Significance
The alteration of cell communication in the immune system of patients with multiple sclerosis (MS) can be analyzed by phosphoproteomics. We performed in vitro assays on immune cells from MS patients and controls and quantified the phosphorylation of several kinases. We found increased phosphorylation of several MAPK kinases in patients, which were modulated by several genetic markers associated with the disease. By using flow cytometry, we detected several kinases altered by phosphorylation in B cells. These findings indicate the activation of cell survival and proliferation (MAPK), and proinflammatory (STAT) pathways in the immune cells of MS patients, primarily in B cells. The changes in the activation of these kinases suggest that these pathways may represent therapeutic targets for modulation by kinase inhibitors.
Keywords: multiple sclerosis, phosphoproteomics, signaling pathways, B cells, autoimmunity
Abstract
Dysregulation of signaling pathways in multiple sclerosis (MS) can be analyzed by phosphoproteomics in peripheral blood mononuclear cells (PBMCs). We performed in vitro kinetic assays on PBMCs in 195 MS patients and 60 matched controls and quantified the phosphorylation of 17 kinases using xMAP assays. Phosphoprotein levels were tested for association with genetic susceptibility by typing 112 single-nucleotide polymorphisms (SNPs) associated with MS susceptibility. We found increased phosphorylation of MP2K1 in MS patients relative to the controls. Moreover, we identified one SNP located in the PHDGH gene and another on IRF8 gene that were associated with MP2K1 phosphorylation levels, providing a first clue on how this MS risk gene may act. The analyses in patients treated with disease-modifying drugs identified the phosphorylation of each receptor’s downstream kinases. Finally, using flow cytometry, we detected in MS patients increased STAT1, STAT3, TF65, and HSPB1 phosphorylation in CD19+ cells. These findings indicate the activation of cell survival and proliferation (MAPK), and proinflammatory (STAT) pathways in the immune cells of MS patients, primarily in B cells. The changes in the activation of these kinases suggest that these pathways may represent therapeutic targets for modulation by kinase inhibitors.
Analyzing signaling pathways in patients with multiple sclerosis (MS) may provide insights into processes likely to drive the immune cell’s response, as well as to influence the effects of drugs (1). Phosphoproteomic analyses provide opportunities to evaluate the activation of signaling cascades, and such studies may help to identify the pathways activated in cells (2). Mass spectrometry is the technique most commonly used to identify new phosphosites, whereas xMAP or flow cytometry are often used to evaluate the phosphorylation of larger numbers of kinases in vitro or ex vivo (3).
In MS, the immune system is chronically activated, leading to specific damage of the CNS (4, 5). Genome-wide association studies (GWAS) studies suggest that the genetic susceptibility to suffer MS is mainly due to polymorphisms in genes associated with the immune system, consistent with an autoimmune pathogenesis (6). In addition, also lifestyle/environmental factors are likely to act through effects on the adaptive arm of immunity, in view of their potent interactions with HLA class II and I genes, which are the restriction elements for CD4+ and CD8+ T cells, respectively (7). Several pathways in the immune system have been implicated with MS pathogenesis, including those driven by the TCR, IL-2, IL-7, TNFα, or NFkβ (1, 8, 9), yet we still lack a comprehensive understanding of how immune pathways are truly involved in MS.
Immunomodulatory drugs significantly affect signaling in their target cells after receptor or target engagement. For this reason, changes in the phosphoprotein network may represent a sensitive readout of drug activity and the response of immune cells to the drug (3). Generating better information as to how MS alters cell signaling will benefit the development of new therapies to combat this disease.
In this study, we assessed the phosphorylation of key kinases and other proteins in signaling pathways associated with MS (1). Phosphorylation was assessed by performing xMAP assays in vitro on PBMCs isolated from a cross-sectional cohort of MS patients and healthy controls (HCs) following perturbation with several stimuli and drugs. Moreover, we genotyped 112 single-nucleotide polymorphisms (SNPs) associated with MS (6) to evaluate the influence of genetic background on the phosphorylation in these pathways. Immune cell subtype was also characterized in a subgroup of patients by flow cytometry.
Results
Phosphoproteomic Signatures in MS Patients.
To search for signaling pathways in the immune system that are differentially activated in patients with MS, we analyzed the phosphorylation of kinases in PBMCs from MS patients and sex- and age-matched HCs. We studied a cross-sectional cohort of 195 MS patients and 60 HCs (see clinical characteristics in SI Appendix, Table S1) recruited in the CombiMS project. We obtained phosphoproteomic information that conformed the quality control (QC) assessment from 169 individuals (132 MS patients and 37 HCs) and genotyping data from 154 of those 169 individuals (122 MS patients and 32 HC; 14 subjects were excluded because genotyping did not pass QC checks). The final cohort was representative of the recruited cohort and between sites (SI Appendix, Table S1).
