Significance
A new phenomenon of constructing distinctive funerary monuments, collectively known as megalithic tombs, emerged around 4500 BCE along the Atlantic façade. The megalithic phenomenon has attracted interest and speculation since medieval times. In particular, the origin, dispersal dynamics, and the role of these constructions within the societies that built them have been debated. We generate genome sequence data from 24 individuals buried in five megaliths and investigate the population history and social dynamics of the groups that buried their dead in megalithic monuments across northwestern Europe in the fourth millennium BCE. Our results show kin relations among the buried individuals and an overrepresentation of males, suggesting that at least some of these funerary monuments were used by patrilineal societies.
Keywords: paleogenomics, population genomics, migration, megalithic tombs
Abstract
Paleogenomic and archaeological studies show that Neolithic lifeways spread from the Fertile Crescent into Europe around 9000 BCE, reaching northwestern Europe by 4000 BCE. Starting around 4500 BCE, a new phenomenon of constructing megalithic monuments, particularly for funerary practices, emerged along the Atlantic façade. While it has been suggested that the emergence of megaliths was associated with the territories of farming communities, the origin and social structure of the groups that erected them has remained largely unknown. We generated genome sequence data from human remains, corresponding to 24 individuals from five megalithic burial sites, encompassing the widespread tradition of megalithic construction in northern and western Europe, and analyzed our results in relation to the existing European paleogenomic data. The various individuals buried in megaliths show genetic affinities with local farming groups within their different chronological contexts. Individuals buried in megaliths display (past) admixture with local hunter-gatherers, similar to that seen in other Neolithic individuals in Europe. In relation to the tomb populations, we find significantly more males than females buried in the megaliths of the British Isles. The genetic data show close kin relationships among the individuals buried within the megaliths, and for the Irish megaliths, we found a kin relation between individuals buried in different megaliths. We also see paternal continuity through time, including the same Y-chromosome haplotypes reoccurring. These observations suggest that the investigated funerary monuments were associated with patrilineal kindred groups. Our genomic investigation provides insight into the people associated with this long-standing megalith funerary tradition, including their social dynamics.
Investigations of the genetic relationships among humans from multiple Neolithic sites across western Eurasia have shown that Neolithic lifeways dispersed across Europe via a large-scale process of migration (1–6) starting from Anatolia and the areas of the Aegean at ca. 7000–6500 (cal) BCE (7–10). In Europe, migrating people and Neolithic lifeways dispersed along two main routes: an inland route (partly along the Danube River) and a route along Mediterranean coastal areas (11–13). Around 4000 BCE, Neolithic farming communities reached the northwestern fringes of Europe, including the British Isles (14, 15) and Scandinavia (1, 2, 16, 17). A marked hunter-gatherer (HG) admixture has been observed in the later farmer groups compared with the Early Neolithic farmers on the continent (2, 10, 12).
During this period of important social and demographic change, a new phenomenon of constructing megalithic monuments emerged, starting around 4500 BCE in France (18), 3700 BCE in the British Isles (14, 19–26), and 3600 in Scandinavia (16, 27). These Neolithic megalithic tombs are concentrated along the Atlantic coastal areas, stretching from the Mediterranean to Scandinavia, including the British Isles and regions in the northern European plain (28), but also in southern France, northern Italy, and on the Islands of Corsica and Sardinia (Fig. 1) (19, 27).
Fig. 1.
Map of Europe with megalithic burial sites (red squares) and nonmegalithic sites (red circles) from this study, and comparative published data from megalithic sites (black squares) sequenced to date in Europe (Dataset S1.3). The date range represents the 95% CI of available samples from these sites, except for La Mina in Spain. Blue shading represents the estimated distribution of early megalithic burials. Bold italic type indicates dates (95% CI) estimated for the start of dolmens and passage grave monuments, based on samples from these contexts. Regular text indicates time interval associated with the earliest cultural material in the megaliths (27, 45).
The emergence of these megaliths was closely associated with the development of farming communities (14, 23, 25, 27, 29), but the origin and the social structure of the groups are largely unknown. The similarities in the construction and design of some types of megaliths (i.e., dolmens and passage graves) from Iberia to southern Scandinavia, Britain, and Ireland is compelling. Interregional interaction has been evidenced in the same period from the dispersal of domesticated resources, raw materials, and artifacts, possibly reflecting shared social and cultural systems as well as shared cosmology of the groups (21, 27, 28, 30). Although it is clear that many megaliths were used for collective burials (27, 29, 31), it has been difficult to evaluate which members of the communities were buried in the tombs. Some assemblages include males, females, juveniles, and children, implying familial burials. Many tombs have poorly preserved human remains and also show secondary usage in later times, complicating assessments. The use of megaliths as burial grounds for the community as a whole would imply some level of shared ideology over vast geographical areas (31, 32). However, it has also been argued that the monumental burials and associated rich material culture reflect the emergence of social differentiation or stratification (33–36; see ref. 37 on segmentally structured societies), with the monuments perhaps symbolizing status and territorial markers (37–40).
