Abstract
Purpose
Isocitrate dehydrogenase (IDH) and 1p19q codeletion status are importantin providing prognostic information as well as prediction of treatment response in gliomas. Accurate determination of the IDH mutation status and 1p19q co-deletion prior to surgery may complement invasive tissue sampling and guide treatment decisions.
Methods
Preoperative MRIs of 538 glioma patients from three institutions were used as a training cohort. Histogram, shape, and texture features were extracted from preoperative MRIs of T1 contrast enhanced and T2-FLAIR sequences. The extracted features were then integrated with age using a random forest algorithm to generate a model predictive of IDH mutation status and 1p19q codeletion. The model was then validated using MRIs from glioma patients in the Cancer Imaging Archive.
Results
Our model predictive of IDH achieved an area under the receiver operating characteristic curve (AUC) of 0.921 in the training cohort and 0.919 in the validation cohort. Age offered the highest predictive value, followed by shape features. Based on the top 15 features, the AUC was 0.917 and 0.916 for the training and validation cohort, respectively. The overall accuracy for 3 group prediction (IDH-wild type, IDH-mutant and 1p19q co-deletion, IDH-mutant and 1p19q non-codeletion) was 78.2% (155 correctly predicted out of 198).
Conclusion
Using machine-learning algorithms, high accuracy was achieved in the prediction of IDH genotype in gliomas and moderate accuracy in a three-group prediction including IDH genotype and 1p19q codeletion.
Keywords: Glioma, Isocitrate dehydrogenase (IDH), 1p19q codeletion, Machine learning, Random forest, MRI
Introduction
Gliomas account for 81% of primary malignant brain tumors and confer significant mortality and morbidity [1]. Patients with glioblastoma, the most common glioma histology, have a median survival of 15–16 months, despite surgery, chemotherapy, and radiation therapy [2]. Although the natural history of low-grade gliomas (LGGs) varies greatly with survival averaging approximately 7 years, LGGs in adults eventually progress to high-grade glioma and premature mortality [3].
IDH1 mutations, specifically involving the amino acid arginine at position 132, were first described in 12% of glioblastomas [4], followed by observation that they are present in 50–80% of LGG patients [5]. Importantly, IDH mutations confers diagnostic and prognostic implications. Gliomas with the IDH1 mutation (or its homolog IDH2) is associated with a significantly more favorable survival outcomes than the IDH1/2 wild-type tumors, independent of histological grade [6, 7]. Due to the prognostic significance of the IDH mutation, the World Health Organization (WHO) updated its classification criteria in 2016 to integrate IDH1/2 status as a molecular parameter for classifying gliomas [8]. IDH mutants are driven by specific epigenetic alterations, which may make them susceptible to therapeutic interventions (such as temozolomide) that are less effective against IDH wild type tumor [9, 10]. This is supported by in vitro experiments, which demonstrated increased radio- and chemo-sensitivity in IDH-mutated cancer cells [11, 12]. Targeted therapy against IDH mutation has been shown to be associated with a favorable safety profile and prolonged stable disease in Phase I study of cholangiocarcinoma [13] and glioma [14]. Codeletion of 1p19q has also been proven to be a prognostic molecular marker for positive tumor response to radiation and chemotherapy and associated with better survival [15–17]. The Cancer Genome Atlas Research Network classified LGGs into three molecular categories: gliomas with IDH mutation and 1p19q codeletion (IDHmut-codel), gliomas with IDH mutation and no 1pI19q codeletion (IDHmut-non-codel) and gliomas with wild-type IDH (IDHwt). IDHwt LGGs are associated with poor outcome. LGGs with an IDH mut-codel are more sensitive to radiation and chemotherapy and associated with longer survival than other types of diffuse gliomas [18].
Although biopsies can be performed at relatively low-risk, an approach using MRI imaging to predict IDH and 1p19q genotype preoperatively is a less expensive and noninvasive alternative. The early identification of IDH and 1p19q status may benefit the prediction of patient’s prognosis and predictive of responsiveness to chemotherapy and radiation.
Several advanced imaging techniques have been shown to predict IDH and/or 1p19q status in gliomas. Most of the previous approaches utilized a single imaging feature or parameter, such as relative cerebral blood volume, sodium, spectroscopy, blood oxygen level-dependence, perfusion and 11C-methionine PET [19–25]. However, inclusion of these advanced imaging sequences such as DWI, PWI, MRI spectroscopy and [18F] fluoroethyltyrosine-PET(FET-PET) images may not be useful or reliable for determining genotype of the gliomas compared with conventional MR images [26, 27]. Besides, many of these imaging acquisitions are not routinely obtained in clinical care. In this study, we strived to develop a method solely employing imaging sequences that would be acquired during standard of care in clinical evaluations.
