Significance
A sequence variant (I148M) in PNPLA3 is a major genetic risk factor for nonalcoholic fatty liver disease. Previously, we showed that PNPLA3(148M) evades ubiquitylation-mediated degradation and accumulates to high levels on lipid droplets (LDs). Here we address how this accumulation is related to steatosis. We generated an active, ubiquitylation-resistant isoform that accumulated on LDs and increased hepatic triglyceride levels when expressed in livers of mice. Conversely, depletion of PNPLA3 resolved the excess hepatic fat accumulation associated with expression of PNPLA3(148M). Our results provide direct evidence that accumulation of PNPLA3 per se causes fatty liver, and that depletion of the protein is a potential strategy for therapeutic intervention.
Keywords: PNPLA3, fatty liver disease, lipid droplet, autophagy, proteasome
Abstract
Fatty liver disease (FLD) is a disorder in which accumulation of triglycerides (TGs) in the liver can lead to inflammation, fibrosis, and cirrhosis. Previously, we identified a variant (I148M) in patatin-like phospholipase domain-containing protein 3 (PNPLA3) that is strongly associated with FLD, but the mechanistic basis for the association remains elusive. Although PNPLA3 has TG hydrolase activity in vitro, inactivation or overexpression of the WT protein in mice does not cause steatosis. In contrast, expression of two catalytically defective forms of PNPLA3 (I148M or S47A) in sucrose-fed mice causes accumulation of both PNPLA3 and TGs on hepatic lipid droplets (LDs). To determine if amassing PNPLA3 on LDs is a cause or consequence of steatosis, we engineered a synthetic isoform of PNPLA3 that uncouples protein accumulation from loss of enzymatic activity. Expression of a ubiquitylation-resistant form of PNPLA3 in mice caused accumulation of PNPLA3 on hepatic LDs and development of FLD. Lowering PNPLA3 levels by either shRNA knockdown or proteolysis-targeting chimera (PROTAC)-mediated degradation reduced liver TG content in mice overexpressing PNPLA3(148M). Taken together, our results show that the steatosis associated with PNPLA3(148M) is caused by accumulation of PNPLA3 on LDs.
Nonalcoholic fatty liver disease (FLD) is a burgeoning metabolic disorder in which features of alcohol-associated liver disease develop in individuals who consume little or no alcohol (1). Accumulation of triglycerides (TGs) in the liver (hepatic steatosis) is the first stage of the disorder. In a subset of individuals, steatosis is associated with an inflammatory response (steatohepatitis) that can progress to cirrhosis and even hepatocellular carcinoma (2). The primary risk factor for nonalcoholic FLD is obesity, but genetic factors play a major role in susceptibility (and resistance) to the disorder (2–5). Previously, we identified a nonsynonymous variant (I148M) in patatin-like phospholipase domain-containing protein 3 (PNPLA3) that is associated with the full spectrum of both alcoholic and nonalcoholic FLD (6–10). Although PNPLA3(148M) is the most common and strongest genetic risk factor for nonalcoholic FLD (3, 11), the mechanism by which this variant increases liver TG levels remains elusive.
In vitro assays using purified recombinant PNPLA3 indicate that the enzyme has TG hydrolase activity that is reduced ∼80% by the I148M substitution (12). Loss of PNPLA3-mediated TG hydrolase activity alone is unlikely to cause hepatic steatosis, since Pnpla3−/− mice do not have increased liver TG levels (13, 14). Overexpression of PNPLA3(148M), but not PNPLA3(WT), results in hepatic steatosis (15). Moreover, 148M knockin (KI) mice (Pnpla3148M/M) develop FLD when challenged with a high-sucrose diet (HSD) (16), which stimulates PNPLA3 expression in liver (17). A similar phenotype is seen in mice expressing a catalytically dead form of the protein (Pnpla347A/A) (16). Thus, the FLD associated with PNPLA3(148M) requires two conditions: (i) the protein must be expressed (since Pnpla3−/− mice do not develop hepatic steatosis), and (ii) the expressed protein must have reduced TG hydrolase activity, since overexpression of catalytically impaired forms of PNPLA3 (I148M and S47A) causes steatosis whereas overexpression of the wild-type (WT) protein does not (15). In KI mice that express PNPLA3(148M) or PNPLA3(47A), the levels of PNPLA3 on hepatic lipid droplets (LDs) are ∼40-fold higher than those in WT mice, despite similar levels of PNPLA3 mRNA in the two lines (16). A similar accumulation of PNPLA3 protein was observed in transgenic mice expressing human PNPLA3(148M) compared with mice expressing the WT transgene (15). The massive increase in PNPLA3(148M and 47A) levels appears to be due to decreased ubiquitylation of the mutant proteins, which leads to decreased proteasomal degradation (18).
Here we ask whether accumulation of PNPLA3(148M or 47A) on LDs is a cause or consequence of the associated steatosis. If accumulation of PNPLA3 per se is causal, then accumulation of the WT protein, which is usually maintained at low levels (16), should result in FLD. Conversely, reduction in the amount of PNPLA3(148M) should eliminate the FLD phenotype. To test this hypothesis, we used two complementary strategies: First, we engineered a form of the WT protein that retained activity but evaded proteasomal degradation, and thus accumulated to high levels on LDs. Expression of this isoform caused hepatic steatosis in WT mice. Second, we reduced levels of PNPLA3(148M) on LDs using two different methods: (i) shRNA to knock down PNPLA3 expression, and (ii) proteolysis-targeting chimera (PROTAC)-mediated degradation of Halo-tagged PNPLA3(148M). Both strategies lowered PNPLA3 levels on LDs and reduced hepatic steatosis.
Results
Inhibition of Autophagy Promotes Accumulation of PNPLA3.
Previously, we showed that PNPLA3(WT) is polyubiquitylated and degraded by proteasomes (18). Treatment with the proteasome inhibitor bortezomib results in an increase in hepatic PNPLA3 in WT mice but not in mice expressing the 148M isoform (18). In contrast, treatment with an inhibitor of macroautophagy, 3-methyladenine (3-MA), failed to alter PNPLA3 levels (18). However, the efficacy of 3-MA as a macroautophagy inhibitor has been disputed (19). Therefore, to further evaluate the role of autophagy in PNPLA3 degradation, we used two alternative inhibitors of the pathway.
