Significance
p180/RRBP1 is an essential factor for high-rate protein synthesis on the endoplasmic reticulum (ER) in terminally differentiated secretory cells. A unique system for selectively enhanced translation has been proposed for p180-associated polyribosomes. Here we provide evidence that SF3b4, an RNA-binding protein in the splicing factor, plays a key and unexpected role in translation control as a cofactor for p180. It appears that abundant expression of both SF3b4 and p180 is critical for increased ER mRNA targeting, and consequently leads to the heavy polyribosome assembly. This discovery could shed light on the pathogenesis of Nager syndrome caused by SF3B4 mutation, as such mutations may result in abnormal collagen biosynthesis, potentially affecting bone morphogenesis in patients with this disease.
Keywords: p180, SF3b4, polyribosome, 5′ UTR, endoplasmic reticulum
Abstract
One of the morphological hallmarks of terminally differentiated secretory cells is highly proliferated membrane of the rough endoplasmic reticulum (ER), but the molecular basis for the high rate of protein biosynthesis in these cells remains poorly documented. An important aspect of ER translational control is the molecular mechanism that supports efficient use of targeted mRNAs in polyribosomes. Here, we identify an enhancement system for ER translation promoted by p180, an integral ER membrane protein we previously reported as an essential factor for the assembly of ER polyribosomes. We provide evidence that association of target mRNAs with p180 is critical for efficient translation, and that SF3b4, an RNA-binding protein in the splicing factor SF3b, functions as a cofactor for p180 at the ER and plays a key role in enhanced translation of secretory proteins. A cis-element in the 5′ untranslated region of collagen and fibronectin genes is important to increase translational efficiency in the presence of p180 and SF3b4. These data demonstrate that a unique system comprising a p180–SF3b4–mRNA complex facilitates the selective assembly of polyribosomes on the ER.
Protein entry into the secretory pathway is initiated at the rough endoplasmic reticulum (ER), which is essential for intracellular transport of proteins to secretory compartments within the cell, and ultimately to the extracellular environment. One of the morphological hallmarks of “professional” secretory cells is the prominent proliferation of rough ER membranes densely occupied by polyribosomes (also known as polysomes). Such prominent proliferation is observed in both tissues (1, 2) and cultured cells (3, 4), but the molecular basis for this enhanced biosynthesis is poorly understood. One important issue regarding translational control on the ER is the molecular mechanism that ensures an efficient mRNA supply. In addition to classical mRNA delivery via the signal recognition particle (SRP)-dependent pathway, recent reports have provided substantial evidence for alternative routes, including translation- and SRP-independent mRNA targeting to the ER (reviewed in refs. 5–7) and ribosome-independent mRNA association with the ER (8, 9). However, little information is available on which pathways are used to ensure efficient biosynthesis in secretory cells. Studies on noncanonical mRNA association with the ER highlight the intrinsic complexity of translation at the ER (9–12).
Another important parameter of translational control on the ER is the degree of polyribosome assembly, which can directly affect the efficiency of translation (3). When ER-associated polyribosomes produce secretory proteins, it is assumed that higher-order coordination between each unit of a ribosome/translocon complex is essential for synchronized translation and subsequent translocation across the membrane. Such highly controlled organization is especially important for heavy polyribosomes encoding large proteins such as collagen. One important factor for heavy polyribosome formation is p180, an ER membrane protein originally identified as a ribosome receptor (13). p180 is abundantly expressed in a variety of professional secretory cells (14, 15) and comprises multiple domains involved in various functions, such as microtubule bundling (16) and association with a ribosomal subunit or translocon (3, 17, 18). Our previous report suggested that mRNAs forming polyribosomes at the ER are segregated into p180-dependent and p180-independent mRNAs (3). mRNAs belonging to the p180-dependent genes include COL1A1 and FN1, and p180 is required to form actively translating polyribosomes. In contrast, p180 depletion does not affect polyribosomes of CANX, MMP2, and TIMP1, which are classified as p180-independent genes (3). It was also shown that p180 depletion selectively inhibits collagen biosynthesis without reducing ER-associated mRNAs (17). However, the exact mechanism underlying specificity remains to be clarified. We previously presumed that an unknown factor or factors mediates the specific interaction between p180 and the target mRNAs (3, 17), in contrast to the direct binding of p180 to mRNAs, as previously reported (19).
Here, we provide evidence that p180 specifically interacts with a spliceosome component, SF3b4, which is a major constituent of the SF3b complex in the U2 small nuclear ribonucleoprotein particle. The specific association of p180 and SF3b4 at the ER appears to provide a very unique platform for up-regulating translation, thereby leading to a high rate of biosynthesis in secretory cells. Recently, haploinsufficiency of the SF3B4 gene was identified as a genetic cause of Nager syndrome, an autosomal genetic disorder resulting in acrofacial dysostosis and limb malformations (20). Our unexpected finding of a function of SF3b4 in collagen biosynthesis may provide crucial insight into the pathogenesis of Nager syndrome.
Results
The C-Terminal Coiled–Coil Domain of p180 Mediates Target mRNA Association to Promote Efficient Protein Biosynthesis.
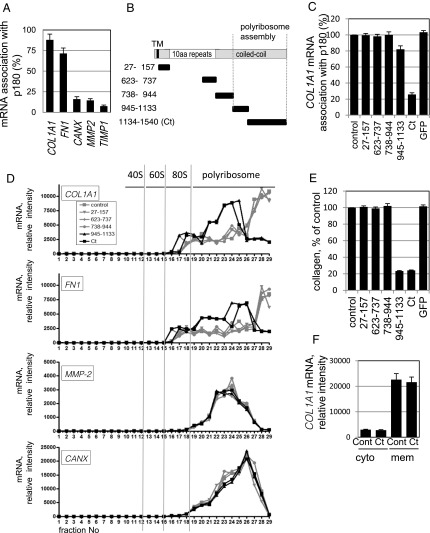
To explore the specific role of p180 in actively translating polyribosomes, mRNA association with p180 was examined using collagen-secreting human embryonic lung fibroblast (HEL) cells (3, 17). Ribosome-stripped membrane fractions were immunoprecipitated with an anti-p180 antibody, and mRNAs in the immunoprecipitates were quantified (3). For collagen and fibronectin mRNAs, the majority of input mRNAs were immunoprecipitated with the anti-p180 antibody (88% and 72% for COL1A1 and FN1, respectively; Fig. 1A), whereas minimal amounts (<0.5% of input) were detected in control Ig fractions. In contrast, <20% of input mRNAs encoding CANX, MMP2, and TIMP1 were recovered with the anti-p180 antibody (Fig. 1A). To identify the domain responsible for mRNA association, a series of truncated p180 polypeptides was overexpressed in the cytosol of HEL cells (Fig. 1B and SI Appendix, Fig. S1A), and the effects on mRNA association with p180 were examined. We found that overexpression of the C-terminal peptide (aa 1,134–1,540, hereafter designated Ct) prevented efficient COL1A1 mRNA association with p180, whereas the other polypeptides, including the N-terminal and repeat domains, had no significant effect (Fig. 1C).
