Abstract
Castration-resistant prostate cancer remains as an incurable disease. Exploiting DNA damage repair defects via inhibition of poly (ADP-ribose) polymerase (PARP) is becoming an attractive therapeutic option. The TOPARP-A clinical trial demonstrated that the PARP inhibitor olaparib may be an effective strategy for treating prostate cancer. However, several unanswered questions regarding the use of olaparib remain: 1) How do we best stratify patients for olaparib treatment? 2) Where do we place olaparib in the treatment sequence paradigm? 3) Is there cross-resistance between olaparib and currently used therapies? Here, we tested putative cross-resistance between current therapies and olaparib in treatment-resistant castration-resistant prostate cancer models. Docetaxel-resistant cells exhibited robust resistance to olaparib which could be attributed to blunted PARP trapping in response to olaparib treatment. Upregulated ABCB1 mediates cross-resistance between taxanes and olaparib, which can be overcome through decreasing ABCB1 expression or inhibiting ABCB1 using elacridar or enzalutamide. We also show that combining olaparib with enzalutamide is more effective in olaparib-sensitive cells than either single agent. Our results demonstrate that cross-resistance between olaparib and other therapies could blunt response to treatment and highlight the need to develop strategies to maximize olaparib efficacy.
Introduction
Castration-resistant prostate cancer (CRPC) remains an incurable disease responsible for significant morbidity and mortality. Recent efforts have added several therapies to the armamentarium for CRPC including the next-generation antiandrogen therapies, enzalutamide and abiraterone, and the taxanes docetaxel and cabazitaxel [1], [2], [3], [4]. Despite these advances, patients still succumb to the disease, highlighting the urgent need for both novel therapies and research to understand the optimal sequencing of all available options for patients.
Inhibition of poly (ADP-ribose) polymerase (PARP) using small molecule PARP inhibitors (PARPis) is quickly emerging as an efficacious treatment option for CRPC [5]. The PARP family consists of 17 members, each of which possesses ADP-ribose transfer function [6], [7]. Adding chains of ADP-ribose is known as poly ADP-ribosylation (PARylation). PARylation can alter the functioning of several different substrates and is involved in numerous cellular processes. A key function of PARP is to detect and initiate repair of single-strand DNA breaks [8]. Inhibition of PARP activity leads to increased DNA repair stress and the creation of double-strand breaks which must be repaired by additional mechanisms such as homologous recombination [7]. In the context of cells with mutations or alterations to DNA-repair proteins, loss of PARP activity can lead to synthetic lethality [9], [10]. PARPi treatment is emerging to exploit this synthetic lethality effect in select tumors with defined DNA-repair defects such as mutations in BRCA1 and BRCA2. While initial research is promising, further research is needed to fully understand PARPi function in varying contexts as this will improve our ability to treat patients.
Several PARPis are now in clinical development with exciting results in varying cancer indications. Notably, the TOPARP-A study tested the PARPi olaparib in the context of metastatic CRPC [11]. Fifty patients were recruited and treated with olaparib along with extensive genomic testing for biomarkers of DNA-repair deficiency. Of 49 evaluable patients, 16 were documented as having had a response. Of those 16 patients, 14 were determined to have a DNA-repair defect, suggesting that biomarker stratification may highlight a subset of patients who will have a response to PARPis. These results indicate the promise of using PARPis in CRPC clinical practice, but questions still abound regarding their use in this indication. It is noted that 2 of the 16 responders were not determined to be biomarker positive, and it was suggested that some patients considered biomarker positive had only single allele alterations which may not be sufficient to induce functional deficiency [7]. This suggests that further work is needed to fully understand response to these drugs. Also, the patient population recruited for this trial had been heavily pretreated with other approved CRPC treatments [11]. One hundred percent of patients had previously received docetaxel, while varying percentages had also been given abiraterone, enzalutamide, and cabazitaxel. It is currently unknown how prior therapeutic exposure may impact response to PARPis, nor is it understood where best to place PARPis or how best to utilize them in the CRPC clinical treatment paradigm. Our previous work demonstrated select cross-resistance between currently used CRPC treatments [12], [13]. Studies to evaluate putative cross-resistance between these therapies and PARPis are lacking.
Due to the promise of using olaparib in CRPC based on the TOPARP-A trial, olaparib received FDA breakthrough therapy designation, paving the way for a possible approval for this indication. Studies to understand how to use and sequence olaparib with other approved therapies are warranted to allow for maximized clinical efficacy. In this study, we assess the ability of olaparib to treat varying models of treatment resistant CRPC to understand putative cross-resistance. We find that taxane resistance induces robust cross-resistance to olaparib and that this is mediated by increased ABCB1 expression. Inhibition of ABCB1 resensitizes resistant cells to treatment. We also show that putative olaparib combination therapies may be highly effective in olaparib-resistant and -sensitive tumors.