To the best of our knowledge, there are limited studies that predict IDH and 1p19q status utilizing standardized imaging methodology and through large sample size from multiple institutions. We hypothesized that a model integrating features from conventional MRI using a machine-learning approach could diagnose IDH mutation and 1p19q codeletion status and identify specific features relevant to the genotype.
Methods
Patient cohort
The training cohort consisted of patients with histologically confirmed diffuse gliomas treated at Hospital of the University of Pennsylvania (HUP), Brigham and Women’s Hospital (BWH), and Massachusetts General Hospital (MGH). Institutional Review Board (IRB) approval was obtained for the training cohort with waiver of consent. The validation cohort consisted of patients with gliomas who have overlapping clinical and molecular data from The Cancer Genome Atlas (TCGA) and presurgical MR imaging data from The Cancer Imaging Archive (TCIA), an imaging sharing resource that houses images corresponding to TCGA patients [28, 29]. Analysis of the TCGAITCIA cohort is exempt from IRB approval under the TCGAITCIA data use agreements (http:IIcancergenome.nih.gov/abouttcgaIpoliciesIinformedconsent). All patients identified met the following criteria: (i) histo-pathologically confirmed primary grade II-IV glioma according to current WHO criteria, (ii) known IDH genotype, and (iii) available preoperative MR imaging consisting of post-contrast axial T1-weighted (T1 post-contrast) and T2-weighted fluid attenuation inversion recovery (FLAIR) images. Patients whose IDH genotype were not confirmed per criteria (see “Tissue Diagnosis and Genotyping” section below) were excluded (N = 93). Our final patient cohort included 227 patients from HUP, 156 patients from BWH, 155 patients from MGH and 206 patients from TCIA.
Tissue diagnosis and genotyping
For the HUP cohort, IDH1R132H mutant status was determined using either immunohistochemistry (IHC) or next-generation sequencing, performed by the Center for Personalized Diagnostics at HUP. For the BWH cohort, IDH1/2 mutations were determined using IHC, mass spectrometry-based mutation genotyping (OncoMap), or capture-based sequencing (OncoPanel), depending on the available genotyping technology at the time of diagnosis. OncoMap was performed by Center for Advanced Molecular Diagnostics of the BWH and OncoPanel was performed by Center for Cancer Genome Discovery of the Dana-Farber Cancer Institute. For the MGH cohort, IDH1/2 status were confirmed either by immunochemistry or next generation sequencing. For this retrospective study, only gliomas with absence of IDH1/2 mutations as determined by full sequencing assay were included in our analyses as IDH-WT gliomas. IDH-mutated gliomas were defined by the presence of mutation as indicated by IHC or sequencing on samples provided to the pathology department at each institution at the time of surgery. IDH1- and IDH2-mutated gliomas were collapsed into one category. For patients in the TCIA cohort, IDH½ mutation data were downloaded from TCGA and IvyGap data portal.
The 1p/19q co-deletion genotype was determined via fluorescence in situ hybridization (FISH) or polymerase chain reaction (PCR) depending on the availability of the hospital. For patients in the TCIA cohort, 1p19q codeletion data were downloaded from TCGA and IvyGap data portal.
Expert tumor segmentation
For the HUP and TCIA cohorts, MR imaging for each patient was loaded into Matrix User (v2.2), and 3D FLAIR tumor regions were manually drawn slice-by-slice in the axial plane for the FLAIR image by a user (H.Z.) followed by editing by a radiologist (H.X.B). For the BWH cohort, FLAIR tumor regions were drawn with the 3D Slicer Editor and Segmentation Wizard module(https://www.slicer.org/wiki/Documentation/Nightly/Extensions/SegmentationWizard) on the FLAIR image (K.C.) and edited by an neuroradiologist (R.Y.H.) [30, 31]. For the MGH cohort, FLAIR tumor regions were drawn by a user (V.K.K., J.T.S.) and edited by a neurosurgery resident (A.B.) on the FLAIR image with 3D Slicer. We derived expert manual tumor segmentations for half of the patients. The entire abnormality was segmented (Fig. 1) on FLAIR image of each patient. We used FLAIR to draw the mask for the predictive model, then this FLAIR mask was applied to both contrast enhanced T1 and FLAIR sequences.
Fig. 1.
