Significance
DNA double-strand breaks (DSBs) are severe insults that lead to different types of genome rearrangements if repaired incorrectly. We explored the genesis of genomic rearrangements mediated by the nonhomologous end-joining pathway of DSB repair. Previous studies suggested that the efficiency of trans repair (i.e., rearrangements) is strongly influenced by the distance between two stretches of DNA incurring DSBs. We systematically investigated the impact of predamage spatial proximity on trans repair frequency in yeast and found no correlation between them, which has important implications for understanding chromosome movement and disease-causing rearrangements.
Keywords: genome rearrangement, translocation, nonhomologous end joining, homologous recombination, single-strand annealing
Abstract
DNA double-strand breaks (DSBs) are serious genomic insults that can lead to chromosomal rearrangements if repaired incorrectly. To gain insight into the nuclear mechanisms contributing to these rearrangements, we developed an assay in yeast to measure cis (same site) vs. trans (different site) repair for the majority process of precise nonhomologous end joining (NHEJ). In the assay, the HO endonuclease gene is placed between two HO cut sites such that HO expression is self-terminated upon induction. We further placed an additional cut site in various genomic loci such that NHEJ in trans led to expression of a LEU2 reporter gene. Consistent with prior reports, cis NHEJ was more efficient than trans NHEJ. However, unlike homologous recombination, where spatial distance between a single DSB and donor locus was previously shown to correlate with repair efficiency, trans NHEJ frequency remained essentially constant regardless of the position of the two DSB loci, even when they were on the same chromosome or when two trans repair events were put in competition. Repair of similar DSBs via single-strand annealing of short terminal direct repeats showed substantially higher repair efficiency and trans repair frequency, but still without a strong correlation of trans repair to genomic position. Our results support a model in which yeast cells mobilize, and perhaps compartmentalize, multiple DSBs in a manner that no longer reflects the predamage position of two broken loci.
DNA double-strand breaks (DSBs) are serious chromosomal lesions that lead to genome instability. Eukaryotic cells primarily use two mechanisms, homologous recombination (HR) and nonhomologous end joining (NHEJ), to repair DSBs, each of which is highly conserved from yeast to humans (1, 2). Whereas HR requires a homologous template to bring about repair, NHEJ executes the direct (re)joining of two DSB ends (2–4). Most often, ends of the same DSB are ligated together in events we refer to as occurring in cis. Alternatively, aberrant ligation of ends from two different DSBs, that is, in trans, leads to gross chromosomal rearrangements, including translocations, inversions, deletions, insertions, and duplications. Joint analysis of most spontaneous rearrangements reveals microhomology at junctions, suggesting that they are often the outcome of NHEJ (3, 5, 6). However, the factors that prevent or promote trans rearrangements in a nucleus with multiple DSBs are poorly understood.
Importantly, cis and trans outcomes at DSBs each represent bona fide DNA repair. Thus, similar to the manner in which Rad52-dependent mechanisms can execute both homologous and ectopic recombination (7–9), cis and trans NHEJ events can both be catalyzed by a common mechanism dependent on the Ku DSB-binding protein and DNA ligase IV (Dnl4 in yeast) (10). Alternatively, different mechanisms might be used for different outcomes. Alternative end-joining (alt-NHEJ) and the closely related microhomology-mediated end-joining (MMEJ) pathways execute inaccurate DSB joining that nearly always deletes bases at the junctions (3, 11–13). A number of reports have revealed a role of alt-NHEJ in the formation of reciprocal translocations in mouse cells (14–17). In contrast, translocation frequency decreased in the absence of c-NHEJ in human cell lines demonstrating species-specific differences in the generation of genomic rearrangements (18). Thus, one key determinant of DSB-dependent mutagenesis might be the extent to which different repair pathways commit DSBs to cis or trans repair.
DSB-dependent mutagenesis might also depend on spatial disposition of lesions in the nucleus, specifically whether DSBs in different locations are equally likely to have their ends joined. Lisby et al. (19) demonstrated that, after formation of multiple DSBs, an average of only two Rad52 foci were observed in Saccharomyces cerevisiae, which suggested that each focus recruits more than one DSB for repair. This suggested that DSBs tend to cluster, that is, are brought together in close proximity for repair. Clustering of DSBs has also been reported in mammalian cells following endonuclease induction or after exposing the cells to ionizing radiation (20–24). It has been argued that DSB clustering is favored and not restricted to chromosomal domains that are in proximity, so that spatial proximity is not sufficient to explain DSB clustering (21).
The factors that influence trans DSB repair outcomes have been studied in several ways. In mammalian cells, Chiarle et al. (22) employed high-throughput genome-wide translocation sequencing and showed that 75% of novel junctions were within 10 kb of a DSB site. In another study, the translocation frequency between the MYC gene with its translocation partners IGH, IGK, and IGL correlated with their nuclear proximity (25). In yeast, the spatial proximity of an ectopic donor sequence directly correlated with DSB repair efficiency by HR (26, 27). In contrast, during single-strand annealing (SSA), the frequency of intrachromosomal and interchromosomal repair was similar when two DSBs were induced (7). In an effort to elucidate the mechanism of reciprocal translocations, Lee et al. (28) demonstrated that the checkpoint kinase Tel1 suppresses NHEJ-mediated translocations, likely by promoting the tethering of cognate DSB ends. Indeed, the fact that most translocations are reciprocal suggests that the two DSB ends remain tethered even when joining occurs in trans (24).
In this work, we examine how the relative position of concurrent DSBs in the yeast nucleus influences trans DSB repair frequency. We focus on precise NHEJ, the major NHEJ outcome, as well as an MMEJ-like SSA event. Following formation of multiple DSBs, cis repair predominated over trans repair, but trans repair efficiency was strikingly independent of the reported predamage spatial proximity of DSB loci in the nucleus. These findings are consistent with DSBs being mobilized and possibly compartmentalized for repair.
Results
Reporter-Based Assay to Detect Reciprocal Translocations.
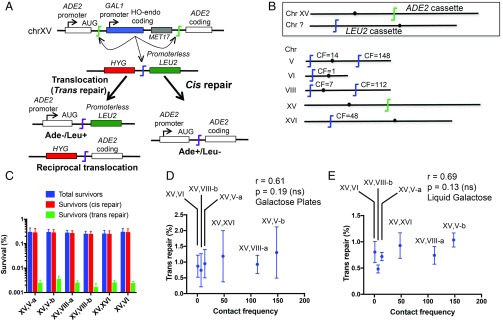
We designed a reporter-based assay that measures cis (same site) vs. trans (different site) repair. The assay is based on a previously established suicide deletion approach wherein the DSB-inducing HO endonuclease is placed between two 36-bp HO cut sites that are flanked by the ADE2 promoter (including the start codon) and coding regions (Fig. 1A and SI Appendix, Table S1) (29). Following galactose induction, HO endonuclease expression is self-terminated upon cutting of the HO cut sites and subsequent DNA repair. The version of the assay used in this study was designed such that precise NHEJ, the most frequent outcome of yeast NHEJ (29), leads to conversion of the initial Ade−/red to the Ade+/white phenotype. Although this NHEJ event arises from two DSBs, we considered it to be cis repair for purposes here due to the close proximity of the sites (see more below). To detect trans repair, we placed a second cut site at different locations in the yeast genome, which contained a promoterless and ATG-less LEU2 coding sequence such that reciprocal repair of the two DSB loci leads to productive coupling of the LEU2 coding region with the ADE2 promoter and thus an Ade−/Leu+ phenotype.