Materials and Methods
Cell Culture and Reagents
C4-2B cells were kindly provided and authenticated by Dr. Leland Chung (Cedars-Sinai Medical Center, Los Angeles, CA). DU145 cells were obtained from the American Type Culture Collection, which uses short tandem repeat profiling for testing and authentication of cell lines. All cell lines are routinely tested for mycoplasma using ABM mycoplasma PCR detection kit (cat. #G238). All experiments with these cell lines and their derivatives were conducted within 6 months of receipt or resuscitation after cryopreservation. Cells were maintained in RPMI 1640 media supplemented with 10% fetal bovine serum, 100 IU penicillin, and 0.1 mg/ml streptomycin. Enzalutamide-resistant C4-2B cells (C4-2B-MDVR), abiraterone-resistant C4-2B cells (C4-2B-AbiR), docetaxel-resistant C4-2B cells (C4-2B-TaxR), and DU145-DTXR cells were characterized and described previously and maintained in complete RPMI 1640 supplemented with 20 μM enzalutamide, 10 μM abiraterone, or 5 nM docetaxel (TaxR and DU145-DTXR), respectively [14], [15], [16]. C4-2B and DU145 parental cells were cultured alongside derivative cell lines during their creation as an appropriate control. TaxR-Control and TaxR-shABCB1 were described previously and maintained in complete RPMI 1640 supplemented with 2 μg/ml puromycin [16]. CabR cells were created through continuous culture of TaxR cells in increasing doses of cabazitaxel over a 3-month period. CabR cells are maintained in complete RPMI 1640 supplemented with 5 nM cabazitaxel. All cells were maintained at 37°C in a humidified incubator with 5% carbon dioxide. Docetaxel (cat. #RS019) was purchased from TSZ CHEM. Enzalutamide (cat. #S1250), olaparib (cat. #S1060), and cabazitaxel (cat. #S3022) were purchased from Selleckchem. Abiraterone Acetate (cat. #X6144) was purchased from AK Scientific, Inc. Elacridar (cat. #143664-11-3) was purchased from Sigma-Aldrich. Puromycin (cat. #BP2956-100) was purchased from ThermoFisher Scientific. Methyl methanesulfonate (MMS; 99%) (cat. #156890050) was purchased from Fisher Scientific.
Cell Growth Assay
Cells were plated at a density of 25,000 cells/well in 24-well plates in complete RPMI 1640 media without any selection agent. After 24 hours, cells were subjected to indicated treatments. All drugs were administered simultaneously in each given assay. Total cells were counted via Coulter counter 72 hours posttreatment. Alternatively, cell proliferation was measured using Cell Counting Kit-8 (cat. #CK04) purchased from Dojindo Laboratories using manufacturer's instructions. Data are displayed as percent of control cell growth − treatment group cell number/control group cell number × 100. All conditions were performed in triplicate. All experiments were performed at least twice.
Colony Formation Assay
Cells were plated at 500 cells/well in 6-well plates in complete RPMI 1640 with no selection agent. Plated cells were subsequently treated 24 hours later as indicated. All drugs were administered simultaneously in each given assay. Colonies formed for 14 days. At the completion of each assay, cell colonies were fixed and stained using the following solution for 20 minutes: 0.05% w/v crystal violet, 1% of 37% formaldehyde, 1% methanol, 1× PBS. After staining, colonies were rinsed, allowed to air dry, and counted. Data are displayed as a percent of control cell colony growth (control is vehicle treatment only). All conditions were performed in duplicate. All experiments were performed at least twice.
Preparation of Whole Cell Lysates and Chromatin-Bound Fraction
Cells were harvested, washed with PBS, and lysed in RIPA buffer supplemented with 0.5 mM EDTA, 1 mM NaV, 10 mM NaF, and 1× Halt Protease Inhibitor Cocktail (cat. #78430) purchased from ThermoFisher Scientific. Protein concentration was determined with Pierce Coomassie Plus (Bradford) Assay Kit (cat. #23236) purchased from ThermoFisher Scientific. Chromatin-bound protein fractions were isolated using the Subcellular Protein Fractionation Kit for Cultured Cells (cat. #78840) purchased from ThermoFisher Scientific and used according to manufacturer's protocol.
Western Blot
Protein extracts were resolved by SDS-PAGE, and indicated primary antibodies were used. For PAR analysis, cells were first treated with indicated doses of olaparib for 30 minutes prior to harvest. For PARP trapping analysis, cells were first treated with 0.01% MMS and indicated doses of olaparib for 4 hours prior to harvest. Antibodies used were as follows: ABCB1 antibody (SC-8313, rabbit-polyclonal, 1:500 dilution) was purchased from Santa Cruz Biotechnology; PARP antibody (CS-9532S, rabbit-monoclonal antibody, 1:1000 dilution) was purchased from Cell Signaling Technology; PAR antibody (4335-MC-100, mouse-monoclonal antibody, 1:1000 dilution) was purchased from Trevigen; Tubulin antibody (T5168, mouse monoclonal antibody, 1:6000 dilution) was purchased from Sigma-Aldrich; GAPDH antibody (MAB374, mouse monoclonal antibody, 1:10000) was purchased from EMD Millipore; and Histone-H3 antibody (cat. #39163) was purchased from Active Motif. Tubulin and Histone-H3 were used to monitor the amounts of samples applied. Proteins were visualized with a chemiluminescence detection system (cat. #WBLUR0500) purchased from Millipore. Densitometry to quantify blots was performed using ImageJ.