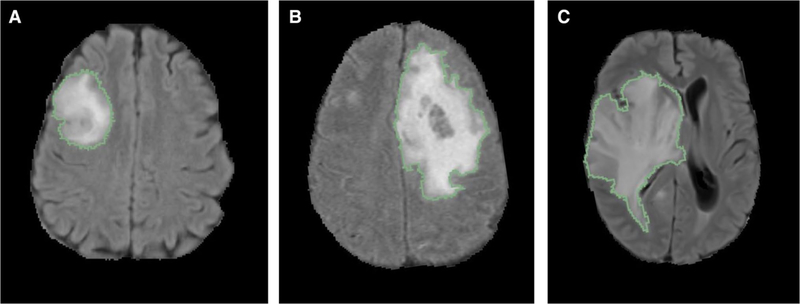
FLAIR tumor segmentation of a a grade II, b grade III, c grade IV. Segmentations shown are overlaid on axial, non-contrasted FLAIR image
Preprocessing
Brain extraction was performed using Robust Learning-Based Brain Extraction [32]. We then normalized the intensity values of each image to have zero mean and unit variance. To account for differences in image resolution, we resampled the images to isotropic 1 mm with linear interpolation. Similarly, the manual tumor segmentations were resampled using nearest neighbor interpolation.
MRI feature extraction
For each glioma case, we extracted 3 categories of features (histogram, shape and texture) from brain and tumor masks to maximize the characterization of the tumor. Eighteen types of histogram features were calculated: variance, skewness, kurtosis, standard deviation, maximum, mean, mean absolute deviation, minimum, range, root mean squared, uniformity, max probability, entropy, and intensity percentiles (10, 25, 50, 75, 90). Ten types of shape features were calculated: volume, number of centroids, surface area, compactness (calculated 2 different ways), solidity, maximum diameter, spherical disproportion, sphericity, and surface area to volume ratio. The ratios of different shape features were then calculated and also inputted as additional shape features. Types of texture features implemented included 8 texture features from the Gray-Level Co-occurrence Matrix (GLCM), 13 texture features from the Gray-Level Run-Length Matrix (GLRLM), 13 texture features from the Gray-Level Size Zone Matrix (GLSZM), and 5 texture features from the Neighborhood Gray-Tone Difference Matrix (NGTDM). Before automated features were calculated, the intensity of MR images was normalized to adjust for any differences in acquisition parameters. The intensity was normalized first by identifying the normal brain for each patient (non-skull and non-tumor regions, including both white matter and gray matter), followed by dividing the image intensity of the tumor region by the interquartile range of intensity values within the normal brain region. The magnetic field strength, resolution, and slice thickness, echo time and repetition time of pre-operative MR images from 4 institutions are attached in Supplementary Figs. 1, 2 and 3.
Random forest classification
The machine-learning procedure was performed using the Statistics and Machine Learning Toolbox (MATLAB 2015a Natick, MA). A random forest algorithm was used to classify patients as either IDH wild type or mutant. Random forest is one of several machine-learning algorithms that have been applied in clinical classification problems. It is especially advantageous when the number of predictor variables greatly exceeds sample size because it is resistant to overfitting [33]. Furthermore, it provides an estimation of generalizability though calculation of out-of-bag error, defined as the mean prediction error in each patient within the training cohort, using only the trees that did not have that patient in the bagged sample [34]. In this study, the training cohort (HUP/BWH/MGH) was used to develop the learning model, while cases in the validation cohort (TCGA) were used to independently evaluate the performance of the final model. The predictive value of each MRI feature for determining IDH genotype and 1p19q codeletion was calculated individually by area under the receiver operating characteristic curve (AUROC) analysis. All the MRI extracted features were used in our training algorithm. The heatmap of Pearson correlations among all these features was attached as Supplementary Fig. 4. The tree depth was set to 64, and the algorithm was set to grow to a total of 4096 trees, a number empirically determined to be a reasonable upper bound in our learning models and before which the training set classification error commonly begins to converge [35]. Finally, the model was tested on the validation cohort using the same model score threshold selected based on AUROC analysis of the training cohort. Figure 2 provides an overview of our MRI processing pipeline.
Fig. 2.
