Significance
A persistent mystery in olfaction is how the combinatorial activation of a family of 400 olfactory receptors (ORs) encodes odor perception. We take advantage of the high frequency of natural OR knockouts in the human genome to tackle a major bottleneck in the field—namely, how an odor is transduced into perceptual characteristics. We demonstrate that loss of function of an individual OR correlates with changes in perceived intensity and pleasantness. This study demonstrates how natural variation can provide important clues to the normal translation of OR activation to odor information and places a constraint on the amount of redundancy in the olfactory code.
Keywords: olfaction, genetic variation, human genome, odor intensity, ancestry
Abstract
Humans use a family of more than 400 olfactory receptors (ORs) to detect odors, but there is currently no model that can predict olfactory perception from receptor activity patterns. Genetic variation in human ORs is abundant and alters receptor function, allowing us to examine the relationship between receptor function and perception. We sequenced the OR repertoire in 332 individuals and examined how genetic variation affected 276 olfactory phenotypes, including the perceived intensity and pleasantness of 68 odorants at two concentrations, detection thresholds of three odorants, and general olfactory acuity. Genetic variation in a single OR was frequently associated with changes in odorant perception, and we validated 10 cases in which in vitro OR function correlated with in vivo odorant perception using a functional assay. In 8 of these 10 cases, reduced receptor function was associated with reduced intensity perception. In addition, we used participant genotypes to quantify genetic ancestry and found that, in combination with single OR genotype, age, and gender, we can explain between 10% and 20% of the perceptual variation in 15 olfactory phenotypes, highlighting the importance of single OR genotype, ancestry, and demographic factors in the variation of olfactory perception.
Understanding how the olfactory system detects odorants and translates their features into perceptual information is one of the fundamental questions in olfaction. Although early color vision researchers were unable to directly observe receptor responses, perceptual deficits caused by genetic variation (i.e., color blindness) helped show that color vision is mediated by three receptors responding to different wavelengths of light (1, 2). Guillot (3), and then Amoore (4), extended this idea to olfaction and proposed that cataloging specific anosmias (the inability to perceive a particular odorant) may provide similar clues linking genes and perception. Early applications of this idea failed, presumably because olfaction relies on hundreds of receptors, and without direct observation of their responses, psychophysical tests could not untangle the fundamental rules of odor coding. However, with the advent of next-generation genome sequencing to profile olfactory receptor (OR) genes and cell-based assays to identify ligands for ORs, receptor variation can now be matched to individuals and receptor responses can be directly observed.
Humans have approximately 400 OR genes that are intact in at least part of the population, but individuals have different repertoires of pseudogenes, copy number variations, and single nucleotide polymorphisms (SNPs) that can alter receptor responses (5–7). While nonfunctional genes are rare in the genome (on average approximately 100 heterozygous and 20 homozygous pseudogenes in an individual), they are significantly enriched in OR genes (8). This provides a useful set of “natural knockouts” to examine the role of a single OR in olfactory perception and distinguish between different hypotheses of how odor information is encoded. For example, the number of ORs activated by an odorant has been proposed to encode both intensity and pleasantness (9, 10). Alternatively, the large set of ORs may redundantly encode odorant representations, such that loss of function of a single OR only rarely has perceptual consequences. Recent work suggests that functional changes in a single receptor can have significant perceptual consequences, but data linking perceptual and genetic variation exist for only five ORs (11–15).
We extended these studies to the entire OR repertoire, under the premise that studies of natural perceptual variation will improve our understanding of the normal translation of OR activity to odor perception. Here we identified associations between genetic variation in 418 OR genes and 276 olfactory phenotypes, used cell-based functional assays to investigate the mechanisms underlying the associations, and examined the contributions of single OR genotype, genetic ancestry, age, and gender to variations in olfactory perception.
Results
We carried out high-throughput sequencing of the entire OR subgenome in a cohort of 332 participants who had been previously phenotyped for their sense of smell. These data include ratings of the perceived intensity and pleasantness of 68 odorants at two concentrations (SI Appendix, Table S1), detection thresholds of three odorants, and overall olfactory acuity (16). Participants rated each stimulus twice, and the median within-subject test-retest correlation was 0.63 for intensity ratings and 0.57 for pleasantness ratings.
High-Throughput Sequencing of the OR Gene Family.