Statistics
All quantitated data are displayed as percent of control mean ± standard deviation. Significance was assessed using a two-tailed, two-sample, equal-variance Student’s t test. A P value of ≤ .05 was accepted as significant.
Results
Olaparib Efficacy Is Blunted in Models of Treatment-Resistant CRPC
While recent clinical work has shown the potential utility of using olaparib to treat advanced CRPC, it is not known where in the treatment sequence this drug should be used, nor has it been determined how previous exposure to different drugs will affect its efficacy [11]. We hypothesized that cross-resistance may exist between olaparib and currently approved CRPC treatments. We have created and characterized several models of CRPC therapeutic resistance using C4-2B as a parental cell line: enzalutamide-resistant C4-2B-MDVR cells (MDVR), abiraterone-resistant C4-2B-AbiR cells (AbiR), and docetaxel-resistant C4-2B-TaxR cells (TaxR) [14], [15], [16]. Using these models, we tested response to olaparib in the context of therapeutic resistance.
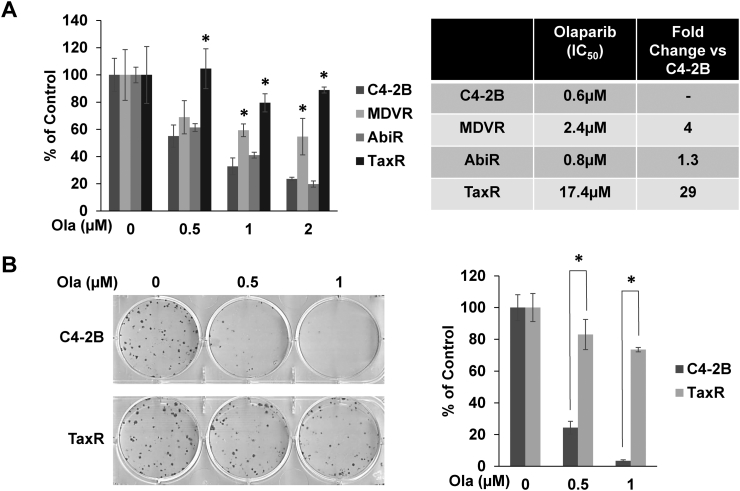
Cell growth assays and determination of IC50s demonstrate differential olaparib responses in varying resistant models versus parental C4-2B cells (Figure 1A). While AbiR cells remain relatively sensitive to olaparib, MDVR cells do exhibit moderate cross-resistance to the treatment. In contrast, TaxR cells exhibit robust insensitivity to olaparib treatment, prompting additional study. To further test TaxR cross-resistance to olaparib, we used colony formation assays (Figure 1B). While olaparib markedly reduced colony-forming ability in C4-2B cells, this effect was largely blunted in TaxR cells. These data suggest that prior exposure to docetaxel may greatly retard responses to subsequent treatment with olaparib.
Figure 1.
Response to olaparib is blunted in models of therapeutic-resistant CRPC. (A) Cell growth assays were used to test response to olaparib in MDVR, AbiR, and TaxR cells versus parental C4-2B cells using indicated doses of olaparib over a 72-hour period. IC50 values were calculated and are represented in the table. (B) Colony formation assays were used to test response to olaparib in TaxR cells versus parental C4-2B cells. Quantification of the assay is shown (right panel). Ola = olaparib, * = P value ≤ .05.
Olaparib PARP Trapping Is Blunted in Docetaxel-Resistant CRPC Cells
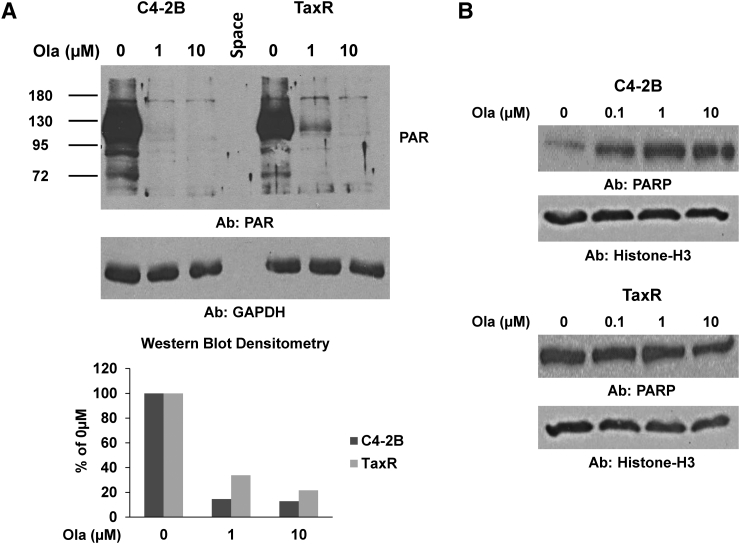
PARP inhibitors such as olaparib are known to exert their effects in two ways: 1) inhibition of PARP catalytic function and 2) trapping of PARP on chromatin leading to DNA damage and apoptosis [17], [18]. To better understand olaparib cross-resistance in TaxR cells, we first tested whether olaparib's inhibition of PARP catalytic activity was altered using Western blots for PAR to assess the relative level of inhibited PARylation (Figure 2A). C4-2B and TaxR cells were treated with indicated doses of olaparib for 30 minutes prior to harvest, and then whole cell lysates were subjected to Western blot for PAR. Quantification of the blots is also shown. Interestingly, we found a similar decrease in PAR levels in response to treatment with increasing doses of olaparib, suggesting no significant difference in olaparib's ability to inhibit PARP catalytic activity at these doses in TaxR cells versus parental C4-2B cells. To assess whether the levels of trapped PARP were altered, we treated C4-2B and TaxR cells with 0.01% MMS to recruit PARP to damaged DNA and subjected each cell line to increasing doses of olaparib to trap PARP at sites of damage [18]. After 4 hours of treatment, we isolated the chromatin-bound cellular fractions and performed Western blots for PARP (Figure 2B). We found that in olaparib-sensitive C4-2B cells, treatment with increasing olaparib concentrations led to increased trapped PARP. However, in TaxR cells, trapped PARP levels were unchanged. These data suggest that olaparib's ability to lock PARP onto DNA is blunted in docetaxel-resistant prostate cancer, and this may be responsible for cross-resistance between these therapies.