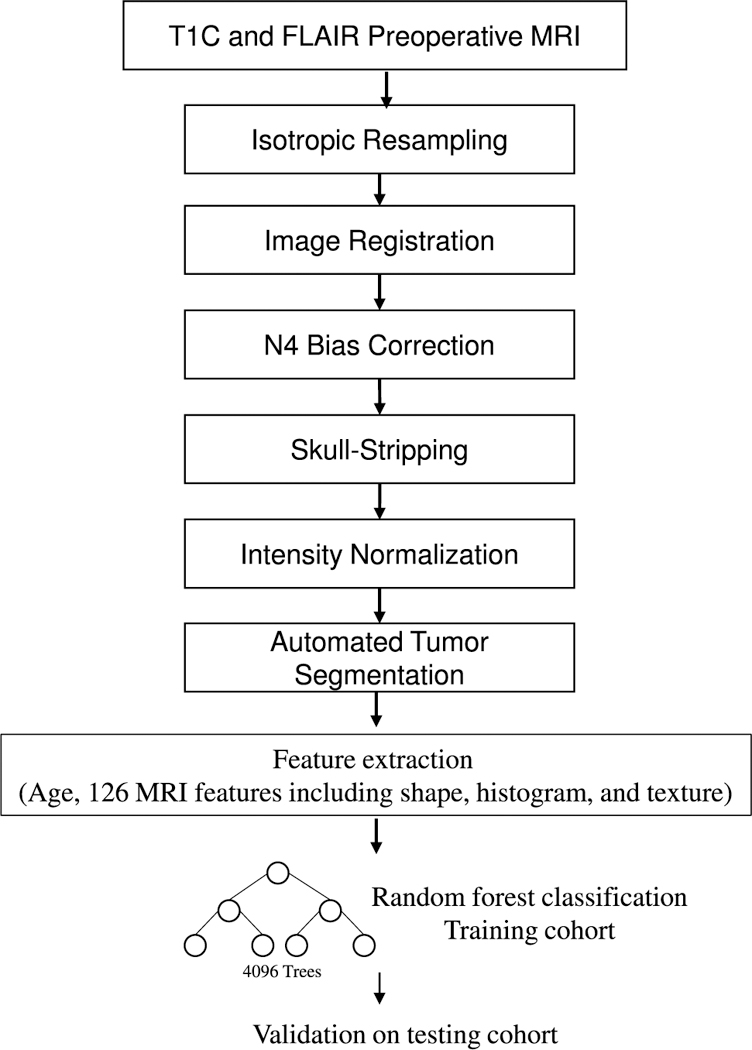
MRI feature extraction and machine-learning pipeline
Features with most significant contributions to the final model were determined by the increase in prediction error if the values of that feature were permuted across the out- of-bag observations (a method of measuring the prediction error of random forests models utilizing bootstrap aggregating (bagging) to sub-sample data samples used for training). This measure was computed for every tree and then averaged over the entire ensemble and divided by the standard deviation over the entire ensemble [36].
Results
Patient characteristics
The median age of the HUP, BWH, MGH, and TCIA cohorts were 53, 48, 52, and 53 years respectively. The percentage of males was 53%, 56%, 60%, and 56%, respectively. The HUP cohort was 22% grade II, 34% grade III, and 42% grade IV. The BWH cohort was 20% grade II, 29% grade III, and 51% grade IV. The MGH cohort was 13% grade II, 36% grade III, and 51% grade IV. The TCIA cohort was 22% grade II, 28% grade III, and 50% grade IV. Collectively, the HUP, BWH, MGH, and TCIA cohorts were 37%, 55%, 48%, and 41% IDH-mutant, respectively. IDH mut-codel gliomas accounted for 10%, 25%, 13% and 12% of HUP, BWH, MGH, and TCIA cohorts (see Table 1).
Table 1.
Summary of patient characteristics
| Cohort | BWH | MGH | HUP | TCIA |
|---|---|---|---|---|
| n | 156 | 155 | 227 | 206 |
| Age, mean: years, (range) | 48;18–85 | 52; 22–86 | 53; 18–88 | 53; 20–84 |
| Sex: n, male (%) | 88 (56%) | 93 (60%) | 122 (53%) | 116 (56%) |
| Grade II, n (%) | 31 (20%) | 19 (13%) | 51 (22%) | 45 (22%) |
| Grade III, n (%) | 46 (29%) | 56 (36%) | 79 (34%) | 58 (28%) |
| Grade IV, n (%) | 79 (51%) | 79 (51%) | 97 (42%) | 103 (50%) |
| IDH mutated, n (%) | 87 (55%) | 74 (48%) | 89 (37%) | 85 (41%) |
| IDH wild type, n (%) | 69(45%) | 81(52%) | 138(63%) | 121(59%) |
| IDH mutant-1p19q codel, n (%) | 39 (25%) | 20 (13%) | 24 (10%) | 25 (12%) |
| IDH mutant-1p19q noncodel, n (%) | 39 (25%) | 37 (24%) | 38 (17%) | 59 (28%) |
| IDH mutant-1p19q unknown, n (%) | 9 (6%) | 17 (11%) | 27 (12%) | 1 (0.4%) |
MRI and clinical features
From each patient’s imaging, a total of 126 features (histogram, shape and texture) were extracted. Together with one clinical feature (age), they were all used in the training algorithm.
IDH genotype prediction
The model (Model 1) using age and imaging features achieved an area under the receiver operating characteristic curve (AUC) of 0.921 (95% CI 0.894–0.941) in the training cohort and 0.919 (95% CI 0.871–0.949) in the validation cohort. Age offered the highest predictive value, followed by shape features. The 15 features that contributed most to our model are shown in Table 2. Based on the top 15 features, a model was generated and achieved the AUC of 0.917 (95% CI 0.889–0.938) in the training cohort and 0.916 (95% CI 0.867–0.947) in the validation cohort, respectively. To assess the impact of imaging features alone, we also built a model excluding age; this model achieved an AUC of 0.870 (95% CI 0.836–0.897) in the training cohort and 0.886 (95% CI 0.827–0.926) in the validation cohort.