The phosphoproteomic assays were designed to study signaling deregulation using dynamic network modeling. This linked study compiled and presented a manually curated immune- and MS-based signaling network (10). After topological analysis, we screened the 70 antibodies for the xMAP assays that maximized coverage of that network, identifying 30 antibodies with good signal-to-noise ratio. We selected a set of 17 assays that were suitable for the multiplex assays. The 17 assays used in the vitro assays were as follows: AKT1, CREB1, FAK1, GSK3A, HSPB1, IKBA, JUN, MK03, MK12, MP2K1, PTPN11, STAT5, STAT1, STAT3, STAT6, TF65, and WNK1 (Fig. 1 and SI Appendix, Table S2). Considering that phosphorylation is a dynamic event that takes place soon after stimulation and that is sensitive to different stimuli, we performed ex vivo assays in PBMCs stimulated with 19 different stimuli known to activate such kinases or that are relevant for MS pathogenesis or therapy, including cytokines, metabolites, and drugs (Fig. 1 and SI Appendix, Table S3). To capture the differences already present at baseline and those that require activation of specific pathways, phosphorylation was analyzed at baseline (unstimulated), and 5 and 25 min after stimulation (stimulated). These time points after stimulation were chosen as they showed the strongest phosphorylation (either 5 or 25 min), as well as the stimulus that produced the most significant differences (SI Appendix, Fig. S1). We checked that no center or batch effect was present in the normalized phosphorylation levels by performing principal component analysis. In addition, we genotyped our cohort for 112 SNPs (6) associated with susceptibility for MS (SI Appendix, Table S4), and these genotypes were used to evaluate the effect of such SNPs on phosphorylation levels (see Dataset S1 for raw genotyping data).
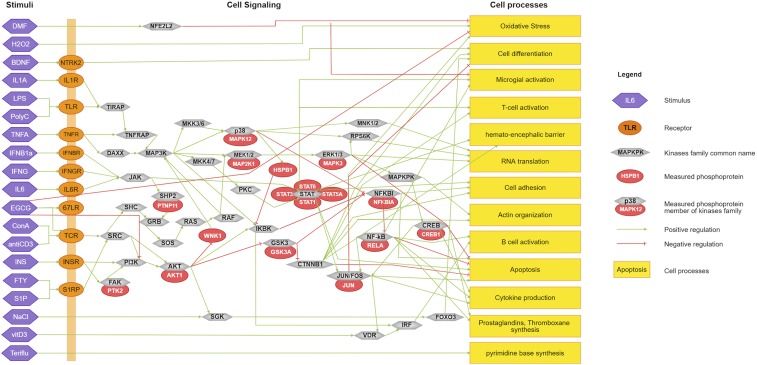
Fig. 1.
The immune signaling network in MS. Shown are the stimuli used for the in vitro assays (purple hexagons on the left), targeting receptors on the cell membrane (orange circles), although some stimuli directly target kinases or pathways (e.g., H2O2 directly induces oxidative stress). Membrane receptors are linked to intracellular kinases (light grey circles) as part of their intracellular signaling networks. The specific phosphoproteins tested in these assays are shown as red circles associated to the master kinase. Finally, kinases influence cellular and molecular processes. Stimulation (e.g., phosphorylation) is indicated by green arrows, whereas inhibitory interactions (e.g., dephosphorylation) is indicated by a red T link.
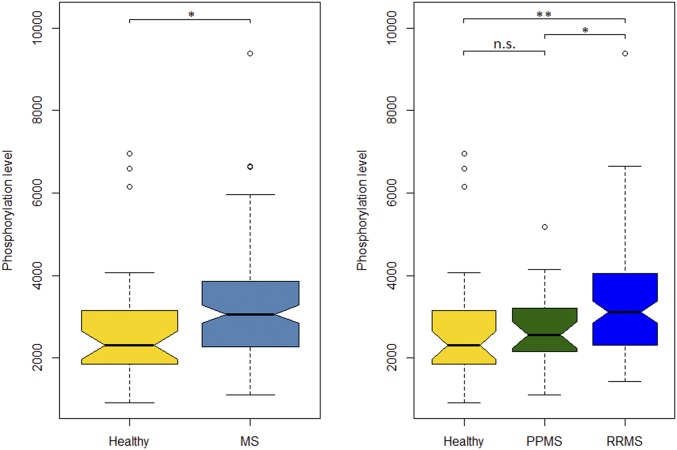
First, we analyzed the baseline differences in phosphoproteins between MS patients and HCs. We found a significant increase in the baseline phosphorylation of MP2K1 in MS patients compared with controls after adjustment for multiple comparisons (false discover rate, FDR). MP2K1 is an element of the MAPK signaling pathway that promotes cell survival and inhibits apoptosis, in particular through the NFkβ cascade. Indeed, this difference remained significant in the relapsing-remitting (RRMS) subgroup after adjusting for multiple comparisons (Wilcoxon test RRMS vs. control P value = 0.0016, FDR = 0.03; Wilcoxon test all MS patients vs. control P = 0.0034, FDR = 0.065: Fig. 2). MKO3, GSK3A, JUN, and STAT3 phosphorylation levels at baseline were also different between MS and controls in the unadjusted analysis, although such differences were not significant after correction for multiple testing (SI Appendix, Table S5). In addition, we found differences in kinase phosphorylation levels when the cells were subjected to distinct stimuli in vitro, although these effects were not significant after adjusting for multiple comparisons.
Fig. 2.