Figure 2.
Assessment of olaparib mechanistic function in TaxR vs. C4-2B cells. (A) C4-2B and TaxR cells were treated with increasing doses of olaparib for 30 minutes. Cells were then harvested and whole cell lysates were subjected to Western blot for PAR (upper panel). GAPDH served as a loading control. Densitometric quantification of the blot is shown (lower panel). PAR levels are normalized to GAPDH expression. (B) C4-2B and TaxR cells were treated with 0.01% MMS and indicated doses of olaparib for 4 hours and then harvested and fractionated to isolate the chromatin-bound fraction. Chromatin-bound fraction was subjected to Western blot for PARP. Histone-H3 served as a loading control. Ola = olaparib, Ab = antibody used.
ABCB1 Mediates Robust Resistance to Olaparib
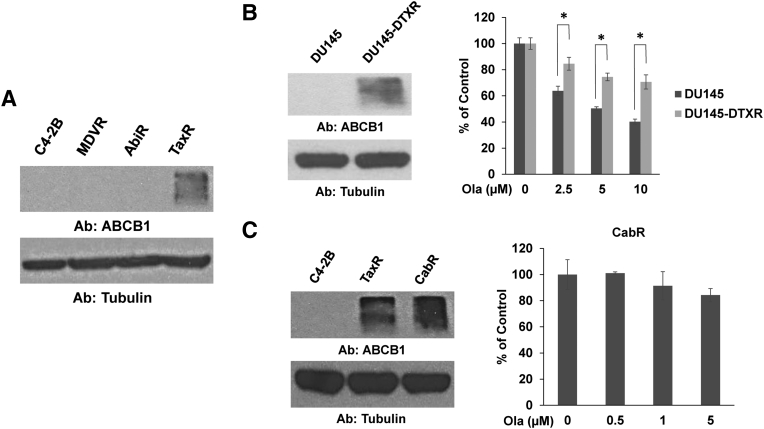
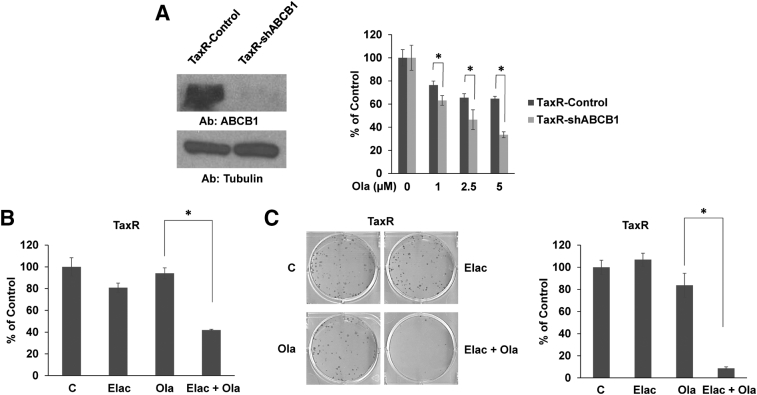
Our data suggest that prior exposure to taxane chemotherapy induces cross-resistance to olaparib. We've shown previously that taxane-resistant TaxR cells become cross-resistant to both docetaxel and cabazitaxel through increased levels of ABCB1 compared to control C4-2B cells [13], [16]. We hypothesize that ABCB1-mediated taxane cross-resistance may additionally induce resistance to olaparib in CRPC cells. Western blot for ABCB1 confirms our previous data showing TaxR cells have increased ABCB1 expression (Figure 3A). Whether ABCB1 confers resistance to olaparib in CRPC is unknown.
Figure 3.