We used Illumina short-read DNA sequencing to analyze target regions consisting of 418 ORs and 256 olfactory-related genes (∼800 kb), obtaining a minimum of 15× coverage for 96% of the targeted bases and identifying 19,535 variants. We validated a subset of these by comparing them with variants identified from Sanger sequencing data for 10 ORs. Eight ORs had >95% concordance between the two sequencing methods, while concordance was <85% for OR10G4 and OR10G9. These ORs share >95% sequence similarity (17), making it difficult to assign genetic variants to the correct genomic location. Therefore, alternate sequencing methods may prove necessary for ORs with high sequence similarity. However, high-throughput sequencing can be expected to accurately identify sequence variants for ORs in which sequencing reads map with high confidence (90% of ORs have a median mapping confidence >99.9%).
Genetic Variation in Single ORs Frequently Associates with Odorant Perception.
We first examined the association between the genotype of each OR and 276 different phenotypes (Fig. 1, SI Appendix, Fig. S1, and Dataset S1). Eight odorant perception phenotypes (including mixtures; 12% of the tested odors) were significantly correlated with variation in a single OR locus [linear model, P < 0.05 following false discovery rate (FDR) correction]. These results indicate that although a given odorant typically activates multiple ORs, variation in a single OR was frequently associated with perceptual features. For these top associations, OR variation tended to associate with perceived intensity (88% of eight significant associations) rather than with perceived pleasantness (P = 0.07 via a binomial test).
Fig. 1.
Top OR-olfactory phenotype associations. Each row represents the association between an OR genomic locus and the perceived intensity (circles) or pleasantness (squares) of an odor at a high (filled) or low (open) concentration (−log10 P values). Associations to the right of the gray line were significant (P < 0.05) following multiple comparisons correction (FDR). The top 50 associations (P = 0.66 following FDR correction; black line), excluding mixtures, were tested in cell culture. Associated ORs for each locus are listed in order of significance.
In Vitro Assays Confirm Genetic Associations.
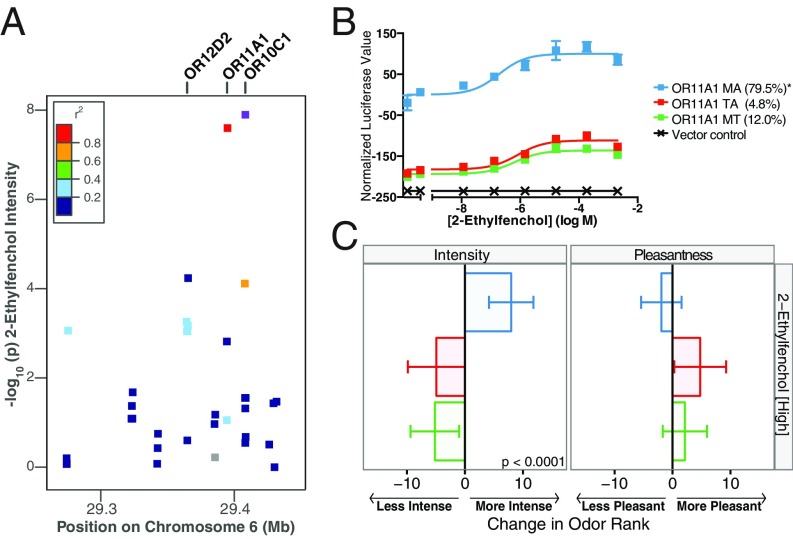
We next used a functional assay to search for a mechanistic explanation for the associations between OR genetic variation and perception. ORs tend to cluster in the genome, and this spatial proximity makes it difficult to discriminate between the causal mutation and nearby SNPs in linkage disequilibrium with the causal mutation (18, 19). To illustrate this point, three different ORs in close proximity on chromosome 6 were significantly associated with the perceived intensity of 2-ethylfenchol: OR11A1, OR12D2, and OR10C1 (Fig. 2A). The most highly associated SNP (Dataset S2), found in OR10C1, is in high linkage disequilibrium with SNPs in OR11A1 and OR12D2, with >60% correlation in the 1000 Genomes Project EUR superpopulation data (20, 21).
Fig. 2.
Functional variation in OR11A1 correlates with the perception of 2-ethylfenchol. (A) Associations (−log10 P values) between SNPs in a locus on chromosome 6 and the perceived intensity of the high concentration of 2-ethylfenchol. The most highly associated SNP is shown in purple, and flanking SNPs are colored according to their linkage disequilibrium with the best-associated SNP, using pairwise r2 values from the 1000 Genomes Project EUR data (20, 21). ORs labeled at the top of the plot were tested in cell culture for their response to 2-ethylfenchol. (B) Response of OR11A1 haplotypes to increasing doses of 2-ethylfenchol. Error bars represent the SEM of three replicates. The y-axis values are normalized to the response of the most responsive haplotype. Letters indicate the amino acids that differ from the hg19 reference haplotype (marked with an asterisk). (C) Changes in perceived intensity and pleasantness rank attributed to a single copy of each OR11A1 haplotype (intensity, P < 0.0001; pleasantness, P > 0.05 following FDR correction).