MP2K1 phosphorylation in PBMCs from MS patients and controls. Phosphorylation of MP2K1 in PBMCs from MS patients and controls, as assessed by xMAP and compared with a Wilcoxon test: *P < 0.05; **adjusted P < 0.05. n.s., not significant.
Then, we analyzed how the MS susceptibility genotype influenced phosphorylation levels (Fig. 3 and SI Appendix, Fig. S2). After FDR correction, we found a significant influence of the TT genotype of the rs666930 SNP (located in the phosphoglycerate dehydrogenase gene —PHGDH) on baseline MKO3 and MAP2K1 phosphorylation in MS patients. Similarly, the CT genotype of the rs35929052 SNP [located on IFN regulatory factor 8 (IRF8) gene, a key target of vitamin D receptor; refs. 11 and 12] had a significant influence on MAP2K1 phosphorylation after stimulation with vitamin D3 (Table 1). We found significant effects of several SNPs on the phosphorylation differences between MS and HCs (ANOVA-adjusted P < 0.05). Twelve kinases were shown to be involved in these effects, including AKT1, FAK1, GSK3A, JUN, MKO3, MP2K1, STAT1, STAT3, STAT5, STAT6, TF65, and WNK1 (SI Appendix, Table S6), although only those ones described above for MKO3 and MP2K1 (Table 1) remained significant for the allele-specific pairwise comparisons.
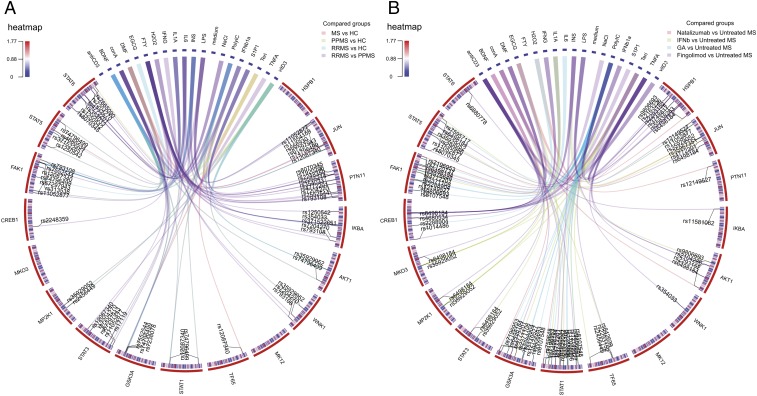
Fig. 3.
The phosphoproteomic signature in MS. The lines in the circle graph show the pairs of stimuli/phosphoproteins found to be significant and the corresponding MS susceptibility genes (ANOVA test for association between kinase levels and SNPs; Benjamini correction for multiple testing). The outer circle shows the 17 phosphoproteins analyzed and the 19 stimuli used, while the inner circle shows the 112 SNPs tested (color coded for the mean allelic distribution) where the SNP indicated by the link is referred to by its letter code. (A) Phosphoproteomic signature in MS, healthy controls, and the RMS and PMS subtypes of MS. (B) Phosphoproteomic signature of the DMDs compared with untreated RRMS patients.
Table 1.
Differential phosphorylation in PBMCs from MS patients relative to healthy controls
| Phosphorylation levels | ||||||
| Group | Stimulus | SNP: allele | MS | HCs | P | Adjusted P |
| MS | ||||||
| MKO3 | Baseline | rs666930: TT | 937 ± 480 | 463 ± 86 | 0.0011 | 0.004 |
| MP2K1 | vitD3 | rs35929052: CT | 0.44 ± 0.42 | −0.24 ± 0.2 | 1.32e-05 | 0.0016 |
| Baseline | rs666930: TT | 3,849 ± 1,794 | 2,041 ± 646 | 0.0019 | 0.04 | |
| RRMS | ||||||
| JUN | INS | rs11554159: GG | 0.12 ± 0.14 | −0.017 ± 0.1 | 0.00022 | 0.027 |
| MP2K1 | vitD3 | rs35929052: CT | 0.48 ± 0.47 | −0.24 ± 0.2 | 6.71e-05 | 0.008 |
The table shows the kinases that were more strongly phosphorylated in MS patients relative to the HCs, indicating the stimulus used in the in vitro assays and the susceptibility SNPs (Benjamin correction for multiple tests). The results are shown as the mean ± SD. Unstimulated basal phosphoprotein levels were compared using a Wilcoxon test, whereas a phosphorylation after stimulation was normalized and compared using a t test. INS, insulin; VitD3, vitamin D3.
Indeed, we assessed whether disease subtype was associated with differential phosphorylation, comparing RRMS and progressive MS with HCs. We found the same differential phosphorylation at baseline for MAP2K1 in RRMS patients relative to HCs but not in PMS (Fig. 2). Regarding the analysis of phosphorylation levels and its association with MS-associated SNPs for disease subtypes, we found significant differences in JUN phosphorylation after insulin stimulation in RRMS individuals with the GG genotype for the rs11554159 SNP, located in the IFN gamma-inducible protein 30 (IFI-30) gene. Similarly, the effect of the CT genotype of the rs35929052 SNP on the phosphorylation MP2K1 following VitD3 stimulation was significantly different between RRMS patients and HCs (Table 1).