Fig. 1.
Spatial proximity and interchromosomal trans NHEJ repair frequency do not correlate. (A) Schematic showing the ADE2 suicide deletion cassette and LEU2 cassette (with MET17 and HYG as their selection markers, respectively) on two different chromosomes, as well as the expressed reporter end products of cis and trans NHEJ repair. (B) Diagram showing the fixed position of the ADE2 cassette on chrXV and variable positions of the LEU2 cassette. The reported contact frequency (CF) between regions ±30 kb around ADE2 and each trans target location are indicated. (C) Genome rearrangement assay (galactose plates) measuring absolute survival for each ADE2-LEU2 pair. (D) Lack of correlation between trans repair and contact frequencies (galactose plate method). (E) Lack of correlation between trans repair and contact frequencies (liquid galactose method). Data are the mean ± SD from four independent experiments. ns, not significant.
Spatial Distance Between DSB Sites Does Not Determine Interchromosomal trans Repair Frequency.
We employed our assay system to explore the impact of genomic and nuclear context on trans repair frequencies. We exploited yeast interlocus contact frequencies obtained from the Hi-C genome-wide cross-linking data of Duan et al. (30), the same data used by Lee et al. (27). We generated six reporter strains by moving the LEU2 DSB cassette to different genomic locations having a range of contact frequencies with the ADE2 locus on chrXV (Fig. 1B). Cells were grown in glucose for 2 d until saturation followed by plating to galactose to score cis and trans repair events. DSB formation was relatively slow in this assay, and the cutting efficiency of the LEU2 cassette proved lower than the ADE2 cassette, but the cell-autonomous appearance of ADE2 product alleles was generally coincident with DSB formation (SI Appendix, Fig. S1).
Only 0.25–0.3% of the total cells survived on the galactose plates (Fig. 1C), compared with ∼1.4% observed when only the ADE2 cassette was present. The trans repair (Ade−/Leu+) frequency ranged from 0.7 to 1.3% of the total survivors in different strains, with the rest being cis repair (Ade+/Leu−) (Fig. 1 C and D), affirming a cis preference of yeast NHEJ (28). As expected, joint analysis of 20 Ade−/Leu+ isolates in one of the strains showed that 39 of the 40 individual repair junctions corresponded to precise NHEJ (SI Appendix, Fig. S2 and Table S1); the remaining junction did not give a PCR product and likely arose by deletion beyond a primer site. Some Ade−/Leu− colonies, typically representing imprecise NHEJ, were also seen but not routinely scored due to their much lower frequency.
Strikingly, no significant difference was observed in the trans repair frequency between strains with different locations of the second DSB, nor was any correlation observed between the contact and trans repair frequencies (Fig. 1D), which suggested that spatial distance between DSB sites does not determine trans repair efficiency. Surprised by these results, we repeated the measurements using a different method, hereafter called “liquid galactose” (as opposed to the “galactose plates” method above). In the liquid galactose method, saturated starting cultures were reinoculated into galactose liquid medium and allowed to grow to saturation again. HO endonuclease induction and NHEJ occur in this liquid phase, before plating to glucose selection plates to count cis and trans repair events. The liquid galactose method is more reproducible but does not allow measurement of absolute survival frequencies. The trans repair frequencies of the six strains followed the same trend with liquid galactose as with galactose plates, except that the values were slightly lower with the former (Fig. 1 D and E). Again, no correlation was observed between contact and trans repair frequencies (Fig. 1E).
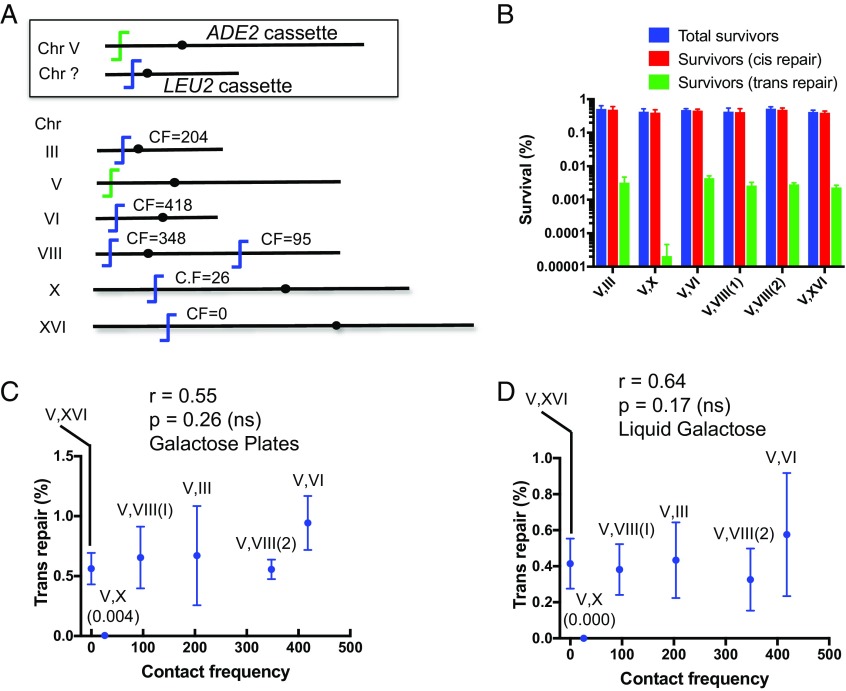
The lack of correlation between contact and trans repair frequencies could somehow be caused by the specific location of the ADE2 suicide deletion cassette. To test this possibility, we moved the suicide deletion cassette from chrXV to chrV near CAN1, 33 kb from the telomere. As above, we generated six different strains by moving the LEU2 cassette to different locations having a range of contact frequencies with CAN1 (Fig. 2A). Notably, the location of the ADE2 cassette at CAN1 and five of the six placements of the LEU2 cassette were within ∼1 kb of loci used by Lee et al. (27) when examining HR. Results with both the galactose plates and liquid galactose methods reaffirmed that the spatial distance between the two DSB loci had no impact on the trans repair frequency (Fig. 2 B–D). One strain showed extremely rare Leu+ colonies, likely due to inviability of trans repair events for unknown reasons.
Fig. 2.
The lack of correlation between proximity and trans NHEJ repair is not a result of ADE2 marker location. (A) Diagram of DSB positions similar to Fig. 1, with the ADE2 cassette now located on chrV. (B) Genome rearrangement assay (galactose plates) measuring absolute survival. (C) Lack of correlation between trans repair and contact frequencies (galactose plate method). (D) Lack of correlation between trans repair and contact frequencies (liquid galactose method). Data are the mean ± SD from four independent experiments. ns, not significant.
As noted above, the suicide deletion cassette contains two cut sites that we treated as a single DSB. To determine whether this configuration influenced the outcome, we mutated the downstream cut site at ADE2 in three different trans reporter strains (SI Appendix, Fig. S3A). Doing so prevents the self-termination of HO endonuclease expression, thereby permitting recleavage of the HO cut site and preventing detection of precise NHEJ repair on galactose plates. For this reason, we performed the genome rearrangement assay in these strains using “transient galactose” method in which saturated glucose cultures were reinoculated into glycerol medium and grown to log phase followed by galactose induction for 3 h. Appropriate dilutions were then plated on glucose plates to count repair events. All strains showed a trans repair frequency similar to their parent strains with two cut sites at ADE2 (SI Appendix, Fig. S3B). Further comparison of the three different induction methods in the different HO cut site combinations showed that the trans repair frequency was always comparable, indicating that the suicide deletion format was not responsible for the lack of correlation of Hi-C contact and trans repair frequencies (SI Appendix, Fig. S3B).