Some scholars hypothesize that the people buried in the megalithic structures were kin related (41–43). Analyses of mitochondrial data (mtDNA) from megalithic burials at Falbygden and Gotland in modern-day Sweden have revealed a large lineage variation, and thus the groups did not seem to have been organized matrilineally (44, 45; however, contra ref. 43). Genomic data are necessary to provide deeper information on kin relations and the social dynamics and general social structure of the societies or groups. However, as genomic data have been available from only a few individuals from megalithic burials, the origin and dispersal dynamics of the funerary practices, as well as the population history of the people that used the burial constructions, have also remained uncertain.
In the present study, we investigated the genetic structure and demographic affinities of people buried within megaliths to shed light on this burial phenomenon, the social dynamics of the people buried in the monuments, and their demographic history. We generated and examined genome sequence data from 24 individuals from five megalith burial sites located in Ireland, the Orkney Isles, and the Island of Gotland in the Baltic Sea dated between ca. 3800 and 2600 cal BCE encompassing wide-ranging examples from the megalithic tradition in northern Europe. The study also incorporated three individuals from nonmegalith contexts from mainland Scotland (3370–3100 cal BCE) and the Czech Republic (4825–4555 cal BCE) (Table 1).
Table 1.
Summary of genetic and archaeological information about the 27 individuals in the study
| Radiocarbon date (95% CI, cal BCE) | Sequence coverage | Haplogroup | Estimated contamination | ||||||||
| Individual | Site | Upper | Lower | nuDNA | mtDNA | Sex | mt | Ychr | mtDNA | 95% CI | Autosomal |
| Primrose 2 | Primrose | 3790 | 3660 | 5.76 | 817.93 | XX | H1+16189 | 0.05 | 0.01–1.22 | 1.283 | |
| Primrose 17 | Primrose | 3780 | 3650 | 0.19 | 49.51 | XY | K1a+195 | I | 0.66 | 0.11–21.63 | 0.049 |
| Primrose 18 | Primrose | 3770 | 3650 | 0.10 | 55.71 | XY | K1a+195 | I | 0.59 | 0.10–18.30 | 0.000 |
| Primrose 12 | Primrose | 3770 | 3650 | 0.25 | 325.42 | XY | W1+119 | I2a2a1a1a2 | 0.09 | 0.01–2.62 | 0.000 |
| Primrose 3 | Primrose | 3770 | 3650 | 0.22 | 125.69 | XY | H1i | I | 5.28 | 1.91–12.50 | 0.000 |
| Primrose 16 | Primrose | 3690 | 3530 | 6.40 | 442.67 | XY | K1a4a1 | I2a2a1a1a | 0.06 | 0.01–1.53 | 0.951 |
| Primrose 10 | Primrose | 3640 | 3520 | 0.23 | 178.60 | XY | K1a+195 | I | 0.17 | 0.03–5.23 | 0.000 |
| Primrose 6 | Primrose | 3640 | 3380 | 0.27 | 1,158.06 | XX | K1a+195 | 0.03 | 0.00–0.84 | 0.000 | |
| Primrose 13 | Primrose | 3630 | 3370 | 4.73 | 675.01 | XY | T2b3c | I2a2a1a1a | 0.03 | 0.01–0.64 | 1.731 |
| Primrose 7 | Primrose | 3510 | 3360 | 0.01 | 43.44 | XY | K1a4a1 | NA | 1.44 | 0.18–14.26 | 0.000 |
| Primrose 9 | Primrose | 3500 | 3360 | 7.10 | 923.93 | XY | U5b2c | I2a2a1a1a | 0.03 | 0.00–0.88 | 1.520 |
| Carrowmore 4 | Carrowmore | 3640 | 3380 | 0.04 | 451.69 | XY | T2c1d1 | I | 0.03 | 0.00–0.72 | 0.100 |
| Midhowe 1 | Midhowe | 3630 | 3370 | 0.27 | 22.00 | XY | H5+16311 | I2a1b | 1.52 | 0.24–44.17 | 1.150 |
| Lairo 1 | Lairo | 3360 | 3100 | 0.22 | 25.08 | XY | U5b2 | I2a1b | 0.96 | 0.16–31.07 | 0.022 |
| Balintore 4 | Balintore | 3370 | 3110 | 1.54 | 168.43 | XX | H1 | 0.18 | 0.03–4.71 | 0.033 | |