Fig. 1.
The C-terminal coiled–coil domain of p180 associates with target mRNAs and promotes efficient protein synthesis. (A) Ribosome-stripped membrane fractions from ascorbate-treated HEL cells were immunoprecipitated with an anti-p180 antibody. The relative amounts of mRNA were analyzed for COL1A1, FN1, CANX, MMP2, and TIMP1 cDNA recovered in the p180 immunoprecipitates; percentages of the input value are depicted. Data represent means ± SD (n = 4). (B) Structures of full-length p180 and a series of truncated proteins are illustrated. TM, transmembrane domain; white box, a highly basic tandem repeat domain; gray box, a C-terminal acidic coiled–coil domain. Each truncated mutant protein contains the indicated amino acid residues of human p180. (C) The series of green fluorescent protein (GFP)-tagged p180 truncated polypeptides shown in B were expressed in HEL cells. Relative mRNA amounts (vs. mock-transfected cells, set as 100%) recovered in p180 immunoprecipitates of the membrane fractions are depicted. (D) Analyses from sucrose density gradient centrifugation of the membrane fractions are shown. Relative amounts of the indicated mRNAs estimated by qPCR analysis are depicted. Total RNA profiles are shown in SI Appendix, Fig. S1B. (E) Collagen secreted from HEL cells overexpressing a series of polypeptides was quantified by MS analysis. The amount of collagen secreted from mock-transfected cells was set as 100%. (F) After overexpression of Ct as in C, the relative amounts of COL1A1 mRNA in the cytosolic and membrane fractions were compared. Data in C, E, and F represent means ± SD (n = 3).
To further examine the role of mRNA-p180 association during active translation, mRNAs were monitored using fractionated polyribosomes after density gradient centrifugation. Ct overexpression induced a prominent shift of COL1A1 and FN1 mRNAs: the extremely heavy polyribosomes containing COL1A1 mRNAs (Fig. 1D, fraction nos. 27–29) shifted to lighter fractions (fraction nos. 23–25), suggesting that Ct perturbed translational initiation. Another polypeptide (aa 945–1,133), which affects polyribosome assembly irrespective of the C-terminal polypeptides (3), showed a similar effect. Parallel analyses revealed that both Ct and the aa 945–1,133 polypeptide impaired the efficiency of type I collagen protein synthesis to <25% that in control cells (Fig. 1E). Similar dominant-negative effects of the two polypeptides were observed for FN1 mRNAs, but not for CANX and MMP2 (Fig. 1D). It is notable that COL1A1 mRNAs remained in the membrane polyribosomes after Ct overexpression (Fig. 1 D and F), in contrast to marked reduction of total membrane polyribosomes (SI Appendix, Fig. S1B).
Identification of a Cofactor for p180.
The very acidic pI (∼4.6) of the Ct domain and the selective manner of the p180-mRNA association led us to predict that an unknown cofactor mediates this specific association. Our findings suggested that this cofactor binds to the Ct domain and recognizes the particular mRNA encoding COL1A1. Therefore, we postulated that transient association of the cofactor with the ER membrane would be perturbed after overexpression of cytoplasmic Ct.
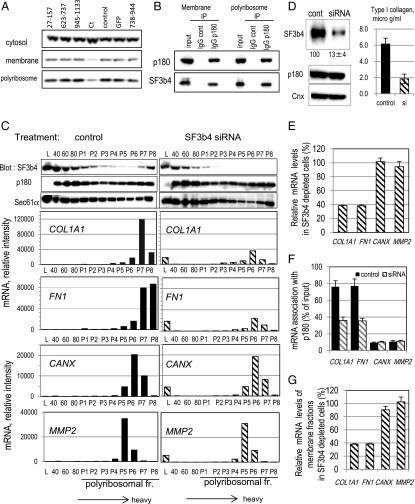
To isolate this candidate Ct-binding protein, we expressed and purified hexahistidine-tagged recombinant Ct in HEL cells (SI Appendix, Fig. S2A). Mass spectrometric analyses of trypsin-digested Ct fractions identified 15 copurified proteins that were completely absent in fractions from mock-transfected cells (SI Appendix, Table S1). Western blotting analyses revealed that five of the 15 proteins were present in membrane fractions (SI Appendix, Table S1). Of the five candidates, overexpression of Ct induced specific release of SF3b4 and eEF1A from the membranes, whereas the other three proteins remained membrane-associated on overexpression of any of the fragments (Fig. 2A and SI Appendix, Fig. S2B). Special attention was paid to SF3b4, a major component of the SF3b complex in the U2 small nuclear ribonucleoprotein particle essential for the general splicing process. SF3b4 has two RNA-recognition motifs (RRMs) in its N terminus, which potentially mediate selective mRNA binding. SF3b4 was clearly present in the membrane polyribosome fraction of collagen-secreting HEL cells (Fig. 2A). Moreover, immunoprecipitation experiments confirmed that SF3b4 specifically associated with p180 in the membrane polyribosome fraction (Fig. 2B and SI Appendix, Fig. S2F). Nuclease treatment did not disturb the interaction (SI Appendix, Fig. S2G).
Fig. 2.
SF3b4 in membrane polyribosomes is associated with p180 and cosediments with COL1A1 mRNA. (A) After overexpression of GFP-tagged truncated polypeptides of p180 in ascorbate-treated HEL cells (Fig. 1), the changes in SF3b4 levels in the cytosolic, membrane, and membrane polyribosome fractions were examined by Western blotting. (B) The membrane or membrane polyribosome fractions were immunoprecipitated with an anti-p180 antibody or control IgG and then analyzed for SF3b4 and p180 by Western blotting. Nuclease treatment did not affect these data (SI Appendix, Fig. S2G). (C) The membrane fractions were subjected to sucrose density gradient centrifugation and analyzed by Western blotting to monitor SF3b4 distribution. HEL cells treated with control siRNA (Left) or siRNA specific for human SF3b4 (Right) were used. Polyribosomal profiles of the respective mRNAs are depicted. 40, 40S subunit; 60, 60S subunit; 80, monosome; L, light fraction; P1–P8, polyribosome fractions. (D) Total cell lysates from HEL cells treated with control siRNA or siRNA specific for human SF3b4 were subjected to Western blotting. Relative amounts estimated by densitometric scanning are shown below the image. The amounts of secreted collagen were quantified by MS analysis. (E) Relative amounts of mRNA in total cell lysates of the SF3b4-depleted cells were quantified by qPCR. The corresponding value in control siRNA-transfected cells was set as 100%. (F) Membrane fractions were immunoprecipitated with an anti-p180 antibody (see also SI Appendix, Fig. S2F for verification of the immunoprecipitates). Relative amounts of mRNA in the p180 immunoprecipitates were quantified, and percentages of the input value are depicted. Black bars, control siRNA; shaded bars, siRNA specific for human SF3b4. (G) Effects of SF3b4 depletion on the amount of membrane mRNA estimated by qPCR are depicted (vs. control siRNA-transfected cells, set as 100%). Data in D–G represent means ± SD (n = 3).