Table 2.
Top 15 features contributing to the predicting model of IDH
| Single var AUC testing cohort |
Measure of importance |
|
|---|---|---|
| Age | 0.8475 | 2.9742 |
| Shape_Volume | 0.8475 | 1.0796 |
| Shape_N umberCentroids | 0.8568 | 0.5361 |
| Shape_MaxAxialDiameter | 0.8085 | 0.4336 |
| Shape_Solidity | 0.7536 | 0.3998 |
| FLAIR_Histogram_Skewness | 0.7697 | 0.3994 |
| Shape_SurfaceArea | 0.7833 | 0.3892 |
| FLAIR_Histogram_Kurtosis | 0.6237 | 0.3281 |
| Shape_Compactness | 0.7681 | 0.3185 |
| FLAIR_Histogram_Range | 0.5816 | 0.3175 |
| FLAIR_Histogram_Entropy | 0.6676 | 0.3107 |
| Shape_SphericalDisproportion | 0.699 | 0.279 |
| FLAIR_Histogram_MaximumProbability | 0.5301 | 0.2642 |
| FLAIR_GLSZM_HGZE | 0.5463 | 0.2371 |
| Shape_Sphericity | 0.6785 | 0.2355 |
1P19q genotype prediction
For IDH mutant subset, we trained a model (Model 2) integrating age to predict 1p/19q status using patients from MGH, BWH, HUP (total N = 197) and tested the model by samples from TCIA (total N = 84). The model achieved a training AUC of 0.685 (95% CI 0.606–0.754) and testing AUC of 0.716 (95% CI 0.577–0.823). Age offered the highest predictive value, followed by histogram features. The 15 features that contributed most to our model are shown in Table 3. Based on the top 15 features, a model was generated and achieved the AUC of 0.670 (95% CI 0.589–0.743) in the training cohort and 0.692 (95% CI 0.543–0.801) in the validation cohort, respectively. To assess the impact of imaging features alone, we also built a model excluding age; this model achieved an AUC of 0.681 (95% CI 0.603–0.751) in the training cohort and 0.690 (95% CI 0.549–0.803) in the validation cohort.
Table 3.
Top 15 features contributing to the predicting model of 1p19q
| Single var AUC testing cohort |
Measure of importance |
|
|---|---|---|
| Age | 0.5986 | 0.0994 |
| T1C_Histogram_Percentile50 | 0.5444 | 0.0945 |
| T1C_Histogram_Percentile75 | 0.6034 | 0.0926 |
| T1C_Histogram_Percentile25 | 0.6719 | 0.0879 |
| T1C_Histogram_Uniformity | 0.5569 | 0.0859 |
| T1C_GLRLM_SRHGE | 0.5505 | 0.0854 |
| T1C_GLRLM_HGRE | 0.5051 | 0.0835 |
| T1C_GLRLM_LRLGE | 0.5763 | 0.075 |
| T1C_Histogram_MeanAbsoluteDeviation | 0.6769 | 0.0716 |
| FLAIR_Histogram_Percentile90 | 0.5614 | 0.0655 |
| FLAIR_GLCM_AutoCorrelation | 0.638 | 0.0654 |
| Shape_Max3DDiameter | 0.5559 | 0.0621 |
| FLAIR_GLSZM_SZLGE | 0.5946 | 0.0534 |
| T1C_GLSZM_HGZE | 0.5614 | 0.0532 |
| T1C_Histogram_Minimum | 0.5925 | 0.0512 |
Three-group prediction
Two separate models (Model 1 and Model 2 as stated above) were applied sequentially to the TCIA testing data set (N = 198, those with both IDH and 1p/19q status available): Model 1 for prediction of IDH status followed by model 2 for prediction of 1p/19q status using IDH mutants (N = 71) predicted by the first model output. This generates 3 predictions: IDHwt, IDHmut-codel, IDHmut-non-codel. The overall accuracy for the 3 group prediction was 78.2% (155 correctly predicted out of 198).