To investigate these associations, we cloned all major haplotypes in this locus and found that only OR11A1 haplotypes responded in the heterologous assay (Fig. 2B). OR haplotypes with lower maximum responses in cell culture were associated with lower perceived intensity ratings (Fig. 2C). These results indicate that of the three ORs found to significantly associate with the perceived intensity of 2-ethylfenchol in our analysis, OR11A1 was the best candidate for a causal receptor at this locus.
Hyporesponsive OR Haplotypes Are Associated with Changes in Perceived Intensity or Pleasantness.
We performed a similar analysis for our top 50 associated OR/odorant phenotype pairs (Fig. 1), relating the response of associated and linked ORs to odorant perception in our participants. After examining linkage disequilibrium at each locus (SI Appendix, Fig. S2) and removing cases in which multiple ORs from a single locus were associated with the same odorant, we identified a total of 36 unique OR loci-odorant associations. Based on the FDR for these top associations, we expected 66% of them to be false positives (22). To discriminate false positives from true positives, we cloned all major haplotypes for associated ORs, along with at least one haplotype for any OR linked to the associated receptor (SNP correlation >0.6, corresponding to a total of 200 clones; Dataset S3), and tested their response to the associated odorant in our heterologous assay. We tested clones that accounted for on average 73% of the haplotypes found in our participant population.
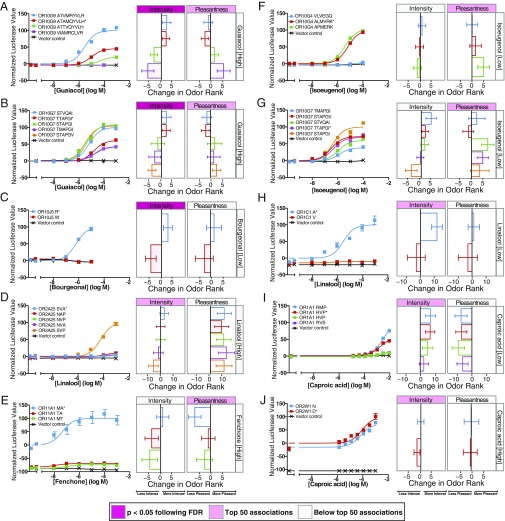
Eleven of these loci (31%) had at least one OR that responded to the associated odorant in cell culture, for a total of 14 responsive ORs (Figs. 2A and 3). Three OR-odorant pairs (OR7D4-androstenone, OR7D4-androstadienone, and OR10G4-guaiacol) were previously identified using this dataset with Sanger sequencing (12, 15) and were confirmed here via Illumina sequencing. In 5 of these 14 cases, subjects with less responsive OR haplotypes rated the perceived intensity of the receptor ligands as less intense compared with subjects with more responsive OR haplotypes (Figs. 2 B and C and 3 C, H, and I), as was also demonstrated by a previously reported correlation between OR7D4 function and the perceived intensity of androstenone (12). In other cases, less responsive OR haplotypes were correlated with greater perceived intensity (Fig. 3D), changes in perceived pleasantness (Fig. 3 E and F), or both (Fig. 3A), as shown by previously reported correlations between OR7D4 and OR10G4 function and the perception of androstadienone (12) and guaiacol (15), respectively. For three ORs, haplotype function did not clearly associate with perception; similar in vitro function was associated with changes in perception (Fig. 3 B and J), or differences in function were associated with little change in perception (Fig. 3G). Overall, in vitro functional variation in a single OR predicted intensity perception, pleasantness perception, or both for 10 different OR-odorant pairs in this dataset, three of which have been reported previously (12, 15).
Fig. 3.
Functional variation in vitro is associated with changes in perceived intensity and pleasantness. Shown are correlations among genetic variation, OR response to odorants in vitro, and perceptual rankings for all OR-odorant associations with at least one responsive OR, excluding previously published OR-odorant pairs (12, 15). In A–J, the Left show the responses of the different OR haplotypes to increasing doses of odorant for OR10G9 and guaiacol (A), OR10G7 and guaiacol (B), OR10J5 and bourgeonal (C), OR2A25 and linalool (D), OR11A1 and fenchone (E), OR10G4 and isoeugenol (F), OR10G7 and isoeugenol (G), OR1C1 and linalool (H), OR1A1 and caproic acid (I), and OR2W1 and caproic acid (J). Error bars represent SEM of three replicates. The y-axis values are normalized to the response of the most responsive haplotype. Haplotypes are listed in order of the associated change in perceived intensity. Letters indicate the amino acids that differ from hg19 reference haplotype (marked with an asterisk). The right panels show the changes in perceived intensity and pleasantness rank attributed to a single copy of the haplotype. Pink labels indicate associations that fall within the top 50, and darker pink labels indicate associations that are significant (P < 0.05) following FDR correction.