To assess the role of SF3b4 in the association of mRNA with p180, polyribosome analyses were performed in combination with knockdown of endogenous SF3b4. Membranes fractionated by density gradient centrifugation were used to observe the distribution of SF3b4 and mRNAs. Surprisingly, SF3b4 protein was enriched in extremely heavy fractions near the bottom of the gradient, and cosedimented with COL1A1 and FN1 mRNAs in control cells (Fig. 2C, Left). Concomitantly, p180 and translocon components were abundantly present in these fractions (Fig. 2C and SI Appendix, Fig. S2E), as reported previously (3). Next, we down-regulated endogenous SF3b4 by siRNA transfection; this markedly decreased collagen secretion to 30% of that in control cells without apparent loss of other membrane markers (Fig. 2D and SI Appendix, Fig. S2C). Unlike Ct overexpression, the total and membrane amounts of COL1A1 and FN1 mRNAs decreased to ∼40% of those in control cells (Fig. 2 C and E), whereas the polyribosomal RNA profiles remained unchanged (SI Appendix, Fig. S2D). The reduced levels of COL1A1 mRNA on SF3b4 knockdown may be caused by disruption of mRNA maturation at the splicing process through loss of nuclear SF3b4. Alternatively, metabolism or nuclear-cytoplasmic shuttling may be disturbed by weak mRNA association with the ER in the absence of SF3b4, consequently leading to the degradation of COL1A1 mRNA. Notably, loss of SF3b4 decreased p180 association with COL1A1 and FN1 mRNAs to <40% of the input mRNAs (Fig. 2F) and resulted in reduced levels of polyribosomal mRNAs for COL1A1 and FN1 (Fig. 2C, Right). In contrast, no significant changes were observed in total and membrane mRNA levels or in the association of p180 with other mRNAs (Fig. 2 E–G), suggesting that SF3b4 depletion did not induce critical damage that perturbed general mRNA biosynthesis. These data suggest an intrinsic role for SF3b4 in efficient translation of p180-associated polyribosomes, thereby leading to a high rate of biosynthesis of the corresponding proteins.
SF3b4 and p180 Cooperatively Enhance Collagen Biosynthesis Dependent on the 5′ Untranslated Region Sequence of COL1A1 mRNA.
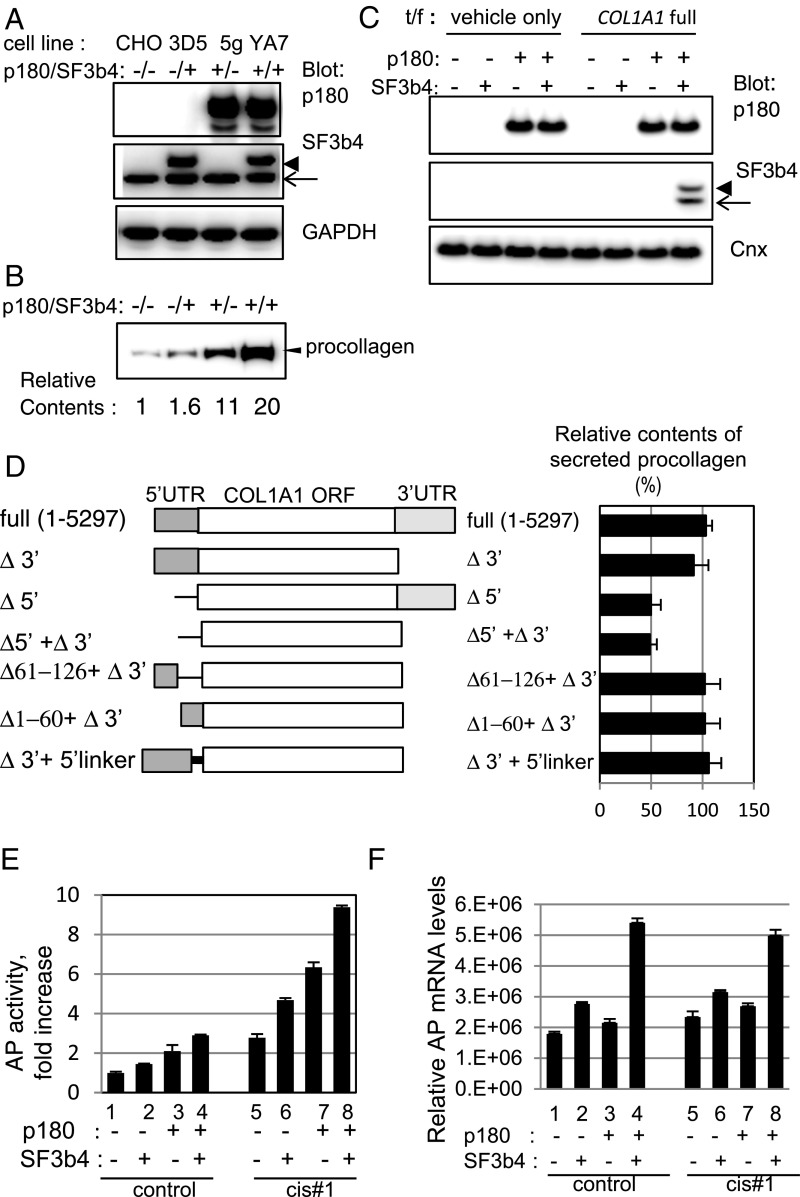
To examine the molecular basis of SF3b4 and p180 cooperation in collagen biosynthesis, double and single stable transfectants were generated in Chinese hamster ovary (CHO) cells lacking endogenous p180 (Fig. 3A), followed by transfection with a plasmid encoding human full-length COL1A1 cDNA. Concomitant overexpression of SF3b4 and p180 led to a 20-fold increase in procollagen secretion (Fig. 3B). COL1A1 mRNA was mainly membrane-associated in the double transfectant (clone YA7), but predominantly cytosolic in the single transfectants, as in the parental cells, although the sum of their cytosolic and membrane levels remained unchanged (SI Appendix, Fig. S3A, lanes 1–4). Importantly, membrane association of SF3b4 only occurred after transfection with COL1A1 (Fig. 3C), suggesting that a specific sequence in COL1A1 mRNA is required for SF3b4 transition to the membrane. Several mutant forms were examined to determine the cis-interacting sequence facilitating efficient collagen biosynthesis (Fig. 3D). Mutated plasmids lacking the 5′ untranslated region (5′ UTR; Δ5′ and Δ5′+Δ3′) decreased procollagen secretion by approximately half, whereas deletion of the 3′ UTR (Δ3′) had no significant effect (Fig. 3D). Interestingly, the polyribosome pattern of Δ5′+Δ3′ shifted to lighter fractions compared with that of cells harboring full-length COL1A1 cDNA (SI Appendix, Fig. S3B). Partial deletion mutants were examined to define effective regions within the 5′ UTR. Unexpectedly, neither of the deletion mutations (Δ1–60 + Δ3′ and Δ61–126 + Δ3′) affected collagen secretion (Fig. 3D). COL1A1 mRNA contains a stem–loop structure in a region spanning from the 5′ UTR to the ORF (21). Because a rigid stem–loop structure is considered to exert inhibitory effects on translation, a mutant gene that inhibited stem–loop formation was examined to rule out this possibility. The mutant gene (Δ 3′ + 5′ linker) did not activate collagen biosynthesis (Fig. 3D). Taken together, the present results suggest that multiple regions within the 5′ UTR (hereafter called cis#1) are important for efficient collagen secretion.