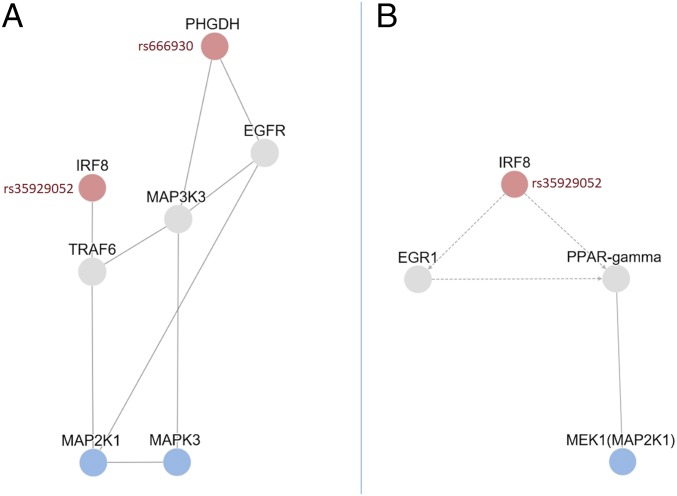
Finally, we assessed the biological relationship of the found genetic and phosphoprotein associations by performing a protein network analysis (see SI Appendix, Supplementary Methods). This pathway analysis indicates that the potential influence of the SNP rs35929052 located in the IRF8 gene on MAP2K1 kinase activity could be mediated by the interaction of IRF-8 with TRAF6 and with changes in the transcription of EGR1 and PPAR-gamma. Moreover, the analysis also indicates that the influence of SNP rs666930 located in the PHGDH gene on MAPK3 can be explained by a physical interaction between PHGDH protein with both EGFR and MAP3K3 proteins (Fig. 4).
Fig. 4.
Protein network analysis of phosphoproteins and SNPs associated with MS. The graph shows the proteins from either genes containing MS-susceptibility SNPs (in red) or phosphokinases (in blue) found associated in our analysis. (A) First-order physical interactions of the studied proteins identified on the iRefIndex and MetaBase/MetaCore database. (B) Transcriptional regulation (dashed lines) and physical interactions (solid lines) of the associated proteins identified using the TieDie algorithm on the background of directed MetaBase network.
Phosphoproteomic Signatures Associated with Immunomodulatory Therapies.
To assess the effects of disease-modifying drugs (DMDs) approved for the treatment of MS on the signaling network in immune cells from patients with MS, we analyzed the phosphoproteomic profile of the 17 kinases in patients treated for more than 1 y with a given DMD and compared it to that in untreated RRMS patients (see SI Appendix, Table S1 for clinical details). We performed the analysis for the most common DMDs at the time of the analysis, namely fingolimod (FTY), natalizumab (NTZ), IFN beta1a s.q. 44 mcg (IFNB1a), and glatiramer acetate (GA). Teriflunomide and dimethyl-fumarate were not included in the analysis because there were insufficient patient numbers treated with these DMDs at our centers by the timing of recruitment.
Regarding the changes in phosphorylation induced by FTY, the phosphorylation of STAT1, MKO3, and PTPN11 after stimulation differed in FTY-treated patients (n = 13) compared with untreated RRMS patients (n = 58: SI Appendix, Table S7). Moreover, we found significant differences between FTY-treated and FTY-untreated RRMS patients after adjustment for genotype, specifically in terms of IKBA phosphorylation for the AG rs11581062 SNP, TF65 phosphorylation for the GG rs2293152 SNP, and CREB1 phosphorylation for the CT rs6498184 SNP (Fig. 3B and SI Appendix, Table S7B). In summary, in FTY-treated patients, we identified several kinases downstream of the S1P receptor that are implicated in the MAPK or NFKβ pathway (e.g., MKO3, IKBA, TF65) as well as several kinases that do not participate in the S1P receptor pathway, yet that were related to pathways involved in the immune response (e.g., STAT1, STAT3, STAT5, STAT6).
Patients treated with NTZ (n = 19) showed differential phosphorylation of STAT3, STAT5, STAT6, and MP2K1 compared with untreated RRMS patients (n = 58: SI Appendix, Table S8). After adjusting for the genotype, we found an association with JUN, AKT1, FAK1, GSK3A, HSPB1, PTN11, STAT1 phosphorylation, and NTZ therapy (Fig. 3B). The VLA4 (ITGA4) receptor interacts with the Ras/MAPK, PI3K, and NFKβ pathways, and our results implicate the activation of several members of the Ras/MAPK (MP2K1, HSPB1) as well as other kinases associated with overall immune activation (STAT3, STAT5, STAT6).
Patients treated with IFNB1a (n = 23) showed distinct STAT1 phosphorylation compared with untreated RRMS patients (n = 58), with a dependence on the AC rs759648 SNP (adjusted P = 0.0088). After adjusting for genetic susceptibility, we observed an association between IFNB therapy and the phosphorylation of JUN, AKT1, FAK1, GSK3A, MKO3, MP2K1, STAT1, STAT3, STAT5, and TF65 (Fig. 3B). Considering that JAK/STAT participate in type 1 IFN receptor pathways, our findings suggest that several kinases in this pathway are likely to be activated (STAT1, STAT3, STAT5), as well as those in other pathways like PI3K, MAPK, or NFKβ, supporting the pleiotropic immunomodulatory activity of IFNB.