Two Interchromosomal trans Repair Events in Competition Occur at Equal Frequency.
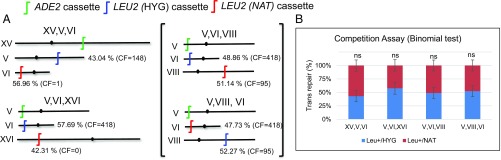
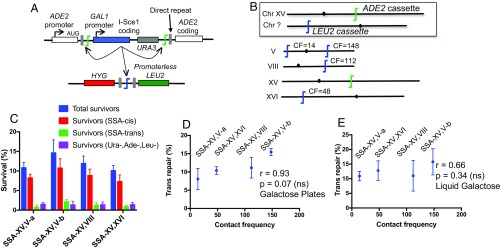
A limitation of strains with only two DSB loci is that two different strains must be compared in order to explore the impact of spatial proximity. To provide a direct and internally controlled comparison, we performed a competition assay by introducing a third HO cut site locus into selected strains, which again carried a promoterless LEU2 coding sequence (Fig. 3A). Trans repair can occur between the ADE2 promoter and the LEU2 coding sequence at either locus (but not both in one cell). Using LEU2 at both trans repair targets limited the number of variables that might influence the results but necessitated that we use PCR to determine the trans target locus used in a given Leu+ survivor. At least 78 Leu+ colonies (∼20 from each independent experiment) were scored for each strain (SI Appendix, Fig. S4). Again using Hi-C data of Duan et al. (30), we chose the positions of the two LEU2 targets such that one was in close proximity to ADE2 while the other was distant (Fig. 3A). Consistent with results above, the trans repair frequencies of the two possible targets were not statistically significantly different from a 50:50 ratio, despite the large difference in their contact frequencies with the common ADE2 rearrangement partner (Fig. 3). To ensure that there was no influence of the HYG and NAT selectable markers used to create the two LEU2 targets, we swapped the LEU2 cassettes in one strain and confirmed that the two competing trans repair frequencies were again comparable (Fig. 3).
Fig. 3.
Two trans NHEJ target alleles are used at equal frequency when placed in competition. (A) Schematic of the position of three HO cut sites in four different strains that place two LEU2 trans target cassettes in competition. Contact frequencies (CFs) are for each LEU2 locus relative to ADE2. Strains V, VI, and VIII, and V, VIII, and VI are the same except that the LEU2 (HYG) and LEU2 (NAT) cassettes were swapped. (B) The relative contribution of LEU2 (HYG) and LEU2 (NAT) cassette usage during Leu+ trans repair was consistent with a 50:50 ratio in all strains. Data are the mean ± SD from four independent experiments. ns, not significant.
Intrachromosomal trans Repair Occurs at a Similar Frequency as Translocation.
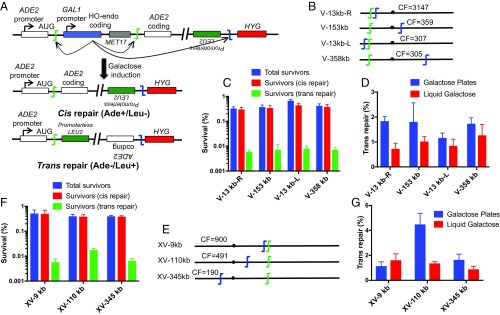
It is well known that chromosomes occupy substantially exclusive territories within nuclei, leading to much higher contact frequencies between loci on the same chromosome than between those on different chromosomes (30, 31). Comparing trans repair frequencies between two intrachromosomal loci vs. two interchromosomal loci should thus be insensitive to any limitations imposed by the quality of our Hi-C reference data (30). It is also an interesting question whether different locations on the same chromosome behave similarly, or whether features such as centromeres impose a DSB repair restriction (32). To this end, we integrated the LEU2 target cassette at four different locations on chrV, which also carried the ADE2 suicide deletion cassette (Fig. 4 A and B). The LEU2 cassettes were oriented such that precise NHEJ led to chromosomal inversions to be compatible with cell growth and survival.
Fig. 4.
Genome rearrangement assay detecting intrachromosomal trans NHEJ repair (inversion). (A) Schematic showing the ADE2 suicide deletion cassette and LEU2 cassette on the same chromosome. As designed, inversion by precise NHEJ led to Leu+ outcomes. (B and E) Relative locations of LEU2 cassette on chrV and XV, respectively (not to scale). Contact frequencies (CFs) are similar to Fig. 1. (C and F) Genome rearrangement assays measuring total survival (galactose plates). (D and G) Trans repair frequency using galactose plates (blue bars) and liquid galactose (red bars). As in Figs. 1–3, trans repair is largely independent of the LEU2 genomic position. Data represent the mean ± SD from four independent experiments.
Several features are notable about the pattern of cis and trans repair frequencies obtained with the inversion strains, whether measured by the galactose plate or liquid galactose methods. First, cis repair again predominated, with trans repair frequencies ranging from 1.2 to 1.8% (Fig. 4 C and D). Moreover, this trans repair frequency was the same as observed with the interchromosomal trans repair targets above (compare Fig. 4 C and D to Fig. 1 C–E and Fig. 2 B–D). Finally, there was no impact of the location of the second DSB site within the same chromosome, including whether it was placed close to the ADE2 DSB cassette (13 kb) or on the opposite chromosome arm with an intervening centromere (Fig. 4B). Similar to the interchromosomal assays, we validated these results by moving the ADE2 suicide deletion and LEU2 cassettes from chrV to chrXV (Fig. 4E). Overall, the same pattern was seen (Fig. 4 F and G). The only notable exception among all strains studied was that, when using the galactose plating method only, the Inv-110 kb strain showed a significantly higher trans repair frequency (Fig. 4G). We suspect this outlier result could be due to a property of this target locus (Discussion).
Influence of Genetic Factors on trans Repair Frequencies.
The overall survival as well as the trans repair efficiency in our assay strains were lower than previously reported (28). The lower trans repair frequency could be due to the relatively inefficient cleavage by HO that we observed for our alleles (SI Appendix, Fig. S1), which very likely reduces the rate of simultaneous DSB formation and biases toward cis repair. However, the cleavage efficiency of the LEU2-marked second DSB was the same regardless of its genomic location (SI Appendix, Fig. S1), so this property cannot easily explain the pattern of trans repair efficiencies we observed with our different alleles.