Increased ABCB1 expression is associated with resistance to olaparib. (A) Whole cell lysates from indicated cell lines were subjected to Western blot for ABCB1. Tubulin served as a loading control. (B) Cell growth assay was used to test response to olaparib in docetaxel-resistant DU145-DTXR cells versus parental DU145 cells. Western blot was used to assess ABCB1 expression in these cell lines. Tubulin served as a loading control. (C) Cell growth assay was used to test response to olaparib in cabazitaxel-resistant CabR cells. Western blot was used to assess ABCB1 expression in C4-2B, TaxR, and CabR cells. Tubulin served as a loading control. Ola = olaparib, Ab = antibody used, * = P value ≤ .05.
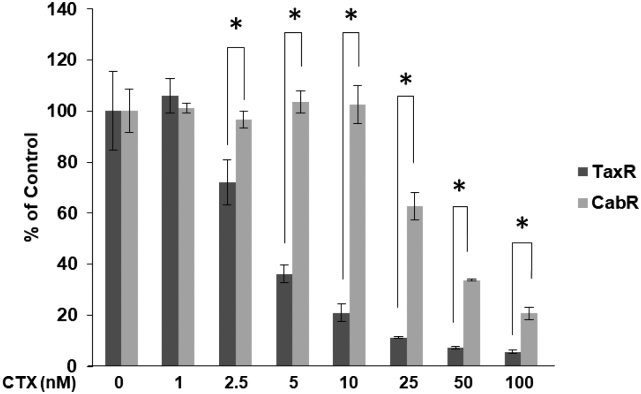
To test the role of ABCB1 in mediating cross-resistance to olaparib, we first used two additional models: 1) a DU145-derived docetaxel-resistant cell line, DU145-DTXR, which we've previously demonstrated also possesses augmented ABCB1 levels versus parental DU145 cells, and 2) a novel cabazitaxel-resistant cell line, CabR, derived from docetaxel-resistant TaxR cells (Supplemental Figure 1) [13], [16]. Western blots confirm increased ABCB1 expression in DU145-DTXR cells versus control cells and also show that CabR cells further increase ABCB1 expression compared to TaxR cells (Figure 3, B-C). Similar to TaxR cells, cell growth assays demonstrate that DU145-DTXR cells exhibit decreased sensitivity to olaparib versus control cells (Figure 3B). We also found very little response to olaparib in CabR cells (Figure 3C). These data suggest that overexpression of ABCB1 is associated with resistance to olaparib in CRPC.
Supplemental Figure 1.
Characterization of novel cabazitaxel-resistant CabR cells. Prolonged exposure to increasing doses of cabazitaxel over a 3-month period to docetaxel-resistant TaxR cells led to the creation of cabazitaxel-resistant CabR cells. A cell growth assay was used to demonstrate the resistance to cabazitaxel in CabR versus TaxR cells. CTX = cabazitaxel, * = P value ≤ .05.
We previously demonstrated that constitutive inhibition of ABCB1 in TaxR cells using a shRNA targeting ABCB1 (TaxR-shABCB1 cells) resensitizes taxane-resistant cells to both docetaxel and cabazitaxel [13], [16]. To test the role of ABCB1 in mediating cross-resistance to olaparib, we assessed response to olaparib treatment using the same TaxR-shABCB1 cells (Figure 4A). Western blot confirms decreased ABCB1 expression in TaxR-shABCB1 versus shGFP expressing TaxR-control cells. Cell growth assays demonstrate that decreased ABCB1 expression leads to increased sensitivity to olaparib. These data taken together suggest that increased ABCB1 expression can induce resistance to olaparib in CRPC.
Figure 4.
Inhibition of ABCB1 expression or function resensitizes TaxR cells to olaparib treatment. (A) Cell growth assay was used to assess response to olaparib in TaxR-shABCB1 cells versus TaxR-Control cells. Western blot was used to assess ABCB1 expression in these cell lines. Tubulin served as a loading control. (B) Cell growth assay was used to test response to elacridar (0.5 μM), olaparib (1 μM), or both in TaxR cells. (C) Colony formation assays were used to test response to elacridar (0.25 μM), olaparib (1 μM), or both in TaxR cells. Quantification of the assay is shown (right panel). C = control (DMSO), Elac = elacridar, Ola = olaparib, Ab = antibody used, * = P value ≤ .05.
Pharmacologic Inhibition of ABCB1 Activity Resensitizes Taxane-Resistant Cells to Olaparib Treatment
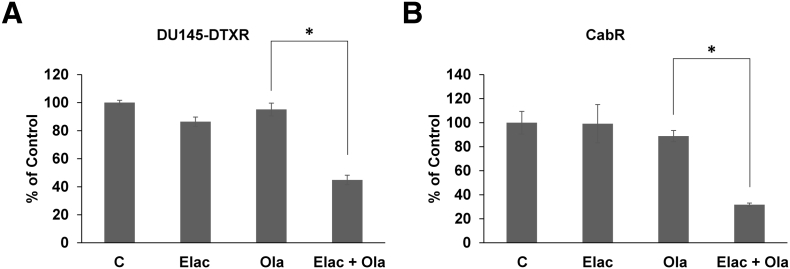
We've demonstrated that increased ABCB1 expression can mediate olaparib cross-resistance in taxane-resistant CRPC cells. Inhibition of ABCB1 using small molecule drugs could provide novel treatment strategies capable of rendering resistant tumors sensitive to olaparib treatment. To test this idea, we used elacridar, a small molecule inhibitor of ABCB1. We've previously shown that elacridar inhibits ABCB1 ATPase activity and greatly diminishes ABCB1 function leading to enhanced sensitivity to taxanes [13], [16], [19]. Cell growth assays demonstrate that combination of elacridar with olaparib is synergistic and resensitizes TaxR cells to olaparib treatment (Figure 4B). Colony formation assays further demonstrate that elacridar is highly efficacious in combination with olaparib (Figure 4C). We also show that elacridar can resensitize both DU145-DTXR and CabR cells to olaparib treatment (Figure 5, A and B). These data suggest that small molecule ABCB1 inhibition could be a viable strategy in combination with olaparib in the setting of ABCB1 mediated taxane resistance.