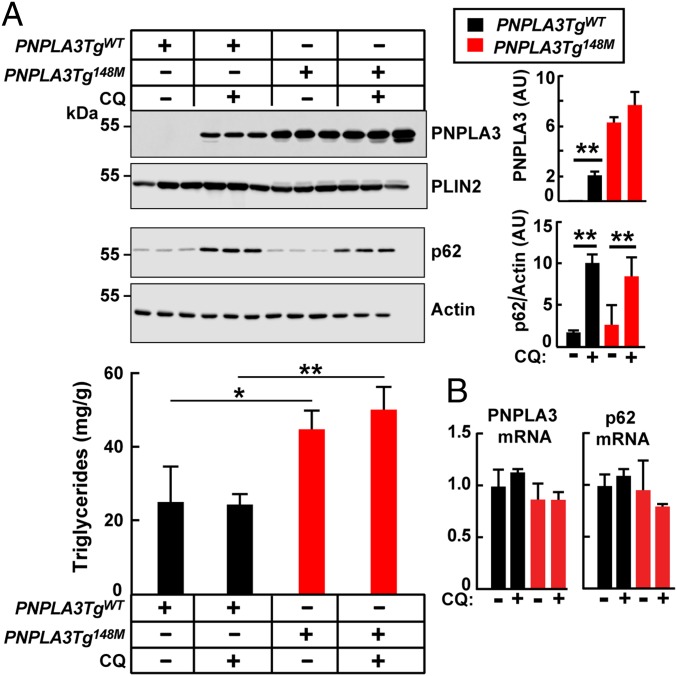
First, we treated transgenic mice expressing either PNPLA3(WT) or PNPLA3(148M) with chloroquine (25 mg/kg) for 3 h (20) (Fig. 1A, Upper). In PNPLA3TgWT mice, chloroquine treatment increased PNPLA3 levels on LDs ∼20-fold compared with vehicle control. The changes in PNPLA3 protein levels were not associated with changes in PNPLA3 mRNA levels (Fig. 1B). In contrast to mice expressing the WT transgene, chloroquine treatment had no effect on PNPLA3 levels in PNPLA3Tg148M mice. As a positive control for chloroquine-mediated inhibition of autophagy, we monitored levels of sequestosome 1 (SQSTM1)/p62, an autophagy substrate (21). Chloroquine treatment resulted in comparable increases in levels of p62 in the two lines of transgenic mice. Despite the increased PNPLA3 levels in the PNPLA3TgWT mice, no change was seen in the level of hepatic TGs (Fig. 1A, Lower).
Fig. 1.
Chloroquine treatment increases levels of hepatic PNPLA3 in mice. (A) Male PNPLA3TgWT and PNPLA3Tg148M mice (n = 4 per group, aged 11 to 12 wk) were fed an HSD for 4 wk and then placed on the synchronization protocol. Chloroquine (CQ) phosphate sodium salt (Sigma) was dissolved in PBS and administered to mice (25 mg/kg) via tail vein injection. Control animals were injected with vehicle alone. Mice were killed 3 h after injection, and liver lysates and LDs were isolated as described in SI Appendix, Methods. A total of 2 μg of LD protein was subjected to immunoblot analysis using an anti-human PNPLA3 mAb (11C5) and a guinea pig anti-mouse perilipin 2 (PLIN2) polyclonal Ab. Liver lysate (25 μg) was immunoblotted with an SQSTM1/p62-specific mAb as described in SI Appendix, Methods. Signals were quantified using a LI-COR imaging system and expressed as arbitrary units (AUs). Liver lipids were measured as described in SI Appendix, Methods. Each bar represents mean ± SEM values. Mean values were compared among lines using Student’s t test. *P < 0.05, **P < 0.01. (B) Chloroquine treatment did not change the mRNA levels of PNPLA3 or p62. Total hepatic mRNA was prepared from the livers of the mice described in A, and real-time PCR quantification was performed as described in SI Appendix, Methods. The experiment was repeated twice and the results were similar.
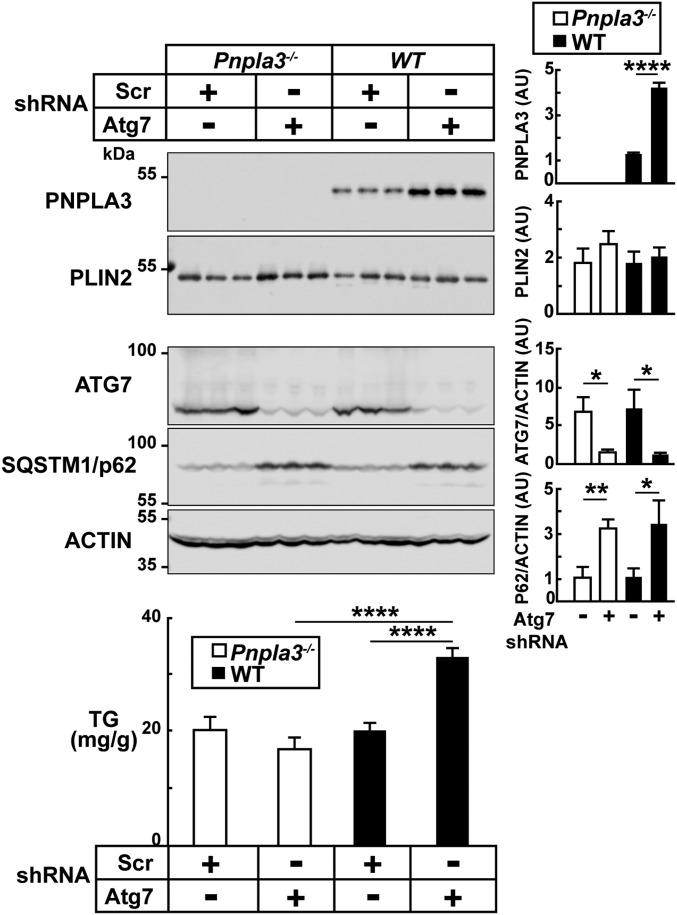
Since chloroquine has effects on protein degradation that are independent of autophagy (22), we inactivated autophagy more specifically by genetically depleting autophagy-related 7 (Atg7), a key component of the autophagy machinery (23). Sucrose-fed Pnpla3−/− (13, 14) and WT mice were infected with recombinant adeno-associated viruses (AAVs) expressing either an shRNA targeting the Atg7 transcript or a scrambled (Scr) shRNA. Levels of ATG7 protein were effectively reduced in mice treated with the Atg7-targeted shRNA (Fig. 2, Middle). Comparable inhibition of autophagy was confirmed by analysis of p62, which increased to a similar extent in the Pnpla3−/− and WT mice. In WT animals, Atg7 knockdown (KD) was associated with a threefold increase in PNPLA3 levels on hepatic LDs (Fig. 2, Top). Hepatic TG content was also increased in the WT mice, but not in the Pnpla3−/− mice (Fig. 2, Bottom). Thus, inhibition of autophagy increased both the levels of PNPLA3 and TGs on hepatic LDs. Moreover, the development of steatosis in association with inactivation of autophagy was dependent on expression of PNPLA3, since it did not occur in the Pnpla3−/− mice.
