Significance
Primary cilia are sensory organelles that detect a wide range of extracellular signals for regulation of cell proliferation. Defects of their assembly/disassembly (primary ciliogenesis) and maintenance are linked to a variety of medical disorders. The signals they detect include mechano-signals from liquid flow, which requires a reversible bending property of the primary cilium axoneme. An accurate 3D structure of primary cilia is essential to fully understand the process and regulation of primary ciliogenesis and clarify the structural basis of the elastic bending property. Here we report complete 3D structural maps and a model for typical epithelial primary cilia, which will serve as the updated structural base for studies of primary cilium assembly and regulation, and mechanosensory function.
Keywords: primary cilium, 3D structure, electron microscopy
Abstract
We report a complete 3D structural model of typical epithelial primary cilia based on structural maps of full-length primary cilia obtained by serial section electron tomography. Our data demonstrate the architecture of primary cilia differs extensively from the commonly acknowledged 9+0 paradigm. The axoneme structure is relatively stable but gradually evolves from base to tip with a decreasing number of microtubule complexes (MtCs) and a reducing diameter. The axonemal MtCs are cross-linked by previously unrecognized fibrous protein networks. Such an architecture explains why primary cilia can elastically withstand liquid flow for mechanosensing. The nine axonemal MtCs in a cilium are found to differ significantly in length indicating intraflagellar transport processes in primary cilia may be more complicated than that reported for motile cilia. The 3D maps of microtubule doublet–singlet transitions generally display longitudinal gaps at the inner junction between the A- and B-tubules, which indicates the inner junction protein is a major player in doublet–singlet transitions. In addition, vesicles releasing from kidney primary cilia were observed in the structural maps, supporting that ciliary vesicles budding may serve as ectosomes for cell–cell communication.
Epithelial primary cilia are hair-like, membrane-enveloped cellular projections that extend into the extracellular space. They are sensors detecting environmental signals for modulation of cellular physiological activities. Due to the importance of these cellular activities, defects in primary cilium assembly/disassembly (ciliogenesis) and maintenance underlie many developmental and organ disorders (1–3). Accurate information of the 3D structure of primary cilia is essential to fully understand the process and regulation of primary ciliogenesis.
In mammalian organs, such as kidney, liver, and pancreas, each epithelial cell carries a single primary cilium as a mechanosensor, which bends reversibly under liquid flow (4–6). The passive elastic bending of primary cilia is believed to modulate epithelial cell proliferation and autophagy, thereby maintaining the normal architecture of mature organ tissues essential to their physiological functions, although the identity of the signaling messenger is controversial (2, 7–13). The elastic bending property relies on the structure of the ciliary microtubule-based framework (axoneme). The 3D architecture of primary cilium axoneme is fundamental to understanding the mechanosensory function of primary cilia in kidney and other cells.
Electron microscopic (EM) studies of both primary cilia and motile cilia started over half century ago (14–18). The primary cilium axoneme has long been described as a “9+0” architecture, with 9 peripheral microtubule doublets of the same length forming a hollow cylindric architecture as the core framework based on some electron micrographs (1, 17–24). The 9+0 model is currently the most accepted structural understanding in the field for primary cilium axoneme. However, there were some early cross-section EM studies, showing deviation of primary cilium axoneme from the 9+0 paradigm (25–29). An accurate 3D structural model derived from 3D structural map has not been established for typical primary cilia.
The 9+0 paradigm stipulates an architecture similar to the array of the 9 peripheral microtubule doublets in the 9+2 motile cilium axoneme. In 9+2 motile cilia, the 9 microtubule doublets are integrated into the cylindric array with radial spokes, nexin links, and dynein arms (14–16, 30–32), but these structural components do not exist in the primary cilium. It has been unclear if there are alternative structural components integrating the 9 microtubule doublets into a stable cylindrical 9+0 axoneme to enable the elastic properties necessary for reversible bending of primary cilia. Microtubules in cells present in singlet (33–36), doublet (37–39), and triplet (40) forms. The latter two forms are unique to cilia and flagella. Although microtubule doublets were found to become singlets at the tip end of cilia or flagella (29, 41, 42), the doublet–singlet transitions remain structurally uncharacterized.
In this work we determined complete 3D structural maps of epithelial primary cilia using serial section electron tomography (SSET) and developed a 3D structural model of typical primary cilia as an updated structural baseline for primary cilium research. We found that key features of primary cilia architecture are distinct from expectations based on the 9+0 paradigm. We also discovered a structural component that cross-links axonemal microtubule complexes (MtCs), and connects MtCs with the ciliary membrane. The 3D structure provides a mechanistic understanding of the elastic bending of primary cilia under liquid flow required for the mechanosensory function. We have also analyzed the 3D structural maps of the doublet–singlet transition regions and gained insights about doublet assembly/disassembly. In addition, vesicles releasing from kidney primary cilia were observed in our structural maps. The updated structural knowledge in this report provides insights into primary cilia structure, assembly, and maintenance.
Results and Discussion
In our pilot studies, we employed cryoelectron tomography to investigate vitreously frozen primary cilia from kidney cells of Lilly Laboratories cell–porcine kidney 1 (LLC-PK1) (43) and mouse inner medullary collecting duct-3 (IMCD3) (44). The results did not agree with the 9+0 structural model. We therefore utilized conventional EM to investigate the primary cilia from the three most commonly used kidney cell lines in primary cilium studies: IMCD3, LLC-PK1, and Madin–Darby canine kidney (MDCK) (45). The data confirmed that their axoneme structures do not follow the 9+0 paradigm.
The cross-section images demonstrated that the axonemal microtubules change from doublets to singlets and most terminate before they reach the ciliary tip. We use the term “microtubule complex” to refer to the microtubule, either in the doublet or singlet form, which originates from a basal body triplet. MtCs need to be clearly tracked through entire serial cross-section sets to develop a detailed understanding of the 3D structure of typical primary cilium axoneme. Most primary cilia are not perfectly perpendicular to the cell surface, having an angle of deflection and regions of bending or tilting (e.g., Movies S7 and S11). Such characteristics make the MtCs in serial sections difficult to identify and track in the serial projection images, and complicate the distinction between doublets, pairs of neighboring singlets, and singlets, particularly in the distal portion of the cilia. As a consequence, serial section datasets are rarely interpretable for the entire full length of the primary cilium. Structural information in 3D is required to clarify the detailed MtC structural changes along the primary cilium.
We used SSET to obtain the 3D structure of the primary cilium by combining electron tomograms of serial sections (46). SSET is probably the only method currently available for obtaining the 3D structural maps at transmission electron microscopy (TEM) resolution for large unique subcellular objects that cannot be imaged by EM if not sectioned into multiple slices. This method has been used to work with serial sections of a few slices, but has never been successfully applied at a scale to dozens of serial sections before this work, because the technical challenge and required experimental effort increase exponentially with the slice number increasing. For SSET, there is always a thin layer of damage introduced by knife cutting between two adjacent sections. The damaged region is very thin and generally tolerable for 3D map interpretation at a large scale, but we carried out SSET for serial cross-sections instead of longitudinal sections to minimize the impact of knife-cuts to the precise lengthwise tracking of MtC structural changes in the density maps.
We focused our effort on primary cilia from IMCD3 cells. The structural map of each primary cilium requires tomographic data collection and integration of dozens (IMCD3 cilia of several micrometers) to hundreds (LLC-PK1 and MDCK cilia of dozens of micrometers) of electron tomograms of uninterrupted serial sections. IMCD3 cells are currently the most commonly used kidney cell line for primary cilia studies and their short cilium lengths (2–5 μm) make them the most amenable to our analysis.
Three-Dimensional Structural Model of the Primary Cilium Axoneme.
Our strategy was to survey the largest number of primary cilia that was practically achievable, and then to select a subset of cilia to obtain structural maps over their full length. Finally, we chose a typical example among the acquired structural maps that shares common features with most other maps. This was used for investigation of local structural features within the cilium and to build the 3D structural model for result interpretation, offering both breadth and depth to our research at a practical level of resource expenditure.