Figure 5.
Elacridar resensitizes DU145-DTXR and CabR cells to olaparib treatment. Cell growth assays were used to test response to elacridar (0.5 μM), olaparib (10 μM for DU145-DTXR or 5 μM for CabR), or a combination of both in (A) DU145-DTXR cells and (B) CabR cells. C = control (DMSO), Elac = elacridar, Ola = olaparib, * = P value ≤ .05.
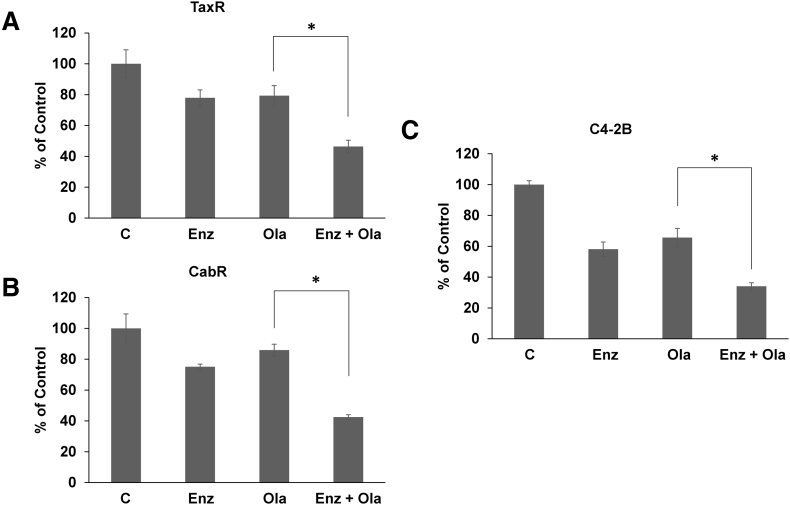
Interestingly, we've previously shown that the antiandrogens bicalutamide and enzalutamide possess secondary function as inhibitors of ABCB1 ATPase activity [19]. Both are capable of decreasing ABCB1 function and resensitizing taxane-resistant cells to taxane treatment [13], [19]. As enzalutamide is currently preferred to treat taxane-resistant CRPC, we sought to test whether treatment with enzalutamide can resensitize taxane-resistant cells to olaparib [20]. Cell growth assays demonstrate that combination treatment with enzalutamide and olaparib is more effective in reducing viability than either treatment alone in both TaxR and CabR cells (Figure 6, A and B). These data suggest that combining olaparib with a current CRPC standard-of-care drug in enzalutamide may be more beneficial than a single agent in the context of taxane resistance.
Figure 6.
Combination of olaparib with enzalutamide is more effective than either single-agent treatment. Cell growth assays were used to test response to enzalutamide, olaparib, or both in (A) TaxR cells (enzalutamide, 10 μM; olaparib, 1 μM), (B) CabR cells (enzalutamide, 10 μM; olaparib, 5 μM), and (C) C4-2B cells (enzalutamide, 20 μM; olaparib, 0.5 μM). C = control (DMSO), Enz = enzalutamide, Ola = olaparib, * = P value ≤ .05.
Olaparib Combined with Enzalutamide Is More Effective than Single-Agent Treatment in Olaparib-Sensitive C4-2B Cells
Lastly, we sought to explore the putative combination therapy of enzalutamide and olaparib in the nonresistant setting. It is hypothesized that next-generation antiandrogens, such as enzalutamide, may produce better responses in combination with PARP inhibitors earlier in CRPC disease progression before the development of resistance. Cell growth assays demonstrate that combination of enzalutamide with olaparib significantly reduces C4-2B cell viability versus either single agent treatment (Figure 6C). Thus, novel combination treatments may provide significant benefit to patients earlier in their disease progression prior to the development of therapeutic resistance.
Discussion
In the present study, we test the efficacy of the PARPi olaparib in models of treatment-resistant CRPC. We find that docetaxel-resistant TaxR cells display robust cross-resistance to olaparib versus parental C4-2B cells. In contrast, AbiR cells remain sensitive to treatment, while MDVR cells are moderately cross-resistant to olaparib. These results suggest that PARPi efficacy may be altered by previous lines of therapy, thus raising the need for further study into rational treatment sequencing and the creation of combination therapies.
