathogens, including protozoa. For example, in response to Toxoplasma, Salmonella typhimurium, and Legionella infection of IECs, GBP1, GBP2, and tripartite motif family proteins are recruited to the pathogen–host interface, specifically the parasitophorous vacuole.33, 34, 35 Their recruitment ultimately promotes parasitophorous vacuole disruption, enhances exposure to host immune responses, and facilitates pathogen clearance.34 Because parasitophorous vacuole formation is a hallmark feature of C parvum interface with host IEC,36, 37 it is probable that up-regulation of these particular genes represents a conserved mechanism of epithelial defense that may be similarly effective against C parvum.
Surprisingly, results of our microarray did not identify a concurrent increase in types I–III IFN expression to explain the IFN-stimulated gene response of piglet intestinal epithelium to C parvum. A similar failure to identify the source of IFN was reported for RNA sequencing data obtained from C parvum–infected human enteroids in which a predominant IFN-stimulated gene response also was observed and speculated to be owing to type I IFN.25 It remains unclear why IFN gene transcription was not observed by microarray in the intestinal epithelium from infected piglets. Our demonstration (by qRT-PCR) of significant IFN gene expression by C parvum–infected IEC monolayers in vitro and using the entire mucosa from C parvum–infected piglets in vivo suggests that the epithelium is sufficient but may not be the primary source for IFN synthesis in vivo. Upon investigating C parvum–infected piglet ileum mucosa for the source of an epithelial-targeted IFN response, we determined that type I IFN (IFN-α and IFN-β) was not up-regulated during peak infection. However, there was significantly induced expression of a novel type III IFN, IFN-λ3 (200-fold). Mucosal expression of type II IFN (IFN-γ), long recognized as an indispensable immune cell–derived cytokine associated with C parvum infection, also was up-regulated, although to a log-fold lesser degree than IFN-λ3. Expression of IFN-λ3 in response to C parvum infection was an unexpected finding, as was the observation at IFN-λ3 could influence the in vivo pathogenesis of a nonviral infection. Accordingly, identification of IFN-λ3 expression in cryptosporidiosis is a remarkable finding. Until recently, IFN-λ was considered to be largely redundant to the cellular effects of type I IFN. However, it is now recognized that IFN-λ receptors are selectively expressed by IEC, resulting in an epithelial-targeted mucosal immune response that plays a decisive role in the clearance of epitheliotropic viruses such as rotavirus, norovirus, and reovirus.6, 7, 22, 38, 39, 40, 41 In light of this knowledge, our findings suggest that C parvum likewise stimulates an epithelial targeted IFN response that is mediated, at least in part, by IFN-λ3.
To test a hypothesis that IFN-λ3 contributes specifically to epithelial defense against C parvum infection, we used a neonatal mouse model of the infection that enabled immunologic manipulation of IFN-λ signaling. The use of IFN-λR1 knockout mice was considered a superior approach for these studies, however, their use was repeatedly precluded by a prescreening diagnosis of norovirus infection.42 In mice, norovirus stimulates a type III IFN response that is critical for mediating viral clearance6 and we expected this would confound interpretation of this study. Consequently, we obtained pregnant C57BL/6 wild-type mice that were established to be specific pathogen-free from murine norovirus as well as other common, and potentially confounding, gastrointestinal co-infections. We subsequently infected the neonatal mouse pups with C parvum and confirmed conserved up-regulation of IFN-λ3 during peak infection. We then infected pups with or without pre-infection systemic immunoneutralization of IFN-λ2/3. Compared with mice treated with an isotype control antibody, mice receiving anti-mouse IFN-λ2/3 immunoneutralizing antibody harbored a significantly larger number of epithelial parasites, developed more severe villous blunting and crypt hyperplasia, and were more likely to shed C parvum and for a longer period of time. These findings strongly support a significant and specific influence of IFN-λ in promoting epithelial defense and possibly resolution of C parvum infection.
In response to viral pathogens, IECs are not only a target of IFN-λ, but also serve as a significant source of the cytokine.22, 41 By using porcine IEC monolayers as a reductionist model of the intestinal epithelium, we identified that C parvum infection alone is sufficient to induce endogenous IFN-λ3 expression and induction of ISG15 by IECs. Paracrine stimulation of ISG15 gene expression by IFN-λ3 was supported by the ability of exogenous rHuIFN-λ3 to similarly stimulate an IEC ISG15 response in the absence of C parvum. Infected IECs did not up-regulate IFN-γ, most likely because immune cells and not IECs are considered the predominant sources of IFN-γ in vivo.43 The only other IFN induced by C parvum–infected IECs in the time frame of these studies was the type I IFN: IFN-β. Others have similarly shown expression of type I IFN (IFN-α or IFN-β) by C parvum–infected IECs, but did not examine expression of IFN-λ.44 It is known that pretreatment of IECs with type I IFN inhibits C parvum development and neonatal mice treated with anti–IFN-α/β neutralizing antibodies have more severe C parvum infection.44 Accordingly, we did not examine the responsiveness of our IEC monolayers to treatment with exogenous type I IFN and it is likely that endogenously produced IFN-β also contributed to the ISG15 response observed in our C parvum–infected IECs. Although type I IFN and IFN-λ have distinct receptor types, both stimulate the same intracellular signaling pathway (signal transducer and activator of transcription 1/2), which results in indistinguishable overlap in target gene expression. Therefore, results of these studies do not negate an important role of type I IFN but identify a novel and significant impact of type III IFN (IFN-λ3) in epithelial defense against C parvum infection. Many studies provide additional context for understanding the differential influence of type I and type III IFN, at least as it pertains to viral infection. For example, restriction of the IFN-λ receptor to IECs in vivo targets the effect of IFN-λ to the epithelium, whereas type I IFN has a ubiquitous receptor expression. Recent studies using enteroid cultures have suggested that epithelial cells may up-regulate both type I and type III IFNs in response to viral infection, but only translate and secrete type III IFN.45 Therefore, recognition that C parvum induces IFN-λ3 gene expression and that epithelial defense mechanisms are dependent on IFN-λ3 has important implications for further development of targeted therapies for the infection.