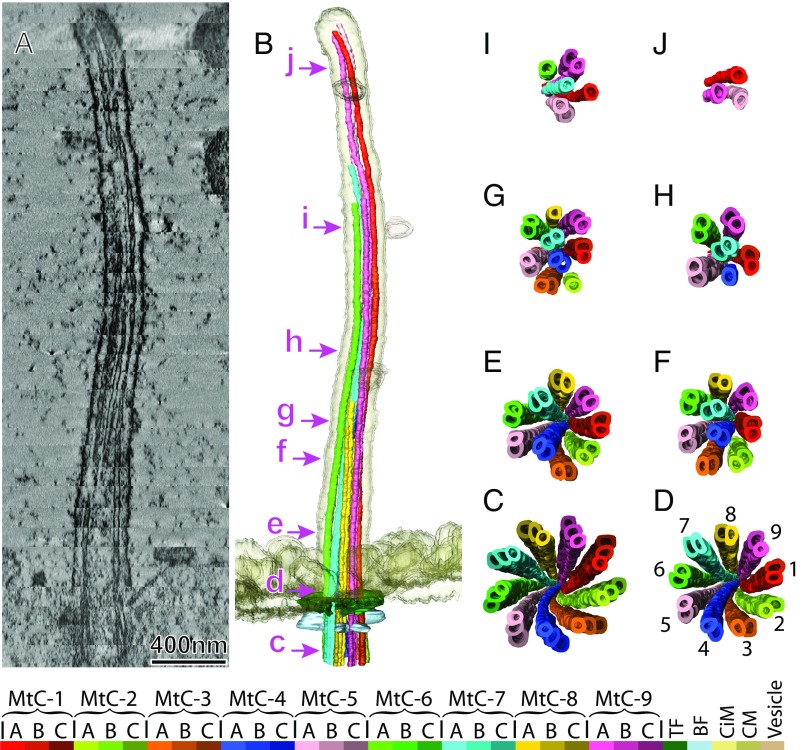
Fig. 1A displays a central longitudinal section of the selected structural map for a typical primary cilium. This structural map was obtained by combining 33 dual-axis electron tomograms of serial cross-sections of a primary cilium on an IMCD3 kidney cell (Movie S1). Looking from the basal region toward the distal end, the microtubule-based axoneme starts at the basal body with nine microtubule triplets in a cylindrical arrangement (Fig. 1C). The nine triplets then become nine doublets (Fig. 1D) as the axoneme initially extends out of the cell surface (position “d” in Fig. 1B). The ninefold symmetry of the microtubule doublets is preserved for a very short length, as displayed in the symmetrized tomo-slice (33 nm) (SI Appendix, Fig. S1B) and in the density map (SI Appendix, Fig. S1A). This is where the structural model matches the widely accepted 9+0 configuration.
Fig. 1.
Overview of the primary cilium structure. The typical primary cilium structural map obtained by combining 33 serial dual-axis tomograms. A longitudinal central slice of 13.4-nm thickness is shown in A (see also Movie S1). The structural model built from the serial tomograms using the Amira package is shown in B. Progressive changes of the microtubule configuration from proximal to distal end of the cilium are demonstrated by the end-on cross-section views shown in C–J that correspond to the locations marked in B (see also Movie S2). The viewing direction is from the basal to the distal end. The color key is shown at the bottom. BF, basal foot; CiM, ciliary membrane; CM, cytoplasmic membrane of the cell; MtC-A, -B, -C, microtubule complex A-, B-, C-tubule; TF, transition fiber.
Two microtubule doublets progressively shift toward the core of the axoneme at the region where the primary cilium starts to extend into the extracellular space (Fig. 1 E and F), and the axoneme becomes a 7+2 configuration with two central microtubule doublets surrounded by seven peripheral microtubule doublets (Fig. 1F). The 7+2 arrangement is the dominant configuration in most primary cilia we investigated. We also noted one occasion in which one microtubule doublet moved inward first, generating an 8+1 configuration (SI Appendix, Fig. S2F), quickly followed by an inward shift of another MtC to give the 7+2 configuration. There were also occasions when the 7+2 configuration was never evident due to one or more MtCs terminating shortly after leaving the basal body. SI Appendix, Fig. S2 D and D′ display such an occasion, in which two MtCs terminate while one microtubule doublet moves toward the core. The 7+2 and 8+1 arrangements were visualized by serial section EM micrographs previously (42). In our 3D maps, the two inward-shifting microtubule doublets were any two nonadjacent peripheral doublets, separated by one, two, or three other doublets (Fig. 1E and SI Appendix, Fig. S2).
Termination of the MtCs happens sequentially rather than simultaneously (Movie S2); in most cases, B-tubules are lost from microtubule doublets as the cilium extends toward the distal end, and subsequently the A-tubules terminate. Moving toward the distal ciliary tips, the 7+2 configuration of the MtCs bundle evolves as the total number of MtCs reduces (Fig. 1 G–J). The evolution of the architecture happens in a gradual and consistent manner. We did not observe significant location changes or portion swapping between the neighboring MtCs, which indicates a structural mechanism is stabilizing the architecture of the MtC bundle (Fig. 1 E–I). The axoneme eventually terminates with a limited complement of microtubule singlets reaching the ciliary tip (Fig. 1J and SI Appendix, Fig. S3; see also the green-lined regions in Fig. 4I and SI Appendix, Fig. S4).
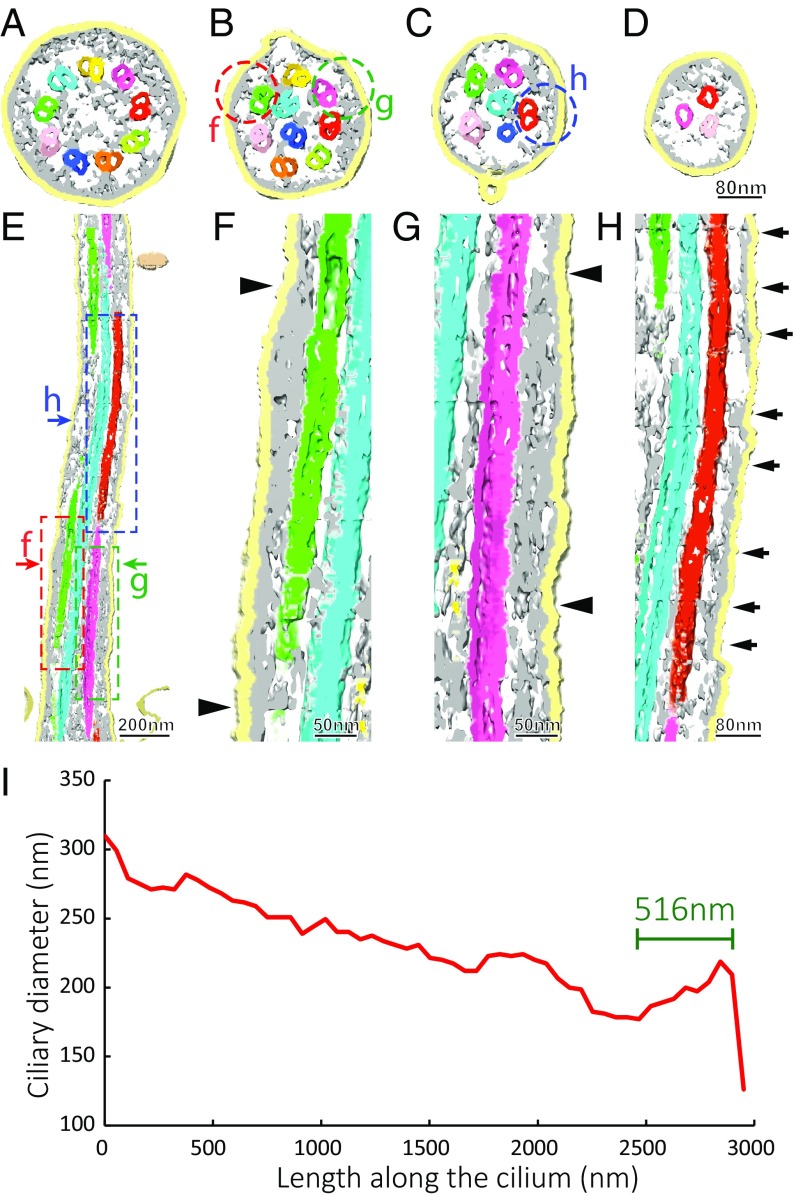
Fig. 4.
Protein networks linking the axonemal MtCs and the ciliary membrane and the ciliary envelope showing a decreasing diameter toward the distal tip. Cross-slices of the 3D density map rendered using the Chimera package are shown in A–D at locations close to the “d,” “f,” “h,” and “i” arrows in Fig. 1B, respectively. The network-like densities are colored in gray, showing a wide distribution along the whole length of the primary cilium. At the basal region (shown in A), the nine doublets are linked to the ciliary membrane by somewhat Y-shaped densities. There are also long filaments connecting the MtCs to each other as shown in A–C. The networks connecting the ciliary membrane and MtCs display two kinds of morphologies as shown in the longitudinal slice in E and magnified in F–H. One is a continuous density as shown between the arrows in F and G, of which the cross-section views are circled in B. The other class of densities are discrete features as in H for which a cross-section view is circled in C. MtCs are colored following the color key in Fig. 1. The diameter of the membrane enveloped primary cilium decreases toward the distal tip, as shown in the profile in I (also see SI Appendix, Fig. S4 for diameter profiles of six additional primary cilia). The last B-tubule terminates 516-nm away from the axoneme tip end, which leaves three microtubule singlets present in the tip region (indicated by a green line in I).