Finally, the phosphorylation of STAT6 in response to insulin differed in patients treated with GA (n = 10) compared with untreated RRMS patients (n = 58: pSTAT6 levels in GA-treated patients 0.206585; untreated patients 0.011394; adjusted P = 0.009). The JUN, CREB1, FAK1, GSK3A, STAT1, STAT5 phosphoproteins were more evident in GA-treated patients after adjustment for SNPs of MS susceptibility (Fig. 3B). Although GA seems to primarily mediate the induction of GA-specific regulatory T cells, it also appears to display broad immunomodulatory effects (13). Thus, the activation of such kinases may be related with the immunomodulatory effects of such cells.
Analysis of Kinase Phosphorylation in Immune Cell Subtypes by Flow Cytometry.
To analyze the cell subtypes responsible for the differences in phosphorylation, we analyzed the phosphoproteins levels in PBMCs by flow cytometry. Phosphorylation was assessed in a representative subgroup of 47 MS patients and 22 HCs from the original cohort (SI Appendix, Table S1). We analyzed the phosphorylation of 7 of the 17 kinases used for the xMAP assays for which cytometry assays passed QC and showed a good signal-to-noise ratio: CREB1, HSPB1, IKBA, MK03, MK12, STAT1, STAT3, STAT5, TF65, and WNK1 (SI Appendix, Table S9 shows the list of antibodies used, and SI Appendix, Table S10 shows the list of ex vivo assays conducted on each disease subgroup). However, MP2K1 was not studied in this cytometry substudy due to the lack of antibodies that passed the QC checks and provided good signal-to-noise ratio. We found significant differences in the phosphorylation of HSPB1 in monocytes (CD33+ cells) and STAT3 in B cells (CD19+ cells) from MS patients relative to HCs. Moreover, there were significant differences in the phosphorylation of STAT1, STAT3, and TF65 in CD19+ cells in PPMS patients compared with the HCs. In terms of the differential phosphorylation related to the use of DMDs, we found significant differential expression of HSPB1 in CD19+ cells in patients treated with FTY (SI Appendix, Table S11).
Discussion
We have performed a comprehensive analysis of the phosphoproteomic changes in immune cells from patients with MS. We found distinct patterns of kinase phosphorylation in patients with MS, mainly involving the MAPK pathway but also affecting the NFkβ and STAT pathways. Such pathways are known to be critical for cell survival and proliferation, cell adhesion and chemotaxis, and the proinflammatory response of the immune cells. The phosphoproteomic data were generated to perform logic network modeling (10). With this approach, proinflammatory and prosurvival pathways were found to be deregulated. Further, these pathways were used for combination therapy prediction and subsequently validated (10). Hence, both studies jointly demonstrate that such differential activation can potentially benefit from new immunomodulatory therapies for MS and other autoimmune diseases using approved and novel kinase inhibitors.
Our studies highlight the prominent role of the MAPK pathway in the peripheral immune system of patients with MS, primarily the ERK subpathway. This importance of the MAPK pathway is illustrated not only by the increased phosphorylation of MP2K1 and its downstream target MK03 but also, by that of the p38 subpathway, as reflected by HSPB1 phosphorylation. MP2K1 (also known as CFC3, MEK1, MKK1, MAPKK1, or PRKMK1) promotes cell survival and inhibits apoptosis, in particular through the NFkβ cascade (14). MAP2K1 is activated through KRAS and BRAF activity (e.g., after EGFR activation), and it phosphorylates ERK kinases and interacts with the C-Raf, phosphatidylethanolamine binding protein 1, MAP2K1IP1, GRB10, MAPK3, MAPK8IP3, MAPK1MP1, and MAP3K1. MK03 (also known as ERK1, MAPK3, or PRKM3) is phosphorylated by MAP2K1, contributing to the prosurvival signaling in this pathway. MK03 is important to induce T cell energy and it acts as a negative regulator of dendritic cells, controlling their capacity to prime T cells toward an inflammatory phenotype (15). HSPB1 (Hsp27) is a small heat shock protein that displays chaperone activity, inhibits apoptosis, and regulates cell development and differentiation (16). HSPB1 is also part of the MAPK pathway, as it is activated by the p38 kinase MK2-3 (although it can also be activated by MK5, PRAK, PKCγ, and PKD) and plays an important role in inflammation (16). Moreover, HSPB1 is overexpressed by astrocytes in MS plaques (17), probably in response to the inflammatory stress that helps protect the CNS and prevent apoptosis (18). Due to the key involvement of the MAPK pathway in cancer (RAS, BRAF, CRAF, MEK1, or MEK2 mutations), MEK1 inhibitors like trametinib or cobimetinib have been approved for the treatment of BRAF-mutated melanoma and new ERK1 inhibitors are being actively sought (19). Therefore, there is an opportunity to test approved MEK1 inhibitors, or new ones under development, for their capacity to modulate the immune response in MS and other autoimmune diseases (14).