Fig. 2.
Genetic depletion of ATG7 in livers of mice increases levels of hepatic PNPLA3 and TGs. Male Pnpla3−/− and WT mice (n = 7 per group, aged 9 to 11 wk) were fed an HSD for 2 wk and then treated with an shRNA targeted against Atg7 or a scrambled shRNA (AAV-Atg7-shRNA or AAV-Scr, respectively). Recombinant AAVs [1.5 × 1011 genome copies (GCs)] were administered via the tail vein, and the mice were maintained on an HSD for another 2 wk. Mice were then switched to the synchronization protocol and killed after 3 d. Hepatic LDs were isolated and processed as described in Methods. Levels of PNPLA3 in LDs were analyzed by immunoblotting using a rabbit anti-mouse PNPLA3 monoclonal antibody (19A6), quantified using a LI-COR imaging system, and expressed as arbitrary units as described in the legend to Fig. 1. PLIN2 was analyzed as a loading control. Levels of ATG7, SQSTM1/p62, and actin were assessed by immunoblot analysis of liver lysates (25 μg per lane). Hepatic TGs were measured as detailed in SI Appendix, Methods. Each bar represents mean ± SEM values. Signal intensities were compared using Student’s t test. *P < 0.05, **P < 0.01, ****P < 0.0001. The experiment was repeated twice and the results were similar.
If the increase in hepatic TGs in this experiment is a consequence of accumulation of PNPLA3 on LDs, why was steatosis seen in mice treated with Atg7 KD but not in those treated with chloroquine (Fig. 1)? One possible explanation is the difference in the time course of the two experiments. The livers of the chloroquine-treated animals were harvested after 3 h of treatment, which is the optimal time to see the effects of chloroquine on autophagy in vivo (20, 24). In contrast, the mice infected with the AAV-Atg7-shRNA were killed after 2 wk. Three hours is insufficient time to develop the steatosis associated with PNPLA3 accumulation on LDs (18).
Disruption of PNPLA3 Ubiquitylation Results in Accumulation of PNPLA3 and TGs on Hepatic LDs.
To determine the site(s) at which PNPLA3 undergoes ubiquitylation, we expressed the patatin domain (1 to 276) [PNPLA3(PAT)] and the C-terminal (C-ter) region (277 to 481) of PNPLA3 in HepG2 cells. Cells were treated with MG132 (10 µM) for 8 h to inhibit proteasome-mediated degradation (17). As reported previously (17), levels of PNPLA3 were increased in lysates from cells treated with the proteasome inhibitor (SI Appendix, Fig. S1). A similar increase in the amount of PNPLA3 was seen in cells expressing either PNPLA3(PAT) or PNPLA3(C-ter), although the C-ter region was poorly expressed in these cells. We concluded from this experiment that lysine residues in both halves of the protein undergo ubiquitylation (SI Appendix, Fig. S1A).
To determine if the steatosis associated with PNPLA3(148M and 47A) requires loss of TG hydrolase activity, or if accumulation of PNPLA3 with an intact catalytic site would have a similar effect, we created an expression construct in which all 19 lysine residues in PNPLA3(WT) were replaced with arginines (PNPLA3:K→R). We predicted that (PNPLA3:K→R) would not be ubiquitylated, and would avoid degradation and accumulate on LDs.
First, we tested if the substitution of arginines for lysines in PNPLA3 inhibited ubiquitylation of the protein (SI Appendix, Fig. S1B). AAV-infected mice expressing PNPLA3(WT), PNPLA3(148M), and PNPLA3(K→R) or vector alone (V) (with a tandem array of three Flag and two Strep epitope tags at the C-ter region) were treated with bortezomib for 5 h. After treatment, PNPLA3 was immunoprecipitated from LDs using an anti-Flag monoclonal antibody (mAb). The immunoprecipitates were then immunoblotted with anti-PNPLA3 and anti-Ub (ubiquitin) Abs (SI Appendix, Fig. S1B). Treatment with bortezomib increased levels of PNPLA3(WT) 20-fold compared with mice treated with vehicle alone (SI Appendix, Fig. S1B, Upper), and resulted in the appearance of a higher molecular-weight band that reacted with the anti-Ub antibody (SI Appendix, Fig. S1B, Lower). Mice expressing PNPLA3(148M) had much higher PNPLA3 levels at baseline. Bortezomib treatment did not further increase either the ubiquitylation or levels of PNPLA3 in the PNPLA3(148M)-expressing mice (SI Appendix, Fig. S1B, Lower). Mice expressing PNPLA3(K→R) had levels of PNPLA3 similar to those of mice expressing PNPLA3(148M). Bortezomib treatment was not associated with any further increase in the levels of PNPLA3(K→R) or with the appearance of higher molecular-weight ubiquitylated forms of the protein (SI Appendix, Fig. S1B, Lower).