In the present study, IECs primed with IFN-λ3 were more resistant to infection by C parvum. The principle mechanism of IFN-λ3 action was to impair parasite invasion of enterocytes. This finding substantiates our observation of an increased burden of epithelial parasitism in mouse pups treated with IFN-λ2/3 immunoneutralizing antibodies. Once C parvum invaded the IEC cells, however, there were no appreciable changes in the ability of the parasites to develop intracellularly. Enterocyte resistance to C parvum also has been described in monolayers pretreated with types I and II IFN, however, given preferential targeting of intestinal epithelial cells by IFN-λ in vivo,7, 22 our results likely have more biological relevance. In addition to enhanced resistance to infection by C parvum, IECs primed with IFN-λ3 were protected from decreases in barrier function resulting from the infection. Barrier protective effects of IFN-λ at the level of the blood-brain barrier, as well as in polarized T84 cells (colonic adenocarcinoma), have been observed in the presence and absence of viral or bacterial injury.8, 9 In contrast, we observed IFN-λ3’s barrier mitigating effect only in the face of C parvum infection. The reason for this observational difference is unknown. Regardless of IFN-λ3’s role in influencing IEC barrier function at homeostasis, our findings now provide further proof that IFN-λ3 defends IECs against pathogen-induced barrier disruption. We probed the paracellular pathway by which C parvum and IFN-λ3 influenced the TEER of IEC monolayers and isolated the barrier defect to a corresponding change in flux of 22Na+, indicating the characteristics of an ion/charge selective pathway. Interestingly, IFN-λ has been shown to promote barrier function in the blood-brain barrier by enhancing junctional localization of the tight junctional protein claudin-5.8 Furthermore, overexpression of claudin-5 in Madin-Darby Bovine Kidney Cells (MDBK) epithelial cells led to an increase in TEER and selectively decreased the relative permeability of Na+ ions.46 In light of these findings, the influence of IFN-λ3 on the expression and localization of claudin-5 during C parvum infection is worth investigating.
In closing, these studies have identified a novel mechanism for intestinal epithelial defense against C parvum involving IFN-λ3. In our studies, IFN-λ3 promoted epithelial defense by not only limiting parasite invasion but also promoting intestinal epithelial barrier function. These findings broaden the scope and depth of our current understanding of IFN-λ’s significance for epithelial defense and provide a mechanistic focus for development of targeted therapies for Cryptosporidium species infection.
Materials and Methods
Piglet Model of Cryptosporidiosis
One-day-old piglets were obtained from the College of Agriculture and Life Sciences. Piglets were transported to the College of Veterinary Medicine and placed into infected or control Biosafety Level 2 isolation facilities and fed a liquid diet (nonmedicated Advance Liqui-Wean; Milk Specialties, Dundee, IL) hourly by an automated delivery device. An inoculum of 108 C parvum oocysts (Bunchgrass Farms, Deary, ID) was given to piglets by orogastric tube at 3 days of age and samples were collected at peak infection 3–5 days later. Before sample collection, each piglet was anesthetized with ketamine (15 mg/kg) and xylazine (0.5 mg/kg) given intramuscularly and they were killed using sodium pentobarbital (200 mg/kg). Sections of ileum mucosa then were collected for epithelial exfoliation (microarray), fixation in 10% neutral buffered formalin (histology), or embedded in optimal cutting temperature media and frozen in liquid nitrogen (in situ hybridization). All studies were approved by the Institutional Animal Care and Use Committee of North Carolina State University.
RNA Microarray
The intestinal epithelium was exfoliated from freshly obtained sections of uninfected control (n = 4) and C parvum–infected (n = 8) porcine ileum mucosa in an oxygenated citrate-phosphate buffer containing 2.5 mmol/L glucose, 10 mmol/L EDTA, 137 mmol/L NaCl, 2.7 mmol/L KCl, 8.0 mmol/L Na2HPO4, and 1.5 mmol/L KH2PO4 (pH 7.4) for 20 minutes at 37°C. To dislodge epithelial cells from the lamina propria, submerged mucosa was grasped with forceps, which then were vibrated by contact with a vortex. Removal of epithelial cells was confirmed by histologic examination. The epithelial cells were pelleted by centrifugation at 200g for 10 minutes at 4°C. Each fraction was placed in 10 volumes of RNAlater or RLT buffer containing mercaptoethanol (Qiagen, Valencia, CA) and stored at -80°C.
Total RNA was isolated and treated with DNase (RNeasy Mini Kit; Qiagen, Germantown, MD) according to the manufacturer's instructions. RNA quantity and quality were determined by capillary electrophoresis (Agilent 2100 Bioanalyzer; Agilent Technologies, Santa Clara, CA). Total RNA (2 μg) was converted into complementary DNA (cDNA) using Reverse Transcriptase (Enzo Biosciences, Farmingdale, NY) and a modified oligo(dT)24 primer that contains T7 promoter sequences (GenSet, Boulder, CO). After first-strand synthesis, residual RNA was degraded by the addition of RNaseH and a double-stranded cDNA molecule was generated using DNA polymerase I and DNA ligase. The cDNA then was purified and concentrated using a phenol:chloroform extraction followed by ethanol precipitation. The cDNA products were incubated with T7 RNA polymerase and biotinylated ribonucleotides using an In Vitro Transcription kit (Affymetrix, Santa Clara, CA). The resultant complementary RNA product was purified using an RNeasy column (Qiagen) and quantified with a spectrophotometer. The complementary RNA target (20 μg) was incubated at 94°C for 35 minutes in fragmentation buffer (Tris, MgOAc, KOAc). The fragmented complementary RNA was diluted in hybridization buffer (2 N -morpholino-ethanesulfonic acid, NaCl, EDTA, Tween 20, herring sperm DNA, acetylated bovine serum albumin) containing biotin-labeled OligoB2 and Eukaryotic Hybridization Controls (Affymetrix). The hybridization cocktail was denatured at 99°C for 5 minutes, incubated at 45°C for 5 minutes, and then injected into an Affymetrix Porcine GeneChip cartridge. The GeneChip array was incubated at 42°C for at least 16 hours in a rotating oven at 60 rpm. GeneChips were washed 91 times with a series of nonstringent (25°C) and stringent (50°C) solutions containing variable amounts of MES, Tween20, and Sodium Chloride-Sodium Phosphate-EDTA. The microarrays then were stained with streptavidin phycoerythrin and the fluorescent signal was amplified using a biotinylated antibody solution. Fluorescent images were detected in a GeneChip Scanner 3000 and expression data were extracted using the GeneChip Command Console Software (AGCC) v 2.0 or later (Affymetrix).