As TaxR cells demonstrated the most striking insensitivity to olaparib, we further explored the mechanism by which cross-resistance exists. Our data support the role of ABCB1 as a significant factor in determining sensitivity to olaparib. We show that two disparate docetaxel-resistant models, TaxR and DU145-DTXR, display augmented levels of ABCB1 and have a poor response to olaparib, which can be overcome by inhibition of ABCB1 expression or function via elacridar or enzalutamide. These data suggest that patients who have developed ABCB1-mediated resistance to taxanes, which are commonly used for the treatment of CRPC, may fare poorly on subsequent olaparib treatment. We also show that a model of cabazitaxel-resistant CRPC in the postdocetaxel setting further augments ABCB1 levels and is highly resistant to olaparib, which can be overcome using pharmacologic ABCB1 inhibition. Thus, even in advanced taxane resistance, blocking ABCB1 activity could be an effective therapeutic strategy. Previous reports have shown that olaparib resistance can be developed through ABCB1 overexpression in breast and ovarian cancers, which could be reversed using ABCB1 inhibitors [21], [22]. These data support our study suggesting ABCB1 inhibition strategies could be beneficial for resistant tumors.
We've previously shown that the antiandrogen drug enzalutamide has a secondary function as an inhibitor of ABCB1 activity [19]. In this study, we show that combination of olaparib with enzalutamide is highly effective over single-agent treatment in taxane-resistant cells, presumably through inhibition of ABCB1 and resensitization to olaparib. As enzalutamide is already approved to treat docetaxel-resistant prostate cancer, our study suggests that combination therapy may be preferred over single-agent treatment [23]. We also show that enzalutamide in combination with olaparib is more effective than either single agent in olaparib-sensitive C4-2B cells. A previous study demonstrated that enzalutamide could reduce the expression of several genes involved in DNA repair and homologous recombination, thus inducing a more sensitive phenotype which provides a rationale for this combination [24]. It has also been demonstrated that PARP promotes AR signaling, which is thought to be mediated by aiding AR access to the chromatin [25]. Thus, inhibition of PARP with a PARPi drug and the addition of an antiandrogen may synergize to inhibit AR signaling and prostate cancer growth and survival. The combination of a PARPi with a taxane has also been shown to work better than single-agent treatment in treatment-naive CRPC [25]. These data taken together suggest that combination therapies of currently used agents with novel PARPis may be more efficacious upfront prior to exposure to any other single treatment.
We've previously provided evidence for cross-resistance between currently available treatments for CRPC [12], [13]. Here, we tested for the potential for cross-resistance between olaparib and these therapies. As PARPis advance through clinical trials toward approval for prostate cancer, our data have important and timely clinical implications. Olaparib has now received FDA breakthrough therapy designation based on results of the TOPARP-A trial. However, it should be noted that the participants of the TOPARP-A trial were heavily pretreated [11]. Interestingly, 100% of the participants of the trial had previously been given docetaxel, 96% had been given abiraterone, 58% had been given cabazitaxel, and 28% had received enzalutamide. While it appears abiraterone has a minimal effect on olaparib treatment, our findings suggest that previous therapeutic exposure to taxanes and enzalutamide may have precluded some patients from responding to olaparib or may have blunted their response. It will be important to conduct further research and clinical trials to understand the optimal sequencing of all available treatments as, currently, olaparib is being studied for those patients who have already had docetaxel and either enzalutamide or abiraterone. In addition to olaparib, rucaparib has also recently received the FDA breakthrough therapy designation based on results of the TRITON2 trial. However, the patient indication is the same regarding previous therapy exposure. Further study of PARPi resistance mechanisms will further our understanding of how best to use these drugs.
In conclusion, our study shows that meaningful cross-resistance may exist between olaparib and currently administered drugs for CRPC. We specifically focus on robust cross-resistance between olaparib and taxanes and show that olaparib resistance may be mediated by increased ABCB1 expression, which can be overcome through inhibition of ABCB1 expression or function. Future research into the PARP family and PARPi function will bring key advances in the field of prostate cancer treatment.
Acknowledgements
This work was supported in part by; National Institutes of Health/National Cancer Institute (USA) grants CA168601 (A.C.G) and CA179970 (A.C.G); Department of Defense (USA) grant PC150229 (A.C.G); and the U.S. Department of Veterans Affairs, Office of Research & Development BL&D (USA) grant I01BX0002653 (A.C.G), and a Research Career Scientist Award (A.C.G).
Footnotes
Conflict of interest: none.