Axoneme structure defines the elastic bending property in liquid flow that is required for mechanosensory functions of primary cilia. We demonstrated that the 3D structure of epithelial primary cilia is not a constant cylindric array of 9 doublets (9+0) of the same length, analogous to the 9-doublet array in a typical 9+2 motile cilium. There have been conventional EM studies indicating a configuration different from the 9+0 model since the 1960s (25–27, 29). The 7+2 or 8+1 arrangements were reported in kidney primary cilia as early as a half century ago (28, 42). However, these observations contradicting the 9+0 model were apparently underappreciated. Most of our knowledge about cilium assembly and maintenance comes from motile cilium research. The generally reported 9+0 configuration of epithelial primary cilia may have arisen from sample preparation deficiencies, combined with the assumption that the axoneme structure of the primary cilium stays the same along its length similar to that of the typical 9+2 motile cilia. Initially in our work, we found that primary cilia broke off easily during specimen preparation particularly when/if osmium was used in the postfixation step. All primary cilia were broken away near location “d” of Fig. 1B, where cilia start to extend out of the cell. The 9+0 configurations would dominate the results if analysis is restricted to the residual basal portion of the truncated cilia. Protection of cilia against such truncation was the major technical challenge of our study. We finally solved the cilia breakage problem with our preparation strategy and are able to routinely obtain serial cross-sections across the full length of primary cilia for cultured kidney cells.
MtCs in a Primary Cilium Differ in Length and Ciliary Diameters Reduce Toward the Tip.
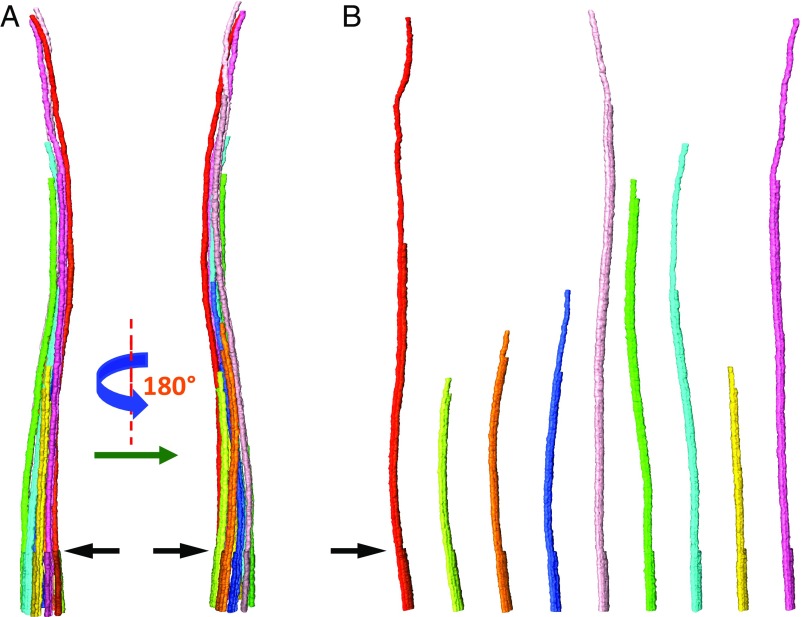
The overall length of each of the nine axonemal MtCs of a cilium is distinct (Fig. 2). From the basal side to the distal end, the B-tubule of the microtubule doublets terminates first for most cases and the A-tubule singlets terminate at a location closer to the ciliary tip (Fig. 3 A–E). In Fig. 2, the individual MtCs in the axoneme model of primary cilia (Fig. 2A) are dissected sequentially, as shown in Fig. 2B and Movie S3. A length profile of the dissected MtCs was generated, as shown in SI Appendix, Fig. S3G, displaying a length range from 0.9 to 3.0 µm. This is a common feature of most of the reconstructed IMCD3 primary cilia (SI Appendix, Fig. S3), except for one case where the cilium is only 0.85-μm long. (SI Appendix, Fig. S3C).
Fig. 2.
Length profile of the MtCs in the axoneme. The axoneme structural model, shown in A, can be disassembled to extract individual MtCs sequentially, as shown in B (also see Movie S3). For each MtC, from the left to the right, the A-tubule and B-tubule are in slightly different colors following the color key in Fig. 1. The black arrows point to where triplets become doublets: location “d” marked in Fig. 1B, is also where the primary cilium starts to extend into extracellular space. The lengths of the nine MtCs within this primary cilium are different. They range from 0.9 to 3.0 µm. Please refer to SI Appendix, Fig. S3G for the length profile of the primary cilium.
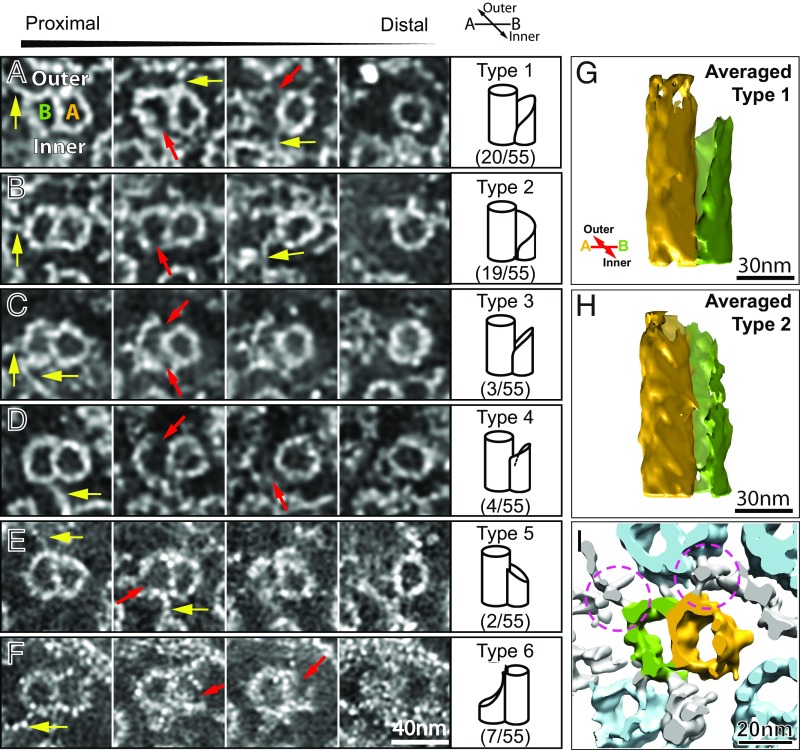
Fig. 3.
Microtubule doublet-to-singlet transition. Six transition types are shown with their end-on views of sequential cross-sectional tomo-slices looking from the proximal end to the distal end (left to right) in A–F. The upside of the doublet images face to the outer side. Red arrows in the tomo-slices point to the gap of the microtubule wall of protofilaments. The last column of the A–F displays schematic illustrations and the population ratios for each type of transition. Type 1 and type 2 dominate the doublet–singlet transition occurrence. Their averaged density maps are displayed in G and H, respectively, which reveals a large gap at the inner junctions (see Movies S4 and S5 of the averaged 3D maps for the type 1 and 2 transition regions, respectively). A-tubules are displayed in yellow, B-tubules in green. The yellow arrows in the tomo-slices, A–F, point to some fibrous structural components (inter-MT links). The volume rendering in I shows the same region of that in A, and demonstrates a fibrous feature of the inter-MT links (indicated by the magenta circles). The inter-MT links connect neighboring MtCs as seen in the magenta circle on the right. They are also found to extend further, as marked by the left magenta circle in I, toward the ciliary membrane, and finally join a dispersed density on the membrane (also see Fig. 4).
To determine if a given length of the MtC is related to its location in the primary cilium basal body, we defined the MtC closest to the daughter centriole as #1 and named the others #2 to #9, following the clockwise numbering system looking from the basal side toward the distal end (16, 47, 48). We found that #1 is a relatively long MtC in the structural maps of six of the seven primary cilia (shown in SI Appendix, Fig. S3 B–G), extending to the ciliary tip. The exceptional case (shown in SI Appendix, Fig. S3A) is inconclusive, because the nonciliated centriole was located beneath the basal body instead of at the side, making the assignment of the first MtC ambiguous.
The diameter of the cilium reduces gradually toward the distal end (Fig. 1 D–J; also see Fig. 4 A–D) as the complement of the MtCs decreases (Fig. 4I). This trend is common among all our reconstructed primary cilia (SI Appendix, Fig. S4). The tip regions of our reconstructed primary cilia only contain microtubule singlets that are the A-tubules of the remaining MtCs (the green lined regions in Fig. 4I and in SI Appendix, Fig. S4).