Impairing Clearance of Ubiquitylation-Resistant PNPLA3 Causes Hepatic TG Accumulation.
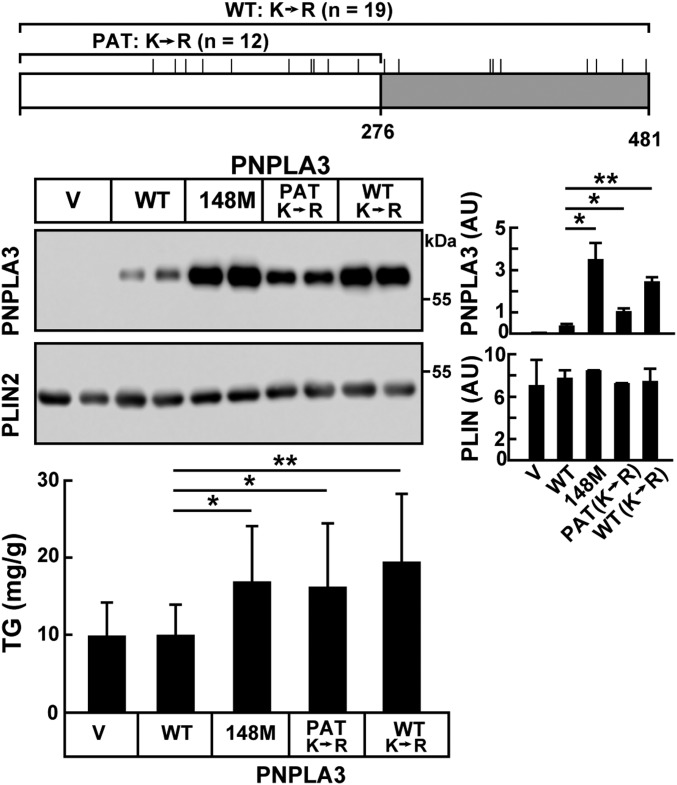
If accumulation of TGs in the liver is due to defective ubiquitylation and clearance of PNPLA3(148M), then AAV-mediated expression of a ubiquitylation-resistant isoform of PNPLA3(WT) [PNPLA3(K→R)] should be associated with fatty liver. Our findings were entirely consistent with this prediction (Fig. 3). We also included in this experiment an expression construct in which only the 12 lysines in the PAT domain were changed to arginines [PNPLA3(PAT:K→R)] (Fig. 3, Top). Mice expressing PNPLA3(PAT:K→R) had levels of PNPLA3 that were intermediate between those of PNPLA3(WT) and PNPLA3(K→R) mice (Fig. 3, Middle). The levels of PNPLA3 on the LDs of the PNPLA3(K→R) mice were similar to those seen in the PNPLA3(148M)-expressing mice.
Fig. 3.
Hepatic TG accumulation in ubiquitylation-resistant PNPLA3. A schematic of PNPLA3 showing the location of the lysine residues that were changed to arginines in either the total protein [PNPLA3(K→R)] (n = 19) or in just the patatin domain [PNPLA3(PAT:K→R)] (n = 12). Male WT mice (n = 7 per group, aged 8 to 11 wk) were infected with AAV (1.25 × 1011 GCs) containing no insert (V) or with AAVs expressing PNPLA3(WT), PNPLA3(148M), PNPLA3(PAT:K→R) (in which the 12 lysines in the patatin domain were replaced with arginines), or [PNPLA3(K→R)] (in which all 19 lysines in the protein were replaced with arginines). Mice were fed an HSD for 3 wk and then placed on a dietary synchronization protocol. After the 3-d protocol, mice were killed and LDs were isolated from the liver. A total of 2 μg of LD protein was subjected to immunoblot analysis with an anti-PNPLA3 mAb (11C5) and an anti-PLIN2 polyclonal Ab (Methods). The signals were quantified using a LI-COR imaging system. The TGs were measured from liver lysates using an enzymatic assay. Each bar represents mean ± SEM values. *P < 0.05, **P < 0.01. The experiment was repeated and the results were similar.
Hepatic TG levels were significantly higher in mice expressing PNPLA3(PAT:K→R) or PNPLA3(K→R) than in those expressing the WT protein (Fig. 3, Lower). These results are consistent with the notion that PNPLA3 accumulation is causally linked to hepatic steatosis. A caveat to this conclusion is that the K→R substitutions may alter the localization or function of PNPLA3.
PNPLA3(K→R) Localizes to LDs and Promotes TG Clearance in Cultured Hepatocytes.
To examine the effect of the lysine substitutions on PNPLA3 localization and activity, we took advantage of our recent observation that coexpression of PNPLA3(WT) and comparative gene identification 58 (CGI-58) in cultured cells depletes them of LDs, whereas coexpression of PNPLA3(47A) and CGI-58 does not (25). CGI-58 is a cofactor of adipocyte triglyceride lipase (also known as PNPLA2), the PNPLA family member that is most closely related to PNPLA3 (26). Since the TG-clearing activity of PNPLA3 in cells is dependent on the integrity of the catalytic serine, this observation provided an assay to assess the effects of the lysine substitutions on PNPLA3 activity. Accordingly, we expressed PNPLA3 alone or together with CGI-58 in cells supplemented with oleic acid (OA) (200 μM) to stimulate formation of LDs (SI Appendix, Fig. S2).
Similar to the expression of either PNPLA3(WT) or PNPLA3(47A), the lysine-less form of PNPLA3 colocalized with LDs (SI Appendix, Fig. S2A). We then expressed the three PNPLA3 constructs together with CGI-58 in cells. When CGI-58 was expressed alone, it was present in a diffuse, nonnuclear pattern. When CGI-58 was expressed together with PNPLA3(WT), no LDs were seen and both PNPLA3 and the cofactor were diffusely distributed in the cytoplasm. In contrast, when CGI-58 was expressed with a catalytically dead form of the protein [PNPLA3(47A)], LDs were present and both PNPLA3 and CGI-58 localized on LDs (SI Appendix, Fig. S2B). Thus, coexpression of CGI-58 with PNPLA3 promoted LD depletion, which was inhibited by substitution of alanine for serine at the active site of the enzyme. Coexpression of CGI-58 with the lysine-less form of PNPLA3 resulted in a distribution of the enzyme that was similar to that seen in cells expressing CGI-58 and PNPLA3(WT) (SI Appendix, Fig. S2B). From this experiment, we concluded that the lysine-less form of PNPLA3 does not alter the localization of PNPLA3 or its effect on TG accumulation in cells.