References
- 1.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324(5928):787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mostaghel EA, Marck BT, Plymate SR, Vessella RL, Balk S, Matsumoto AM, Nelson PS, Montgomery RB. Resistance to CYP17A1 inhibition with abiraterone in castration-resistant prostate cancer: induction of steroidogenesis and androgen receptor splice variants. Clin Cancer Res. 2011;17(18):5913–5925. doi: 10.1158/1078-0432.CCR-11-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, Oudard S, Theodore C, James ND, Turesson I. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351(15):1502–1512. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 4.de Bono JS, Oudard S, Ozguroglu M, Hansen S, Machiels JP, Kocak I, Gravis G, Bodrogi I, Mackenzie MJ, Shen L. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: a randomised open-label trial. Lancet. 2010;376(9747):1147–1154. doi: 10.1016/S0140-6736(10)61389-X. [DOI] [PubMed] [Google Scholar]
- 5.Palmbos PL, Hussain MH. Targeting PARP in Prostate Cancer: Novelty, Pitfalls, and Promise. Oncology (Williston Park) 2016;30(5):377–385. [PubMed] [Google Scholar]
- 6.Morales J, Li L, Fattah FJ, Dong Y, Bey EA, Patel M, Gao J, Boothman DA. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr. 2014;24(1):15–28. doi: 10.1615/critreveukaryotgeneexpr.2013006875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramakrishnan Geethakumari P, Schiewer MJ, Knudsen KE, Kelly WK. PARP Inhibitors in Prostate Cancer. Curr Treat Options in Oncol. 2017;18(6):37. doi: 10.1007/s11864-017-0480-2. [DOI] [PubMed] [Google Scholar]
- 8.del Rivero J, Kohn EC. PARP Inhibitors: The Cornerstone of DNA Repair-Targeted Therapies. Oncology (Williston Park) 2017;31(4):265–273. [PubMed] [Google Scholar]
- 9.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 10.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 11.Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, Nava Rodrigues D, Robinson D, Omlin A, Tunariu N. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med. 2015;373(18):1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lombard AP, Liu L, Cucchiara V, Liu C, Armstrong CM, Zhao R, Yang JC, Lou W, Evans CP, Gao AC. Intra versus Inter Cross-resistance Determines Treatment Sequence between Taxane and AR-Targeting Therapies in Advanced Prostate Cancer. Mol Cancer Ther. 2018;17(10):2197–2205. doi: 10.1158/1535-7163.MCT-17-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lombard AP, Liu C, Armstrong CM, Cucchiara V, Gu X, Lou W, Evans CP, Gao AC. ABCB1 Mediates Cabazitaxel-Docetaxel Cross-Resistance in Advanced Prostate Cancer. Mol Cancer Ther. 2017;16(10):2257–2266. doi: 10.1158/1535-7163.MCT-17-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, Lou W, Zhu Y, Nadiminty N, Schwartz CT, Evans CP, Gao AC. Niclosamide inhibits androgen receptor variants expression and overcomes enzalutamide resistance in castration-resistant prostate cancer. Clin Cancer Res. 2014;20(12):3198–3210. doi: 10.1158/1078-0432.CCR-13-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C, Armstrong C, Zhu Y, Lou W, Gao AC. Niclosamide enhances abiraterone treatment via inhibition of androgen receptor variants in castration resistant prostate cancer. Oncotarget. 2016;7(22):32210–32220. doi: 10.18632/oncotarget.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y, Liu C, Nadiminty N, Lou W, Tummala R, Evans CP, Gao AC. Inhibition of ABCB1 expression overcomes acquired docetaxel resistance in prostate cancer. Mol Cancer Ther. 2013;12(9):1829–1836. doi: 10.1158/1535-7163.MCT-13-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72(21):5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murai J, Huang SY, Renaud A, Zhang Y, Ji J, Takeda S, Morris J, Teicher B, Doroshow JH, Pommier Y. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 2014;13(2):433–443. doi: 10.1158/1535-7163.MCT-13-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Y, Liu C, Armstrong C, Lou W, Sandher A, Gao AC. Antiandrogens Inhibit ABCB1 Efflux and ATPase Activity and Reverse Docetaxel Resistance in Advanced Prostate Cancer. Clin Cancer Res. 2015;21(18):4133–4142. doi: 10.1158/1078-0432.CCR-15-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shore ND, Chowdhury S, Villers A, Klotz L, Siemens DR, Phung S van Os, Hasabou N, Wang F, Bhattacharya S. Efficacy and safety of enzalutamide versus bicalutamide for patients with metastatic prostate cancer (TERRAIN): a randomised, double-blind, phase 2 study. Lancet Oncol. 2016;17(2):153–163. doi: 10.1016/S1470-2045(15)00518-5. [DOI] [PubMed] [Google Scholar]
- 21.Rottenberg S, Jaspers JE, Kersbergen A, van der Burg E, Nygren AO, Zander SA, Derksen PW, de Bruin M, Zevenhoven J, Lau A. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008;105(44):17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaidyanathan A, Sawers L, Gannon AL, Chakravarty P, Scott AL, Bray SE, Ferguson MJ, Smith G. ABCB1 (MDR1) induction defines a common resistance mechanism in paclitaxel- and olaparib-resistant ovarian cancer cells. Br J Cancer. 2016;115(4):431–441. doi: 10.1038/bjc.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367(13):1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Karanika S, Yang G, Wang J, Park S, Broom BM, Manyam GC, Wu W, Luo Y, Basourakos S. Androgen receptor inhibitor-induced "BRCAness" and PARP inhibition are synthetically lethal for castration-resistant prostate cancer. Sci Signal. 2017;10(480) doi: 10.1126/scisignal.aam7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiewer MJ, Goodwin JF, Han S, Brenner JC, Augello MA, Dean JL, Liu F, Planck JL, Ravindranathan P, Chinnaiyan AM. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2(12):1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]