Interestingly, the overall architecture of the peripheral microtubules displays a slight right-handed twist at the basal region, particularly before the number of the microtubules decreases (regions “d” to “g” in Fig. 1B and Fig. 1 D–G). For short cilia, the twist is evident for the whole cilium length (SI Appendix, Fig. S5).
The closely packed MtC bundle shown in Figs. 1 and 2A has a thicker basal region and a thinner distal region, in contrast to the hollow cylinder model of nine peripheral MtCs, for which the axoneme diameter remains relatively constant. Generally, the basal regions of primary cilia exposed to liquid flow are subjected to a much larger torque than the distal regions. The architecture of the primary cilium axoneme, shown in Fig. 2, provides a strong backbone for elastic bending in a liquid flow (6). A thicker basal region with more MtCs is expected to confer higher flexural rigidity for primary cilia. This should aid in both resisting the larger bending torque at the base and in subsequently to regaining the orientation perpendicular to the epithelial. Assuming laminar flow in a tube, the thinner distal region provides a greater bending response to fluid shear at the tip, while reducing the area confronting the flow, thereby reducing the bending stress. The right-handed twist of the MtC bundle (SI Appendix, Fig. S5) is a natural extension of the basal body, for which a right-handed twist was previously reported (49). This twist will also help the axoneme structure withstand overall flow stress.
It is a surprising finding that the lengths of the nine MtCs differ significantly within a primary cilium. This indicates possible divergence in assembly and maintenance processes between those of primary and motile cilia. For example, we know that intraflagellar transport (IFT) is responsible for the transport of proteins between the base and the distal end using MtCs as the transportation track (50, 51). The differences in MtC configuration and length suggest some dissimilarities between IFT processes in motile and primary cilia. In motile cilia, the anterograde IFT trains move precursor proteins along the doublet B-tubules from the ciliary base to the doublet tips, which are at the cilium tip (52). IFT trains reconfigure for retrograde transport on the A-tubules from the ciliary tip to the ciliary base (53). Therefore, collisions between the anterograde and retrograde trains are avoided. A dynamic balancing of the relative rates of anterograde and retrograde transport is proposed to control the protein polymerization and turnover at the ciliary tip and thereby regulates motile ciliary growth and final length (54). This balance-point model explains well the length regulation of motile cilia, in which all microtubule doublets have a same or similar length. However, this model cannot explain the significant length differences among the nine MtCs within the same primary cilium, unless one postulates that the nine MtCs in a single cilium do not share the same IFT activity levels.
When anterograde IFT trains reach the end of a doublet B-tubule, which is far from the primary cilium tip, there are two possible options for the next step in the movement: (i) changing track to a longer doublet B-tubule or (ii) transitioning into retrograde IFT trains in the middle of the primary cilium. The published study on the dynamics of IFT-88 trains showed that almost all of the anterograde IFT trains reach the distal tip regions before they turn into retrograde IFT trains (55). In addition, sometimes there are short pauses in the anterograde movement of IFT trains toward the tip, as shown in the kymograph (figure S3A in ref. 55). It is plausible that the pauses are associated with a track switch event where the anterograde IFT trains reach the end of a short B-tubule and then change to a longer doublet B-tubule. Such kinds of pauses are seldomly seen in the movement of retrograde IFT trains in primary cilia, which is a logical consequence of the longer A-tubules extending to the tip, providing a direct track back to the ciliary base.
In the primary cilium structure, we generally find only extended singlet A-tubules in the cilium tip region, particularly for the relatively long cilia: all B-tubules terminate away from the tip, at distances that can exceed 500 nm (shown in Fig. 4I and SI Appendix, Fig. S4). This indicates that anterograde IFT trains may not directly reach the ciliary tip, because they generally travel via the B-tubules, which avoid collisions with retrograde IFT trains traveling along the A-tubules (52). An alternative mechanism in Caenorhabditis elegans has OSM-3 kinesin-driving anterograde IFT trains along singlet A-tubules (56). However, knockout in mice of Kif17, a mammalian analog of OSM-3, did not yield a phenotype for the rod photoreceptor outer segment (57). This indicates that Kif17 is unlikely a kinesin motor responsible for driving IFT trains toward the distal end at the tip region through A-tubules in mammalian primary cilia. Diffusion has been found to play an important role for molecular trafficking in primary cilia (55) and likely contributes to protein trafficking near the ciliary tip region.
Doublet–Singlet Transitions.
To analyze how microtubule doublets transition to microtubule singlets, the density maps at the transitional regions for MtCs in seven primary cilia (63 MtCs in all) were investigated. Six transition types were identified and defined based on structural characteristics. Fig. 3 A–F displays sequential cross-slices of the tomograms viewing from the proximal to distal end for the six transition types. The inner and outer sides were identified by tracking the doublets from the proximal end near the basal bodies. The doublet–singlet transition mostly occurred with the incomplete B-tubule terminating first (shown in Table 1: types 1–5 in addition to some type-unassigned cases in which the B-tubules clearly terminate first but were difficult to define further). A-tubule terminated first for a small portion of transitions (11%; type 6).
Table 1.
Profiles of doublet-singlet transitions in the IMCD3 primary cilia
| Class | No. | Population percentile (%) | Transition length average (nm) |
| B-tubule terminates first | |||
| Type 1 | 20 | 31.75 | 39.67 ± 13.37 |
| Type 2 | 19 | 30.16 | 53.28 ± 12.43 |
| Type 3 | 3 | 4.76 | 38.41 ± 5.42 |
| Type 4 | 4 | 6.35 | 35.02 ± 10.76 |
| Type 5 | 2 | 3.17 | 38.23 ± 0.66 |
| Unidentifiable | 8 | 12.70 | |
| Subtotal | 56 | 88.89 | 44.15 ± 13.82 |
| A-tubule terminates first | |||
| Type 6 | 7 | 11.11 | |
| Total | 63 | 100 |
The type 1 and type 2 transitions predominate over 70% of the population for the 55 transitions that are type-identifiable (Table 1). At the proximal portion of the transition region, both type 1 and type 2 transitions start with a large gap at the inner side as indicated by the red arrows in the second column of Fig. 3 A and B, which implies a breakage of the inner junction between the A- and B-tubules of the microtubule doublet (37–39, 58). Toward the distal region, type 1 develops another breakage at the outer junction between the A- and B-tubules before the B-tubule terminates (Fig. 3A). In contrast, type 2 does not display an outer-junction breakage. Rather, the distal end of the B-tubule terminates completely at the connection location (Fig. 3B). The transition characteristics described above are clearly recognizable in the averaged subvolumes of the type 1 and type 2 as shown in Fig. 3 G and H, respectively (also see Movies S4 and S5). The average length of the transition region (the distance from the proximal end of tubule disruption to the region where the tubule terminates) is about 40 nm and 53 nm for type 1 and type 2, respectively (Table 1).
There are also other transition types with a small number of occurrences (Fig. 3 C–F and Table 1). Type 3 shares some similarity to that of type 1, but the gaps at the inner and outer junctions start their development simultaneously (Fig. 3C). For type 4, the termination of the B-tubule starts with an outer-junction breakage (Fig. 3D). Occasionally, termination of B-tubule starts with protofilament damage instead of loss of the connection between the A- and B-tubule (type 5) (Fig. 3E).
For the cases where the A-tubules terminate first (type 6), the disruption of the A-tubules always begins with loss of some protofilaments on the side away from the B-tubule (indicated by the red arrow in the second column of Fig. 3F). The protofilaments that seal B-tubule, referred to as the “partition” (37, 58, 59), always maintain until the B-tubule terminates. The B-tubule always terminates shortly after the A-tubule ends, and a cluster of biological material (five of seven, 71.4%) or sometimes fiber-like densities are found at the end of the terminated B-tubule (two of seven, 28.6%) (last column in Fig. 3F).
In the primary cilium axoneme, the microtubule doublets often transform into singlets by termination of the B-tubule. Most doublet–singlet transitions display a longitudinal gap or breakage at the inner junction between the A- and B-tubules (types 1 and 2 in Fig. 3 G and H). Based on cryoelectron tomographic studies of isolated microtubule doublets, the A- and B-tubule protofilaments at the inner-junction region are loosely connected by a large linker protein density (later referred to as MIP3) with a 16-nm periodicity (37, 39, 58). It indicates that the inner junction is less compact and possibly weaker than that of the outer junction for the doublet (37, 58). Meanwhile, additional density was found at the inner junction of the doublet in the intact motile cilium axoneme (38). In our density maps of the doublet–singlet transitions, the large gap between the last B-tubule protofilament and A-tubule at the inner side (shown in Fig. 3 G and H) indicates that the inner-junction proteins play an important role in doublet–singlet transition.