Fig. 3.
The 5′ UTR sequence in COL1A1 mRNA is important for enhanced translation in a stable cell line coexpressing p180 and SF3b4. (A) Protein expression levels in established CHO stable cell lines expressing p180 (clone 5g), SF3b4 (clone 3D5), or both proteins (clone YA7) were analyzed. p180 was not detected in control CHO and 3D5 cells. An arrow indicates positions for endogenous SF3b4, whereas an arrowhead indicates that of recombinant myc-tagged SF3b4; the same applies hereafter. (B) The four indicated cell lines were transfected with an expression vector encoding full-length procollagen cDNA of the COL1A1 gene. Secretion levels of procollagen were analyzed by Western blotting. Relative amounts estimated by densitometric scanning are shown below the image. (C) Membrane fractions were prepared from cells transfected with a vector encoding full-length COL1A1 or empty vector as indicated, and the p180 and SF3b4 levels in each fraction were compared. (D) Schematic structures of full-length and truncated mutants of human COL1A1 cDNA (Left). A mutant with a 13-nt (aagcttcgaattc) linker inserted at the initiation codon (5′-linker) was used to prevent stem–loop formation (21). Bar graphs (Right) represent relative procollagen secretion levels (%) in 3-d culture media from YA7 cells. (E) Cells were transfected with control reporter plasmid (lanes 1–4) or reporter plasmid containing cis#1 upstream of the ORF encoding AP (lanes 5–8). Relative secreted AP activity is shown (vs. the value in lane 1, set as 1). (F) Relative AP mRNA levels of the membrane fractions of the transfected cells are depicted.
To examine the general activity of cis#1, this element was inserted alone into a reporter plasmid encoding secreted alkaline phosphatase (AP). Addition of cis#1 upstream of the initiation codon resulted in an approximately threefold increase in AP activity (Fig. 3E, lanes 1 and 5). The highest enhancement was observed in YA7; approximately ninefold higher than that in control CHO cells lacking cis#1 (lanes 1 and 8). Membrane AP mRNA was increased in YA7 cells irrespective of cis#1 insertion (Fig. 3F and SI Appendix, Fig. S3C).
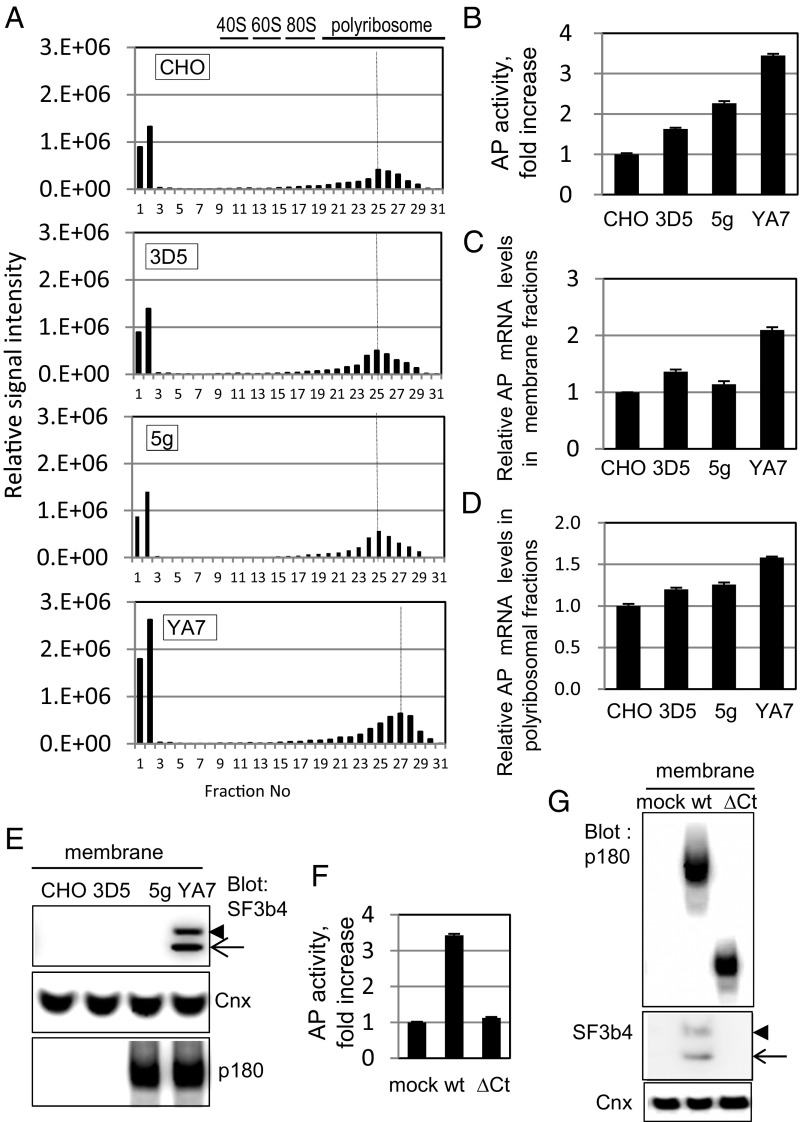
The molecular basis of efficient secretion in the presence of cis#1 element (Fig. 3E, lane 8) was further examined by comparing polyribosome profiles among SF3b4- or p180-overexpressing cells (clones 3D5 or 5g, respectively) and YA7 (Fig. 4 A–E). A significant peak shift to heavy fractions occurred only in YA7, and not in 3D5 or 5g (Fig. 4A). Consistently, a marked increase in AP activity (Fig. 4B) was observed only in YA7 cells, whereas mRNA levels of the membrane and the polyribosomal fractions increased only moderately (Fig. 4 C and D). It is notable that membrane recruitment of SF3b4 was found only in YA7, and not in 3D5 (Fig. 4E), although their cytosolic SF3b4 levels were similar (SI Appendix, Fig. S3D).
Fig. 4.