| Midhowe 2 | Midhowe | 3360 | 3100 | 0.25 | 29.38 | XY | K1a+195 | I | 0.75 | 0.14–23.16 | 0.281 |
| Ansarve 5 | Ansarve | 3500a | 3130* | 0.13 | 114.73 | XX | K1a2b* | 0.21 | 0.04–7.79 | 0.000 | |
| Ansarve 3 | Ansarve | 3490a | 3110* | 0.14 | 300.87 | XX | T2b8* | 0.04 | 0.01–1.02 | 0.046 | |
| Ansarve 8 | Ansarve | 3340a | 3030* | 1.94 | 1,462.38 | XY | J1c5* | I2a1b1a1† | 0.01 | 0.00–0.14 | 0.441 |
| Ansarve 14 | Ansarve | 3330a | 2950* | 2.58 | 431.47 | XY | J1c5* | I2a1b1a1† | 0.02 | 0.00–0.41 | 0.525 |
| Ansarve 17 | Ansarve | 3330a | 2930* | 6.80 | 491.04 | XY | HV0a* | I2a1b1a1† | 0.06 | 0.01–2.06 | 1.461 |
| Ansarve 6 | Ansarve | 3090a | 2920* | 0.0027 | 137.06 | XY | J1c8a* | NA | 0.06 | 0.01–1.70 | NA |
| Ansarve 7 | Ansarve | 3010a | 2890* | 0.0014 | 24.54 | XY | K2b1a* | NA | 0.33 | 0.06–8.90 | NA |
| Ansarve 9 | Ansarve | 2880a | 2630* | 0.0009 | 26.73 | XX | K2b1a* | 0.29 | 0.05–6.99 | NA | |
| Ansarve 16 | Ansarve | 2810a | 2580* | 0.33 | 23.17 | XY | H7d* | I2a1b† | 1.60 | 0.27–46.97 | 0.004 |
| Kolin6 | Kolin | 4910 | 4740 | 1.51 | 218.40 | XX | H+16129 | 0.10 | 0.02–2.23 | 2.639 | |
| Kolin2 | Kolin | 4650 | 4460 | 0.10 | 42.39 | XX | W1+119 | 0.37 | 0.06–10.83 | 0.068 | |
Results
We present genome data from 27 individuals excavated from European Neolithic contexts, of whom 24 were buried in megaliths; Primrose Grange (n = 11) and Carrowmore (n = 1) in Ireland; Lairo (n = 1) and Midhowe (n = 2) in the Orkney Islands, Scotland; and Ansarve (n = 9) in the island of Gotland, Sweden (16, 45, 46) (Table 1 and SI Appendix, section S2). Individuals from the Scottish “short cist” burial Balintore (n = 1) and the Czech Republic Kolin Rondel site (n = 2) (46), associated with the Stroked Pottery culture, were also investigated. These individuals were all radiocarbon-dated to between 4825 and 2580 cal BCE (Table 1). We compared our data with genetic data previously generated from 36 individuals from 16 megalithic sites (Fig. 1 and Dataset S1.3), as well as with farmer groups of nonmegalithic contexts (Dataset S1.3), to investigate the population history of people buried in megaliths.
The individuals buried in these megaliths from the British Isles and Scandinavia show an ancestry similar to other contemporaneous farmer groups (Fig. 2A), with a majority of their ancestry related to early Neolithic farmers and a partial admixture component related to European Mesolithic HGs (Fig. 2B) (1, 2, 5–7, 10, 16, 46).
Fig. 2.
(A) PCA of 429 present-day west Europeans (gray dots) with previously published Western HG (WHGs), Atlantic coast and Central European Neolithic farmer samples (filled symbols), and the samples from the present study (shaded symbols) projected onto the first two principal components (more details in SI Appendix, Section S11.1). (B) Inferred ancestry components (assuming seven clusters) of ancient individuals (Methods and SI Appendix, Section S11.2). All individuals to the left of Yoruba are prehistoric individuals, all of which are shotgun-sequenced unless marked with “CP” for SNP capture data. In the label names, the following letters indicate an archaeological context: CA, Chalcolithic; EN, Early Neolithic; N, Neolithic; MN, Middle Neolithic; LN, Late Neolithic. The LN individuals from Portugal come from different sites (key provided in Dataset S1.3).