The MtCs in the primary cilia axoneme are stable acetylated microtubules and should not exhibit highly dynamic length changes despite their significant differences in length. However, the plus ends of the A-tubules and B-tubules are not static, and may extend or disassemble as suggested by the balance point model (54, 60). If so, the transition types shown in Fig. 3 may represent either growing or shrinking stages. It is remarkable that the sequential cross-slices from the proximal to the distal end of the transition type 2 class (Fig. 3B) resemble the sequential disassembly steps of the microtubule doublet in motile axoneme during Sarkosyl solubilization treatment (figure 19 in ref. 59). In contrast, the structural morphology of the type 1 transition, in which the protofilament sheet segment protrudes from the distal end of the B-tubule (Fig. 3G), is compatible with the growing end of microtubule polymerization model (61, 62). The majority of doublet–singlet transition cases display an open gap at the inner junction, which extends much farther than that of the outer junction. Considering that the inner-junction gap must be present for both growth and shortening of B-tubules, the inner-junction proteins must join the assembly more slowly than the outer-junction proteins during B-tubule growth for doublet assembly, and leave the assembly ahead of others when the B-tubule disassociate. It is therefore a rate-controlling factor for doublet assembly and an initiating factor for doublet disassembly.
For the cases where A-tubule terminates first (type 6), the leftover A-tubule partition protofilaments seal the opening of the B-tubule. The partition was reported as being relatively stable during chemical disruption of the motile cilium axoneme (figure 19 in ref. 59). These partition protofilaments can protrude far out of the end of chemically disrupted doublets (63, 64). It was suggested that the proteins in the intraluminal side of microtubules help integrate these protofilaments (31, 35, 39, 58, 65). However, for cases of the transition type 6, the partition protofilaments do not extend far and terminate with the B-tubule protofilaments immediately after the A-tubules terminate.
Protein Networks Link MtCs to MtCs, and MtCs to the Ciliary Membrane.
In the structural maps, we discovered protein densities interconnecting MtCs to each other and to the ciliary membrane through the entire length of the cilium (colored gray in Fig. 4). These densities are also visible in Fig. 3 A–F, as indicated by the yellow arrows and their fibrous characteristic is visible in the 3D volume-rending image (colored gray in Fig. 3I). The fibrous characteristic of these densities is maintained when linking to neighboring MtCs, as shown in Fig. 3I (we describe these as inter-MT links). The inter-MT links can also extend toward the ciliary membrane to link MtCs to the membrane; however, the densities generally spread into a dispersive feature when they reach the ciliary membrane (Fig. 4 A–D).
The basal region near where the cilium starts to protrude (location “d” in Fig. 1B) follows a ninefold symmetry and displays a morphology of MtC–membrane connections similar to the Y-shape linkers or “champagne-glass” structures reported in motile cilia (66) and primary cilia (26) (Fig. 4A and SI Appendix, Fig. S1). They are believed to be associated with the “necklace” structures in the ciliary membrane of motile cilia (66). There are also filamentous connections that link neighboring doublets at this location (Fig. 4A), but a regular structure resembling the stellate pattern reported for the basal region of motile cilia was not observed (48). These filamentous connections are invisible after applying the ninefold rotational symmetry, indicating that they lack a regular structure with a ninefold symmetry (SI Appendix, Fig. S1).
Moving away from the basal region, the microtubule–membrane connections display either discrete or continuous morphologies along the MtCs. The discrete densities are separated from each other and sometimes display distinct locations about 88-nm (±13 nm) apart (indicated in Fig. 4H by arrows). These discrete connections show fibrous characteristics like the inter-MT links (Fig. 3I), but they branch out into the dispersive densities on the membrane (Fig. 4H and the blue “h” region in Fig. 4C). In contrast, the continuous densities cover some distance along the MtCs beneath the membrane, and they generally do not display fibrous characteristics. Their long and continuous nature resembles the train-like densities of IFT complexes in Chlamydomonas (51, 52, 67). They are relatively thick and sometimes cause the ciliary membrane to bulge slightly (e.g., between arrowheads in Fig. 4 F and G). However, we were unable to identify the previously reported structural features of individual IFT complexes (52, 67), probably due to the missing pyramid in the Fourier space that is parallel to these density features. In the structural map, the continuous densities are found to be associated with both the A-tubule and B-tubule of microtubule doublets. These are likely to represent IFT complexes.
As we discussed in previous sections, the closely packed structure of the MtC bundle generally presents a 7+2 format but evolves along the length of primary cilium with a reducing number of MtCs. The progressive evolution and rearrangement is orderly and proceeds in a relatively stable manner, as shown in Fig. 1 C–G, rather than being a stochastic process. Inter-MT links are found cross-linking adjacent to both MtCs, and MtCs to the ciliary membrane. These features likely augment axonemal-structural stability, thereby strengthening the primary cilia to withstand the liquid flow and perform reversible elastic bending.
Membrane Vesicles Are Released from the Primary Ciliary Membrane.
The structural morphology of our 3D maps provided a direct visualization of vesicle budding from primary cilia. The ciliary membrane was well preserved in our structural maps, and was found to contain protrusions and vesicles, as shown in Fig. 5. The overall morphology of the ciliary membrane in Fig. 5 suggests stages of vesicle release, including initiation of protrusion (Fig. 5E), budding out (Fig. 5B), and completion of separation (Fig. 5 C and D).
Fig. 5.
Membrane vesicles along the surface of primary cilia. A ciliary membrane model (A) was generated by segmentation of the 3D structural map using the Amira package. Protrusions and vesicles on the ciliary membrane are highlighted by arrows in A, and the corresponding cross-sectional views are displayed in B–E. A double-membrane vesicle was found as shown in D. The morphologies of the vesicles indicate they are released from the ciliary membrane.
It was proposed that signaling molecules may be released with ciliary membrane budding (68, 69), and vesicles released from motile cilia/flagella and neuronal primary cilia are proposed to be ectosomes for cell–cell communication (70, 71). Furthermore, receptors were reported to be released with membrane shedding of at the tip end of primary cilia (72). The direct visualization of budding vesicles in the structural maps shown in Fig. 5 confirmed that vesicle budding from the ciliary membrane occurs in primary cilia of kidney cells. In addition, the data demonstrate that the vesicles may be released from regions away from the ciliary tip end.
Conclusions
The architecture of the primary cilia differs extensively from the commonly acknowledged 9+0 paradigm. The local axonemal architecture of primary cilia is relatively stable, but gradually evolves from the base to the tip with decreasing numbers of the MtCs. This causes a reducing diameter for the axonemal architecture from the base to the tip, compatible with a bending property transitioning from rigid to relatively flexible. The axonemal MtCs in primary cilia are cross-linked by previously unrecognized fibrous protein networks. They presumably amend mechanical properties of the primary cilium axoneme and support the elastic bending under liquid flow required for the mechanosensory function. The nine axonemal MtCs of a primary cilium differ significantly in length, indicating that IFT behaviors in primary cilia must be more complicated than those reported for motile cilia. Anterograde IFT may need to switch from one MtC to another to continue travel toward the ciliary tip. A-tubule singlets dominant the framework at the tip regions of primary cilia, which is compatible with a mechanism where diffusion, in addition to IFT, plays an important role for molecular trafficking. The inner-junction gap is a remarkable feature in most of the 3D maps of microtubule doublet–singlet transition regions, indicating the inner-junction proteins could be a controlling factor in doublet–singlet transition. Ciliary vesicles may serve as ectosomes for cell–cell communication.
Materials and Methods
Cell Culture and Cilia Growth.
For this work, we examined the structure of primary cilia with an IMCD3 cell line stably transfected with a GFP-fusion-SSTR3 [a gift of the Nachury laboratory at Stanford University, Stanford, CA (73)], an MDCK cell line with YFP-fusion-Smo [a gift from the Lippincott-Schwartz laboratory at NIH, Bethesda, MD (74)], and a LLC-PK1 cell line with a GFP-fusion-SSTR3 that we generated. IMCD3 cells and LLC-PK1 cells were cultured in DMEM with 5% FBS and 10% FBS, respectively. MDCK cells were cultured in Eagle’s MEM (EMEM) with 7% FBS. Cells were seeded inside the high pressure freezing (HPF) planchets (part no. 242; Wohlwend) that were carbon-coated in advance, and incubated at 37 °C under 5% CO2. IMCD3 and LLC-PK1 cells were serum-free in DMEM (without serum for 2 d) and MDCK cells in serum-free EMEM to induce cilia, and then evaluated for cilia by light microscopy.
Primary Cilia Protection for Making EM Specimens.