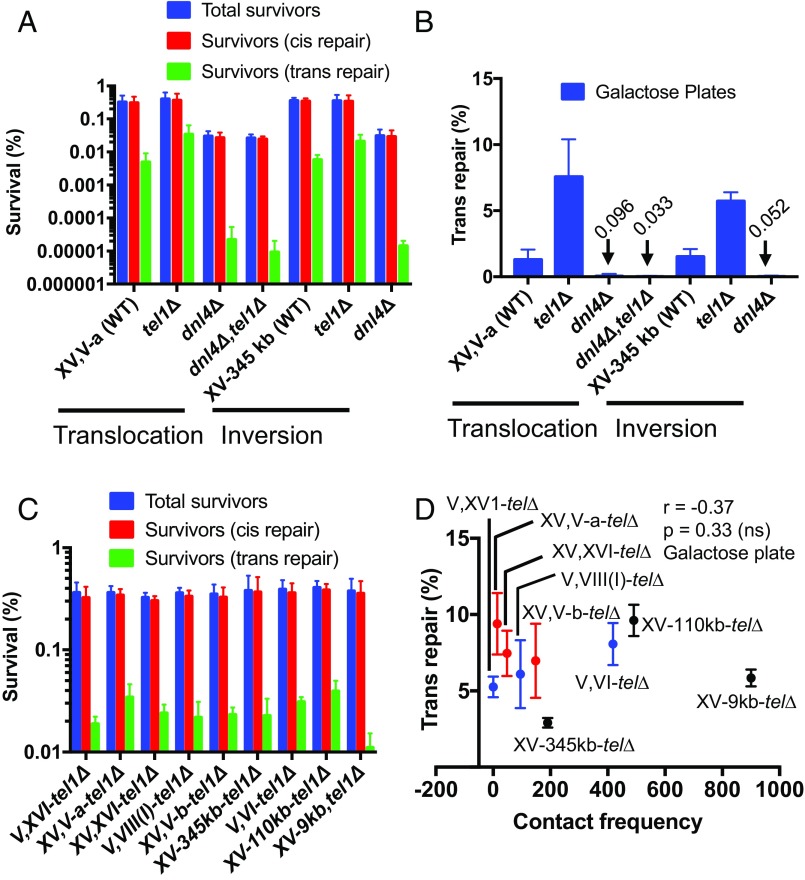
To explore this point further, we deleted TEL1 from our assay strains to reexamine the allele pattern in the context of higher trans repair efficiencies, since Tel1 has been shown to suppress interchromosomal NHEJ (28). We first made two tel1∆ strains, one from each of our interchromosomal and intrachromosomal allele sets. As expected, TEL1 deletion enhanced the trans repair frequency by threefold to fivefold (Fig. 5 A and B). DNL4 deletion abrogated trans repair regardless of TEL1 status, reaffirming that tel1∆ promotes trans repair by a Dnl4-dependent mechanism (Fig. 5 A and B) (28). We next examined a larger series of nine total tel1∆ strains that sampled multiple interchromosomal and intrachromosomal DSB allele combinations. As was observed above with TEL1 wild-type strains, no correlation was observed between contact and trans repair frequencies in the absence of Tel1 (Fig. 5 C and D). In fact, the lowest trans repair efficiency was obtained with the intrachromosomal XV-345 kb-tel1∆ reporter strain.
Fig. 5.
Deletion of TEL1 increases trans repair frequency independently of spatial proximity (A) Genome rearrangement assay measuring survival in interchromosomal as well as intrachromosomal repair assays in the indicted wild-type and mutant strains (galactose plates). (B) Trans repair frequency in wild-type and mutant strains. Absolute survival (C) and trans repair frequency (D) in the interchromosomal and intrachromosomal repair assay strains. Red (ADE2 cassette on chrXV) and blue (ADE2 cassette on chrV) circles represent data obtained from interchromosomal assay strains, while the black circles (ADE2 cassette on chrXV) represent data from intrachromosomal assay strains. Data are the mean ± SD from four independent experiments. ns, not significant.
In yeast, increased mobility of DSBs is thought to facilitate HR and requires Rad9, Rad51, and Mec1 (33, 34). To ascertain the potential role of DSB mobility during precise NHEJ repair, we deleted RAD9 and RAD51 from our interchromosomal and intrachromosomal trans reporter strains. Neither deletion significantly changed the trans repair frequency (SI Appendix, Fig. S5). The implications of these results are discussed below.
SSA via Short Terminal Direct Repeats Is also Independent of Proximity.
The insensitivity of trans NHEJ to genomic location described above was different from the reported correlation between donor location and HR efficiency in yeast (26, 27). Notably, in these earlier HR studies, only one of the two interrogated loci had suffered a DSB, since the donor allele remained intact. We wondered which pattern would prevail if two DSBs were allowed to repair through an HR-like mechanism. SSA is a Rad52-dependent subpathway of HR in which DSBs ends are subjected to 5′ resection followed by annealing of the 3′ terminated strands. To measure trans repair efficiency by SSA, we adapted a previously described version of our suicide deletion reporter assay in which SSA occurs between terminal 28-bp direct repeats on either side of the endonuclease cut sites, here using the endonuclease I–SceI instead of HO (Fig. 6A) (29). We generated four strains with a range of contact frequencies between the two DSB loci (Fig. 6B). As expected from prior studies with a single DSB locus (29), the total survival frequency increased with SSA, ranging from 10 to 15% compared with 0.25 to 0.50% for NHEJ (Fig. 6C). The trans repair frequency also increased, ranging from 8 to 15% for SSA strains compared with 1 to 2% for NHEJ (Fig. 6 D and E), indicating that cis repair via short resected direct repeats is not favored as strongly as during precise NHEJ. Overall, little to no correlation was observed between spatial proximity and trans repair frequency by SSA (a shallow linear trend was seen only with the galactose plate method that was not statistically significant), suggesting that, as with NHEJ, repair is independent of spatial proximity when two DSBs are repaired by a Rad52-dependent mechanism (Fig. 6 D and E) (29).
Fig. 6.
Spatial proximity and interchromosomal trans SSA repair frequency do not correlate. (A) Schematic of single-strand annealing (SSA) assay showing ADE2 suicide deletion cassette and LEU2 cassettes with I-SceI cut sites flanked by 28-bp direct repeats. (B) Diagram showing the fixed position of the ADE2 cassette on chrXV and variable positions of the LEU2 cassette. (C) Absolute survival (galactose plates). Non-SSA outcomes (e.g., precise NHEJ) were scored as colonies that had successfully deleted the URA-marked suicide deletion cassette to become Ura− but that had not converted to either the Ade+ (cis SSA) or Leu+ (trans SSA) phenotypes. (D) Lack of correlation between trans repair and contact frequencies (galactose plate method). (E) Lack of correlation between trans repair and contact frequencies (liquid galactose method). Data represent the mean ± SD from four independent experiments. ns, not significant.
We also repeated the experiment of Lee et al. (27) in our strains in which the HR repair of a single DSB locus is measured using a variety of donor alleles. We first introduced a 2-kb ade2 donor sequence at a subset of the same loci used for the second DSB in our NHEJ assays. This donor supported HR repair of the closely spaced suicide deletion DSBs and included a MluI restriction site to distinguish NHEJ and HR outcomes. The final set of 11 strains had the DSB allele at either ADE2 (SI Appendix, Fig. S6A) or CAN1 (SI Appendix, Fig. S6B) and included both interchromosomal and intrachromosomal locus combinations. Cells were grown in glycerol to log phase before plating to galactose to measure HR efficiency. Overall, HR was markedly more efficient than NHEJ, with as much as ∼74% of cells repairing the suicide deletion allele (SI Appendix, Fig. S7 A and B), as confirmed by MluI digestion of the repaired allele from 16 independent colonies (SI Appendix, Fig. S8A). However, we did not observe a linear correlation between contact and HR frequencies as reported by Agmon et al. (26) and Lee et al. (27). Instead, we noted a possible threshold pattern, only when the suicide deletion allele was located at CAN1, in which the donor with the lowest contact frequency of zero had an HR frequency of ∼38% while all other donors had indistinguishable HR frequencies of ∼70% (SI Appendix, Fig. S7B).