Discussion
Several studies have reported an association between radiographic appearance and genotypes of gliomas [37–40]. Metellus et al. showed that IDH wild type in grade II gliomas were associated with an infiltrative pattern on MRI [41]. Qi et al. demonstrated that IDH mutants have a characteristic appearance on imaging, including sharp tumor margins and homogenous signal intensity [42]. Through MR imaging features of 175 LGGs reviewed by using Visually AcceSAble Rembrandt Images(VASARI), Park et al. built a model predictive of IDH1 mutation with AUC of 0.859 and 0.788 in the discovery and validation sets [43]. However, one major limitation in these studies is that they requires manual rating thus rater variability is inevitable. Using clinical and imaging features in a random forest model to predict IDH genotype in 120 high-grade gliomas, Zhang et al. reported accuracies of 86% in the training cohort and 89% in the validation cohort derived from a single-institution data set [44]. Yu et al. built a radiomic model to predict IDH mutation in grade II gliomas using only T2-weighted imaging and reported an accuracy of 80% in the primary cohort and 83% on the validation cohort [45]. In a recent study, Hao et al. used routine preoperative MRIs of 165 patients from the TCGA/TCIA to generate texture features predicting IDH mutation and 1p19q status with an accuracy of 0.86 and 0.96 within the same data set [46]. Akkus et al. presented machine intelligence to predict 1p19q status of 159 LGGs from post-contrast T1- and T2-weighted MR images using convolutional neural networks (CNN) and achieved accuracy of 87.7% [47]. However, none of these studies has attempted to predict genotype of gliomas in a combined cohort of diffuse gliomas from multi-institutional data and all of them lack external independent validation.
In the current study, using data from multiple institutions, a random forest model integrating preoperative multimodal automated imaging features and age was constructed to predict IDH and 1p19q genotype in grade II-IV gliomas. This model relied only on standard-of-care preoperative MRI including contrast enhanced T1-weighted sequence and T2 FLAIR sequence. The model predictive of IDH achieved high area under the receiver operating characteristic curve (AUC) of 0.921 in the training cohort and 0.919 in the validation cohort. For model predicting 3 group prediction (IDHwt, IDHmut-codel, IDHmut-non-codel), however, the overall accuracy was moderate (78.2%). For model predictive of 1p19q, the AUC was 0.685 in training set and 0.716 in validation set. We believe that this is likely due to sample size and should improve with larger data set available in the future.
While WHO grade information was available, we did not include this data in our model in order to understand the predictive accuracy based on pre-operative data alone. The only non-imaging feature included in the final model is patient age which is available to the radiologists preoperative and remains a strong feature correlating with IDH mutation and 1p19q status.
The predictive potential of individual imaging features was assessed in univariate and multivariate analysis, which was outperformed by the model constructed using machine learning methods. This demonstrates the ability of machine learning techniques to synergistically integrate features to discover distinct imaging phenotypes. In addition, the predictive model based on clinical and automated features derived from multi-institutional data was successfully validated on an independent multicenter patient cohort from the TCGA/TCIA, demonstrating the generalizability of our model.
The random forest method was chosen over other machine learning methods because of its high stability, generalizability, and predictive performance compared with other popular machine-learning classification methods. In a radiomic study by Chintan et al., 12 popular machine-learning classification methods arising from 12 classifier families (Bagging(BAG), Bayesian (BY), Boosting (BST), Decision trees (DT), Discriminant analysis (DA), Generalized linear models (GLM), Multiple adaptive regression splines (MARS), Nearest neighbors (NN), Neural networks (Nnet), Partial least square and principle component regression (PLSR), Random forests (RF), and Support vector machines (SVM)) were evaluated and compared in terms of their predictive performance and stability against data perturbation. Random forests (RSD = 3.52%, AUC = 0.66 ± 0.03) was proven to have the highest prognostic performance with high stability against data perturbation. The current study has the largest sample size and independent validation of results among all studies investigating different machine-learning modeling methods for radiomics based clinical predictions [48]. Moreover, RF has displayed high predictive performance in several other biomedical and other domain applications as well [49].
The features that contributed the most to our automated model predictive of IDH were predominantly age and shape features. Among the 15 features that contributed the most to the model, 9 were derived from the shape features. The high predictive values of shape features reflected the differences in growth pattern between IDH mutants and wild type. Unlike in our model, texture features were the top-performing features in the prediction of IDH of high grade gliomas in the study by Zhang et al. [24]. This may be caused by the difference of tumor grade ratio enrolled between two studies (70.8% of tumors are WHO grade IV gliomas in their study while 47.4% in our study population). Although the precise relationship between automated imaging features and the biology of IDH mutations remains unclear, our results provided quantitative insight into intratumor heterogeneity and behavior of tumor margins.
Even though our study constructed a model using conventional MRI imaging that are routinely obtained pre-operatively to predict IDH status with a high accuracy, we acknowledge several limitations to our study. Due to the retrospective study design where only known gliomas were selected, our model cannot readily be applied to situations where other tumor types as well as non-tumor mimickers are also common in the study population. The current model is most applicable for patients who have high likelihood of primary glioma based on standard radiographic evaluation. A more general model using data set from other lesion types will be needed to further improve generalizability. Our study also did not utilize advanced MR modalities such as perfusion MRI and MR-spectroscopy, which have been demonstrated to have potential in the prediction of IDH genotype [19, 21, 23]. Future studies combining conventional and advanced imaging may further improve the accuracy of noninvasive IDH status prediction. Furthermore, our work to continue to enlarge our sample size is ongoing. We believe that the accuracy of 1p19q prediction should improve when larger data set is available. Finally, although our combined model achieved successful prediction of IDH status, the biological significance of the individual features remains unknown and require further evaluation.