To further explore the demographic history of the individuals buried in the megaliths, we investigated the genetic affinities among sets of individuals and groups, using an f3 outgroup test for groups of individuals buried in megalithic or nonmegalithic contexts, as well as between individuals from Atlantic coastal and inland Neolithic sites (SI Appendix, section S11.3 and Fig. S19). These analyses showed genetic associations between individuals from the same/similar geographic region and time period (Fig. 2A and SI Appendix, Figs. S16 and S17). However, some tests (SI Appendix, Fig. S19) indicated similar trends as shown in our principal component analysis (PCA) and previous studies (5, 11, 15, 47, 48) and suggested a demic connection among western European Neolithic groups to the exclusion of central European Neolithic groups, as well as a connection between the British Isles and Iberian groups (SI Appendix, section S11.4 and Figs. S20–S22). These results were not driven by greater levels of HG ancestry among the populations at the fringes of the Neolithic expansion (11, 12, 15, 16) (SI Appendix, section S11.4).
Interestingly, we also found a significant farmer-specific genetic affinity between the British Isles Neolithic populations and the Scandinavian populations (Ansarve and Gökhem; Fig. 1) to the exclusion of central European farmers (SI Appendix, Figs. S21 and S22). This observation is compatible with a further migration of farming groups along the European Atlantic coast, as has been suggested by the archaeological record (21, 49, 50).
We found that significantly more males than females were buried in the British Isles megaliths (31 of 42 randomly sampled individuals; P = 0.0014, binomial test) and at the Primrose megalith alone (9 of 11; P = 0.032) (SI Appendix, section S8). However, other megalithic tombs with at least four individuals investigated, including Ansarve (6 of 9; P = 0.25), Gökhem (1 of 4; P = 0.93), La Mina (2 of 4; P = 0.68), Holm of Papa Westray (2 of 4; P = 0.68), and Isbister (Tomb of the Eagles) (8 of 10; P = 0.054), did not show the same striking pattern, nor did nonmegalithic burials from the British Isles (15) (nonmegalithic burials: 6 of 10; P = 0.27, cave burials: 10 of 15; P = 0.27, both nonmegalithic and cave burials: 16 of 25; P = 0.11). Overall, genetic data from all individuals from megalithic contexts suggest a higher male-to-female ratio in these burial chambers (41 of 60; P = 0.0031) (SI Appendix, Table S3), although the tendency is similar (but not significant) for nonmegalithic burials (SI Appendix, section S8).
We found greater macrohaplogroup mtDNA diversity than Y-chromosomal (YDNA) diversity. Whereas mtDNA lineages from megalith burials harbor haplogroups K, H, HV, V, U5b, T, and J (among others), males from megalith burials belong almost exclusively to YDNA haplogroup I, more specifically to the I2a sublineage, which has a time to most recent common ancestor of ∼15000 BCE (51). This pattern of uniparental marker diversity is found not only among individuals buried in megaliths, but also in other farmer groups from the fourth millennium BCE, which display similar patterns of uniparental marker diversity (SI Appendix, Figs. S6 and S23) (10, 15, 48, 52). Some mtDNA lineages appear to be overrepresented at megalithic sites, with information from more than six individuals, including Primrose (n = 11; K1a+195 and K1a4a1 at 36% and 18% frequency, respectively), Ansarve (n = 9; J1c5 and K2b1a at ∼20% frequency), and Isbister (n = 10; K1a+195 at 20% frequency). Males from the present study belonged to YDNA haplogroup I, and those who could be resolved beyond this level were characterized as belonging to the I2a2a or I2a1b branch. Four of the 10 Primrose/Carrowmore males (Primrose 9, 12, 13, and 16) could be further resolved to the former sublineage, while the two Scottish males and the four Ansarve males could be further placed in the latter branch (Table 1 and SI Appendix, section S7).