Fluorophore-labeled cDNA was prepared and hybridized to Affymetrix Porcine GeneChip microarrays (20,201 genes) by a commercial laboratory (Expression Analysis, Durham, NC). All GeneChips were scaled to a median intensity setting of 500. The arrays were first normalized with a Loess normalization technique using JMP Genomics (SAS, Cary, NC), and analysis of gene expression changes was performed via analysis of variance. False-discovery rates were calculated according to the methods of Storey47 and a false-discovery threshold of 0.05 was adopted to identify differentially expressed genes. A 2-fold change in gene expression from the control cells was the minimum change to be considered for further analysis. Lists of differentially expressed genes were analyzed further using Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA). Determination of enriched biological processes was determined by entering up-regulated genes into the Protein ANalysis THrough Evolutionary Relationships (PANTHER) database.
Quantitative Histopathology
Formalin-fixed sections of ileum mucosa were paraffin-embedded, sectioned at a thickness of 5 μm, and stained with H&E. For each piglet, the average crypt depth and villus height was calculated from measurements of 5 well-oriented, villus-crypt units using a brightfield microscope and ocular micrometer. The percentage of infection was determined by dividing the number of infected villus enterocytes by the total number of villus enterocytes.
Fluorescence In Situ Hybridization
Custom probes designed for hybridization with porcine ISG15 were purchased (Affymetrix). Probes were hybridized to formalin-fixed and paraffin-embedded, 5-μm sections of porcine ileum mucosa (ViewRNA in situ hybridization kit, catalog QVC0001; Affymetrix) using the manufacturer protocol for 1-plex hybridization. Slides were deparaffinized using xylene (30 minutes) and heat and protease K pretreated (10 and 30 minutes, respectively). After initial hybridization, signal amplification was performed using pre-amplifier and amplifier probes using a slide hybridization and denaturation system (ThermoBrite, catalog 23-021-580; Leica Microsystems, Wetzlar, Germany). To detect the ISG15 target, slides were incubated with fast-red chromagen (1:30 dilution for 45 minutes), washed, and coverslipped using mounting media containing DAPI (VECTASHIED antifade mounting medium with DAPI, catalog H-1200-10; Vector Labs, Burlingame, CA). Slides were imaged using a fluorescence microscope (Leica DB 5000B with LASX software; Leica Microsystems). Rat ubiquitin probes were hybridized to formalin-fixed and paraffin-embedded rat kidney slides as a hybridization control.
Immunoneutralization of IFN-λ2/3 in Neonatal Mouse Pups Infected With Cryptosporidiosis
Conventional (specific pathogen-free, which includes common gastrointestinal pathogens such as norovirus, reovirus, rotavirus, Citrobacter rodentium, Salmonella, Helicobacter, and Giardia) C57BL/6 pregnant dams (embryonic days 13–15) were purchased (Charles River Laboratories, Durham, NC) and housed in accordance with Biosafety Level 2 guidelines. Litters were assigned to 1 of 4 treatment groups, as follows: (1) uninfected control mice treated with isotype (rat IgG) antibody, (2) uninfected control mice treated with rat anti-mouse IFN-λ2/3 immunoneutralizing antibody, (3) C parvum–infected mice treated with isotype (rat IgG) antibody, and (4) C parvum–infected mice treated with rat anti-mouse IFN-λ2/3 immunoneutralizing antibody. Each mouse pup was administered either 15 μg of IFN-λ2/3 neutralizing antibody (Monoclonal Rat IgG2B clone 244716, catalog MAB17892; R&D Systems, Minneapolis, MN) or 15 μg of rat IgG isotype control antibody (Monoclonal Rat IgG2B clone 141945, catalog MAB0061; R&D Systems) by intraperitoneal injection on days -1, 0, and 3 of infection. The dose was calculated based on the quantity required to achieve 100% immunoneutralization of IFN-λ2/3 in vitro (ED100) and estimated blood volume of pups on day 10 of infection (day 14 of life). Pups were infected orogastrically with 2 × 105 oocysts of C parvum in 10 μL of phosphate-buffered saline (PBS) using a 25-mm, plastic gavage needle (Petsurgical, Westlake Village, CA). During the 10-day course of infection, pups were weighed daily. Pups were euthanized using isofluorane anesthesia followed by decapitation. Immediately after euthanasia the gastrointestinal tract was removed from the abdominal cavity. A section of the small intestine extending from the distal jejunum to the proximal ileum was snap-frozen for RNA extraction. The ileocecocolic region of the intestine was formalin-fixed and paraffin-embedded, sectioned at a thickness of 5 μm, and stained with H&E before quantifying the burden of epithelial infection and histomorphometry using light microscopy (Accu-scope 3025; Olympus Corporation, Tokyo, Japan). Digital images of well-oriented individual villus-crypt units were captured (cellSens software; Olympus) at a magnification of 400×. The total number of epithelial cell–associated Cryptosporidium parasites were counted for each of 10 well-oriented villus-crypt units. Villus height and crypt depth measurements were obtained from each villus-crypt unit using ImageJ software (National Institutes of Health, Bethesda, MD).