In conclusion, using machine-learning algorithms, high accuracy was achieved in the prediction of IDH genotype in gliomas and moderate accuracy in a three-group prediction including IDH genotype and 1p19q codeletion. Our model may have the potential to serve as a noninvasive tool that complements invasive tissue sampling and guiding patient management at an earlier stage of disease.
Supplementary Material
Acknowledgements
This work was supported by the Natural Science Foundation of China (81301988 to L.Y.), Shen-ghua Yuying Project of Central South University to L.Y. and the Natural Science Foundation of Hunan Province for Young Scientists, China (Grant No: 2018JJ3709 to Li Yang). This project was supported by a training grant from the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health under Award Number 5T32EB1680 to K. Chang. This project was supported by the RSNA research fellow Grant to H.X.B (RF1802), SIR Foundation resident research grant to H.X.B, and Research Fund for International Young Scientist by the National Natural Science Foundation of China (818580410556 to H.X.B.). This research was carried out in whole or in part at the Athinoula A. Martinos Center for Biomedical Imaging at the Massachusetts General Hospital, using resources provided by the Center for Functional Neuroimaging Technologies, P41EB015896, a P41 Biotechnology Resource Grant supported by the National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health.
Funding This work was supported by the Natural Science Foundation of China (81301988 to L.Y.), Shenghua Yuying Project of Central South University to L.Y. Project supported by the Natural Science Foundation of Hunan Province for Young Scientists, China (Grant No: 2018JJ3709 to Li Yang). This project was supported by a training grant from the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health under Award Number 5T32EB1680 to K. Chang. This project was supported by the RSNA research fellow grant to H.X.B (RF1802), SIR Foundation resident research grant to H.X.B, and Research Fund for International Young Scientist by the National Natural Science Foundation of China (818580410556 to H.X.B.).
Footnotes
Data availability The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Compliance with ethical standards
Ethical approval For this type of study, formal consent is not required.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s11060–019-03096–0) contains supplementary material, which is available to authorized users.
Extended author information available on the last page of the article
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Ostrom QT et al. (2014) The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol 16(7):896–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi WL, Beroukhim R (2014) Beating the odds: extreme long-term survival with glioblastoma. Neuro Oncol 16(9):1159–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Recht LD, Lew R, Smith TW (1992) Suspected low-grade glioma: is deferring treatment safe? Ann Neurol 31(4):431–436 [DOI] [PubMed] [Google Scholar]
- 4.Parsons DW et al. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321(5897):1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckel-Passow JE et al. (2015) Glioma Groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 372(26):2499–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartmann C et al. (2010) Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol 120(6):707–718 [DOI] [PubMed] [Google Scholar]
- 7.Houillier C et al. (2010) IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology 75(17):1560–1566 [DOI] [PubMed] [Google Scholar]
- 8.Louis DN et al. (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820 [DOI] [PubMed] [Google Scholar]
- 9.Weller M et al. (2013) Molecular neuro-oncology in clinical practice: a new horizon. Lancet Oncol 14(9):e370–e379 [DOI] [PubMed] [Google Scholar]
- 10.SongTao Q et al. (2012) IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci 103(2):269–273 [DOI] [PubMed] [Google Scholar]
- 11.Molenaar RJ et al. (2015) Radioprotection of IDH1-mutated cancer cells by the IDH1-mutant inhibitor AGI-5198. Cancer Res 75(22):4790–4802 [DOI] [PubMed] [Google Scholar]
- 12.Mohrenz IV et al. (2013) Isocitrate dehydrogenase 1 mutant R132H sensitizes glioma cells to BCNU-induced oxidative stress and cell death. Apoptosis 18(11):1416–1425 [DOI] [PubMed] [Google Scholar]
- 13.Lowery MA et al. (2017) Phase I study of AG-120, an IDH1 mutant enzyme inhibitor: results from the cholangiocarcinoma dose escalation and expansion cohorts. J Clin Oncol 35(15_suppl):4015–4015 [Google Scholar]
- 14.Mellinghoff IK et al. (2016) ACTR-46. AG120, a first-in-class mutant IDH1 inhibitor in patients with recurrent or progressive IDH1 mutant glioma: results from the phase 1 glioma expansion cohorts. Oxford University Press, Oxford [Google Scholar]