If accumulation of PNPLA3(148M) on LDs causes the development of steatosis, then reducing levels of PNPLA3 in the liver should reverse the fatty liver in mice expressing PNPLA3(148M). We tested this prediction using two approaches.
Pnpla3 Knockdown Resolves Fatty Liver in Pnpla3148M/M Mice.
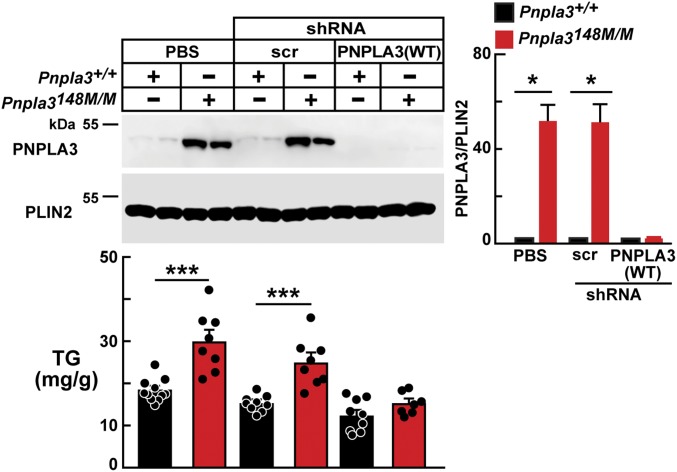
To test the hypothesis that reducing Pnpla3 expression would alleviate fatty liver in the Pnpla3148M/M mice, we used AAVs to express an shRNA targeting Pnpla3 in the livers of mice. We used a 2-wk high-fructose diet to elicit accumulation of PNPLA3 and an FLD phenotype (SI Appendix, Fig. S3). Expression of Pnpla3 shRNA in fructose-fed mice completely abrogated expression of PNPLA3 (Fig. 4) and was associated with a reduction in hepatic TG levels that was not seen in mice infected with the Scr shRNA or in the PBS-treated control groups (Fig. 4). These results suggest that the accumulation of PNPLA3 per se causes the steatosis associated with the PNPLA3(148M) variant. Importantly, this experiment also implies that interventions that effectively lower PNPLA3 levels will reverse steatosis in individuals with the PNPLA3(148M) variant.
Fig. 4.
Pnpla3 knockdown ameliorates fatty liver in the Pnpla3148M/M mice. Male Pnpla3+/+ and Pnpla3148M/M mice (n = 7 per group, aged 7 to 9 wk) were injected with AAV-expressing anti-Pnpla3 shRNA (2.25 × 1011 GCs). The mice were fed a high-fructose diet for 2 wk before dietary synchronization. Livers were harvested and LDs were prepared as described in SI Appendix, Methods. LD proteins (4 μg) were analyzed by immunoblotting using an anti-rabbit mAb (19A6) and guinea pig anti-mouse PLIN2 polyclonal Ab (Methods). Each bar represents mean ± SEM values. *P < 0.05, ***P < 0.001. The experiment was repeated and the results were similar.
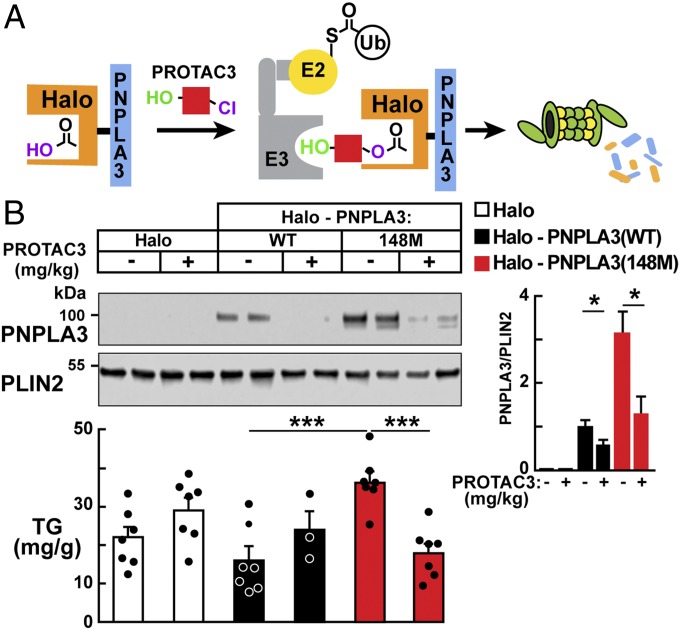
PROTAC-Mediated Degradation of Halo-PNPLA3(148M) Ameliorates Fatty Liver.
We used an independent strategy to lower hepatic PNPLA3 levels posttranslationally. In this experiment, we used a heterobifunctional proteolysis-targeting chimera (PROTAC3) comprising a hydroxyproline derivative that has high affinity for the E3 ligase VHL, coupled to a chloroalkane, which binds covalently to a modified bacterial dehalogenase (Halo). The Halo tag is expressed as a fusion protein with PNPLA3 (Fig. 5A) (27, 28). PROTAC3 would be anticipated to recruit VHL to Halo-PNPLA3(148M) and thereby promote ubiquitylation and degradation of the mutant protein. Levels of hepatic PNPLA3 (SI Appendix, Fig. S4A) and TGs (SI Appendix, Fig. S4C) were significantly higher in mice expressing Halo-PNPLA3(148M) than in those expressing Halo-PNPLA3(WT). Thus, the Halo tag did not interfere with the accumulation of PNPLA3(148M) or the associated FLD.
Fig. 5.