MS genetic susceptibility also seems to be associated with signal pathway activation as we found several SNPs associated with altered kinase phosphorylation in immune cells. At present we lack sufficient understanding of how genetic polymorphisms regulate protein phosphorylation in a direct or indirect manner, although it is well known that mutations in specific kinases alter their activity and the phosphorylation of their downstream targets (20). Convincing data has appeared with regard to the TYK2 gene polymorphism where a protective variant reduces cytokine signaling, without compromising the defense against infections (21). Up to 10% of the human genome encodes for proteins that modulate phosphorylation or other types of posttranslational protein modifications. Mutations in ligands, receptors, and adaptors affect protein phosphorylation indirectly, thereby suppressing or enhancing the activation of signaling networks (20). Indeed, recent GWAS have shown association signals between SNPs at loci linked to genes encoding kinases and related proteins, and numerous complex and common disease phenotypes (22), including the association of SNPs of TyK2, RPS6KB1, MAPK1, MAPK3, RELA, NFKβ1, STAT3, or STAT4 with MS (6, 8, 21, 23–26). These polymorphisms may be implicated in the direct regulation of signaling pathways or they may serve as markers of regulatory elements that segregate with MS. We here provide preliminary evidences suggesting that MS risk gene SNPs may be associated functionally to kinase signaling.
The selection of SNPs used in this study was based on the GWAS of 2013 (6), but more recent available studies have increased the number of SNPs. In addition, fine-mapping the SNPs associated with phosphoprotein levels would help to characterize the biological basis of the statistical association identified here. For this reason, new studies with high genetic coverage would be required to further characterize the influence of genetic susceptibility in the signaling pathways involved in MS pathogenesis (9).
The effects recorded preferentially on B cells are of great interest, since the depletion of CD20+ cells has demonstrated unexpected high efficacy in MS (27). Several mechanisms for the effect of B cells have been presented, like the B cells capacity to present antigens, their cytokine production, and development into antibody producing cells (28). We here suggest one more potential piece of evidence in the puzzle: a preferential activation of kinases in B cells.
We identified altered phosphorylation of several kinases downstream of the S1P receptor that are involved in the MAPK or NFKβ pathways in patients treated with FTY. Similarly, we found alterations in the phosphorylation of proteins in the VLA4 receptor pathways in patients treated with NTZ, such as ras/MAPK, or the activation of several STATs that participate in type I IFN pathways. Nevertheless, we also observed altered phosphorylation in other pathways. Considering the main mechanism of action of some drugs (FTY and NTZ) is preventing cell migration, it remains unclear to which extent our findings are a direct consequence of the activation of drug-targeted pathways or a reflection of the immune system adaptation to changes in immune cells dynamics. However, the cross-sectional design of our study and the influence of such drugs in PBMC’s composition prevents the establishment of a definite causality between drug use and kinase activation, and for this reason prospective validation studies are required to confirm such findings. Indeed, longitudinal studies are also required for defining the role of these kinases in the response to therapy. Anyway, phosphoproteomics is evolving as the method of choice in some areas of the drug discovery process, and it has also become more suitable for the discovery of novel targets or biomarkers (3).
This study has several limitations, such as the use of a medium throughput approach such as xMAP, or flow cytometry with limited number of assays available with a good signal-to-noise ratio. Indeed, we have measured only levels of phosphorylated proteins, without solving whether this was due to increased phosphorylation or by higher abundance of the kinase or both. However, the kinases we have included in the analysis are known to participate in pathways previously associated with MS and autoimmunity, supporting the biological relevance of our results (1). The selection of the stimuli was based in the literature pointing for T cell activation, and for this reason, specific activators of B cell response were not included. Our analysis simplified the signaling process to the analysis of phosphorylation events but not other posttranslational changes, employing few time-points and conditions. Moreover, our study was cross-sectional, whereas longitudinal analysis will be required to define the response to each of the DMDs at the clinical level and being used for identifying the role of several pathways and kinases in the response to therapy. In addition, we performed many statistical tests and obtained results adjusted for statistical multitesting across the entire study, although this approach does not allow us to exclude false positive results. Indeed, the use of multiple testing corrections decreases the power of the study for detecting true associations and, for example, only MP2K1 remained significant in the comparison between patients and controls. However, despite such limitations, our study provides evidence of the specific activation of the MAPK pathway in the immune cells of MS patients, which may promote the development of MAPK-targeted therapies for MS.
Methods
See SI Appendix, Supplementary Information Text for extended method explanations.
Subjects.
We recruited 255 subjects, 195 patients with MS and 60 healthy controls matched for age and sex with the RRMS group, from four MS centers (SI Appendix, Table S1): Hospital Clinic of Barcelona (n = 69); Karolinska Institute (n = 64); University of Zurich (n = 40), and Charité University (n = 82). Patients fulfilled the McDonald 2005 criteria (29), and their disease subtype was defined using Lublin criteria (30). Patients were allowed to receive any therapy and in the previous 6 mo they had not required any adjustment to their therapy.
xMAP Assays.
xMAP assays were performed blinded at ProtAtOnce (Athens, Greece). We optimized the assays from a list of 70 candidates based in QC checks and the signal-to-noise analysis and obtained a final list of 17 phosphoproteins with optimized assays (SI Appendix, Table S2). We used a set of 19 stimuli that included proinflammatory and prooxidant stimuli; immunomodulatory stimuli; neuroprotectants and antioxidants; and DMDs (SI Appendix, Table S3). Such stimuli are known to activate several pathways known being associated with MS pathogenesis (e.g., MAPK, NFKβ, or STAT) or trigger the activation of DMDs receptors (Fig. 1). The samples were collected at the baseline (time 0) and 5 and 25 min after stimulation. All of the data were normalized after reading the signals. Changes in phosphorylation for each protein and each patient were calculated with respect to the control conditions (31). The phosphorylation of each protein in response to stimulation was defined as the log2 of the response to the stimulus relative to the response to the medium.