Combining the YDNA lineages and the radiocarbon dates of the individuals, a possible scenario of paternal continuity is observed for the Primrose and Ansarve megaliths. From the Primrose site, Primrose 9, 13, and 16, separated in time by at least 1 generation and possibly up to 12 generations, display the I2a2a1a1a haplotype. In addition, the Primrose 3, 10, and 17 individuals were inferred to harbor variants common to the I2a2 lineage, although with low coverage support (SI Appendix, section S7). A similar scenario is observed for the Ansarve megalith, with the individuals Ansarve 8, 14, and 17, separated by at most a few generations, carrying haplotype I2a1b1a. Ansarve 16, dated to at least 100 y younger, shares variants along the I2a1b lineage (Table 1 and SI Appendix, section S7).
The high frequency of the HG-derived I2a male lineages among megalith as well as nonmegalith individuals (SI Appendix, section S11.6) suggests a male sex-biased admixture process between the farmer and the HG groups (2, 12, 53, 54), but when this admixture occurred is unclear. To characterize the extent of sex-biased admixture between HGs and the individuals of the megalithic contexts, we assessed the affinity of all individuals buried in megaliths with sufficient genetic data, to an Early Neolithic farmer or a HG ancestry on the autosomes and the X chromosome using f4-statistics (SI Appendix, section S11.5). Higher levels of HG admixture on the autosomes than on the X chromosome implies a greater genetic contribution of male HGs than female HGs to these individuals, suggesting an HG male sex bias admixture. We find that in general, megalith groups do not harbor higher levels of HG ancestry on the autosomes compared with on the X chromosome (SI Appendix, Table S7 and Dataset S1.6), but the Scottish_MN farmers of this study showed a tendency toward an HG male-sex biased admixture in the recent past. The Scandinavian (Ansarve and Gökhem) individuals displayed an HG admixture for both the autosomes and the X chromosome (SI Appendix, Table S7), suggesting a scenario of more recent admixture with HGs in northern Europe.
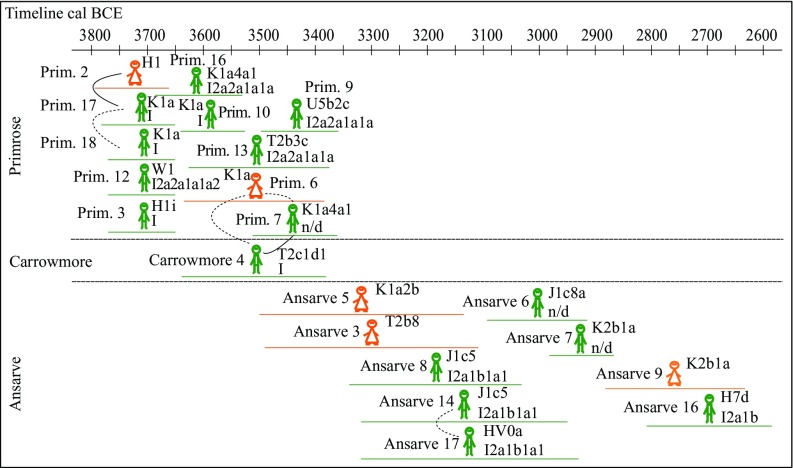
Using READ (Relationship Estimation from Ancient DNA) software (55), we inferred six kin relationships among the megalith individuals of this study: five relations among the Irish megaliths (two first-degree and three second-degree connections) and a second-degree relation in the Ansarve tomb (Fig. 3 and SI Appendix, section S10). First-degree relationships are characterized by either parent-offspring or a full sibling relationship, second-degree kin connections are represented by half-siblings, grandparent-grandchild, aunt/uncle-niece/nephew, and double cousins. Combining the READ predictions, uniparental lineages, radiocarbon dating, and age at death if available for those individuals who could be assessed, we inferred the potential familial relationships (Fig. 3 and SI Appendix, sections S2, S6, S7, and S10). Among the Irish megaliths, we observed two potential familial structures (SI Appendix, Fig. S10). The first is composed of three individuals from Primrose Grange (Tomb 1; individuals Primrose 2, 17, and 18), which overlap broadly in time (Fig. 3). Primrose 2 and 17 were predicted to be related in the first degree, representing a father-daughter relationship. Primrose 17 and 18 were predicted to be second-degree relatives (harboring the same mtDNA lineage but with possibly different YDNA haplogroups) and thus could have been half-siblings or double cousins. However, the YDNA prediction is hindered by low coverage and few informative markers, and thus a grandfather-grandson or uncle-nephew relationship cannot be fully excluded.
Fig. 3.
Kinship relationships in the Primrose, Carrowmore, and Ansarve burials. Solid line, first degree; dashed line, second degree. Males are displayed in green; females, in orange. The MtDNA and YDNA haplogroups are presented to the right of the figures. Bars underneath figures represent calibrated dating, with 95% CI (details in Table 1 and SI Appendix, Table S1).