PROTAC-mediated degradation of Halo-PNPLA3(148M) reduces liver TGs. (A) A schematic of the experiment. Adapted with permission from ref. 28. Copyright 2015 American Chemical Society. (B) Recombinant AAVs were used to express Halo-tagged PNPLA3 constructs under the control of a liver-specific promoter (thyroxine-binding globulin). Female mice (n = 3 to 7 per group, aged 12 wk) were injected with AAVs (1.25 × 1011 GCs) expressing Halo alone, Halo-PNPLA3(WT), or Halo-PNPLA3(148M). After 2 wk on an HSD, the mice were treated with vehicle alone (2.5% DMSO in 0.9% NaCl) or PROTAC3 (4.8 mg/kg) for 2 wk (three doses per wk) and then killed after 3 d of dietary synchronization. Livers were harvested for lipid analysis and for isolation of LDs as described in SI Appendix, Methods. Bars represent mean ± SEM values. *P < 0.05, ***P < 0.001.
To test the efficacy of this strategy, we treated QBI-293A cells expressing the Halo-tagged PNPLA3 constructs with increasing concentrations of PROTAC3. PROTAC3 treatment significantly reduced PNPLA3 levels, even at the lowest concentration (5 μM) used (SI Appendix, Fig. S5). These findings are consistent with the assumption that PROTAC3 links Halo-tagged PNPLA3 with the E3 ligase, resulting in ubiquitylation and degradation of the fusion protein.
We then tested the effect of PROTAC3 treatment (4.8 mg/kg, three times per wk for 2 wk) in mice expressing PNPLA3(WT) and PNPLA3(148M). PNPLA3 was significantly lower in both groups of PROTAC-treated mice compared with mice treated with vehicle alone (Fig. 5), but did not affect the levels of transcripts encoding Halo-PNPLA3(WT) or Halo-PNPLA3(148M) (SI Appendix, Fig. S4B). In the PNPLA3(148M)-expressing mice, treatment was associated with a significant reduction in hepatic TG levels. Thus, two different methodologies—shRNA- and PROTAC-mediated degradation—that reduced levels of PNPLA3(148M) ameliorated the FLD associated with expression of the variant protein.
Discussion
The major conclusion of this paper is that the fatty liver phenotype associated with PNPLA3(148M) is caused by an accumulation of PNPLA3 on LDs and that reduction of PNPLA3 levels in the liver reverses the steatosis. Expression of a recombinant isoform of PNPLA3 that resisted ubiquitylation and retained PNPLA3 activity in cultured cells resulted in accumulation of PNPLA3 on LDs and the development of hepatic steatosis. Conversely, ablation of PNPLA3 expression in the liver attenuated the hepatic steatosis associated with the PNPLA3(148M) variant. Hepatic steatosis was improved irrespective of whether PNPLA3 was depleted by shRNA-mediated KD or by promoting proteasomal degradation of the protein using PROTAC3. Taken together, these findings strongly support the hypothesis that PNPLA3(148M) promotes hepatic steatosis by accumulating on hepatic LDs, and that preventing this accumulation would effectively ameliorate PNPLA3(148M)-associated FLD.
Ubiquitin plays a key role in both of the major intracellular pathways of protein degradation: the ubiquitin proteasome system and autophagy. We provide evidence here that both pathways contribute to the degradation of PNPLA3. Previously, we found that the autophagy inhibitor 3-methyladenine did not promote accumulation of PNPLA3 in mice, and proposed that PNPLA3 was not a target of autophagy-mediated breakdown (18). Since it now has become clear that the effects of 3-MA on autophagy can be highly variable (19), we undertook additional experiments to assess the role of autophagy in PNPLA3 degradation. We used two different methods to inhibit autophagy: chloroquine, which inhibits lysosome activity (29), and knockdown of ATG7, an essential component of the autophagy pathway (23). Both inhibitors increased hepatic levels of PNPLA3(WT) in mice (Figs. 1 and 2). Thus, autophagy, in addition to proteasome-mediated degradation, plays a role in the turnover of PNPLA3. The relative contributions of the two pathways cannot be determined from the studies performed here, and are likely dependent on multiple factors. Moreover, we cannot exclude the possibility that chaperone-mediated autophagy, which specifically targets individual proteins to lysosomes independent of vesicular trafficking (30), also contributes to PNPLA3 degradation.
A recent report by Negoita et al. (31) showed that autophagic flux and lipophagy were lower in HepG2 cells expressing PNPLA3(148M) than in cells expressing the WT protein. They also reported that PNPLA3 bound to LC3, and proposed that PNPLA3 might play an essential role in lipophagy in hepatocytes. This hypothesis is difficult to reconcile with the observation that mice lacking PNPLA3 expression do not have elevated levels of TGs in the liver (Fig. 2). Further studies will be required to elucidate the relationship between PNPLA3, autophagy, and hepatic TG metabolism.
In contrast to PNPLA3(WT), which was increased by an order of magnitude both by bortezomib and by chloroquine, levels of the PNPLA3(148M) isoform were not significantly increased by either inhibitor (Fig. 1 and SI Appendix, Fig. S1). We show here that the resistance to the effect of bortezomib is coupled to a failure of ubiquitylation of the 148M isoform. Treatment with bortezomib was associated with the appearance of higher molecular-weight Ub-immunoreactive forms of PNPLA3(WT) but not PNPLA3(148M) (SI Appendix, Fig. S1). These data are consistent with the notion that the I148M substitution renders PNPLA3 resistant to ubiquitylation, thus impairing its degradation by either the autophagy or the proteasome degradation pathway.
The resistance of PNPLA3(148M) to ubiquitylation suggested that replacing ubiquitylated residues (typically lysines) with nonubiquitylated residues in PNPLA3(WT) would allow us to test whether accumulation of PNPLA3 per se leads to steatosis. Accordingly, we replaced all 19 of the lysine residues in the WT protein with arginines. The resulting construct [PNPLA3(K→R)] localized to LDs and resembled the effect of expression of the wild-type protein on LDs when coexpressed with CGI-58 (SI Appendix, Fig. S2). When expressed in the livers of mice, the lysine-less protein accumulated to levels as high as those seen with PNPLA3(148M) (Fig. 3). Moreover, the increased levels of PNPLA3(K→R) were associated with steatosis of similar magnitude to that seen in the PNPLA3(148M)-expressing mice.