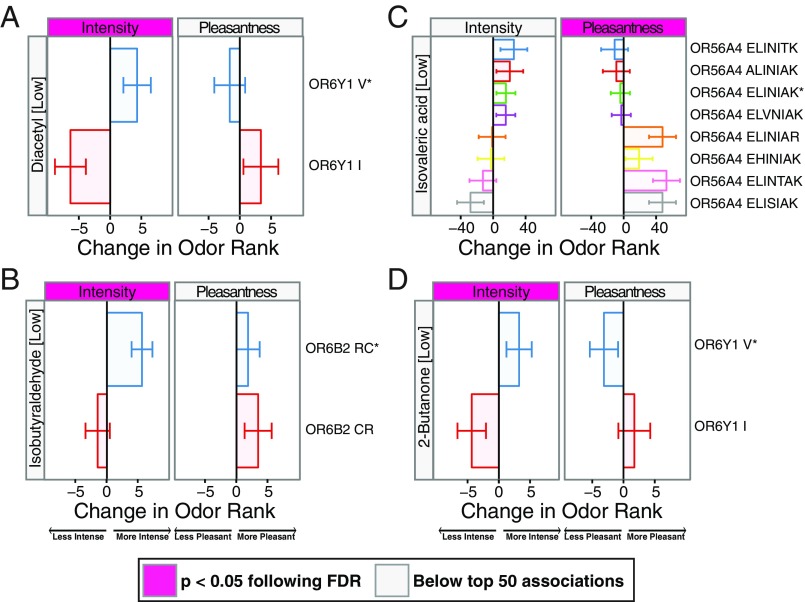
Finally, in four cases, although phenotypes were significantly associated with genetic variation in a single OR locus (P < 0.05 following multiple comparisons correction), we were unable to identify an OR that responded to the associated odorant in cell culture (Fig. 4). Not all ORs have been functionally expressed in in vitro assay systems (23). To determine whether our inability to identify a causal OR was due to technical limitations, we modified the ORs in Fig. 4 using conserved residues in rodent and primate orthologs as a guide and tested their responses to the associated odorant. We produced a consensus version of OR6Y1 by changing nine different amino acids to match the most common amino acid at that position in 10 orthologous rodent and primate receptors (SI Appendix, Fig. S3A). The consensus OR6Y1 responded to diacetyl but not to 2-butanone, providing support for OR6Y1 as the causal receptor for the association with diacetyl (SI Appendix, Fig. S3B).
Fig. 4.
Causal ORs were not identified for all significant associations. Significant associations between genetic variation and perceptual rankings for which we were unable to identify a causal receptor. The changes in perceived intensity and pleasantness rank attributed to a single copy of the haplotype are shown for OR6Y1 and diacetyl (A), OR6B2 and isobutyraldehyde (B), OR56A4 and isovaleric acid (C), and OR6Y1 and 2-butanone (D). Letters indicate the amino acids that differ from hg19 reference sequence (marked with an asterisk). Pink labels indicate associations that fall within the top 50.
Genetic and Demographic Influences on Human Odorant Perception.
We then determined how much phenotypic variance in odorant perception could be explained by four genetic and demographic factors: the genotype of the single OR that explained the greatest variance for each phenotype, genetic ancestry, gender, and age (Dataset S4). All four factors were significant contributors to different olfactory phenotypes, most significantly single OR genotype and genetic ancestry.
Genetic ancestry was quantified by plotting participants according to the first two principal components (PCs) calculated from genotype data (SI Appendix, Fig. S4 A and B). To search for an explanation for the associations with genetic ancestry, we examined the correlation between each PC and odor perception (Dataset S5). Examining participants’ self-reported ancestry demonstrated that PC1 separated Caucasians and Asians from African Americans. PC1 explained >4% of the variance in six different phenotypes (P < 0.01 following FDR correction) (SI Appendix, Fig. S4C), most significantly the perceived pleasantness of vanillin (r = 0.28, P < 0.0001 following FDR correction), which self-reported Caucasians and Asians rated as more pleasant than African Americans did (SI Appendix, Fig. S4D). PC2 separated Caucasians and African Americans from Asians (self-reported), had less effect on olfactory phenotypes (P > 0.05; SI Appendix, Fig. S4E), and explained at most 4% of the variance in spearmint perception (r = −0.20, P = 0.054 following FDR correction), which self-reported Asians tended to rate as more pleasant than African Americans and Caucasians did (SI Appendix, Fig. S4F).