Given the low cutting efficiency of the suicide deletion cassette and the fact that two DSBs are made at this one locus, we further introduced a single HO cut site into a nucleosome-free region of the ILV1 promoter on chrV. This site cleaves very efficiently, with more than 90% DSB formation by 30 min (35). We generated seven different strains with a 2-kb ILV1 wild-type donor sequence, again distributed around interchromosomal and intrachromosomal genomic loci with a range of ILV1 contact frequencies (SI Appendix, Fig. S6C). Here, we again observed a possible threshold pattern where a strain with a very low contact frequency had a significantly lower HR efficiency of ∼22% (SI Appendix, Figs. S7C and S8B) (Discussion).
Discussion
Four assay systems have been used to study site-directed translocations in yeast (28, 36–38). Our system allowed efficient detection of precise NHEJ by linking DSB formation to termination of HO expression, as well as distinction of cis and trans repair by reporter selection. We generated 12 strains with different combinations of DSB loci to measure interchromosomal trans NHEJ (reciprocal translocation), and 7 different strains to measure intrachromosomal trans NHEJ (inversion), employing 2 different loci for the ADE2 DSB cassette and 14 loci for the LEU2 trans repair target. Both cis and trans repair junctions arose by NHEJ as validated by joint sequencing and dependence on DNL4.
Despite testing many strains with a large range of predamage contact frequencies, based on the same Hi-C data (30) used in a similar study of HR donor proximity (27), we observed no significant differences in trans repair frequency in essentially any wild-type strains (Figs. 1 and 2). These monotonous results do not reflect an inability to detect altered trans repair frequencies because TEL1 deletion increased both interchromosomal and intrachromosomal trans repair, as reported by Lee et al. (28). However, trans repair frequencies were again not correlated with spatial proximity in tel1∆ strains (Fig. 5). While we relied on published contact frequencies, there is abundant evidence that intrachromosomal locus pairs have much higher contact frequencies than interchromosomal pairs due to chromosomal territoriality (30, 39), yet the trans repair frequency remained constant (Fig. 4). Competition assays with three DSB loci further demonstrated that the insensitivity of trans repair to DSB placement was not a result of comparing different strains (Fig. 3). Finally, mutating one HO cut site in the ADE2 locus established that results were not an artifact of the suicide deletion format (SI Appendix, Fig. S3).
In total, we believe our results reflect an underlying biology that NHEJ trans repair frequency is not strongly a function of locus predamage spatial proximity in yeast. Any model invoked to explain this phenomenon (Fig. 7) must also explain the fact that while trans repair did not vary it was a less efficient repair mechanism. Importantly, DSBs at different loci must be roughly simultaneous for trans repair to occur, so we are very likely underestimating the true potential for trans NHEJ given the low HO cleavage efficiency at our alleles (SI Appendix, Fig. S1). Nevertheless, cis repair preference has been observed in other studies and is an expected finding (24, 28).
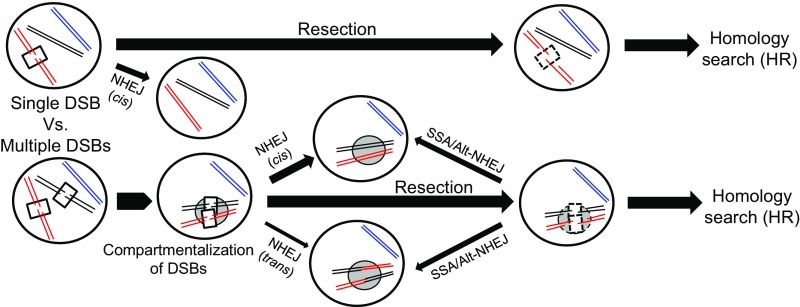
Fig. 7.
Relationship between spatial proximity, HR, and NHEJ. One potential model to explain a lack of special proximity effects for NHEJ based on DSB movement and, specifically, clustering of multiple DSBs in a common repair compartment. See Discussion.
Importantly, the pattern we observed with NHEJ is different from the strong influence of donor proximity reported for ectopic gene conversion by HR (26, 27). This difference might result from mechanistic differences between NHEJ and HR, or from the fact that gene conversion involves only one DSB. We note that we did not fully recapitulate the prior HR findings. Instead of a linear correlation, we noted the potential of a threshold effect for HR donor proximity (SI Appendix, Fig. S7 B and C). This difference in result patterns could be due to the smaller number of allele combinations used in our study compared with Lee et al. (27) or, perhaps more likely, influences of different strain backgrounds that might also be evident in NHEJ outcomes.
Our data are consistent with a prior study showing that SSA, which involves two DSBs and an HR-like Rad52-dependent mechanism, is largely independent of the predamage proximity of the two loci in the same manner as we observed for NHEJ (Fig. 6) (7). The fact that SSA appears to be more like NHEJ in this regard suggests that the difference between NHEJ and HR most likely results from the fact that two broken alleles must interact during NHEJ. Notably, SSA, especially via short terminal repeats, is reminiscent of alt-NHEJ/MMEJ known to catalyze translocations in mammals (11–13). Unlike gene conversion, NHEJ, SSA, and alt-NHEJ are Rad51 independent, emphasizing that the temporal sequence of DSB repair must be considered; NHEJ obligatorily occurs before 5′ resection, while SSA occurs after resection but potentially before Rad51 filament formation and a donor search (3, 10).
A model to explain our results (Fig. 7) must invoke widespread locus mobility after DSB formation (33, 34) and perhaps clustering of DSBs in restricted subnuclear “repair compartments” (19, 20, 23). An extreme possibility is that DSB ends separate early and search the nucleus until they find a repair partner. However, this would not explain cis preference, since separated ends would likely join at random. Alternatively, DSB ends pairs might become mobile as a unit, and indeed, indirect evidence from studies of cis vs. trans HR suggests that DSB ends remain associated (40). DSB mobility could also be more directed, driving DSB ends to common nuclear locations while still held together from the initial break (41), as facilitated by Tel1-dependent tethering (28). NHEJ that commenced in such a repair compartment could still obey cis preference due to tethering but show no bias of trans repair based on predamage locations of DSB loci due to their interim movement. The reported collection of DSBs at the nuclear periphery would fulfill these requirements; specifically, DSBs have been shown to move to the nuclear pore complex in G1 in a Rad51-independent manner, consistent with trans NHEJ occurring at these locations (42).
Lisby et al. (19) reported that two endonuclease-induced Rad52 foci become colocalized in S. cerevisiae due to DSB movement. Importantly, such clustering likely occurs on a different spatiotemporal framework, that is, at later times (and possibly at different nuclear locations), than the movement and clustering we infer for NHEJ. Rad52 functions exclusively during HR (43–45) so that DSBs monitored by Rad52 signal are likely resected and past the NHEJ repair phase. Although no one has visualized DSBs in yeast known to be in the NHEJ phase, various findings in mammalian systems support the idea of a transition from a rapid NHEJ clustering to a later HR stage. These include observations that DSB clustering appears in mammalian cells 10–30 min after irradiation (20, 23) and that dynamics of the NHEJ-promoting 53BP1 and HR-promoting BRCA1 proteins support the notion of a regulated transition from NHEJ to HR (46, 47).