The finding that expression of lysine-less PNPLA3 is associated with steatosis strongly supports the notion that the accumulation of PNPLA3, rather than the inhibition of its enzymatic activity, causes the steatosis associated with PNPLA3(148M) and PNPLA3(47A). If accumulation of PNPLA3 per se causes steatosis, then depletion of the accumulated protein should decrease liver fat content. The finding that reducing PNPLA3 levels using two independent methods normalized hepatic TG levels supports this notion.
Our results have important therapeutic implications. Multiple strategies could be used to reduce PNPLA3 levels in individuals expressing PNPLA3(148M). PNPLA3 is a direct target gene of sterol regulatory element-binding protein 1c (SREBP-1c), a transcription factor that also up-regulates fatty acid synthesis (32). Thus, strategies to inactivate SREBP-1c would be expected to reduce expression levels of PNPLA3 and alleviate the associated steatosis. SREBP-1c could be inactivated directly with RNAi (33), or indirectly by inactivating sterol regulatory element-binding protein cleavage activator protein, which is required for activation of SREBP-1c (34). An alternative strategy would be to target the mRNA of PNPLA3 directly using RNAi or antisense oligonucleotides (35). While this manuscript was in review, Lindén et al. (36) reported on siRNA-mediated knockdown of PNPLA3 in the livers of mice that had been fed two different diets. In mice fed a high-sucrose diet, PNPLA3 inactivation resulted in a genotype-specific reduction in hepatic TGs, whereas in mice fed a high-fat, high-fructose, high-cholesterol diet, the hepatic TG levels were decreased independent of PNPLA3 genotype.
Finally, it may be possible to accelerate degradation of PNPLA3 by promoting its association with an E3 ligase, as implemented using the PROTAC system. Thus, reduction of hepatic TG levels by targeting PNPLA3 represents a potentially useful therapeutic approach in nonalcoholic FLD in individuals harboring the PNPLA3(148M) variant.
Methods
Mice.
Knockin mice in which the isoleucine at codon 148 of PNPLA3 was changed to a methionine (Pnpla3148M/M) and transgenic mice expressing human PNPLA3(WT) and PNPLA3(148M) (PNPLA3TgWT and PNPLA3Tg148M) were developed and maintained as described in SI Appendix, Methods. All animal experiments were performed with the approval of the Institutional Animal Care and Research Advisory Committee of the University of Texas Southwestern Medical Center.
Antibodies.
The antibodies used for immunoblot analysis and immunofluorescence in this study are described in SI Appendix, Methods.
Plasmids.
The plasmids used for immunoblot analysis and immunofluorescence microscopy are described in SI Appendix, Methods.
Liver Chemistry.
Lipids were extracted from liver aliquots (100 mg) (37). Cholesterol and TGs were measured using enzymatic assays (Infinity, Thermo Electron, and Wako, respectively). Values were normalized to sample weight.
Messenger RNA Expression.
Total RNA was prepared from mouse tissues as described (38). Levels of transcripts were measured using real-time PCR with all reactions performed thrice. The relative amounts of the mRNA transcripts of interest were compared with those of mouse 36B4 (internal control) and expressed using the comparative threshold cycle (Ct) method (38).
Adeno-Associated Viruses.
Recombinant AAV constructs expressing an shRNA targeting PNPLA3 were generated in-house. The AAV-PNPLA3-shRNA constructs were made using shRNA sequences that targeted exons 4 to 7 of mouse Pnpla3 (11) as described in SI Appendix, Methods. All other AAVs were purchased from Vector Biolabs.
Bortezomib Treatment.
Mice were fed an HSD ad libitum for 4 wk, and then synchronized and treated with bortezomib (1 mg/kg) as described in SI Appendix, Methods.
Chloroquine Treatment.
Mice fed an HSD for 4 wk were synchronized and then treated with chloroquine (25 mg/kg) as described in SI Appendix, Methods.
PROTAC3 Treatment.
Mice were fed an HSD for 2 wk and then treated with PROTAC3 (Promega; CS2072A01; 479 μg per vial) as described (39). Briefly, PROTAC3 in DMSO (60 μL) was diluted 1:20 (vol/vol) in saline and injected (4.8 mg/kg in 200 μL) via the tail vein three times per wk for 2 wk. Mice were maintained on an HSD until the last 3 d of the experiment, when the diet was synchronized as described above. Livers were harvested and LDs were isolated (40).
Immunoblotting.
Liver tissue (50 to 100 mg) was homogenized in RIPA buffer (150 mM NaCl, 1.0% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0) supplemented with protease inhibitors (Protease Inhibitor Mixture; Roche). Lysates and LDs were prepared and immunoblotting was performed as previously described (41). Bands were visualized using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) and quantified using the LI-COR Odyssey System.
Immunofluorescence Microscopy.
QBI-293A cells were cultured in complete medium (DMEM with high glucose, 5% FCS), transfected, and analyzed by immunofluorescence microscopy as described in SI Appendix, Methods.
Isolation of LDs and Immunoprecipitation of PNPLA3.
LDs were isolated as previously described (16). Immunoblot analysis was performed as described in SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Serena Banfi, Tommy Hyatt, Christina Zhao, Liangcai Nie, Fang Xu, Maritza Thomas, Lizbeth Solis, Jeffrey Cormier, and Lisa Beatty for technical support, and Amanda Lowe, Nancy Heard, and Chelsea Burroughs for administrative support. S.B.R. was supported by a postdoctoral research fellowship award from the American Liver Foundation and a Junior Faculty Research Award from the National Lipid Association. J.C.C. and H.H.H. were supported by grants from the National Institutes of Health (R01 DK090066 and P01 HL200948).
Footnotes
The authors declare no conflicts of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901974116/-/DCSupplemental.