Cytometry.
Patient’s samples were washed three times and stained with the antibodies (SI Appendix, Table S6). Four subtypes of immune cells were identified and gated: CD4+ cells, CD8+ cells, B cells (CD19), and monocytes (CD33).
Genotyping.
Genotyping was performed on DNA samples collected from the subjects, assessing SNPs previously validated as associated with MS (6). The final list includes 112 SNPs, including 1 SNP associated with HLA-DRB1*1501 (SI Appendix, Table S4).
Statistical and Bioinformatic Analysis.
We compared pairs of groups using a Wilcoxon test (for baseline) or a t test (for responses to stimuli) using R software. Genotype to phosphoprotein levels associations were tested using two-way ANOVA, and with each SNP as a first independent factor and the patient group as a second factor. The FDR was always corrected using the Benjamini–Hochberg procedure (reported as significant when FDR < 0.05). Protein network analysis is described in SI Appendix, Supplementary Information Text.
Ethics Statement.
This investigation has been conducted according to Declaration of Helsinki principles. The study was approved by the Ethics Committee of the Hospital Clinic of Barcelona, Hospital San Martino of Genova, Charite University of Berlin, and University of Oslo. Patients were recruited by neurologists after they had provided their signed informed consent before inclusion in the study.
Data and Materials Availability.
The phosphoproteomic dataset can be found at https://github.com/saezlab/combiMS, together with the code allowing network modeling of signaling pathways (10).
Supplementary Material
Acknowledgments
We thank Mark Sefton for editorial assistance. This study was supported by the European Commission (CombiMS project) under Grant Agreement 305397 (FP7/2007-2013); Sys4MS project (Horizon2020: Eracosysmed: ID-43); the Instituto de Salud Carlos III (Fondo Europeo de Desarrollo Regional funds “Otra forma de hacer Europa” Grant AC15-00052); and Centres de Recerca de Catalunya Programme/Generalitat de Catalunya.
Footnotes
Conflict of interest statement: D.M. is an employee of ProtATonce; T.O. received honoraria for lectures and/or participation on advisory boards, as well as unrestricted multiple sclerosis research grants from Allmiral, Astrazeneca, Biogen, Genzyme, Merck, and Novartis; R.M. received grants and personal fees from Biogen Idec, personal fees from Genzyme Sanofi Aventis, grants and personal fees from Novartis, and personal fees from Merck Serono, Bionamics, all for work unrelated to that submitted; R.M. received honoraria for lectures and/or participation on advisory boards, as well as unrestricted multiple sclerosis research grants from Biogen, Genzyme, Merck, Celgene, Roche, Novartis, Neuway, and CellProtect, all for work unrelated to that submitted; F.P. received research grants and personal compensation from Alexion, Bayer, Chugai, Novartis, Merck, Teva, Sanofi, Genzyme, Biogen, and MedImmune; L.G.A is the founder and shareholder at ProtATonce; P.V. holds stock in and has received consultancy payments from Bionure Farma SL, QMenta Inc, Health Engineering SL, Spire Therapeutics Inc, and Spire Bioventures Inc.
This article is a PNAS Direct Submission.
Data deposition: Raw data have been deposited at the MultipleMS consortium (https://www.multiplems.eu). The phosphoproteomic dataset can be found on GitHub at https://github.com/saezlab/combiMS.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818347116/-/DCSupplemental.
References
- 1.Kotelnikova E, et al. Signaling networks in MS: A systems-based approach to developing new pharmacological therapies. Mult Scler. 2015;21:138–146. doi: 10.1177/1352458514543339. [DOI] [PubMed] [Google Scholar]
- 2.Stasyk T, Huber LA. Mapping in vivo signal transduction defects by phosphoproteomics. Trends Mol Med. 2012;18:43–51. doi: 10.1016/j.molmed.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Morris MK, Chi A, Melas IN, Alexopoulos LG. Phosphoproteomics in drug discovery. Drug Discov Today. 2014;19:425–432. doi: 10.1016/j.drudis.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Ransohoff RM, Hafler DA, Lucchinetti CF. Multiple sclerosis-a quiet revolution. Nat Rev Neurol. 2015;11:134–142. doi: 10.1038/nrneurol.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotelnikova E, et al. Dynamics and heterogeneity of brain damage in multiple sclerosis. PLOS Comput Biol. 2017;13:e1005757. doi: 10.1371/journal.pcbi.1005757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beecham AH, et al. International Multiple Sclerosis Genetics Consortium (IMSGC); Wellcome Trust Case Control Consortium 2 (WTCCC2); International IBD Genetics Consortium (IIBDGC) Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olsson T, Barcellos LF, Alfredsson L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat Rev Neurol. 2017;13:25–36. doi: 10.1038/nrneurol.2016.187. [DOI] [PubMed] [Google Scholar]
- 8.Housley WJ, et al. Genetic variants associated with autoimmunity drive NFκB signaling and responses to inflammatory stimuli. Sci Transl Med. 2015;7:291ra93. doi: 10.1126/scitranslmed.aaa9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu G, et al. Integrating genome-wide association studies and gene expression data highlights dysregulated multiple sclerosis risk pathways. Mult Scler. 2017;23:205–212. doi: 10.1177/1352458516649038. [DOI] [PubMed] [Google Scholar]
- 10.Bernardo-Faura M, et al. 2019. Prediction of combination therapies based on topological modeling of the immune signaling network in Multiple Sclerosis. bioRxiv:10.1101/541458.