Assessment of molecular basis of efficient secretion in the presence of cis#1 element. (A–E) The four indicated cell lines were transfected with reporter plasmid containing cis#1 upstream of the ORF encoding AP as in Fig. 3E. (A) Polyribosomal profiles of AP cDNA of the membrane fractions of the indicated cells after sucrose density gradient centrifugation analysis are shown. (B) Relative secreted AP activity of the transfected cells is shown. (C and D) Relative AP mRNA levels of the membrane and the membrane polyribosomal fractions of the transfected cells are depicted, respectively. (E) Western blotting analysis of cytosolic and membrane fractions of the indicated cell lines are shown. Membrane localization of SF3b4 occurred in YA7 cells but not in 3D5 cells. (F) Expression plasmids of either the wild-type or deletion mutant (ΔCt) of p180 were transfected into 3D5 cells, and relative secreted AP activity in medium from the cells is shown. (G) Membrane fractions of the cells shown in F were analyzed by Western blotting. Data in B, C, D, and F represent means ± SD (n = 3).
We next examined the role of the p180 Ct domain on the ER association of SF3b4. Overexpression of wild-type p180 in 3D5 cells resulted in marked enhancement of AP activity (Fig. 4F) and SF3b4 transit to the membrane (Fig. 4G). In contrast, a p180 truncation mutant of the Ct domain (ΔCt) failed to increase secretion or membrane association of SF3b4 (Fig. 4 F and G), indicating that the Ct domain plays a critical role in SF3b4 localization to the ER.
Identification of 5′ UTR Motif Sequences Required for Efficient Protein Biosynthesis.
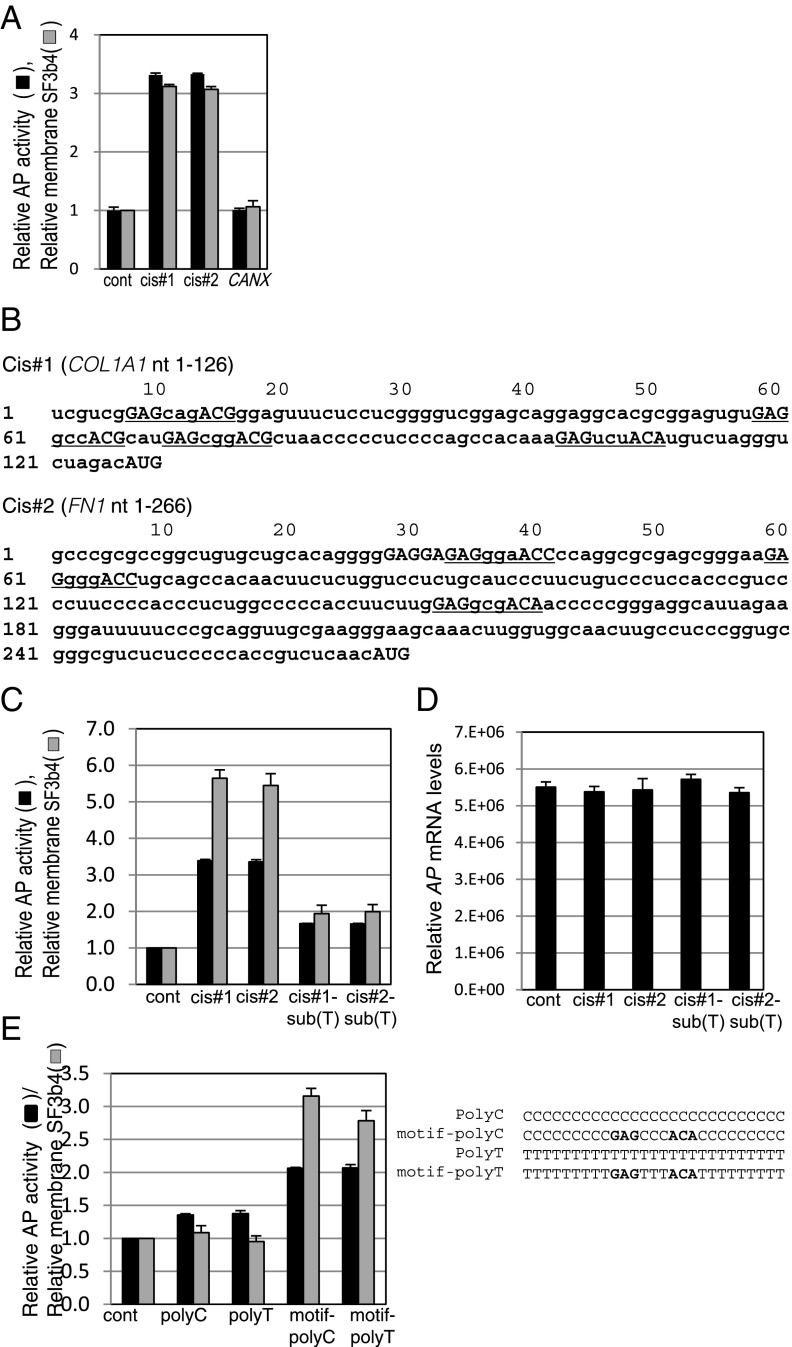
We also found another cis-acting sequence in human FN1 5′ UTR (266-mer, hereafter designated cis#2) that had an enhancing effect similar to cis#1, whereas no enhancement was seen with the CANX 5′ UTR (Fig. 5A). Consistently, increased membrane localization of SF3b4 was seen in the cells transfected with the reporter plasmids bearing cis#1 and cis#2 (Fig. 5A).
Fig. 5.
Identification of a motif sequence responsible for enhanced biosynthesis. (A) YA7 cells were transfected with reporter plasmids bearing the 5′ UTR sequences of COL1A1 (cis#1), FN1 (cis#2), and CANX. Relative secreted AP activities in medium with the reporter plasmids are shown. (B) The cis#1 and cis#2 sequences originated from human COL1A1 and FN1 genes. Sequences of the identified motif GAG-(X)3-ACA/G/C are underlined. (C) YA7 cells were transfected with reporter plasmids bearing mutated cis#1 and cis#2. Relative secreted AP activities (black bars) as in A or membrane SF3b4 levels (gray bars) are shown. See SI Appendix, Fig. S4A for the sequences. (D) Relative AP mRNA levels of membrane fractions of the transfected cells shown in C are depicted. Likewise, those of the cytosol and membrane fractions are shown in SI Appendix, Fig. S4C. (E) Reporter plasmids comprising polyC and polyT or the motif containing polyC and polyT were used to compare relative AP activity and degree of membrane localization of SF3b4, as in C. All data of relative AP activity in Fig. 5 represent means ± SD (n = 3 or 4), and the values obtained from control samples using empty reporter plasmid were set as 1.