References
- 1.Ludwig J, Viggiano TR, McGill DB, Oh BJ. Nonalcoholic steatohepatitis: Mayo Clinic experiences with a hitherto unnamed disease. Mayo Clin Proc. 1980;55:434–438. [PubMed] [Google Scholar]
- 2.Younossi ZM, et al. Changes in the prevalence of the most common causes of chronic liver diseases in the United States from 1988 to 2008. Clin Gastroenterol Hepatol. 2011;9:524–530.e1. doi: 10.1016/j.cgh.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 3.Speliotes EK, et al. NASH CRN; GIANT Consortium; MAGIC Investigators; GOLD Consortium Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buch S, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet. 2015;47:1443–1448. doi: 10.1038/ng.3417. [DOI] [PubMed] [Google Scholar]
- 5.Viitasalo A, et al. Association of MBOAT7 gene variant with plasma ALT levels in children: The PANIC study. Pediatr Res. 2016;80:651–655. doi: 10.1038/pr.2016.139. [DOI] [PubMed] [Google Scholar]
- 6.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: Old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sookoian S, et al. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res. 2009;50:2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valenti L, Dongiovanni P, Ginanni Corradini S, Burza MA, Romeo S. PNPLA3 I148M variant and hepatocellular carcinoma: A common genetic variant for a rare disease. Dig Liver Dis. 2013;45:619–624. doi: 10.1016/j.dld.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Romeo S, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian C, Stokowski RP, Kershenobich D, Ballinger DG, Hinds DA. Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet. 2010;42:21–23. doi: 10.1038/ng.488. [DOI] [PubMed] [Google Scholar]
- 11.Kozlitina J, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Cohen JC, Hobbs HH. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J Biol Chem. 2011;286:37085–37093. doi: 10.1074/jbc.M111.290114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basantani MK, et al. Pnpla3/adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J Lipid Res. 2011;52:318–329. doi: 10.1194/jlr.M011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W, Chang B, Li L, Chan L. Patatin-like phospholipase domain-containing 3/adiponutrin deficiency in mice is not associated with fatty liver disease. Hepatology. 2010;52:1134–1142. doi: 10.1002/hep.23812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JZ, et al. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J Clin Invest. 2012;122:4130–4144. doi: 10.1172/JCI65179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smagris E, et al. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology. 2015;61:108–118. doi: 10.1002/hep.27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, et al. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc Natl Acad Sci USA. 2010;107:7892–7897. doi: 10.1073/pnas.1003585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.BasuRay S, Smagris E, Cohen JC, Hobbs HH. The PNPLA3 variant associated with fatty liver disease (I148M) accumulates on lipid droplets by evading ubiquitylation. Hepatology. 2017;66:1111–1124. doi: 10.1002/hep.29273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu YT, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurney MA, et al. Measuring cardiac autophagic flux in vitro and in vivo. Methods Mol Biol. 2015;1219:187–197. doi: 10.1007/978-1-4939-1661-0_14. [DOI] [PubMed] [Google Scholar]
- 21.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 22.Myeku N, Figueiredo-Pereira ME. Dynamics of the degradation of ubiquitinated proteins by proteasomes and autophagy: Association with sequestosome 1/p62. J Biol Chem. 2011;286:22426–22440. doi: 10.1074/jbc.M110.149252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 24.Kurdi A, et al. Continuous administration of the mTORC1 inhibitor everolimus induces tolerance and decreases autophagy in mice. Br J Pharmacol. 2016;173:3359–3371. doi: 10.1111/bph.13626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Kory N, Cohen JC, Hobbs HH. PNPLA3, CGI-58, and inhibition of hepatic triglyceride hydrolysis in mice. Hepatology. February 25, 2019 doi: 10.1002/hep.30583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmermann R, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 27.Sakamoto KM, et al. Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc Natl Acad Sci. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buckley DL, et al. HaloPROTACS: Use of small molecule PROTACs to induce degradation of HaloTag fusion proteins. ACS Chem Biol. 2015;10:1831–1837. doi: 10.1021/acschembio.5b00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovács AL, Seglen PO. Inhibition of hepatocytic protein degradation by inducers of autophagosome accumulation. Acta Biol Med Ger. 1982;41:125–130. [PubMed] [Google Scholar]
- 30.Juste YR, Cuervo AM. Analysis of chaperone-mediated autophagy. Methods Mol Biol. 2019;1880:703–727. doi: 10.1007/978-1-4939-8873-0_47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Negoita F, et al. PNPLA3 variant M148 causes resistance to starvation-mediated lipid droplet autophagy in human hepatocytes. J Cell Biochem. 2019;120:343–356. doi: 10.1002/jcb.27378. [DOI] [PubMed] [Google Scholar]
- 32.Horton JD, Goldstein JL, Brown MS. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr Opin Chem Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Brown MS, Goldstein JL. Cholesterol feedback: From Schoenheimer’s bottle to Scap’s MELADL. J Lipid Res. 2009;50:S15–S27. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crooke ST, Witztum JL, Bennett CF, Baker BF. RNA-targeted therapeutics. Cell Metab. 2018;27:714–739. doi: 10.1016/j.cmet.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Lindén D, et al. Pnpla3 silencing with antisense oligonucleotides ameliorates nonalcoholic steatohepatitis and fibrosis in Pnpla3 I148M knock-in mice. Mol Metab. 2019;22:49–61. doi: 10.1016/j.molmet.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 38.Engelking LJ, et al. Overexpression of Insig-1 in the livers of transgenic mice inhibits SREBP processing and reduces insulin-stimulated lipogenesis. J Clin Invest. 2004;113:1168–1175. doi: 10.1172/JCI20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raina K, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci USA. 2016;113:7124–7129. doi: 10.1073/pnas.1521738113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding Y, et al. Isolating lipid droplets from multiple species. Nat Protoc. 2013;8:43–51. doi: 10.1038/nprot.2012.142. [DOI] [PubMed] [Google Scholar]
- 41.Smagris E, Gilyard S, BasuRay S, Cohen JC, Hobbs HH. Inactivation of Tm6sf2, a gene defective in fatty liver disease, impairs lipidation but not secretion of very low density lipoproteins. J Biol Chem. 2016;291:10659–10676. doi: 10.1074/jbc.M116.719955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.