We reason that either a small fraction of DSB ends were never tethered properly and become substrates for trans repair, or that trans repair becomes enabled as the tethering machinery loses its grip on DSB end pairs. Such a progressive transition is perhaps inevitable as resection initiates, but even before resection ends might become less well associated, as suggested by augmentation of this effect in tel1 mutant strains (28). Observations with SSA further support the notion of tethering release. As with NHEJ, SSA assays showed poor correlation between spatial proximity and trans repair frequency. Moreover, in our strains, which used a short repeat at the DSB ends, the SSA trans repair frequency (8–15%) was much higher than for NHEJ (1–2%). Nevertheless, our trans repair frequency was lower than the roughly equal cis and trans repair SSA frequencies obtained by Haber and Leung (7) using large repeats several kilobases apart. Thus, we suggest that the loss of DSB end tethering and ensuing potential for trans rejoining may proceed in progressive degrees of release and resection. However, any trans repair that does occur is promiscuous with respect to predamage positions because the DSB loci have moved.
The final transition at a DSB is to initiate Rad51-dependent HR (48, 49). Importantly, the Rad51 filament formed on resected DSB ends changes their dynamics, with the homology search during HR being facilitated by increased DSB mobility that requires Rad51, Rad9, and Mec1 (33, 34). In our study, neither rad9∆ and rad51∆ mutations conferred a significant change in the trans repair frequency (SI Appendix, Fig. S5), consistent with our model in which DSB movement and clustering occur before the later arrival of Rad51. It is thus intriguing that the spatial proximity of DSB and donor loci determines the rate of HR in yeast (26, 27). Our data suggest that a DSB could move from its predamage location during an early NHEJ phase, which might be expected to break down an HR donor preference based on proximity. One possibility is that NHEJ and HR are not sequential in a cell but occur in a parallel fashion in different cells, for example, as a function of cell cycle stage. Thus, NHEJ and HR outcomes might reflect entirely different search processes. Alternatively, movement of DSBs may depend on whether one vs. two or more DSBs are present, an idea supported by observations that a single DSB did not alter cis interactions in G1 chromosomes (50). Thus, a single DSB may not drive movement in the same manner as multiple DSBs, implying that DSBs emanate a signal leading to their movement and clustering. These ideas are indirectly supported by observations that yeast cells with one, but not two, DSBs can adapt to checkpoint signals and undergo cell division with persistent damage (51) and that the yeast recombination enhancer leads to physical association of HML and MAT loci only after a DSB is induced at MAT (52).
In contrast to our findings in yeast, studies of NHEJ-driven genomic rearrangements in mammalian cells often reveal a strong effect of proximity on DSB clustering and relative trans repair frequencies. Clustering occurred more frequently between DSBs on the same chromosome (21). Roukos et al. (24) showed that translocations are preferentially formed between prepositioned proximal DSBs, while Chiarle et al. (22) showed that most translocations arising from a localized DSB resulted from intrachromosomal NHEJ. The translocation frequency of MYC with IGH, IGK, and IGL has been seen to correlate with their spatial proximity (25) similar to other oncogenic translocations (53). While there are many differences between these studies, we believe the most potent effect results from the fact that the yeast nuclear volume is ∼80 times smaller than that of mammalian nuclei (54). It is possible that a movement-driven breakdown of predamage proximity is also true in mammalian cells, but only within a territory substantially smaller than the nucleus, whereas a similar movement space exposes the entire yeast genome to similar opportunities for trans repair.
Our study leaves open the possibility that as yet underappreciated genomic and nuclear factors could influence trans NHEJ outcomes. While major chromosomal features such as centromeres do not appear to act as barriers to trans NHEJ, one strain (XV-110 kb inversion) did show approximately threefold higher trans repair than all others. We presume this is due to an unknown local effect that will require many more locus pairs to meaningfully discern. In mammalian cells, DSB clustering and hence translocations are preferentially targeted to transcriptionally active regions (21, 22), and similar epigenetic phenomena could be important in yeast. We positioned our second cut site in intergenic regions in all but one strain. Moreover, both DSB cassettes were always flanked by the same sequences. While these design features helped isolate the influence of proximity, they limited the ability of this study to reveal the influence of histones and transcription on repair outcomes.
If our model is correct, it predicts a mechanism of DSB movement that is independent of resection, compatible with persistent tethering, and stimulated by multiple simultaneous DSBs. Recent reports suggest that DSB clustering in mammalian cell is dependent on microtubule and actin-related networks (21, 55–57). In the former case, the LINC complex promotes DSB mobility and repair by mediating an interaction between cytoplasmic microtubules and chromatin (56). In contrast, DNA damage stimulates the formation of intranuclear actin filaments that alter mobility or organization of chromatin (55, 57, 58). In each case, the essential movement is initiated by the DSB. What is less clear is whether initial DSB movement needs to be directional. There is precedence for directional movement of DSBs, albeit not in the context of NHEJ. In Escherichia coli, RecA bundles channel DSB ends to facilitate the long-range homology search (59). Similarly, TRF1-Fok1 induces directional telomere movement and intertelomere association (60). F-actin and myosins have been implicated in the directed motion of heterochromatic DSBs to the nuclear periphery (58). To our knowledge, early directional movement of DSBs has not been demonstrated for NHEJ, but it remains a parsimonious explanation for the absence of predamage proximity effects in trans repair. Notably, WASP binds all DSBs but activates ARP2/3 only at DSBs undergoing HR (57). It seems an interesting possibility that a different set of factors may be recruited by WASP or as-yet-unidentified protein for clustering of DSBs destined to be repaired by NHEJ.
Materials and Methods
Strains, Media, and Allele Construction.
The genotypes of all strains used in this study are listed in SI Appendix, Table S2. All cultures were grown at 30 °C. Rich medium was YPA (1% yeast extract, 2% bacto-peptone, 40 μg/mL adenine) plus 2% dextrose (YPAD) or 3% glycerol (YPAGly). Synthetic defined (SD) medium was as described (61) containing 2% glucose or, when specified, 2% galactose. Suicide deletion cassettes were previously described (29). LEU2 trans target cassettes contained 1,192 bp of LEU2 sequence from second codon through the stop codon plus 100 bp of the terminator region, fused by PCR to either the HYG or NAT markers with the 36-bp HO cut site (insufficient to support HR repair) embedded between the marker and Leu2 coding region. All allele constructions were performed by transforming yeast with appropriate PCR products bearing 45-bp tails homologous to the target locus, purifying marker-positive colonies, and verifying correct insertions by PCR.
Genome Rearrangement Assays Measuring NHEJ, SSA, and HR Repair.