The median percentage of total phenotypic variance that we could explain using single OR genotype, genetic ancestry, gender, and age was 4.77%. Fig. 5 illustrates the relative contribution of all four factors to the 25 phenotypes for which we could explain the most variance (P < 0.0001). The test-retest correlation for a phenotype provided an upper bound for the amount of perceptual variance that we could explain (shown in gray in Fig. 5) (24). For some perceptual phenotypes, our model accounted for >80% of the explainable variance, such as the perceived intensity of diacetyl, for which single OR genotype is the main contributor, and the perceived intensity of nonyl aldehyde, for which age is the main explanatory variable. For other phenotypes, such as the perceived intensity of 2-ethylfenchol and linalool, OR genotype, genetic ancestry, age, and gender accounted for <50% of the explainable variance. These results indicate that while genetic variation in a single OR is an important contributor to perceptual variance, methods that consider multiple ORs may allow us to account for more of the explainable variance in a particular phenotype.
Fig. 5.
Contributions of single OR genotype, genetic ancestry, age, and gender to odorant perception. Shown are the relative contributions of these four genetic and demographic factors to the 25 phenotypes for which we can explain the greatest variance (P < 0.0001 following FDR correction). Covariates that significantly alter the linear model are shown in full color (P < 0.05), as determined by one-way ANOVA comparing the complete model (with all four covariates) with a model excluding the covariate. Gray bars indicate the total explainable variance for each phenotype, as determined by its test-retest value. Bold type indicates a high odorant concentration, and regular type indicates a low odorant concentration.
Discussion
In this large-scale study of the relationships among OR genetic variation, OR activation, and odorant perception, we examined 36 different OR loci-odorant pairs in cell culture, demonstrated that at least one OR from 11 of these loci responded to the associated odorant, and found that OR response in vitro matched the perceived intensity or pleasantness of the associated odorant for 10 different OR-odorant pairs. Three of these pairs have been reported previously (12, 15), and here we describe seven pairs in which genetic variation in a single OR predicts intensity or pleasantness perception.
Our results have important implications for olfactory coding. First, although most odors activate multiple ORs, functional variation in only a single OR altered perception for 13% of 68 odors. This increases the number of known cases directly linking OR activation to odorant perception from 6 to 13 (11–15) and places a constraint on the amount of redundancy in the combinatorial code. These cases provide valuable tools for exploring and manipulating the olfactory code using agonists or antagonists and for examining the contributions of activation of individual ORs to the coding of odorant information, much as characterization of OR responses to ligands in the empty neuron system of Drosophila melanogaster (25) has allowed researchers in the field to choose rationally diverse odorant sets (26) and manipulate specific subpopulations of ORs to dissect olfactory coding (27, 28).
Second, loss of function in an OR was correlated with a decrease in the perceived intensity of the ligand in 8 out of 10 cases in this dataset, in accordance with previous studies (12, 14, 15). The specificity of the effect is inconsistent with a proposed model in which the bulk of ORs encode odorant identity and a subset of broadly tuned receptors encode odorant intensity (29). The repertoire of activated ORs changes with odorant concentration, and thus a genetic variation in an OR will be perceptually relevant only at certain concentrations. We tested two concentrations of each odorant and found that perceived intensity was more strongly associated with genetic variation in a particular OR at one of the concentrations. This finding is consistent with work showing that different receptors in D. melanogaster are necessary for perception in different concentration ranges (30).
Third, loss of function in an OR was correlated with both diminished and enhanced perception of pleasantness for different odors. In most cases, OR variation was correlated with changes in perception of both intensity and pleasantness, although the effect on the latter was usually smaller (below the top 50 associations). In only 1 case out of 10 was variation in OR function related to changes in perceived pleasantness with no associated changes in perceived intensity (isoeugenol in Fig. 3F). One possibility is that the change in intensity is driving pleasantness; however, without knowing how pleasantness changes across a full range of concentrations, we could not test this hypothesis here.