The RRM domains of SF3b4 are highly conserved among species. For example, the sequence similarity of the N-terminal RRM domain is 95% between humans and worms (22). A previous in vitro screening study revealed that the RRM2 domain of Caenorhabditis elegans SF3b4 bound to RNA containing the octamer motif CGUGUGAG (23). Alignment searches of cis#1 identified at least two sets of similar, but not identical, sequences to the octamer motif. Further extensive manual searching, together with preliminary reporter assays, identified a candidate motif, GAG-(X)3-ACA/G/C (X represents any nucleotide), found at four sites in cis#1 (Fig. 5B, underlined sequences). Notably, three sets of this motif were also found in cis#2 (underlined sequences), but not in the 5′ UTR regions of genes encoding CANX, MMP2, and TIMP1. To verify the importance of this newly identified motif in enhanced secretion, mutated cis#1 and cis#2 lacking all the motif sequences [cis#1-sub (T) and cis#2-sub (T), respectively] were examined.(Fig. 5C). These mutated reporter genes significantly impaired the enhancement of AP activity and SF3b4 localization to the ER (Fig. 5C) without reduction of the membrane mRNA levels (Fig. 5D). The enhancement activity of the motif was further confirmed using an embedded 9-mer (GAGCCCACA or GAGTTTACA) within polymeric C or T sequences (Fig. 5E), suggesting that the defined sequence is sufficient for enhanced secretion. Although the previously identified C. elegans octamer (23) contains GAG and partially overlaps with the newly identified motif, it only resulted in a moderate increase in AP activity and failed to enhance SF3b4 transition to the membrane (SI Appendix, Fig. S4B).
Discussion
The Splicing Factor Component SF3b4, Which Interacts with p180, Has a Function in Translational Control on the ER.
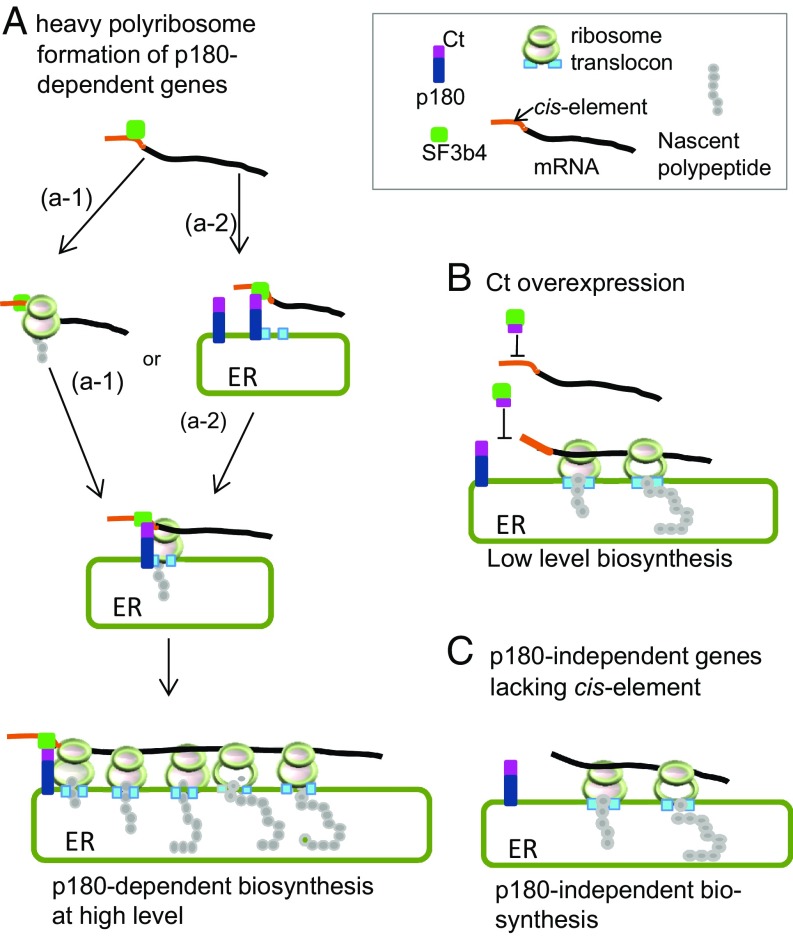
p180 is an essential factor for high-rate protein synthesis of collagen and fibronectin on the ER because of its multiple binding abilities to ribosomes/polyribosomes and to the Sec61 translocon (3). Here we identified a critical cofactor of p180 that plays a key role in a mechanism for the translational control of secretory proteins. Unexpectedly, the cofactor was identified to be SF3b4, a component of SF3b in spliceosomal U2 small nuclear ribonucleoprotein particle. We demonstrated that abundant expression of both SF3b4 and p180 is required for enhanced mRNA association with ER membrane and for enhanced heavy polyribosome assembly of specific mRNA, thereby leading to high-rate biosynthesis of the secreted proteins (Fig. 6). This is a surprising discovery of a distinct function for SF3b4 in translational control other than its well-established role in pre-mRNA splicing.
Fig. 6.
Model for p180-SF3b4-cis-element-dependent polyribosome assembly that facilitates efficient translation. (A) An mRNA containing the cis-element in its 5′ UTR is targeted to p180 on the ER via the bridging factor SF3b4 mediated by Ct domain of p180. p180-SF3b4 interaction facilitates the assembly of heavy polyribosomes that confer high-rate protein synthesis. Either a SRP-dependent pathway (a-1) or a SRP-independent pathway (a-2) is the targeting route for mRNAs toward the ER. (B) In the presence of the cytoplasmic Ct fragment, mRNA containing the cis-element does not interact with p180, thereby preventing heavy polyribosome formation. (C) The translational efficiency of mRNAs devoid of the 5′ UTR cis-element remains constant irrespective of manipulation of SF3b4 or p180.
We also demonstrated that a 5′ UTR cis-element comprising the motif GAG-(X)3-ACA/G/C is required for both ER localization of SF3b4 and enhanced biosynthesis in a p180-dependent manner. Hence, the enhanced translation of secretory proteins appears to involve three factors: p180, SF3b4, and the cis-element in 5′ UTR (Fig. 6). Although the underlying mechanism remains to be elucidated, ER-localized SF3b4 and p180 cooperatively induce targeting and anchoring of the mRNA at the ER, consequently leading to the enhanced translation initiation for mRNAs bearing the cis-element. Previously reported 5′ UTR elements that can modulate translation are normally negative modulators (24). The appropriate model system coupled with sophisticated analyses enabled us to identify both 5′ UTR cis-acting element and a trans-acting factor (i.e., SF3b4) that can upregulate translation of ER-localized mRNAs in eukaryotic cells. It remains to be clarified whether the identified motif sequence is bound directly by SF3b4 or indirectly by a complex containing SF3b4.
ER-mRNA Anchorage Is Mechanistically Independent from the Process Required for Enhanced Translation of ER-Associated Polyribosomes.