NHEJ repair was measured by three different methods. In the galactose plate method, a single colony was inoculated into SD-Met medium and allowed to grow for 2 d. Appropriate dilutions of these saturated cultures were plated on SD galactose medium such that 50–300 colonies were counted on each plate to score for cis as well as trans repair events, which places all measurements well above the practical limit of quantification. As a control, cells were also plated on SD glucose complete plates. Colonies were counted after 4 d. Absolute survival was calculated by dividing the number of colonies on galactose plates by that on glucose plates. cis repair events were identified by counting Ade+/white colonies, and rare sectored colonies, on galactose complete plates. Trans repair frequency was measured by dividing the number of colonies on galactose-Leu plates by that on galactose complete plates. In the liquid galactose method, cells from initial saturated cultures were reinoculated into SD galactose complete liquid medium and grown for 2 d until saturation. Appropriate dilutions were plated on SD glucose medium. The trans repair frequency was calculated by dividing the number of colonies on SD-Leu plates by that on SD complete plates. In the transient galactose method, a single colony was inoculated in YPAD liquid medium and grown overnight. Cultures were reinoculated in a fresh liquid YPA-glycerol to a starting OD600 of 0.04–0.05 and allowed to grow until exponential phase (OD600 of 0.6). Galactose was added to a final concentration of 2% and cultures incubated for 3 h. Cells were spun down and resuspended in YPAD for 15 min and then plated on SD glucose medium, followed by calculations as for the liquid galactose method.
To measure SSA repair, galactose plate and liquid galactose methods were employed as above, except that the single colony was inoculated into SD-Ura instead of SD-Met, and colonies on galactose-Leu plates were counted after 5 d of growth while the colonies on glucose-Leu plates were counted after 4 d. HR repair was measured by glycerol–galactose method. In this method, cells from the overnight grown precultures were reinoculated in a fresh liquid YPA-glycerol to a starting OD600 of 0.04–0.05 and allowed to grow until exponential phase. Appropriate dilutions of cells were plated on SD glucose and SD selective galactose medium plates. HR repair efficiency was calculated as for the galactose plate method.
Competition Assay.
The competition assay used the galactose plate method described above. After outgrowth, 80 Leu+ colonies (20 from each independent experiment) were analyzed to ascertain the relative contribution from the LEU2 (HYG) and LEU2 (NAT) cassettes. Each colony was subjected to two PCRs containing one common primer positioned in the ADE2 promoter and a specific second primer positioned in sequence flanking the LEU2 coding segment that was unique to each chromosomal trans target location. The ADE2 common primer was OW620 (CTTGACTAGCGCACTACCAG, chrXV) or OW3650 (ACTCACCCGGAAACCACACAG, chrV) and the specific primers were OW3765 (AAAGACCTTGGGGAATATACCTG, chrV), OW3773 (GCTACCAAACGTCATGGACAAG, chrVI), OW3767 (CTACAGTCACAACCCAGTTCATC, chrVIII), or OW3771 (AGGTGAAAGCCAGTAGTAAAAGTG, chrXVI), as appropriate.
Analysis of DSB Repair Outcomes.
The nature of NHEJ repair events was verified by amplifying junctions by PCR, followed by gel electrophoresis and Sanger sequencing, using appropriate combinations of the following primers: OW3671 (TTTACCTTTTGATGCGGAATTGAC, ADE2 promoter), OW3667 (ATATCAAGAGGATTGGAAAAGGAGC, ADE2 coding), OW3666 (AGCACCTAACAAAACGGCATC, LEU2 coding), and OW3665 (GGACCGATGGCTGTGTAGAAG, HYG coding).
The restriction digestion analysis of the HR repair events in the strains carrying ADE2 cassette was done by PCR amplifying the DNA flanking the repaired sites using the primers OW3671 and OW3667. The amplified DNA was digested with MluI restriction enzyme followed by gel electrophoresis. In the strains with ILV1-HOcs, DNA was PCR amplified from the HR repaired colonies using primers OW4159 (CGCTTTTCCGCCTCTGTTAATTC) and OW4160 (AACCACTTGTTGGGCGACTC). The amplified DNA was digested using PsiI restriction enzyme followed by gel electrophoresis.
Calculation of Contact Frequencies.
For maximal consistency with prior studies, all contact frequencies were extracted from the yeast genomic Hi-C map reported by Duan et al. (30), specifically from the experiments using HindIII-digested genomic DNA at a false-discovery rate of 1% [the source data for Lee et al. (27)]. Moreover, like Lee et al. (27), we used contacts between regions ±30 kb around each ADE2 suicide deletion query locus and ±20 kb around each LEU2 trans target locus. We first combined SI appendix, tables S5 and S6, from Duan et al. (30) (files nature08973-s2.xls and nature08973-s3.xls), containing intrachromosomal and interchromosomal data, respectively. We next empirically determined that the data were derived from yeast genome version sacCer2 based on the reported restriction fragment midpoints, to accurately localize the positions of all query and target loci. We finally summed the measured contacts between all query and target fragments within the stated genomic windows. Contacts to restriction fragments contained entirely within a query or target region were counted fully, while contacts to restriction fragments overlapping the edges of a region were weighted by their fractional overlap with the region.
DSB-Monitoring Assay.
DSB and repair product formation were monitored as described (35). Overnight cultures were typically reinoculated in YPA-glycerol and grown to a final OD600 of 0.3–0.5 followed by addition of galactose to induce endonuclease expression. Alternatively, cultures were pregrown in SD-Met medium for 2 d as described above for colony counting experiments before transfer to YPA galactose. Samples were drawn at various time points, genomic DNA was extracted, and qPCR was performed to measure the cutting efficiency using the following sets of primers: (i) ADE2 cassette first cut site breakage, OW3613 (GGTGCGTAAAATCGTTGGATCTC), OW3614 (AGCGTATTACTGAAAGTTCCAAAG); (ii) ADE2 cassette second cut site breakage, OW3617 (TCTCTGTTGGTATCGAATTTATTGATG), OW3618 (TGTCCCCCTCCTAATATACCAACTG); (iii) ADE2 cassette repair product, OW3613 and OW3618; (iv) LEU2 cassette, OW3660 (GCTTCGGCTGTGATTTCTTGAC), OW3661 (TGCCCAGATGCGAAGTTAAGTG); and (v) control allele (ACT1), OW3058 (AGAGTTGCCCCAGAAGAACA) and OW3059 (GGCTTGGATGGAAACGTAGA).
Supplementary Material
Acknowledgments
We thank Mr. Dominic Bazzano for assistance in completing SI Appendix, Fig. S1. This work was supported by National Institutes of Health Grant GM120767 (to T.E.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818595116/-/DCSupplemental.