RNA Extraction and Real-Time PCR Analysis
For RNA extraction from tissue, snap-frozen samples of ileum mucosa (piglets) or total distal jejunum/proximal ileum (mice) were placed in ice-cold lysis buffer, homogenized, and sonicated to disrupt the tissue. For RNA extraction from cell culture monolayers, media was removed and monolayers were washed 3 times with PBS, lysis buffer was applied directly to monolayers, and cells were scraped off inserts with a sterile cell lifter. Total RNA was extracted using a commercial kit (Ambion PureLink Mini kit, catalog 12183025; Thermo Fisher Scientific, Raleigh, NC) including DNAse treatment (Turbo DNA free kit, catalog AM1907; Thermo Fischer, Waltham, MA). RNA concentration and purity were quantified by spectrophotometry (Nanodrop ND-1000; Thermo Fisher Scientific, Waltham, MA). A total of 1 μg of total RNA was converted to single-stranded cDNA by reverse transcription (high-capacity cDNA reverse transcription kit, catalog 4368814; Thermo Fisher Scientific, Waltham, MA). After reverse transcription, quantitative PCR was performed in a 15 μL reaction volume with 50 nmol/L of each primer, 60 ng cDNA, and 2 × SYBR green Mastermix (catalog 04707516001; Roche Diagnostics, Basel, Switzerland) using a Roche Lightcycler 480 for 50 cycles at 95°C for 15 seconds and 60°C for 1 minute after an initial incubation for 10 minutes at 95°C. PCR products were additionally examined after electrophoresis in 1.5% agarose gels stained with GelRed DNA stain (catalog 41003; Biotium, Freemont, CA). The identity of amplicons was confirmed by expected size, melting temperature, and commercial sequencing (GeneWiz, South Plainfied, NJ). Expression of the following genes were targeted: C parvum glycoprotein 40/15 (Cp gp40/15) porcine IFN-α (all isoforms, poIFN-α-all), porcine IFN-α isoforms 7/11 (poIFN-α-7/11), porcine IFN α isoform 9 (poIFN-α9), porcine IFN-β (poIFN-β), porcine IFN-γ (poIFN-γ), porcine IFN-λ1 (poIFN-λ1), porcine IFN-λ3 (poIFN-λ3), porcine IFN-λ4 (poIFN-λ4), porcine IFN-λ–receptor 1 (poIFN-λR1), porcine ISG15 (poISG15), murine IFN-λ2/3 (muIFN-λ2/3), murine IFN-β (muIFN-β), and murine ISG15 (muISG15). All primers were purchased from Integrated DNA technologies (Skokie, IL). Primer sequence data and references of origin are shown in Table 1. Primers were designed either by the authors or as previously described.
Table 1.
Specific Primer Sequences and Sources Used in Real-Time RT-PCR and Their Resulting Amplification Product Sizes and Melting Temperatures
| mRNA | Forward, 5’ to 3’ | Reverse, 5’ to 3’ | Product size | Melting temperature, C° | Reference |
|---|---|---|---|---|---|
| Cp gp40/15 | TCATTTGTAATGTGGTTCGGAGAA | AGGGTAAAGGCAAACAAATCG | 326 | 82.5–83.0 | 48 |
| Porcine-IFNα (all) | GGCTCTGGTGCATGAGATGC | CAGCCAGGATGGAGTCCTCC | 197 | 88.45–88.80 | 15 |
| Porcine-IFNα 7/11 | GGGACTTTGGATCCCCTCAT | GTGGAGGAAGAGAAGGATG | 369 | 89.0–89.5 | 49 |
| Porcine-IFNα9 | GTGCTGCTCAGCTGCAAG | AGTCCTCCTCCAGCAGGGGC | 384 | 87.5–88.5 | 49 |
| Porcine-IFNβ | ATGTCAGAAGCTCCTGGGACAGTT | AGGTCATCCATCTGCCCATCAAGT | 246 | 82.20–82.70 | 15 |
| Porcine-IFNγ | GTTTTTCTGGCTCTTACTGC | CTTCCGCTTTCTTAGGTTAG | 410 | 86.3–87.0 | 50 |
| Porcine-IFNλ1 | ACATCCACGTCGAACTTCAGGCTT | TCAGATGTGCAAGTCTCCACTGGT | 209 | 87.5–88.0 | 49 |
| Porcine-IFNλ3 | AAGAGGGCCAAGGATGCCTTTGAA | AGGCGGAAGAGGTTGAACATGACA | 374 | 92.9–93.3 | 49 |
| Porcine-IFNλ4 | GTCACAGAGCTGACTCGCCT | TCACAGACAAGGCCCCGAAT | 110 | 89.2–89.5 | Authors |
| Porcine-ISG15 | GATGCTGGGAGGCAAGGA | CAGGATGCTCAGTGGGTCTCT | 229 | 90.0–90.3 | Authors |
| Porcine-IFNλR1 | CGGTGGCAAAGAGTGAAAAT | GATCTCCTCTGTCCGGGTGA | 249 | 86.7–86.8 | Authors |
| Porcine-Cyclophilin | CCGTCGATGGCGAGCCC | CCCGTATGCTTCAGGATAAAA | 250 | 82.5–84.5 | 51 |
| Murine-IFNL2/3 | AGCTGCAGGCCTTCAAAAAG | TGGGAGTGAATGTGGCTCAG | 244 | 88.5–89.1 | 7 |
| Murine-ISG15 | GAGCTAGAGCCTGCAGCAAT | TTCTGGGCAATCTGCTTCTT | 122 | 84.3–84.5 | 7 |
| Murine-IFNβ | GGAGATGACGGAGAAGATGC | CCCAGTGCTGGAGAAATTGT | 103 | 79.9–80.6 | 52 |
| Murine-GAPDH | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAG | 87 | 84.2–84.4 | 7 |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
All qPCR reactions were performed in technical triplicate and averaged to generate a single cycle threshold value (Ct). Each reaction was confirmed to generate a sigmoidal amplification plot and a single melt curve at the target temperature to be considered a positive amplification. Each sample underwent additional amplification of a housekeeping gene, also performed in triplicate and averaged, for use in generating a normalized Ct value (ΔCt). For porcine-origin samples, cyclophilin was used as the housekeeping gene. For murine-origin samples, glyceraldehyde-3-phosphate dehydrogenase was used as the housekeeping gene. To compare fold-change differences in expression between, as well as within, experimental conditions, a representative sample was chosen from the comparator condition (eg, uninfected, vehicle-treated, or initial time point group) and used as a reference to calculate a ΔΔCt for each sample. Fold-change differences in gene expression between samples and the comparator were calculated using the 2-ΔΔCt method. This approach was used because it enables an objective look at the variability between samples in the control/reference group and works well for studies in which observations between groups are unpaired.