- 15.Erdem-Eraslan L et al. (2013) Intrinsic molecular subtypes of glioma are prognostic and predict benefit from adjuvant procarbazine, lomustine, and vincristine chemotherapy in combination with other prognostic factors in anaplastic oligodendroglial brain tumors: a report from EORTC study 26951. J Clin Oncol 31(3):328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cairncross G et al. (2013) Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol 31(3):337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang L et al. (2018) Reduced expression of DNA repair genes and chemosensitivity in 1p19q codeleted lower-grade gliomas. J N euro-Oncol. 10.1007/s11060-018-2915-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brat DJ et al. (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372(26):2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kickingereder P et al. (2015) IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci Rep 5:16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biller A et al. (2016) Improved brain tumor classification by sodium MR imaging: prediction of IDH mutation status and tumor progression. AJNR Am J Neuroradiol 37(1):66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pope WB et al. (2012) Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol 107(1):197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andronesi OC et al. (2012) Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med 4(116):116ra4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi C et al. (2012) 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med 18(4):624–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stadlbauer A et al. (2017) MR imaging-derived oxygen metabolism and neovascularization characterization for grading and IDH gene mutation detection of gliomas. Radiology 283(3):799–809 [DOI] [PubMed] [Google Scholar]
- 25.Iwadate Y et al. (2016) Molecular imaging of 1p/19q deletion in oligodendroglial tumours with 11C-methionine positron emission tomography. J Neurol Neurosurg Psychiatry 87:1016–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fellah S et al. (2013) Multimodal MR imaging (diffusion, perfusion, and spectroscopy): is it possible to distinguish oligodendroglial tumor grade and 1p/19q codeletion in the pretherapeutic diagnosis? Am J Neuroradiol 34(7):1326–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansen NL et al. (2012) Prediction of oligodendroglial histology and LOH 1p/19q using dynamic [18F] FET-PET imaging in intracranial WHO grade II and III gliomas. Neuro-oncology 14(12):1473–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark K et al. (2013) The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26(6):1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. https://wiki.cancerimagingarchive.net/display/Public/TCGA-LGG#0e9decd5f9664ff990f9a9d7e5c43631.
- 30.Chang K et al. (2016) Multimodal imaging patterns predict survival in recurrent glioblastoma patients treated with bevacizumab. Neuro Oncol 18(12):1680–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fedorov A et al. (2012) 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 30(9):1323–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iglesias JE et al. (2011) Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans Med Imaging 30(9):1617–1634 [DOI] [PubMed] [Google Scholar]
- 33.Breiman L (2001) Random forests. Mach Learn 45(1):5–32 [Google Scholar]
- 34.James G et al. (2013) An introduction to statistical learning, vol 112 Springer, New York [Google Scholar]
- 35.Hanley JA, McNeil BJ (1983) A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology 148(3):839–843 [DOI] [PubMed] [Google Scholar]
- 36.Eoli M et al. (2006) Reclassification of oligoastrocytomas by loss of heterozygosity studies. Int J Cancer 119(1):84–90 [DOI] [PubMed] [Google Scholar]
- 37.Megyesi JF et al. (2004) Imaging correlates of molecular signatures in oligodendrogliomas. Clin Cancer Res 10(13):4303–4306 [DOI] [PubMed] [Google Scholar]
- 38.Jenkinson MD et al. (2006) Histological growth patterns and genotype in oligodendroglial tumours: correlation with MRI features. Brain 129(Pt 7):1884–1891 [DOI] [PubMed] [Google Scholar]
- 39.Brown R et al. (2008) The use of magnetic resonance imaging to noninvasively detect genetic signatures in oligodendroglioma. Clin Cancer Res 14(8):2357–2362 [DOI] [PubMed] [Google Scholar]
- 40.Liu C et al. (2012) Towards MIB-1 and p53 detection in glioma magnetic resonance image: a novel computational image analysis method. Phys Med Biol 57(24):8393–8404 [DOI] [PubMed] [Google Scholar]
- 41.Metellus P et al. (2010) Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol 120(6):719–729 [DOI] [PubMed] [Google Scholar]
- 42.Qi S et al. (2014) Isocitrate dehydrogenase mutation is associated with tumor location and magnetic resonance imaging characteristics in astrocytic neoplasms. Oncol Lett 7(6):1895–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park Y et al. (2018) Prediction of IDH1-mutation and 1p/19q-codeletion status using preoperative MR imaging phenotypes in lower grade gliomas. Am J Neuroradiol 39(1):37–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang B et al. (2017) Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol 19(1):109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu J et al. (2017) Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur Radiol 27(8):3509–3522 [DOI] [PubMed] [Google Scholar]
- 46.Zhou H et al. (2017) MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol 19(6):862–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akkus Z et al. (2017) Predicting deletion of chromosomal arms 1p/19q in low-grade gliomas from MR images using machine intelligence. J Digit Imaging 30(4):469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parmar C et al. (2015) Machine learning methods for quantitative radiomic biomarkers. Sci Rep 5:13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandez-Delgado M et al. (2014) Do we need hundreds of classifiers to solve real world classification problems? J Mach Learn Res 15(1):3133–3181 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