Fourth, we found that genetic variation in a single receptor had a greater effect on intensity and pleasantness than on detection threshold. Previous studies focused on the correlations between OR genetic variation and differences in detection threshold (11, 13, 31), but in our dataset, no single OR was associated with the detection threshold of vanillin, isovaleric acid, or pentadecalactone at a significance level that warranted testing in cell culture. Poor genotyping frequency prevented us from testing a published association between OR11H7 and isovaleric acid detection thresholds (11) (SI Appendix, Fig. S1); however, the OR56A4 genotype was significantly associated with isovaleric acid’s perceived pleasantness, but not with its detection threshold. Similarly, the OR7D4 genotype explains more of the variance in the perceived intensity of androstenone than its detection threshold (12). Differences in phenotype measurements also may have accounted for our failure to find a previously identified association between the OR2J3 genotype and cis-3-hexenol perception, as the original study examined the detection threshold and here we measured perceived intensity (13) (SI Appendix, Fig. S1).
Although we did not confirm our associations in a second population, cell-based assays provide additional evidence of their validity. The high linkage disequilibrium that characterizes many OR loci makes identifying causal ORs difficult using association analyses (18). To overcome these limitations, we used a cell-based assay to identify the response of the associated and linked ORs to odorants. Because heterologous assays have identified ligands for only 12% of the roughly 400 intact human ORs (32), we expected to identify a causal OR for 12% of our associations. Given that the FDR for our top 50 associations was 66%, among the 36 OR-odorant pairs that we examined, we expected to find 12 true positives and 24 false positives. In fact, we found 11 cases in which the associated OR-odorant pair was active in vitro, which was much higher than the expected 12% (approximately one case) based on previous work in heterologous assays.
These results suggest that the receptors relevant to perception are enriched in the set of receptors that respond in the in vitro assay. ORs that are functional in vitro are also more likely to be found in the set of receptors expressed in the human olfactory epithelium (OE) (33). Furthermore, deorphanized (33) or perceptually relevant ORs (11–15) are expressed at 1.5-fold higher levels than other intact ORs (34) (P < 0.0001 for both via a binomial test) (Figs. 2 and 3). One possibility is that a large fraction of intact human ORs are nonfunctional in vivo and not expressed in the OE. In summary, our results indicate that cell-based assays are a useful proxy for identifying behaviorally relevant ORs that are expressed in the OE and whose activation can be directly tied to perception.
Despite these successes, there are certainly cases in which in vitro results do not predict perception. In vitro assays lack critical components of the OE, including proteins in the mucous layer that transport and modify odorant molecules (23). Our cell-based assays were unable to identify a responsive OR for some loci for which we have strong association data (Fig. 4). In one instance, a modified version of OR6Y1 derived from rodent and primate orthologs responded to diacetyl (SI Appendix, Fig. S3), supporting the idea that although the human receptor did not function properly in the in vitro assay, it is the causal receptor that underlies this association. Examining cases in which the in vitro assay does not match perceptual outcomes may provide guidance on how to improve these assays in the future.
Several individual olfactory phenotypes were significantly influenced by ancestry, a common confounding factor in association studies that incorporate different subpopulations (35). Here we quantified ancestry using genetic data, extending a previous examination of the effect of self-reported ancestry on odor perception in this participant cohort (16) by bypassing self-reporting and allowing us to incorporate subjects who self-reported their ancestry as “other.” From these data alone, we were unable to determine whether these differences in odorant perception were due to unknown genetic, cultural, or social factors that cosegregate with ancestry; nonetheless, this work demonstrates the importance of considering ancestry when studying odorant perception in diverse populations. Overall genetic similarity also failed to predict overall perceptual similarity, as the correlations that we identified are obscured by the fact that the vast majority of genetic differences have little or no influence on perception. In a careful search for specific associations, models incorporating OR genotype, ancestry, age, and gender accounted for >70% of the explainable variance (test-retest correlation) for some olfactory phenotypes (guaiacol, diacetyl, and nonyl aldehyde) and <50% of the explainable variance for others (2-ethylfenchol, linalool, and androstadienone). These results indicate that considering the contribution of multiple ORs may be useful in explaining more of the variance for some olfactory phenotypes.
Although we know that the OR gene family is characterized by a large amount of genetic and functional variation, the combinatorial nature of the olfactory code and our limited knowledge of OR-odorant pairs makes it difficult to translate this variation to differences in perception. Here we focused on the perceptual consequences of loss of function of individual ORs, demonstrating that intensity and pleasantness coding for some odors is not redundant, and that loss of function in a receptor reduces the perceived intensity of that receptor’s ligand. Similar studies on colorblindness, a condition in which genetic variation alters color perception, helped determine the tuning of the three photoreceptors to different wavelengths of light. Deciphering the quantitative representation of color by photoreceptors allowed us to digitize color information so that it can be sent and stored without degradation, as well as to develop representations of color space that outline how wavelengths of light can be combined to make novel colors. Understanding how the olfactory receptors encode odors should lead to similar advances in olfaction—namely, digitizing odors and identifying agonists or antagonists of receptors that can produce any desired olfactory percept from a small set of primary odors, if such a set exists.