Although the cis-element in 5′ UTR appears to be essential for highly enhanced translation of p180-associated polyribosomes on the ER, this observation does not exclude any possible targeting routes of mRNAs toward the ER (Fig. 6), including the SRP-dependent pathway (Fig. 6A, a-1) and SRP-independent pathway (a-2) (25). In general, mRNA transit to the ER can be controlled in multiple ways irrespective of cis-element (8, 9, 12). Our data also show that the cis-element was not necessarily required for mRNA anchorage at the ER, but rather that concomitant overexpression of p180 and SF3b4 is important (SI Appendix, Fig. S3 A and C). In contrast, reporter genes that lack the cis-element failed to enhance biosynthesis despite high levels of ER-localized mRNA (Fig. 3 E and F). However, in the presence of the cis-element, ER localization of neither mRNA nor SF3b4 was induced when cells lacked p180 (Figs. 3C and 4 C and E). These findings suggest that abundant levels of SF3b4 are necessary to lead to an increased rate of translation initiation of the mRNAs.
Although the underlying mechanism is unknown, the observed increase in ER-localized mRNAs lacking the cis-element might be a result of low levels of SF3b4 binding to similar motif sequences residing in the reporter gene, and indeed there are five motifs in the protein-coding region. Alternatively, there may be other mRNA receptors on the ER, which might have a domain similar to the Ct domain of p180. In addition, recently it was reported that translocon components act as mRNA receptors at the ER (26). The apparent discrepancy between our present findings and the mRNA receptor function previously proposed for p180 at the ER (27) might be a result of different expression levels of SF3b4 and p180 resulting from differences in cell types or culture conditions.
A p180-SF3b4-Dependent System That Confers Enhanced Translation of ER Polyribosomes.
SF3b4 is ubiquitously expressed in the nucleus, where it functions as a component of the splicing factor. It is therefore assumed that its ordinary expression levels outside the nucleus are very low, which could be why overexpression of SF3b4 is necessary to exert its auxiliary function at the ER in CHO cells. We demonstrated that the translation efficiency of polyribosomes on the ER is defined by the association status with p180 (Fig. 1), and also that the Ct domain of p180 is required for SF3b4 association with p180 and high-rate biosynthesis (Fig. 4 F and G). Although these findings appear to suggest a critical role for interaction of the Ct domain with SF3b4 in translation control of mRNAs, it remains to be elucidated whether and/or how specific interaction of p180 and SF3b4 enables the increased ribosome loading to specific mRNAs comprising the cis-element.
We found notable features in the 5′ UTR of the p180-dependent genes (i.e., both the COL1A1 and FN1 genes lack a Kozak consensus sequence), whereas this sequence is conserved in the p180-independent genes (e.g., CANX, MMP2, and TIMP1; SI Appendix, Table S2). High expression levels of collagen and fibronectin proteins could be ascribed to an enhancing system, using the p180-SF3b4-mRNA complex to compensate for the lack of a canonical Kozak sequence.
The Critical Role of SF3b4 in Collagen Biosynthesis Has Profound Medical Implications.
The observation of the cosedimentation of SF3b4 with polyribosomal COL1A1 mRNA suggests an intrinsic role for SF3b4 in the translation of collagen mRNA. Recently, haploinsufficiency in SF3B4 gene was identified as a major genetic cause of Nager syndrome (20). This syndrome is characterized by poor craniofacial formation, but the molecular basis underlying the dysplasia caused by SF3B4 haploinsufficiency remains to be elucidated. Our unexpected finding of a role for SF3b4 in collagen biosynthesis may provide a crucial insight into the pathogenesis of this disease. Because collagen is essential for bone morphogenesis, our current findings may provide the missing link between SF3B4 haploinsufficiency and the appearance of craniofacial malformations in patients with this syndrome. Although SF3b4 is ubiquitously expressed in most cell types, its endogenous levels appear to be a limiting factor for adequate protein translation on the ER, as discussed earlier. This is consistent with our proposed scenario that reduced SF3b4 levels caused by haploinsufficiency may be associated with impaired collagen secretion in patients with Nager syndrome. Analyses of collagens in fibroblasts from these patients will provide a vital clue for elucidating the mechanisms of skeletal malformation.
In conclusion, we here identified a mechanism for a selective assembly system of p180-associated polyribosomes on the ER operated by highly organized translational machinery. This system, which we have termed spERt (selective polyribosome assembly on the ER with three factors) Technology, is likely applicable to the production of recombinant proteins in cultured cells. ER-bound mRNAs are known to be translated more efficiently than cytosolic-localized mRNAs (9, 28). Our findings provide insight into a proposed dynamic function for the ER that contributes to global posttranscriptional gene regulation.
Materials and Methods
Construction of Expression Plasmids, Cell Culture, Establishment of Cell Lines, and Quantification of Collagen.
The construction of expression plasmids and cell culture conditions are described in SI Appendix. The generation of CHO cells overexpressing p180 (clone 5g) was described previously (16). Parental CHO and 5g cells were transfected with SF3b4-expression plasmid. After selection with the antibiotic G418 (400 µg/mL) and subsequent limiting dilution, cell lines expressing SF3b4 (clone 3D5) and p180/SF3b4 (clone YA7) were established. The concentration of type I collagen was quantified by LC-MS in multiple reaction monitoring mode and normalized using stable-isotope-labeled collagen (29).
Sequential Detergent Extraction of Cytosolic and Membrane Fractions, Ribosome Stripping, Polyribosome Analyses, and Immunoprecipitation.
The preparation of cytosolic and membrane fractions by sequential detergent extraction, ribosome stripping by in situ EDTA treatment, polyribosome analysis by velocity sedimentation, and qPCR of mRNAs (encoding COL1A1, FN1, MMP2, TIMP1 and CANX) were performed as described previously (17). Membrane fractions or ribosome-stripped membranes were incubated at 4 °C for 1 h with anti-rabbit IgG-conjugated magnetic beads (Dynabeads; Veritas) that had been preincubated with an anti-p180 antibody or control rabbit IgG. After washing, the captured complexes were analyzed for proteins by Western blotting, and mRNAs were quantified by real-time PCR, as described previously (17). In some experiments, polyribosomes were fractionated by a gradient fractionator (Biocomp Instruments) equipped with a UV monitor (AC5200; ATTO).
Affinity Column Chromatography of Histidine-Tagged Ct and Proteome Analysis.
Hexahistidine-tagged Ct protein was overexpressed in HEL cells. Cytosolic fractions were subjected to affinity chromatography using TALON (Clontech), as described in SI Appendix. After extensive washing, most of the hexahistidine-tagged Ct protein was eluted in fractions E1, E2, and E3 (SI Appendix, Fig. S2A), whereas no signal was detected in mock-transfected cells. The proteins of E1, E2, and E3 fractions were identified (SI Appendix, Table S1) by LC-MS after trypsin digestion, as described previously (30) (SI Appendix, Materials and Methods).
Supplementary Material
Acknowledgments
We are grateful to Dr. Hiroaki Imataka, from the Graduate School of Engineering, University of Hyogo, for helpful discussions and to Dr. Yuko Ushiki-Kaku, Ms. Yoshiko Yoshioka, Dr. Kazunori Mizuno, and all other members of the Nippi laboratory for technical support and helpful discussion. This work was supported in part by grants from the Japan Science Technology Agency (Projects for Technological Development, Research Center Network for Realization of Regenerative Medicine).