Cells with primary cilia were rinsed in the planchets with PBS for 10 min at 37 °C, and then fixed with 2.5% glutaraldehyde in 37 °C PBS buffer for 0.5 h, and at room temperature for 0.5 h. The cells in planchets were rinsed with PBS once for 10 min followed by soaking in 0.05% poly-lysine (Sigma) solution in PBS for 3 min at room temperature. After a PBS rinse, a drop of concentrated Escherichia coli JM109 cell suspension (prefixed with 2.5% glutaraldehyde and washed with PBS) was added into the planchet. After another fixation with 2.5% glutaraldehyde for 1 h and a rinse with PBS for 10 min, a drop of 10% BSA solution in ddH2O was added. After 20 min, a drop of 1 M DL-DTT (Sigma) in ddH2O was introduced. After 20 min, the planchets would appear to be covered with a layer of flocculent-matrix. The planchets were then placed into 2.5% glutaraldehyde for 1 h.
HPF and Freeze Substitution.
The HPF and freezing substitution were performed as described previously (75, 76) with minor modifications. After rinsing three times with PBS, each planchet containing pretreated IMCD3 cells was covered with a second planchet to form a sandwich, which was subjected to HPF at 300 bar in a high-pressure freezer (Balzers HPM010; Liechtenstein).
The frozen planchets were placed in 1-mL tubes with the precooled (−140 °C) substitution solution: 0.05% uranyl acetate and 1% osmium tetroxide in acetone containing 2% water. Programmed steps in a freeze-substitution device (Leica AFS2) were then applied: −90 °C, 48 h; −60 °C, 12 h; −20 °C, 12 h; 0 °C, 2 h, with 5 °C temperature increment per hour between steps. The substitution solution was replaced by pure acetone with temperature increased from 0 °C to room temperature (six changes over 3–4 h). The samples were then infiltrated with graded resin (SPI-PON 812; SPI-CHEM) in acetone, 10%, 20%, 50%, 75%, 90%, and 100% (3×) over 3 d. Specimens in resin were then polymerized at 60 °C for 2 d.
Room-Temperature Processing for EM Specimen Preparation.
No major structural differences significant to our investigation were noted comparing the freeze-substituted specimens and the samples prepared using the room-temperature process. The microtubule arrangement in the latter specimens is more distinctive than that in the former. Therefore, data for the reported structural model was generated using data from the room-temperature–processed specimens.
The planchets with pretreated IMCD3 cells were rinsed with PBS (3 × 10 min), and were postfixed, dehydrated, and infiltrated at room temperature following previously reported methods with minor modifications (52, 77, 78). In brief, cells were postfixed with 0.125% potassium ferricyanide and 1% osmium tetroxide (Sigma) for 1 h. After washing with ddH2O (3 × 10 min), the cells were dehydrated with a graded ethanol series (30%, 50%, 70%, 90%, and 95% in water and 10 min per step). Pure ethanol was then applied (3 × 10 min). After substitution with pure acetone (3 × 10 min), the samples were infiltrated with a graded series of resin (SPI-PON 812; SPI-CHEM) in acetone (25%, 50%, 75%) within 24 h. After infiltration with pure resin (three times within 24 h), the specimens in resin were polymerized at 60 °C for 2 d.
Scanning EM Sample Preparation and Imaging.
The preservation of primary cilia was examined using scanning EM (SEM) to test the effects of fixation with glutaraldehyde and postfixation with osmium tetroxide, to identify and solve the cilium truncation problem during TEM sample preparation. Cilia truncation was found to occur after postfixation.
For SEM, cell specimens of the planchets were dehydrated and critical point-dried using a Samdri-795 (Tousimis). The samples were then coated with gold using a Cressington 108 sputter coater (Ted Pella). The prepared SEM specimens were examined on a dual beam SEM (NEON 40EsB; Zeiss) operated at 1.5 kV. Images were taken using the signals of secondary electrons and secondary ions that is provided by a SESI detector.
Serial Sectioning and Staining.
The resin blocks containing the specimens were trimmed and serial 120- to 150-nm cross-sections were prepared using a UC6 microtome (Leica). The sections containing specimens were stained with 2% uranyl acetate and Reynolds’ lead citrate solution (77, 79) after collection onto individual slot copper grids coated with Formvar film. These semithin sections were used for electron tomographic data collection.
Electron Tomography.
Primary cilia were located in each serial section from the basal body to the tip with a multispecimen holder (Gatan) in a JEOL 1400 TEM (JEOL) operating at 100 keV. Colloidal gold beads (10 nm; Sigma) were added to both sides of the sections as fiducial markers before tomographic data collection. Electron tomographic imaging was carried out with a Philips Tecnai F20 EM operating at 200 keV using Serial EM software (80). The dual-axis tilt series images were captured on a 4k × 4k charge-coupled device detector (TVIPS TemCam-F415 and TemCam-F416) with the angle ranging from −65° to +65°, with 1° increments.
The tomographic tilting serials were processed and reconstructed with IMOD software (81). Briefly, the 2D tilt-projections were preprocessed to remove X-ray hot spots and make the rough alignment, and then fine alignment was conducted, utilizing the fiducial markers. The SIRT reconstructions were carried out using Tomo3D (82). The reconstructed a-axis and b-axis tomograms of each serial cross-sections were combined using IMOD (81).
Merging the Serial Tomograms for the Complete 3D Structural Maps.
Initial rough combination.
All of the tomograms obtained from each cross-section of the serial sections for the whole primary cilia were combined using the IMOD function “Join Serial Tomograms” (81). Briefly, the density volumes of primary cilium segments were roughly trimmed out with margins from the dual-axis tomogram series. The orientations of these subtomograms were determined based on the x-, y-tilt angles obtained using the “slicer” function of IMOD. The serial tomograms were roughly combined first, after the orientations were corrected, to form a raw tomogram stack.
Refined combination.
The top and bottom slices of each subtomogram in the tomogram series were carefully examined, and the slices that did not contain density of biological materials were trimmed away. These newly generated serial tomograms were recombined, and more detailed features close to the interfaces were used as tracers to achieve the refined joining. In a second round, the interfaces between tomograms were checked carefully. If there was a big shift within the neighboring tomograms, we would trim and separate the joined tomogram at the interface to form two tomograms and rejoin together using more detailed features in the structural maps. The refinement process was considered complete after the entire combined tomograms had an internally consistent appearance. During the whole joining process, translation, rotation, and magnification adjustment were allowed. The default full-linear transformation option was not used avoiding artifacts and structural distortion.
Three-dimensional masks were generated using Amira for the aligned tomograms of each primary cilium. The individual tomograms for each cilium were scaled to the same mean and SD based on density statistics within the corresponding mask. The scaled tomograms for a specific cilium were then joined together to generate its 3D structural map.
The complete structural map for each primary cilium often requires combining dozens of serial tomograms. Direct calculation to combine them together at once was extremely slow and often caused our most advanced image processing workstation to hang. Therefore, we divided the serial tomograms into a number of groups, with each group consisting of ∼10 individual tomograms. We carried out the combination for each group first, and then combined the resulted 3D maps together to obtain the final complete structural maps.
Density Map Analysis.
The file size of the combined 3D structural maps for each cilium may reach over 10 GB. Therefore, the combined structural maps were trimmed, using IMOD tools, to eliminate the structural features outside of the primary cilia that were useful for serial tomographic data combination, but were not part of primary cilia. Bsoft was used to bin and change the data to byte type to reduce the file size before segmentation and visualization (83, 84).
The segmentation of MtCs and membranes and structural model building was carried out using the 3D interactive tool in Amira software (Thermo Fisher). The movies were generated with Amira software using the “Demo Maker” and “Movie Maker” functions. University of California, San Francisco Chimera was used to render and display the densities in cilia (85), in combination with masks generated using Amira software.
For diameter and length measurement, Fiji (ImageJ) software (86) and IMOD (81) were used. Before the measurement, the voxel size was calibrated using 13-protofilament microtubules with known diameter (35). Lengths of the MtCs were measured directly. Diameters along the primary cilia were calculated based on the measured circumferences of the membrane circle shown in the cross-sectional slices of the primary cilia in the tomograms. The interval of the circumference calculation in longitudinal direction is ∼50 nm.
For this work, we located and traced 56 primary cilia of IMCD3 cells in successfully prepared serial sections under electron microscopes. Of them, we successfully collected complete EM micrographs of serial sections for 11 primary cilia. Among these cilia, we successfully completed electron tomographic studies for seven: three single-axis electron tomographic studies (shown in Movies S6, S10, and S11) and four dual-axis studies (Movies S1 and S7–S9) of serial cross-sections. In total, 160 tomographic datasets (78 dual-axis tomograms and 82 single-axis tomograms) were successfully obtained, reconstructed, and merged to generate the seven 3D structural maps of these primary cilia. Movies S1 and S6–S11 display a slicing through of the tomograms, revealing the structural evolution of the MtC architecture along the cilium length. These seven structural maps enabled investigation and measurement of structural features for the microtubule-based axonemes and helped us to develop an understanding on some common structural features for the primary cilia. To make the structural presentation straight-forward, we have chosen one typical structural map of a dual-axis tomogram of a primary cilium of 3-μm length to build a detailed 3D structural model. This 3D model is used to present the common structural features of IMCD3 primary cilia.