- 11.Ramagopalan SV, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: Associations with disease and evolution. Genome Res. 2010;20:1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horiuchi M, Itoh A, Pleasure D, Ozato K, Itoh T. Cooperative contributions of interferon regulatory factor 1 (IRF1) and IRF8 to interferon-γ-mediated cytotoxic effects on oligodendroglial progenitor cells. J Neuroinflammation. 2011;8:8. doi: 10.1186/1742-2094-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lalive PH, et al. Glatiramer acetate in the treatment of multiple sclerosis: Emerging concepts regarding its mechanism of action. CNS Drugs. 2011;25:401–414. doi: 10.2165/11588120-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arthur JS, Ley SC. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol. 2013;13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 15.Bendix I, et al. MAPK3 deficiency drives autoimmunity via DC arming. Eur J Immunol. 2010;40:1486–1495. doi: 10.1002/eji.200939930. [DOI] [PubMed] [Google Scholar]
- 16.Arrigo AP. Mammalian HspB1 (Hsp27) is a molecular sensor linked to the physiology and environment of the cell. Cell Stress Chaperones. 2017;22:517–529. doi: 10.1007/s12192-017-0765-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peferoen LA, et al. Small heat shock proteins are induced during multiple sclerosis lesion development in white but not grey matter. Acta Neuropathol Commun. 2015;3:87. doi: 10.1186/s40478-015-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brownell SE, Becker RA, Steinman L. The protective and therapeutic function of small heat shock proteins in neurological diseases. Front Immunol. 2012;3:74. doi: 10.3389/fimmu.2012.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: The long and winding road. Nat Rev Cancer. 2015;15:577–592. doi: 10.1038/nrc4000. [DOI] [PubMed] [Google Scholar]
- 20.Cohen P. Immune diseases caused by mutations in kinases and components of the ubiquitin system. Nat Immunol. 2014;15:521–529. doi: 10.1038/ni.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dendrou CA, et al. Resolving TYK2 locus genotype-to-phenotype differences in autoimmunity. Sci Transl Med. 2016;8:363ra149. doi: 10.1126/scitranslmed.aag1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lahiry P, Torkamani A, Schork NJ, Hegele RA. Kinase mutations in human disease: Interpreting genotype-phenotype relationships. Nat Rev Genet. 2010;11:60–74. doi: 10.1038/nrg2707. [DOI] [PubMed] [Google Scholar]
- 23.Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene) Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41:824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 24.Lill CM, et al. International Multiple Sclerosis Genetics Consortium MANBA, CXCR5, SOX8, RPS6KB1 and ZBTB46 are genetic risk loci for multiple sclerosis. Brain. 2013;136:1778–1782. doi: 10.1093/brain/awt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isobe N, et al. International Multiple Sclerosis Genetics Consortium An ImmunoChip study of multiple sclerosis risk in African Americans. Brain. 2015;138:1518–1530. doi: 10.1093/brain/awv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International Multiple Sclerosis Genetics Consortium Network-based multiple sclerosis pathway analysis with GWAS data from 15,000 cases and 30,000 controls. Am J Hum Genet. 2013;92:854–865. doi: 10.1016/j.ajhg.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hauser SL, et al. OPERA I and OPERA II Clinical Investigators Ocrelizumab versus interferon beta-1a in relapsing multiple sclerosis. N Engl J Med. 2017;376:221–234. doi: 10.1056/NEJMoa1601277. [DOI] [PubMed] [Google Scholar]
- 28.Li R, Patterson KR, Bar-Or A. Reassessing B cell contributions in multiple sclerosis. Nat Immunol. 2018;19:696–707. doi: 10.1038/s41590-018-0135-x. [DOI] [PubMed] [Google Scholar]
- 29.Polman CH, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald criteria”. Ann Neurol. 2005;58:840–846. doi: 10.1002/ana.20703. [DOI] [PubMed] [Google Scholar]
- 30.Lublin FD, et al. Defining the clinical course of multiple sclerosis: The 2013 revisions. Neurology. 2014;83:278–286. doi: 10.1212/WNL.0000000000000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen YA, Eschrich SA. Computational methods and opportunities for phosphorylation network medicine. Transl Cancer Res. 2014;3:266–278. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.