Polarized Intestinal Epithelial Cell Culture Response to Cryptosporidiosis
Nontransformed porcine jejunal epithelial cells (IPEC-J2) were grown in co-culture media that included Dulbecco’s minimal essential medium:nutrient mixture F-12 (with L-glutamine and 15 mmol/L HEPES, catalog 10-092-CM; Corning, Corning, NY) supplemented with 5 μg/mL each of insulin, transferrin, and selenium (ITS Premix universal culture supplement, catalog 354350; Corning), epidermal growth factor (5 ng/mL, catalog 354052; Corning), penicillin (50,000 IU/mL), streptomycin (50,000 mg/mL) (100× penicillin-streptomycin solution, catalog 30002CI; Corning), and 5% porcine serum (catalog 26250084; Thermo Fischer, Waltham, MA), and incubated at 37°C in 5% CO2. Cells were seeded onto permeable polycarbonate filters (0.4-μm pore size, either 0.6 cm2 or 4.67cm2; catalog PIHP01250 and PIHP03050, respectively; Millipore Sigma, Burlington, MA) and cultured until confluent (TEER, ≥2000 Ω × 0.6 cm2 or ≥1300 Ω × 4.67 cm2). TEER was measured using an EVOM2 epithelial voltohmmeter with chopstick electrodes (World Precision Instruments, Sarasota, FL). For immunofluorescence assessment of C parvum burden, IPEC-J2 cells were seeded onto 8-well chamber slides (Nunc Lab-Tek II, catalog 154534; Thermo Fisher, Waltham, MA) and grown to confluence over a period of 3–4 days before use. IPEC-J2 cells were used at passages 38–50. Media was changed every 3–4 days.
Infection of IPEC-J2 Cells With C parvum
C parvum oocysts (Bunchgrass Farms) were pelleted by centrifugation (13,000 × g for 3 minutes) and treated with 10 mmol/L HCl (catalog 13-1700 SAJ; Millipore Sigma) in PBS for 60 minutes in a 37°C water bath The oocysts then were pelleted by centrifugation to remove the HCl, reconstituted with 0.2 mmol/L sodium taurodeoxycholate and 22 mmol/L NaHCO3 (catalog T0875 and S6014, respectively; Millipore Sigma) in PBS, and incubated for 30 minutes in a 37°C water bath to excyst infective sporozoites. The excysted oocysts were centrifuged and reconstituted in IPEC-J2 culture media. The number of excysted oocysts was counted using a hemacytometer to establish a final concentration of excysted oocysts. A multiplicity of infection of 2:1 of excysted oocysts to IPEC-J2 cells was used to inoculate monolayers. Based on established counts of approximately 5 × 105 IPEC-J2 cells per 30 mm (4.2 cm2) insert at confluence, each monolayer was infected with 1 × 106 excysted oocysts.
Stimulation of IPEC-J2 Cells With IFN-λ3
Monolayers of IPEC-J2 cells were treated at the time of confluence with 100–2500 U/mL of rHuIFN-λ3 in PBS (catalog 11730-1; PBL Assay Science, Piscataway, NJ). For determination of effects on C parvum infection, cells were pretreated with rHuIFN-λ3 beginning 12 hours before infection of the monolayer and sustained within the culture medium throughout the course of infection.
Crystal Violet Cytotoxicity Assay
Uninfected control and C parvum–infected IPEC-J2 cells were grown to confluence on 24-well polystyrene plates. At designated postinfection time intervals, epithelial monolayers were gently washed with PBS to remove detached epithelial cells, fixed with 2% paraformaldehyde in PBS for 15 minutes at room temperature, washed with Hank’s balanced salt solution, and stained with 100 μL of 0.13% crystal violet solution dissolved in a 5:2 (vol/vol) ethanol-paraformaldehyde solution. Monolayers were washed twice with dH20 and allowed to air dry. Stained cells were solubilized in 100 μL 1% sodium dodecyl sulfate in 50% ethanol and transferred to 96-well plates. The intensity of staining was quantified using a spectrophotometer at a wavelength of 570 nmol/L, with a reference wavelength of 650 nmol/L to account for optical interference.
Epithelial Permeability Assays
IPEC-J2 monolayers were grown to confluence on 12-mm diameter, 0.4-μm pore size polycarbonate filters. A subset of monolayers was infected with C parvum, and the permeability of uninfected control and C parvum–infected monolayers was measured at 24 and 48 hours after infection by concurrent transepithelial passage of 3 permeability probes: creatinine, fluorescein isothiocyanate–4 kilodalton dextran, and rhodamine–70 kilodalton dextran (catalog C4255, 46944, and R9379; Millipore Sigma). Probes were prepared in cell culture media at a final concentration of 11 mg/mL. At the beginning of each flux period, 100 μL of the flux solution was added to the apical side of each insert to give a final volume and concentration of 500 μL and 2.2 mg/mL, respectively. After 2 hours of incubation, basolateral recovery of each probe was measured in a plate reader (Synergy HT; BioTek, Winooski, VT) using freshly prepared standards. Fluorescence of fluorescein and rhodamine B were measured at 490 and 555 nm using emission wavelengths of 520 and 585 nm, respectively. Creatinine was measured separately using a colorimetric assay (catalog MAK080; Millipore Sigma).
Epithelial Permeability to 22Na+
Basolateral to apical isotopic flux studies of 22Na+ (1 μCi/mL, catalog NEZ081100UC; Perkin Elmer, Waltham, MA) was performed in uninfected control and C parvum–infected IPEC-J2 monolayers with or without pretreatment with 1000 U/mL of rHuIFN-λ3 12 hours before infection. Barrier function (TEER) was measured at time points of 6, 12, 24, 36, and 48 hours over the course of infection. At 24 hours after infection, a 50-μL aliquot of 22Na+ working stock was added to a 450 μL existing volume of basolateral media. After a flux period of 2 hours, paired 200-μL samples were obtained from both the apical and basolateral compartments. Sample emissions were counted using a gamma counter (Wallac Wizard 1470-020; Perkin Elmer) and basolateral to apical flux of Na+ was expressed as μEq × cm2 per hour.