References
- 1.Jackson SP. Detecting, signalling and repairing DNA double-strand breaks. Biochem Soc Trans. 2001;29:655–661. doi: 10.1042/0300-5127:0290655. [DOI] [PubMed] [Google Scholar]
- 2.Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiruvella KK, Liang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol. 2013;5:a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson TE, Grawunder U, Lieber MR. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 5.Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst) 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 6.Shaw CJ, Lupski JR. Non-recurrent 17p11.2 deletions are generated by homologous and non-homologous mechanisms. Hum Genet. 2005;116:1–7. doi: 10.1007/s00439-004-1204-9. [DOI] [PubMed] [Google Scholar]
- 7.Haber JE, Leung WY. Lack of chromosome territoriality in yeast: Promiscuous rejoining of broken chromosome ends. Proc Natl Acad Sci USA. 1996;93:13949–13954. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CS, Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Microbiol Spectr. 2015;3:MDNA3-0013-2014. doi: 10.1128/microbiolspec.MDNA3-0013-2014. [DOI] [PubMed] [Google Scholar]
- 9.Mehta A, Beach A, Haber JE. Homology requirements and competition between gene conversion and break-induced replication during double-strand break repair. Mol Cell. 2017;65:515–526 e3. doi: 10.1016/j.molcel.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haber JE. Transpositions and translocations induced by site-specific double-strand breaks in budding yeast. DNA Repair (Amst) 2006;5:998–1009. doi: 10.1016/j.dnarep.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 11.Boboila C, et al. Robust chromosomal DNA repair via alternative end-joining in the absence of X-ray repair cross-complementing protein 1 (XRCC1) Proc Natl Acad Sci USA. 2012;109:2473–2478. doi: 10.1073/pnas.1121470109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Della-Maria J, et al. Human Mre11/human Rad50/Nbs1 and DNA ligase IIIalpha/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J Biol Chem. 2011;286:33845–33853. doi: 10.1074/jbc.M111.274159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simsek D, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Difilippantonio MJ, et al. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J Exp Med. 2002;196:469–480. doi: 10.1084/jem.20020851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007;9:978–981. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu C, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 18.Ghezraoui H, et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell. 2014;55:829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- 20.Aten JA, et al. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 21.Aymard F, et al. Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes. Nat Struct Mol Biol. 2017;24:353–361. doi: 10.1038/nsmb.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumaier T, et al. Evidence for formation of DNA repair centers and dose-response nonlinearity in human cells. Proc Natl Acad Sci USA. 2012;109:443–448. doi: 10.1073/pnas.1117849108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roukos V, et al. Spatial dynamics of chromosome translocations in living cells. Science. 2013;341:660–664. doi: 10.1126/science.1237150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet. 2003;34:287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- 26.Agmon N, Liefshitz B, Zimmer C, Fabre E, Kupiec M. Effect of nuclear architecture on the efficiency of double-strand break repair. Nat Cell Biol. 2013;15:694–699. doi: 10.1038/ncb2745. [DOI] [PubMed] [Google Scholar]
- 27.Lee CS, et al. Chromosome position determines the success of double-strand break repair. Proc Natl Acad Sci USA. 2016;113:E146–E154. doi: 10.1073/pnas.1523660113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee K, Zhang Y, Lee SE. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature. 2008;454:543–546. doi: 10.1038/nature07054. [DOI] [PubMed] [Google Scholar]
- 29.Karathanasis E, Wilson TE. Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics. 2002;161:1015–1027. doi: 10.1093/genetics/161.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berger AB, et al. High-resolution statistical mapping reveals gene territories in live yeast. Nat Methods. 2008;5:1031–1037. doi: 10.1038/nmeth.1266. [DOI] [PubMed] [Google Scholar]
- 32.Wang RW, Lee CS, Haber JE. Position effects influencing intrachromosomal repair of a double-strand break in budding yeast. PLoS One. 2017;12:e0180994. doi: 10.1371/journal.pone.0180994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat Cell Biol. 2012;14:502–509. doi: 10.1038/ncb2465. [DOI] [PubMed] [Google Scholar]
- 34.Miné-Hattab J, Rothstein R. Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol. 2012;14:510–517. doi: 10.1038/ncb2472. [DOI] [PubMed] [Google Scholar]
- 35.Liang Z, Sunder S, Nallasivam S, Wilson TE. Overhang polarity of chromosomal double-strand breaks impacts kinetics and fidelity of yeast non-homologous end joining. Nucleic Acids Res. 2016;44:2769–2781. doi: 10.1093/nar/gkw013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruiz JF, Pardo B, Sastre-Moreno G, Aguilera A, Blanco L. Yeast pol4 promotes tel1-regulated chromosomal translocations. PLoS Genet. 2013;9:e1003656. doi: 10.1371/journal.pgen.1003656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villarreal DD, et al. Microhomology directs diverse DNA break repair pathways and chromosomal translocations. PLoS Genet. 2012;8:e1003026. doi: 10.1371/journal.pgen.1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu X, Gabriel A. Reciprocal translocations in Saccharomyces cerevisiae formed by nonhomologous end joining. Genetics. 2004;166:741–751. doi: 10.1093/genetics/166.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain S, Sugawara N, Haber JE. Role of double-strand break end-tethering during gene conversion in Saccharomyces cerevisiae. PLoS Genet. 2016;12:e1005976. doi: 10.1371/journal.pgen.1005976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao S, Honey S, Futcher B, Grollman AP. The non-homologous end-joining pathway of S. cerevisiae works effectively in G1-phase cells, and religates cognate ends correctly and non-randomly. DNA Repair (Amst) 2016;42:1–10. doi: 10.1016/j.dnarep.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horigome C, et al. SWR1 and INO80 chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Mol Cell. 2014;55:626–639. doi: 10.1016/j.molcel.2014.06.027. [DOI] [PubMed] [Google Scholar]
- 43.Essers J, et al. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 2002;21:2030–2037. doi: 10.1093/emboj/21.8.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Li M, Lee EY, Maizels N. Localization and dynamic relocalization of mammalian Rad52 during the cell cycle and in response to DNA damage. Curr Biol. 1999;9:975–978. doi: 10.1016/s0960-9822(99)80427-8. [DOI] [PubMed] [Google Scholar]
- 46.Chapman JR, Sossick AJ, Boulton SJ, Jackson SP. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J Cell Sci. 2012;125:3529–3534. doi: 10.1242/jcs.105353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mok MT, Henderson BR. A comparison of BRCA1 nuclear localization with 14 DNA damage response proteins and domains: Identification of specific differences between BRCA1 and 53BP1 at DNA damage-induced foci. Cell Signal. 2010;22:47–56. doi: 10.1016/j.cellsig.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 48.Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 49.Wolner B, van Komen S, Sung P, Peterson CL. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol Cell. 2003;12:221–232. doi: 10.1016/s1097-2765(03)00242-9. [DOI] [PubMed] [Google Scholar]
- 50.Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee SE, et al. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 52.Avşaroğlu B, Bronk G, Li K, Haber JE, Kondev J. Chromosome-refolding model of mating-type switching in yeast. Proc Natl Acad Sci USA. 2016;113:E6929–E6938. doi: 10.1073/pnas.1607103113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parada LA, McQueen PG, Munson PJ, Misteli T. Conservation of relative chromosome positioning in normal and cancer cells. Curr Biol. 2002;12:1692–1697. doi: 10.1016/s0960-9822(02)01166-1. [DOI] [PubMed] [Google Scholar]
- 54.Miné-Hattab J, Rothstein R. DNA in motion during double-strand break repair. Trends Cell Biol. 2013;23:529–536. doi: 10.1016/j.tcb.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belin BJ, Lee T, Mullins RD. DNA damage induces nuclear actin filament assembly by Formin-2 and Spire-1/2 that promotes efficient DNA repair. eLife. 2015;4:e07735. doi: 10.7554/eLife.07735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lottersberger F, Karssemeijer RA, Dimitrova N, de Lange T. 53BP1 and the LINC complex promote microtubule-dependent DSB mobility and DNA repair. Cell. 2015;163:880–893. doi: 10.1016/j.cell.2015.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schrank BR, et al. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature. 2018;559:61–66. doi: 10.1038/s41586-018-0237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caridi CP, et al. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature. 2018;559:54–60. doi: 10.1038/s41586-018-0242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lesterlin C, Ball G, Schermelleh L, Sherratt DJ. RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature. 2014;506:249–253. doi: 10.1038/nature12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cho NW, Dilley RL, Lampson MA, Greenberg RA. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell. 2014;159:108–121. doi: 10.1016/j.cell.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson TE, Lieber MR. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J Biol Chem. 1999;274:23599–23609. doi: 10.1074/jbc.274.33.23599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.