Subtomogram Alignment and Averaging.
The regions where microtubule doublets change to microtubule singlets were visually identified using IMOD (81), and their coordinates in the structural maps were manually recorded. Subtomograms of these regions were extracted for careful visual inspection, and then were classified and assigned into different types. The subtomograms of the same transition type (for type 1 or type 2) were aligned and averaged using functions provided in EMAN2 package (87). For the ninefold symmetry enforcement of the microtubule-doublet array near the basal body, the density map was rotated 40° each along the refined rotational axis before averaging. The resultant averaged 3D map-rendering and movie-making were conducted using the Chimera package (85, 88).
Data Availability.
The 3D structural map of the primary cilium used for model building has been deposited in the Electron Microscopy Data Bank under accession no. EMD-9022 (https://www.ebi.ac.uk/). The 3D structural maps of the primary cilia for SI Appendix, Figs. S3 A–F and S4 A–F have also been deposited under accession nos. EMD-20061 and EMD-20064–EMD-20068.
Supplementary Material
Acknowledgments
We thank Dr. M. Nachury at Stanford University for the stable GFP-fusion-SSTR3 IMCD3 cells; Dr. G. Pazours laboratory at the University of Massachusetts Medical School for the transiently transfected GFP-fusion-SSTR3 IMCD3 cells; Drs. C. Ott and J. Lippincott-Schwartz at the National Institute of Child Health and Human Development for YFP-fusion-Smo MDCK cells; the Wadsworth Center tissue culture core for LLC-PK1 cells; Drs. Alexey Khodjakov and Michael Koonce for their advice and help on kidney cell culture growth and maintenance, and valuable discussions on cytoskeleton structures; Dr. Jadranka Loncarek at the National Cancer Institute for valuable discussion and suggestions about LM imaging for primary cilium diameters; and Drs. Carmen Mannella and Roxanne Girardin for critical reading of the manuscript and valuable suggestions. We acknowledge Wadsworth Center’s support of the 3D-EM Facility and EM core. This work is supported by National Institutes of Health Grant GM101026 (to H.S.). S.S.B. was supported by National Science Foundation Grant PLR1341612.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The 3D structural maps of the primary cilia in this study are available in the Electron Microscopy Data Bank, https://www.ebi.ac.uk/ (accession nos. EMD-9022, EMD-20061, EMD-20064–EMD-20068).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821064116/-/DCSupplemental.
References
- 1.Brown JM, Witman GB. Cilia and diseases. Bioscience. 2014;64:1126–1137. doi: 10.1093/biosci/biu174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pazour GJ, et al. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun DA, Hildebrandt F. Ciliopathies. Cold Spring Harb Perspect Biol. 2016;9:a028191. doi: 10.1101/cshperspect.a028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poole CA, Flint MH, Beaumont BW. Analysis of the morphology and function of primary cilia in connective tissues: A cellular cybernetic probe? Cell Motil. 1985;5:175–193. doi: 10.1002/cm.970050302. [DOI] [PubMed] [Google Scholar]
- 5.Roth KE, Rieder CL, Bowser SS. Flexible-substratum technique for viewing cells from the side: Some in vivo properties of primary (9+0) cilia in cultured kidney epithelia. J Cell Sci. 1988;89:457–466. doi: 10.1242/jcs.89.4.457. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz EA, Leonard ML, Bizios R, Bowser SS. Analysis and modeling of the primary cilium bending response to fluid shear. Am J Physiol. 1997;272:F132–F138. doi: 10.1152/ajprenal.1997.272.1.F132. [DOI] [PubMed] [Google Scholar]
- 7.Praetorius HA, Spring KR. Bending the MDCK cell primary cilium increases intracellular calcium. J Membr Biol. 2001;184:71–79. doi: 10.1007/s00232-001-0075-4. [DOI] [PubMed] [Google Scholar]
- 8.Yoder BK, Hou X, Guay-Woodford LM. The polycystic kidney disease proteins, polycystin-1, polycystin-2, polaris, and cystin, are co-localized in renal cilia. J Am Soc Nephrol. 2002;13:2508–2516. doi: 10.1097/01.asn.0000029587.47950.25. [DOI] [PubMed] [Google Scholar]
- 9.Nauli SM, et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- 10.Takacs Z, Proikas-Cezanne T. Primary cilia mechanosensing triggers autophagy-regulated cell volume control. Nat Cell Biol. 2016;18:591–592. doi: 10.1038/ncb3366. [DOI] [PubMed] [Google Scholar]
- 11.Pampliega O, Cuervo AM. Autophagy and primary cilia: Dual interplay. Curr Opin Cell Biol. 2016;39:1–7. doi: 10.1016/j.ceb.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delling M, et al. Primary cilia are not calcium-responsive mechanosensors. Nature. 2016;531:656–660. doi: 10.1038/nature17426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delling M, DeCaen PG, Doerner JF, Febvay S, Clapham DE. Primary cilia are specialized calcium signalling organelles. Nature. 2013;504:311–314. doi: 10.1038/nature12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fawcett DW. The study of epithelial cilia and sperm flagella with the electron microscope. Laryngoscope. 1954;64:557–567. doi: 10.1288/00005537-195407000-00002. [DOI] [PubMed] [Google Scholar]
- 15.Afzelius B. Electron microscopy of the sperm tail; results obtained with a new fixative. J Biophys Biochem Cytol. 1959;5:269–278. doi: 10.1083/jcb.5.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibbons IR. The relationship between the fine structure and direction of beat in gill cilia of a lamellibranch mollusc. J Biophys Biochem Cytol. 1961;11:179–205. doi: 10.1083/jcb.11.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnes BG. Ciliated secretory cells in the pars distalis of the mouse hypophysis. J Ultrastruct Res. 1961;5:453–467. doi: 10.1016/s0022-5320(61)80019-1. [DOI] [PubMed] [Google Scholar]
- 18.Rieder CL, Jensen CG, Jensen LC. The resorption of primary cilia during mitosis in a vertebrate (PtK1) cell line. J Ultrastruct Res. 1979;68:173–185. doi: 10.1016/s0022-5320(79)90152-7. [DOI] [PubMed] [Google Scholar]
- 19.Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- 20.Laib JA, Marin JA, Bloodgood RA, Guilford WH. The reciprocal coordination and mechanics of molecular motors in living cells. Proc Natl Acad Sci USA. 2009;106:3190–3195. doi: 10.1073/pnas.0809849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satir P. CILIA: Before and after. Cilia. 2017;6:1. doi: 10.1186/s13630-017-0046-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei Q, Ling K, Hu J. The essential roles of transition fibers in the context of cilia. Curr Opin Cell Biol. 2015;35:98–105. doi: 10.1016/j.ceb.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang Y, Meng D, Zhu B, Pan J. Mechanism of ciliary disassembly. Cell Mol Life Sci. 2016;73:1787–1802. doi: 10.1007/s00018-016-2148-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bloodgood RA. From central to rudimentary to primary: The history of an underappreciated organelle whose time has come.The primary cilium. In: Sloboda RD, editor. Methods in Cell Biology. Vol 94. Academic; New York: 2009. pp. 2–52. [DOI] [PubMed] [Google Scholar]
- 25.Cohen E, Meininger V. Ultrastructural analysis of primary cilium in the embryonic nervous tissue of mouse. Int J Dev Neurosci. 1987;5:43–51. doi: 10.1016/0736-5748(87)90047-5. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto M, Kataoka K. Electron microscopic observation of the primary cilium in the pancreatic islets. Arch Histol Jpn. 1986;49:449–457. doi: 10.1679/aohc.49.449. [DOI] [PubMed] [Google Scholar]
- 27.Wheatley DN. Cells with two cilia in the rat adenohypophysis. J Anat. 1967;101:479–485. [PMC free article] [PubMed] [Google Scholar]
- 28.Gluenz E, et al. Beyond 9+0: Noncanonical axoneme structures characterize sensory cilia from protists to humans. FASEB J. 2010;24:3117–3121. doi: 10.1096/fj.09-151381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagiwara H, Ohwada N, Aoki T, Suzuki T, Takata K. The primary cilia of secretory cells in the human oviduct mucosa. Med Mol Morphol. 2008;41:193–198. doi: 10.1007/s00795-008-0421-z. [DOI] [PubMed] [Google Scholar]
- 30.Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J Cell Biol. 2008;183:923–932. doi: 10.1083/jcb.200808050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicastro D, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 32.Pigino G, et al. Cryoelectron tomography of radial spokes in cilia and flagella. J Cell Biol. 2011;195:673–687. doi: 10.1083/jcb.201106125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- 34.Li H, DeRosier DJ, Nicholson WV, Nogales E, Downing KH. Microtubule structure at 8 A resolution. Structure. 2002;10:1317–1328. doi: 10.1016/s0969-2126(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 35.Sui H, Downing KH. Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure. 2010;18:1022–1031. doi: 10.1016/j.str.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alushin GM, et al. High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell. 2014;157:1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sui H, Downing KH. Molecular architecture of axonemal microtubule doublets revealed by cryo-electron tomography. Nature. 2006;442:475–478. doi: 10.1038/nature04816. [DOI] [PubMed] [Google Scholar]