Determination of IFN-λ3 Effects on C parvum Burden and Intracellular Development in Epithelial Monolayers
IPEC-J2 cells were cultured to confluency (3–4 days) in 8-well chamber slides and treated or not with 1000 U/mL of rHuIFN-λ3 for 12 hours before infection with C parvum or vehicle (0.1% bovine serum albumin in PBS). At 4, 6, 12, 24, and 48 hours after infection, slides were washed 3 times with PBS and fixed with 4% paraformaldehyde for 10 minutes at room temperature. After fixation, cells were washed 3 times with PBS, incubated in a blocking solution of 1% bovine serum albumin in PBS for 30 minutes, and permeabilized for 5 minutes with 0.5% Triton X-100 (Millipore Sigma) in PBS at room temperature and then washed once with PBS for 30 seconds. To visualize epithelial invasion sites of C parvum organisms, slides were incubated with a red fluorescent phalloidin conjugate (acti-stain 555 phalloidin, catalog PHDH1-A; Cytoskeleton, Inc, Denver, CO) at a concentration of 100 nmol/L at room temperature for 30 minutes. The monolayers then were washed 3 times with PBS. For simultaneous visualization of C parvum organisms, monolayers subsequently were incubated at room temperature for 45 minutes with a FITC-labeled rat anti-Cryptosporidium parvum polyclonal antibody (1× Sporo-Glo, catalog A600FLR; Waterborne, Inc, New Orleans, LA). Finally, monolayers were washed 3 times with PBS, partially air-dried, and then cover slipped with mounting media containing DAPI (Vectashield antifade mounting medium with DAPI; Vector Labs). Monolayers were imaged using a fluorescence microscope (Leica DMB5000B with LASX software; Leica Microsystems). Quantification of C parvum burden was determined by counting the total number of FITC-labeled parasites that also were associated with characteristic F-actin invasion sites within ten 400× fields per infected well. For characterization of life stages present, attached parasites were measured using LASX software (Leica Microsystems) in 5 individual 1000× fields per well and categorized as either sporozoite (∼4 × 1 μm, elongate, eccentric F-actin invasion site), early trophozoite (≤1 × 1 to 2 × 2 μm, round, centralized F-actin invasion site), trophozoite (2 ×2 to 3 × 3 μm, round, centralized F-actin invasion site), or meront (<3 × 3 cm, round, often with multiple nuclei visible).
Data Analysis
Data were tested for normality using a Shapiro–Wilk test and equal variance using Levene’s median test. (SigmaStat; Jandel Scientific, San Rafael, CA). Parametric data were analyzed using the Student t test and 1-way analysis of variance. Nonparametric data were analyzed using Kruskal–Wallis rank-sum tests. Differences in the proportion of observations between groups was analyzed using chi-square analysis of contingency tables using a Yates continuity correction. Descriptive data are represented as means ± SD. For all analyses, P ≤ .05 was considered significant. All authors had access to the study data and reviewed and approved the final manuscript.
Acknowledgments
The authors thank Stephen Stauffer for technical support, Steven Nagar for assistance with immunofluorescence imaging, Dr Maria Correa for help with statistical analysis, and North Carolina State University Laboratory Animal Resources personnel for their excellent care of the piglets and mice used in this study.
Footnotes
Author contributions Jody L. Gookin, Sylvia H. Ferguson, Derek M. Foster, and Barbara Sherry were responsible for the study concept and design; Jody L. Gookin, Sylvia H. Ferguson, Derek M. Foster, and Dahlia M. Nielsen acquired data; Sylvia H. Ferguson and Jody L. Gookin drafted the manuscript; Jody L. Gookin, Sylvia H. Ferguson, Derek M. Foster, and Dahlia M. Nielsen performed the statistical analysis; Jody L. Gookin and Sylvia H. Ferguson obtained funding; Jody L. Gookin, Sylvia H. Ferguson, and Derek M. Foster provided study supervision; and all co-authors analyzed and interpreted data and revised the manuscript for important intellectual content.
Conflicts of interest The authors disclose no conflicts.
Funding Research reported in this publication was supported by grants from the Comparative Medicine Institute at North Carolina State University, a Multidisciplinary Research and Professional Development grant from North Carolina State University, North Carolina State Appropriated Funds through the Office of the Associate Dean of Research and Graduate Studies in the College of Veterinary Medicine at North Carolina State University, and a Ruth L. Kirschstein National Research Service Award through the National Institutes of Health (grant T32OD011130) and as part of North Carolina State University’s Comparative Medicine and Translational Research Training Program.
References
- 1.Scallan E., Hoekstra R.M., Angulo F.J., Tauxe R.V., Widdowson M.A., Roy S.L., Jones J.L., Griffin P.M. Foodborne illness acquired in the United States--major pathogens. Emerg Infect Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotloff K.L., Nataro J.P., Blackwelder W.C., Nasrin D., Farag T.H., Panchalingam S., Wu Y., Sow S.O., Sur D., Breiman R.F., Faruque A.S., Zaidi A.K., Saha D., Alonso P.L., Tamboura B., Sanogo D., Onwuchekwa U., Manna B., Ramamurthy T., Kanungo S., Ochieng J.B., Omore R., Oundo J.O., Hossain A., Das S.K., Ahmed S., Qureshi S., Quadri F., Adegbola R.A., Antonio M., Hossain M.J., Akinsola A., Mandomando I., Nhampossa T., Acacio S., Biswas K., O'Reilly C.E., Mintz E.D., Berkeley L.Y., Muhsen K., Sommerfelt H., Robins-Browne R.M., Levine M.M. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 3.Sow S.O., Muhsen K., Nasrin D., Blackwelder W.C., Wu Y.K., Farag T.H., Panchalingam S., Sur D., Zaidi A.K.M., Faruque A.S.G., Saha D., Adegbola R., Alonso P.L., Breiman R.F., Bassat Q., Tamboura B., Sanogo D., Onwuchekwa U., Manna B., Ramamurthy T., Kanungo S., Ahmed S., Qureshi S., Quadri F., Hossain A., Das S.K., Antonio M., Hossain M.J., Mandomando I., Nhampossa T., Acacio S., Omore R., Oundo J.O., Ochieng J.B., Mintz E.D., O'Reilly C.E., Berkeley L.Y., Livio S., Tennant S.M., Sommerfelt H., Nataro J.P., Ziv-Baran T., Robins-Browne R.M., Mishcherkin V., Zhang J.X., Liu J., Houpt E.R., Kotloff K.L., Levine M.M. The burden of cryptosporidium diarrheal disease among children < 24 months of age in moderate/high mortality regions of Sub-Saharan Africa and South Asia, utilizing data from the Global Enteric Multicenter Study (GEMS) PLoS Negl Trop Dis. 2016;10:e0004729. doi: 10.1371/journal.pntd.0004729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., Donnelly R.P. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 5.Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T.E., Kuestner R., Garrigues U., Birks C., Roraback J., Ostrander C., Dong D., Shin J., Presnell S., Fox B., Haldeman B., Cooper E., Taft D., Gilbert T., Grant F.J., Tackett M., Krivan W., McKnight G., Clegg C., Foster D., Klucher K.M. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 6.Nice T.J., Baldridge M.T., McCune B.T., Norman J.M., Lazear H.M., Artyomov M., Diamond M.S., Virgin H.W. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015;347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pott J., Mahlakoiv T., Mordstein M., Duerr C.U., Michiels T., Stockinger S., Staeheli P., Hornef M.W. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazear H.M., Daniels B.P., Pinto A.K., Huang A.C., Vick S.C., Doyle S.E., Gale M., Jr., Klein R.S., Diamond M.S. Interferon-lambda restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Sci Transl Med. 2015;7:284ra59. doi: 10.1126/scitranslmed.aaa4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Odendall C., Voak A.A., Kagan J.C. Type III IFNs are commonly induced by bacteria-sensing TLRs and reinforce epithelial barriers during infection. J Immunol. 2017;199:3270–3279. doi: 10.4049/jimmunol.1700250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Argenzio R.A., Liacos J.A., Levy M.L., Meuten D.J., Lecce J.G., Powell D.W. Villous atrophy, crypt hyperplasia, cellular infiltration, and impaired glucose-Na absorption in enteric cryptosporidiosis of pigs. Gastroenterology. 1990;98:1129–1140. doi: 10.1016/0016-5085(90)90325-u. [DOI] [PubMed] [Google Scholar]
- 11.Lebreton A., Lakisic G., Job V., Fritsch L., Tham T.N., Camejo A., Mattei P.J., Regnault B., Nahori M.A., Cabanes D., Gautreau A., Ait-Si-Ali S., Dessen A., Cossart P., Bierne H. A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science. 2011;331:1319–1321. doi: 10.1126/science.1200120. [DOI] [PubMed] [Google Scholar]
- 12.Bierne H., Travier L., Mahlakoiv T., Tailleux L., Subtil A., Lebreton A., Paliwal A., Gicquel B., Staeheli P., Lecuit M., Cossart P. Activation of type III interferon genes by pathogenic bacteria in infected epithelial cells and mouse placenta. PLoS One. 2012;7:e39080. doi: 10.1371/journal.pone.0039080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Travar M., Vucic M., Petkovic M. Interferon lambda-2 levels in sputum of patients with pulmonary Mycobacterium tuberculosis infection. Scand J Immunol. 2014;80:43–49. doi: 10.1111/sji.12178. [DOI] [PubMed] [Google Scholar]
- 14.Bouzid M., Hunter P.R., Chalmers R.M., Tyler K.M. Cryptosporidium pathogenicity and virulence. Clin Microbiol Rev. 2013;26:115–134. doi: 10.1128/CMR.00076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sang Y., Rowland R.R., Hesse R.A., Blecha F. Differential expression and activity of the porcine type I interferon family. Physiol Genomics. 2010;42:248–258. doi: 10.1152/physiolgenomics.00198.2009. [DOI] [PubMed] [Google Scholar]
- 16.Elliott D.A., Coleman D.J., Lane M.A., May R.C., Machesky L.M., Clark D.P. Cryptosporidium parvum infection requires host cell actin polymerization. Infect Immun. 2001;69:5940–5942. doi: 10.1128/IAI.69.9.5940-5942.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott D.A., Clark D.P. Cryptosporidium parvum induces host cell actin accumulation at the host-parasite interface. Infect Immun. 2000;68:2315–2322. doi: 10.1128/iai.68.4.2315-2322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai P.Y., Zhang B., He W.Q., Zha J.M., Odenwald M.A., Singh G., Tamura A., Shen L., Sailer A., Yeruva S., Kuo W.T., Fu Y.X., Tsukita S., Turner J.R. IL-22 upregulates epithelial claudin-2 to drive diarrhea and enteric pathogen clearance. Cell Host Microbe. 2017;21:671–681 e4. doi: 10.1016/j.chom.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colegio O.R., Van Itallie C.M., McCrea H.J., Rahner C., Anderson J.M. Claudins create charge-selective channels in the paracellular pathway between epithelial cells. Am J Physiol Cell Physiol. 2002;283:C142–C147. doi: 10.1152/ajpcell.00038.2002. [DOI] [PubMed] [Google Scholar]
- 20.Shen L., Weber C.R., Raleigh D.R., Yu D., Turner J.R. Tight junction pore and leak pathways: a dynamic duo. Annu Rev Physiol. 2011;73:283–309. doi: 10.1146/annurev-physiol-012110-142150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner J.R. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 22.Mahlakoiv T., Hernandez P., Gronke K., Diefenbach A., Staeheli P. Leukocyte-derived IFN-alpha/beta and epithelial IFN-lambda constitute a compartmentalized mucosal defense system that restricts enteric virus infections. PLoS Pathog. 2015;11:e1004782. doi: 10.1371/journal.ppat.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen T.S., Prince A.S. Bacterial pathogens activate a common inflammatory pathway through IFN lambda regulation of PDCD4. PLoS Pathog. 2013;9:e1003682. doi: 10.1371/journal.ppat.1003682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Hara S.P., Bogert P.S.T., Trussoni C.E., Chen X.M., LaRusso N.F. Tlr4 promotes cryptosporidium parvum clearance in a mouse model of biliary cryptosporidiosis. J Parasitol. 2011;97:813–821. doi: 10.1645/GE-2703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heo I., Dutta D., Schaefer D.A., Iakobachvili N., Artegiani B., Sachs N., Boonekamp K.E., Bowden G., Hendrickx A.P.A., Willems R.J.L., Peters P.J., Riggs M.W., O'Connor R., Clevers H. Modelling cryptosporidium infection in human small intestinal and lung organoids. Nat Microbiol. 2018;3:814–823. doi: 10.1038/s41564-018-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuang Z., Seo E.J., Leis J. Mechanism of inhibition of retrovirus release from cells by interferon-induced gene ISG15. J Virol. 2011;85:7153–7161. doi: 10.1128/JVI.02610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pincetic A., Kuang Z.Z., Seo E.J., Leis J. The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J Virol. 2010;84:4725–4736. doi: 10.1128/JVI.02478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okumura A., Lu G.S., Pitha-Rowe I., Pitha P.M. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci U S A. 2006;103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okumura A., Pitha P.M., Harty R.N. ISG15 inhibit Ebola VP40VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci U S A. 2008;105:3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malakhova O.A., Zhang D.E. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J Biol Chem. 2008;283:8783–8787. doi: 10.1074/jbc.C800030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vargas-Inchaustegui D.A., Xin L., Soong L. Leishmania braziliensis infection induces dendritic cell activation, ISG15 transcription, and the generation of protective immune responses. J Immunol. 2008;180:7537–7545. doi: 10.4049/jimmunol.180.11.7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oura C.A., McKellar S., Swan D.G., Okan E., Shiels B.R. Infection of bovine cells by the protozoan parasite Theileria annulata modulates expression of the ISGylation system. Cell Microbiol. 2006;8:276–288. doi: 10.1111/j.1462-5822.2005.00620.x. [DOI] [PubMed] [Google Scholar]