Materials and Methods
Detailed descriptions of psychophysical testing, sequencing sample preparation and genotyping, association analysis, OR cloning, and the luciferase assay are provided in SI Appendix, Materials and Methods.
Psychophysical Testing.
Collection of psychophysical data was previously reported by Keller et al. (12, 16) and approved by The Rockefeller University’s Institutional Review Board. A total of 391 subjects rated the intensity and pleasantness of 66 odors at two concentrations and two solvents on a scale of 1–7. Odor ratings were ranked within each subject, such that the odorant with the highest-rated intensity or pleasantness for a concentration was ranked as 68, and the odorant with the lowest-rated intensity was ranked as 1.
Sequencing Sample Preparation and Genotyping.
Paired-end sequencing was carried out in 332 participants using an Illumina GAIIx with a read length of 2 × 75 base pairs, and variants were identified using a custom-made pipeline that followed the current best practices recommended for variant detection by the Broad Institute (36, 37). A custom-written R script (38) was used to translate the phased variant call file into 836 full-length haplotypes (418 ORs × a maternal haplotype and a paternal haplotype) for each subject.
Association Analysis.
Genetic associations were analyzed using multiple linear regression to regress OR haplotype count (0, 1, or 2) of individual ORs against all 276 phenotype measurements and incorporating the first two PCs calculated from all subject genetic data, age (in years), or gender as covariates. P values were corrected for multiple comparisons using the FDR (22).
Luciferase Assay.
The Promega Dual-Glo Luciferase Assay System was used to measure in vitro OR activity as described previously (39, 40). Data were analyzed with GraphPad Prism 6.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R03 DC011373 and R01 DC013339 (to J.D.M.); T32 DC000014 and F32 DC014202 (to C.T.); and DC005782, DC012095, and DC014423 (to H.M.); and São Paulo Research Foundation Grant 2017/00726-2 (to M.H.N.). A portion of the work was performed at the Monell Chemosensory Receptor Signaling Core and Genotyping and DNA/RNA Analysis Core, which are supported in part by National Institute on Deafness and Other Communication Disorders Core Grant P30 DC011735. The collection of psychophysical data was supported by Clinical and Translational Science Award UL1 TR000043 from the National Center for Advancing Translational Sciences. L.B.V. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: J.D.M is on the scientific advisory board of Aromyx and receives compensation for these activities.
This article is a PNAS Direct Submission.
Data deposition: The data have been deposited in the database of Genotypes and Phenotypes (dbGaP), www.ncbi.nlm.nih.gov/gap/ (accession no. phs001757.v1.p1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804106115/-/DCSupplemental.
References
- 1.Rushton WA. A cone pigment in the protanope. J Physiol. 1963;168:345–359. doi: 10.1113/jphysiol.1963.sp007196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathans J, Piantanida T, Eddy R, Shows T, Hogness D. Molecular genetics of inherited variation in human color vision. Science. 1986;232:203–210. doi: 10.1126/science.3485310. [DOI] [PubMed] [Google Scholar]
- 3.Guillot M. Physiologie des sensations: Anosmies partielles et odeurs fondamentales. CR Hebd Acad Sci. 1948;226:1307–1309. [Google Scholar]
- 4.Amoore JE. Specific anosmia: A clue to the olfactory code. Nature. 1967;214:1095–1098. doi: 10.1038/2141095a0. [DOI] [PubMed] [Google Scholar]
- 5.Menashe I, Man O, Lancet D, Gilad Y. Different noses for different people. Nat Genet. 2003;34:143–144. doi: 10.1038/ng1160. [DOI] [PubMed] [Google Scholar]
- 6.Hasin-Brumshtein Y, Lancet D, Olender T. Human olfaction: From genomic variation to phenotypic diversity. Trends Genet. 2009;25:178–184. doi: 10.1016/j.tig.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Olender T, et al. Personal receptor repertoires: Olfaction as a model. BMC Genomics. 2012;13:414. doi: 10.1186/1471-2164-13-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacArthur DG, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubin BD, Katz LC. Optical imaging of odorant representations in the mammalian olfactory bulb. Neuron. 1999;23:499–511. doi: 10.1016/s0896-6273(00)80803-x. [DOI] [PubMed] [Google Scholar]