Footnotes
Conflict of interest statement: T.U., Y.T., and K.O.-G. are coinventors of patent applications based on this work. T.U., Y.T., S.H., and K.O.-G. are employees of Nippi, Inc., an applicant of the patents (JP58488 and other related patent applications, including in the European Union and the United States). Nippi, Inc. is going to develop application of a new engineering technology (spERt Technology) based on ideas described in this study, which is likely to up-regulate productivity of recombinant proteins. Our development of spERt Technology has not influenced any conclusions of this study.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901742116/-/DCSupplemental.
References
- 1.Palade G, editor. A Small Particulate Component of the Cytoplasm. Yale Univ Press; New Haven, CT: 1958. pp. 283–304. [Google Scholar]
- 2.Ross R, Benditt EP. Wound healing and collagen formation. IV. Distortion of ribosomal patterns of fibroblasts in scurvy. J Cell Biol. 1964;22:365–389. doi: 10.1083/jcb.22.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueno T, Kaneko K, Sata T, Hattori S, Ogawa-Goto K. Regulation of polysome assembly on the endoplasmic reticulum by a coiled-coil protein, p180. Nucleic Acids Res. 2012;40:3006–3017. doi: 10.1093/nar/gkr1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen AK, Bourne CM. Shape of large bound polysomes in cultured fibroblasts and thyroid epithelial cells. Anat Rec. 1999;255:116–129. doi: 10.1002/(SICI)1097-0185(19990601)255:2<116::AID-AR2>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 5.Hermesh O, Jansen RP. Take the (RN)A-train: Localization of mRNA to the endoplasmic reticulum. Biochim Biophys Acta. 2013;1833:2519–2525. doi: 10.1016/j.bbamcr.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Pichon X, et al. RNA binding protein/RNA element interactions and the control of translation. Curr Protein Pept Sci. 2012;13:294–304. doi: 10.2174/138920312801619475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraut-Cohen J, Gerst JE. Addressing mRNAs to the ER: Cis sequences act up! Trends Biochem Sci. 2010;35:459–469. doi: 10.1016/j.tibs.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Pyhtila B, et al. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA. 2008;14:445–453. doi: 10.1261/rna.721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reid DW, Nicchitta CV. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem. 2012;287:5518–5527. doi: 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diehn M, Eisen MB, Botstein D, Brown PO. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- 11.Lerner RS, et al. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Q, Jagannathan S, Reid DW, Zheng T, Nicchitta CV. Hierarchical regulation of mRNA partitioning between the cytoplasm and the endoplasmic reticulum of mammalian cells. Mol Biol Cell. 2011;22:2646–2658. doi: 10.1091/mbc.E11-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savitz AJ, Meyer DI. Identification of a ribosome receptor in the rough endoplasmic reticulum. Nature. 1990;346:540–544. doi: 10.1038/346540a0. [DOI] [PubMed] [Google Scholar]
- 14.Ueno T, et al. Expansion of the trans-Golgi network following activated collagen secretion is supported by a coiled-coil microtubule-bundling protein, p180, on the ER. Exp Cell Res. 2010;316:329–340. doi: 10.1016/j.yexcr.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 15.Langley R, et al. Identification of multiple forms of 180-kDa ribosome receptor in human cells. DNA Cell Biol. 1998;17:449–460. doi: 10.1089/dna.1998.17.449. [DOI] [PubMed] [Google Scholar]
- 16.Ogawa-Goto K, et al. p180 is involved in the interaction between the endoplasmic reticulum and microtubules through a novel microtubule-binding and bundling domain. Mol Biol Cell. 2007;18:3741–3751. doi: 10.1091/mbc.E06-12-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueno T, et al. Enhancement of procollagen biosynthesis by p180 through augmented ribosome association on the endoplasmic reticulum in response to stimulated secretion. J Biol Chem. 2010;285:29941–29950. doi: 10.1074/jbc.M109.094607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker F, et al. Expression of the 180-kD ribosome receptor induces membrane proliferation and increased secretory activity in yeast. J Cell Biol. 1999;146:273–284. doi: 10.1083/jcb.146.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui XA, Zhang H, Palazzo AF. p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. PLoS Biol. 2012;10:e1001336. doi: 10.1371/journal.pbio.1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernier FP, et al. FORGE Canada Consortium Haploinsufficiency of SF3B4, a component of the pre-mRNA spliceosomal complex, causes Nager syndrome. Am J Hum Genet. 2012;90:925–933. doi: 10.1016/j.ajhg.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stefanovic B, Schnabl B, Brenner DA. Inhibition of collagen alpha 1(I) expression by the 5′ stem-loop as a molecular decoy. J Biol Chem. 2002;277:18229–18237. doi: 10.1074/jbc.M108065200. [DOI] [PubMed] [Google Scholar]
- 22.Igel H, Wells S, Perriman R, Ares M., Jr Conservation of structure and subunit interactions in yeast homologues of splicing factor 3b (SF3b) subunits. RNA. 1998;4:1–10. [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka Y, Ohta A, Terashima K, Sakamoto H. Polycistronic expression and RNA-binding specificity of the C. elegans homologue of the spliceosome-associated protein SAP49. J Biochem. 1997;121:739–745. doi: 10.1093/oxfordjournals.jbchem.a021648. [DOI] [PubMed] [Google Scholar]
- 24.Leppek K, Das R, Barna M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol. 2018;19:158–174. doi: 10.1038/nrm.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aviram N, Schuldiner M. Embracing the void–How much do we really know about targeting and translocation to the endoplasmic reticulum? Curr Opin Cell Biol. 2014;29:8–17. doi: 10.1016/j.ceb.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Jagannathan S, et al. Multifunctional roles for the protein translocation machinery in RNA anchoring to the endoplasmic reticulum. J Biol Chem. 2014;289:25907–25924. doi: 10.1074/jbc.M114.580688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui XA, Zhang Y, Hong SJ, Palazzo AF. Identification of a region within the placental alkaline phosphatase mRNA that mediates p180-dependent targeting to the endoplasmic reticulum. J Biol Chem. 2013;288:29633–29641. doi: 10.1074/jbc.M113.482505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephens SB, Nicchitta CV. Divergent regulation of protein synthesis in the cytosol and endoplasmic reticulum compartments of mammalian cells. Mol Biol Cell. 2008;19:623–632. doi: 10.1091/mbc.E07-07-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taga Y, Kusubata M, Ogawa-Goto K, Hattori S. Stable isotope-labeled collagen: A novel and versatile tool for quantitative collagen analyses using mass spectrometry. J Proteome Res. 2014;13:3671–3678. doi: 10.1021/pr500213a. [DOI] [PubMed] [Google Scholar]
- 30.Taga Y, Kusubata M, Ogawa-Goto K, Hattori S. Development of a novel method for analyzing collagen O-glycosylations by hydrazide chemistry. Mol Cell Proteomics. 2012;11:010397. doi: 10.1074/mcp.M111.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.