- 33.Foltz C., Napolitano A., Khan R., Clough B., Hirst E.M., Frickel E.M. TRIM21 is critical for survival of Toxoplasma gondii infection and localises to GBP-positive parasite vacuoles. Sci Rep. 2017;7:5209. doi: 10.1038/s41598-017-05487-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meunier E., Dick M.S., Dreier R.F., Schurmann N., Kenzelmann Broz D., Warming S., Roose-Girma M., Bumann D., Kayagaki N., Takeda K., Yamamoto M., Broz P. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–370. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- 35.Feeley E.M., Pilla-Moffett D.M., Zwack E.E., Piro A.S., Finethy R., Kolb J.P., Martinez J., Brodsky I.E., Coers J. Galectin-3 directs antimicrobial guanylate binding proteins to vacuoles furnished with bacterial secretion systems. Proc Natl Acad Sci U S A. 2017;114:E1698–E1706. doi: 10.1073/pnas.1615771114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang B.Q., Chen X.M., LaRusso N.F. Cryptosporidium parvum attachment to and internalization by human biliary epithelia in vitro: a morphologic study. J Parasitol. 2004;90:212–221. doi: 10.1645/GE-3204. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H.L., Guo F.G., Zhu G. Cryptosporidium lactate dehydrogenase is associated with the parasitophorous vacuole membrane and is a potential target for developing therapeutics. PLoS Pathog. 2015;11:e1005250. doi: 10.1371/journal.ppat.1005250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mordstein M., Pott J., Mahlakoiv T., Neugebauer E., Dirt V., Drosten C., Hornef M.W., Michiels T., Staeheli P. A non-redundant role of IFN-lambda in antiviral defense of the intestinal tract. Cytokine. 2010;52:48. [Google Scholar]
- 39.Mordstein M., Neugebauer E., Ditt V., Jessen B., Rieger T., Falcone V., Sorgeloos F., Ehl S., Mayer D., Kochs G., Schwemmle M., Gunther S., Drosten C., Michiels T., Staeheli P. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol. 2010;84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baldridge M.T., Lee S., Brown J.J., McAllister N., Urbanek K., Dermody T.S., Nice T.J., Virgin H.W. Expression of Ifnlr1 on intestinal epithelial cells is critical to the antiviral effects of interferon lambda against norovirus and reovirus. J Virol. 2017;91 doi: 10.1128/JVI.02079-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hernandez P.P., Mahlakoiv T., Yang I., Schwierzeck V., Nguyen N., Guendel F., Gronke K., Ryffel B., Hoelscher C., Dumoutier L., Renauld J.C., Suerbaum S., Staeheli P., Diefenbach A. Interferon-lambda and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol. 2015;16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doom C.M., Turula H.M., Hill A.B. Investigation of the impact of the common animal facility contaminant murine norovirus on experimental murine cytomegalovirus infection. Virology. 2009;392:153–161. doi: 10.1016/j.virol.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leav B.A., Yoshida M., Rogers K., Cohen S., Godiwala N., Blumberg R.S., Ward H. An early intestinal mucosal source of gamma interferon is associated with resistance to and control of Cryptosporidium parvum infection in mice. Infect Immun. 2005;73:8425–8428. doi: 10.1128/IAI.73.12.8425-8428.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barakat F.M., McDonald V., Foster G.R., Tovey M.G., Korbel D.S. Cryptosporidium parvum infection rapidly induces a protective innate immune response involving type I interferon. J Infect Dis. 2009;200:1548–1555. doi: 10.1086/644601. [DOI] [PubMed] [Google Scholar]
- 45.Pervolaraki K., Stanifer M.L., Munchau S., Renn L.A., Albrecht D., Kurzhals S., Senis E., Grimm D., Schroder-Braunstein J., Rabin R.L., Boulant S. Type I and type III interferons display different dependency on mitogen-activated protein kinases to mount an antiviral state in the human gut. Front Immunol. 2017;8:459. doi: 10.3389/fimmu.2017.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen H., Watry D.D., Marcondes M.C., Fox H.S. Selective decrease in paracellular conductance of tight junctions: role of the first extracellular domain of claudin-5. Mol Cell Biol. 2004;24:8408–8417. doi: 10.1128/MCB.24.19.8408-8417.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Storey J.D. A direct approach to false discovery rates. J R Stat Soc B. 2002;64:479–498. [Google Scholar]
- 48.Godiwala N.T., Vandewalle A., Ward H.D., Leav B.A. Quantification of in vitro and in vivo Cryptosporidium parvum infection by using real-time PCR. Appl Environ Microbiol. 2006;72:4484–4488. doi: 10.1128/AEM.00189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sang Y., Rowland R.R., Blecha F. Molecular characterization and antiviral analyses of porcine type III interferons. J Interferon Cytokine Res. 2010;30:801–807. doi: 10.1089/jir.2010.0016. [DOI] [PubMed] [Google Scholar]
- 50.Dijkmans R., Vandenbroeck K., Beuken E., Billiau A. Sequence of the porcine interferon-gamma (IFN-gamma) gene. Nucleic Acids Res. 1990;18:4259. doi: 10.1093/nar/18.14.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gookin J.L., Duckett L.L., Armstrong M.U., Stauffer S.H., Finnegan C.P., Murtaugh M.P., Argenzio R.A. Nitric oxide synthase stimulates prostaglandin synthesis and barrier function in C. parvum-infected porcine ileum. Am J Physiol Gastrointest Liver Physiol. 2004;287:G571–G581. doi: 10.1152/ajpgi.00413.2003. [DOI] [PubMed] [Google Scholar]
- 52.Stewart M.J., Smoak K., Blum M.A., Sherry B. Basal and reovirus-induced beta interferon (IFN-beta) and IFN-beta-stimulated gene expression are cell type specific in the cardiac protective response. J Virol. 2005;79:2979–2987. doi: 10.1128/JVI.79.5.2979-2987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]