- 10.Haddad R, et al. Global features of neural activity in the olfactory system form a parallel code that predicts olfactory behavior and perception. J Neurosci. 2010;30:9017–9026. doi: 10.1523/JNEUROSCI.0398-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menashe I, et al. Genetic elucidation of human hyperosmia to isovaleric acid. PLoS Biol. 2007;5:e284. doi: 10.1371/journal.pbio.0050284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keller A, Zhuang H, Chi Q, Vosshall LB, Matsunami H. Genetic variation in a human odorant receptor alters odour perception. Nature. 2007;449:468–472. doi: 10.1038/nature06162. [DOI] [PubMed] [Google Scholar]
- 13.McRae JF, et al. Genetic variation in the odorant receptor OR2J3 is associated with the ability to detect the “grassy”-smelling odor, cis-3-hexen-1-ol. Chem Senses. 2012;37:585–593. doi: 10.1093/chemse/bjs049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaeger SR, et al. A Mendelian trait for olfactory sensitivity affects odor experience and food selection. Curr Biol. 2013;23:1601–1605. doi: 10.1016/j.cub.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 15.Mainland JD, et al. The missense of smell: Functional variability in the human odorant receptor repertoire. Nat Neurosci. 2014;17:114–120. doi: 10.1038/nn.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller A, Hempstead M, Gomez IA, Gilbert AN, Vosshall LB. An olfactory demography of a diverse metropolitan population. BMC Neurosci. 2012;13:122. doi: 10.1186/1471-2202-13-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18.Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- 19.Clarke GM, et al. Basic statistical analysis in genetic case-control studies. Nat Protoc. 2011;6:121–133. doi: 10.1038/nprot.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruim RJ, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abecasis GR, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073, and erratum (2011) 473:544. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 23.Peterlin Z, Firestein S, Rogers ME. The state of the art of odorant receptor deorphanization: A report from the orphanage. J Gen Physiol. 2014;143:527–542. doi: 10.1085/jgp.201311151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller A, et al. Predicting human olfactory perception from chemical features of odor molecules. Science. 2017;355:820–826. doi: 10.1126/science.aal2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 26.Haddad R, et al. A metric for odorant comparison. Nat Methods. 2008;5:425–429. doi: 10.1038/nmeth.1197. [DOI] [PubMed] [Google Scholar]
- 27.Bhandawat V, Olsen SR, Gouwens NW, Schlief ML, Wilson RI. Sensory processing in the Drosophila antennal lobe increases reliability and separability of ensemble odor representations. Nat Neurosci. 2007;10:1474–1482. doi: 10.1038/nn1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen SR, Wilson RI. Lateral presynaptic inhibition mediates gain control in an olfactory circuit. Nature. 2008;452:956–960. doi: 10.1038/nature06864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Firestein S. How the olfactory system makes sense of scents. Nature. 2001;413:211–218. doi: 10.1038/35093026. [DOI] [PubMed] [Google Scholar]
- 30.Asahina K, Louis M, Piccinotti S, Vosshall LB. A circuit supporting concentration-invariant odor perception in Drosophila. J Biol. 2009;8:9. doi: 10.1186/jbiol108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McRae JF, et al. Identification of regions associated with variation in sensitivity to food-related odors in the human genome. Curr Biol. 2013;23:1596–1600. doi: 10.1016/j.cub.2013.07.031. [DOI] [PubMed] [Google Scholar]
- 32.de March CA, Ryu S, Sicard G, Moon C, Golebiowski J. Structure-odour relationships reviewed in the postgenomic era. Flavour Fragrance J. 2015;30:342–361. [Google Scholar]
- 33.Verbeurgt C, et al. Profiling of olfactory receptor gene expression in whole human olfactory mucosa. PLoS One. 2014;9:e96333. doi: 10.1371/journal.pone.0096333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olender T, et al. The human olfactory transcriptome. BMC Genomics. 2016;17:619. doi: 10.1186/s12864-016-2960-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Der Auwera GA, et al. 2013. From FastQ data to high-confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43;11.10.1–11.10.33.
- 37.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R Development Core Team 2008. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna), Version 3.3.3.
- 39.Zhuang H, Matsunami H. Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat Protoc. 2008;3:1402–1413. doi: 10.1038/nprot.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trimmer C, Snyder LL, Mainland JD. High-throughput analysis of mammalian olfactory receptors: Measurement of receptor activation via luciferase activity. J Vis Exp, 2014:e51640. doi: 10.3791/51640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.