- 38.Nicastro D, et al. Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc Natl Acad Sci USA. 2011;108:E845–E853. doi: 10.1073/pnas.1106178108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ichikawa M, et al. Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins. Nat Commun. 2017;8:15035. doi: 10.1038/ncomms15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li S, Fernandez JJ, Marshall WF, Agard DA. Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J. 2012;31:552–562. doi: 10.1038/emboj.2011.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds MJ, et al. The developmental process of the growing motile ciliary tip region. Sci Rep. 2018;8:7977. doi: 10.1038/s41598-018-26111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flood PR, Totland GK. Substructure of solitary cilia in mouse kidney. Cell Tissue Res. 1977;183:281–290. doi: 10.1007/BF00226625. [DOI] [PubMed] [Google Scholar]
- 43.Hull RN, Cherry WR, Weaver GW. The origin and characteristics of a pig kidney cell strain, LLC-PK. In Vitro. 1976;12:670–677. doi: 10.1007/BF02797469. [DOI] [PubMed] [Google Scholar]
- 44.Rauchman MI, Nigam SK, Delpire E, Gullans SR. An osmotically tolerant inner medullary collecting duct cell line from an SV40 transgenic mouse. Am J Physiol. 1993;265:F416–F424. doi: 10.1152/ajprenal.1993.265.3.F416. [DOI] [PubMed] [Google Scholar]
- 45.Huang K, et al. Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J Cell Biol. 2007;179:501–514. doi: 10.1083/jcb.200704069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soto GE, et al. Serial section electron tomography: A method for three-dimensional reconstruction of large structures. Neuroimage. 1994;1:230–243. doi: 10.1006/nimg.1994.1008. [DOI] [PubMed] [Google Scholar]
- 47.Satir P. Studies on cilia. 3. Further studies on the cilium tip and a “sliding filament” model of ciliary motility. J Cell Biol. 1968;39:77–94. doi: 10.1083/jcb.39.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoops HJ, Witman GB. Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J Cell Biol. 1983;97:902–908. doi: 10.1083/jcb.97.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson RG. The three-dimensional structure of the basal body from the rhesus monkey oviduct. J Cell Biol. 1972;54:246–265. doi: 10.1083/jcb.54.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 51.Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stepanek L, Pigino G. Microtubule doublets are double-track railways for intraflagellar transport trains. Science. 2016;352:721–724. doi: 10.1126/science.aaf4594. [DOI] [PubMed] [Google Scholar]
- 53.Chien A, et al. Dynamics of the IFT machinery at the ciliary tip. eLife. 2017;6:e28606. doi: 10.7554/eLife.28606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marshall WF. Cellular length control systems. Annu Rev Cell Dev Biol. 2004;20:677–693. doi: 10.1146/annurev.cellbio.20.012103.094437. [DOI] [PubMed] [Google Scholar]
- 55.Ye F, Nager AR, Nachury MV. BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J Cell Biol. 2018;217:1847–1868. doi: 10.1083/jcb.201709041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Snow JJ, et al. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- 57.Jiang L, et al. Kinesin family 17 (osmotic avoidance abnormal-3) is dispensable for photoreceptor morphology and function. FASEB J. 2015;29:4866–4880. doi: 10.1096/fj.15-275677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Downing KH, Sui H. Structural insights into microtubule doublet interactions in axonemes. Curr Opin Struct Biol. 2007;17:253–259. doi: 10.1016/j.sbi.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 59.Witman GB, Carlson K, Berliner J, Rosenbaum JL. Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J Cell Biol. 1972;54:507–539. doi: 10.1083/jcb.54.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marshall WF, Qin H, Rodrigo Brenni M, Rosenbaum JL. Flagellar length control system: Testing a simple model based on intraflagellar transport and turnover. Mol Biol Cell. 2005;16:270–278. doi: 10.1091/mbc.E04-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Downing KH, Nogales E, Grant JJ. Methods in Enzymology. Vol 483. Academic; New York: 2010. Cryoelectron microscopy applications in the study of tubulin structure, microtubule architecture, dynamics and assemblies, and interaction of microtubules with motors; pp. 121–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nogales E, Wang HW. Structural intermediates in microtubule assembly and disassembly: How and why? Curr Opin Cell Biol. 2006;18:179–184. doi: 10.1016/j.ceb.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 63.Meza I, Huang B, Bryan J. Chemical heterogeneity of protofilaments forming the outer doublets from sea urchin flagella. Exp Cell Res. 1972;74:535–540. doi: 10.1016/0014-4827(72)90413-2. [DOI] [PubMed] [Google Scholar]
- 64.Linck RW, Norrander JM. Protofilament ribbon compartments of ciliary and flagellar microtubules. Protist. 2003;154:299–311. doi: 10.1078/143446103322454086. [DOI] [PubMed] [Google Scholar]
- 65.Amos LA. Articulated tubes. Structure. 2010;18:892–894. doi: 10.1016/j.str.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 66.Gilula NB, Satir P. The ciliary necklace. A ciliary membrane specialization. J Cell Biol. 1972;53:494–509. doi: 10.1083/jcb.53.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pigino G, et al. Electron-tomographic analysis of intraflagellar transport particle trains in situ. J Cell Biol. 2009;187:135–148. doi: 10.1083/jcb.200905103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bloodgood RA. The future of ciliary and flagellar membrane research. Mol Biol Cell. 2012;23:2407–2411. doi: 10.1091/mbc.E12-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baldari CT, Rosenbaum J. Intraflagellar transport: It’s not just for cilia anymore. Curr Opin Cell Biol. 2010;22:75–80. doi: 10.1016/j.ceb.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wood CR, Rosenbaum JL. Ciliary ectosomes: Transmissions from the cell’s antenna. Trends Cell Biol. 2015;25:276–285. doi: 10.1016/j.tcb.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol. 2014;16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nager AR, et al. An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell. 2017;168:252–263.e14. doi: 10.1016/j.cell.2016.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ye F, et al. Single molecule imaging reveals a major role for diffusion in the exploration of ciliary space by signaling receptors. eLife. 2013;2:e00654. doi: 10.7554/eLife.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hassounah NB, Bunch TA, McDermott KM. Molecular pathways: The role of primary cilia in cancer progression and therapeutics with a focus on Hedgehog signaling. Clin Cancer Res. 2012;18:2429–2435. doi: 10.1158/1078-0432.CCR-11-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He W, et al. A freeze substitution fixation-based gold enlarging technique for EM studies of endocytosed nanogold-labeled molecules. J Struct Biol. 2007;160:103–113. doi: 10.1016/j.jsb.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McDonald KL, Webb RI. Freeze substitution in 3 hours or less. J Microsc. 2011;243:227–233. doi: 10.1111/j.1365-2818.2011.03526.x. [DOI] [PubMed] [Google Scholar]
- 77.Martin A, Hedinger C, Häberlin-Jakob M, Walt H. Structure and motility of primary cilia in the follicular epithelium of the human thyroid. Virchows Arch B Cell Pathol Incl Mol Pathol. 1988;55:159–166. doi: 10.1007/BF02896572. [DOI] [PubMed] [Google Scholar]
- 78.Pigino G, et al. Electron tomography of IFT particles. Methods Enzymol. 2013;524:325–342. doi: 10.1016/B978-0-12-397945-2.00018-4. [DOI] [PubMed] [Google Scholar]
- 79.Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 81.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 82.Agulleiro JI, Fernandez JJ. Tomo3D 2.0—Exploitation of advanced vector extensions (AVX) for 3D reconstruction. J Struct Biol. 2015;189:147–152. doi: 10.1016/j.jsb.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 83.Heymann JB. Bsoft: Image and molecular processing in electron microscopy. J Struct Biol. 2001;133:156–169. doi: 10.1006/jsbi.2001.4339. [DOI] [PubMed] [Google Scholar]
- 84.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 85.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 86.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 88.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 3D structural map of the primary cilium used for model building has been deposited in the Electron Microscopy Data Bank under accession no. EMD-9022 (https://www.ebi.ac.uk/). The 3D structural maps of the primary cilia for SI Appendix, Figs. S3 A–F and S4 A–F have also been deposited under accession nos. EMD-20061 and EMD-20064–EMD-20068.