The other Irish putative pedigree structure was integrated by two individuals from Tomb 1 (Primrose 6 and 7) and one individual from Carrowmore 4 (from the Listhogil Tomb at the Carrowmore site in close vicinity), who harbored different mtDNA lineages. While the 95% CI dating range of Primrose 6 and Carrowmore 4 overlap, Primrose 7 might be slightly younger than the other two individuals. The Carrowmore 4 and Primrose 7 males were inferred to be at least second-degree related (3.14 SE below the expected value for two unrelated individuals), and the best prediction was a first-degree relation (1.79 SE below the value for a second-degree relation, although not statistically significant at the 95% level; SI Appendix, section S10). If a first-degree relation is assumed, then the sole possible kin connection is a father-son relationship, because the individuals are not maternally linked. In the case of a second-degree relationship, any paternally related second-degree familial connection is possible. The other two READ-predicted second-degree kin relationships in the Irish burials (Primrose 6-Primrose 7 and Primrose 6-Carrowmore 4; 1.04 SE and 0.50 SE below the threshold for an unrelated pair, respectively) involved a familial connection of the male individuals to Primrose 6 (female).
Within the Ansarve megalith, we identify a second-degree relationship between the contemporaneous males Ansarve 14 and Ansarve 17 (Fig. 3 and SI Appendix, section S10). Both males have the same YDNA haplotype but different mtDNA lineages, suggesting that they could be related through any second-degree paternal kin relationship. Morphologically, Ansarve 14 was predicted to be an adult, and Ansarve 17 was predicted to be a juvenile (SI Appendix, section S2). Such observations might favor a grandfather-grandson or uncle-nephew relatedness over half-siblings or double cousins; however, the latter alternatives are still compatible with the data (SI Appendix, Fig. S12). READ analyses from other megalith burials where genetic data from at least four individuals were available per site (Gökhem, La Mina, Isbister, and Holm of Papa Westray; Fig. 1) did not reveal any evidence of genetic kinship relations. However, such observations may be hindered by the limited number of individuals investigated or by low genome coverage, which decreases the power to infer kinship (SI Appendix, section S10).
Discussion
The genetic variation and characteristics of individuals buried in megalithic tombs, and also from individuals buried according to other traditions, suggest that the megalithic tradition was linked to socially stratified Neolithic farmer societies, with the genetic data suggesting close connections between Neolithic populations in Atlantic Europe (5, 15, 48) (Fig. 2 and SI Appendix, Figs. S19–S22). Here we provide evidence of a genetic connection among Scandinavian, British, and Irish Neolithic populations. This signal is weaker than the signals observed between the Iberian Peninsula and the British Isles, however (5, 11, 15, 47, 48) (Dataset S1.3), suggesting that migration between the British Isles and Scandinavia along the Atlantic coast was less frequent than that between Iberia and the British Isles (SI Appendix, section S11.4).
The I2 YDNA lineages that are very common among European Mesolithic HGs (2, 3, 15, 56, 57) are distinctly different from the YDNA lineages of the European Early Neolithic farmer groups (8–10), but frequent in the farmer groups of the fourth millennium BCE (2, 3, 8–10, 15, 56, 57), suggesting a male HG admixture over time. The megalith individuals do not show higher levels of HG ancestry on the autosomes than on the X chromosome, but the Scottish_MN group shows a tendency toward a male-biased HG admixture in farmer groups, similar to previous observations (58). For the Scandinavian farmer groups, in contrast to the other megalith groups, we found an HG admixture for both the autosomes and the X chromosome. When these findings are considered together, it appears as if the social dynamics between HGs and Neolithic farmer groups, and thus the genetic admixture with HGs, differed somewhat in different geographic regions—an observation consistent with a combination of previous male sex bias admixture events occurring on the continent and more recent regional encounters with HG groups with a less pronounced sex-biased admixture.
These observations imply that the groups that erected and used the megalithic burial structures were stable and stratified, but probably not isolated farmer societies (37, 41). The genetic connection of the individuals from the Primrose Grange and Carrowmore burials, spatially distanced by only 2 km and in contemporaneous use, suggests that transgenerational patrilineal structured societies could have expanded geographically, possibly leaving a (local) genetic fingerprint related to the social dynamics of the group. Such a scenario of forming patrilineal kin groups and intergroup competition during the Neolithic could explain the inferred Y-chromosome bottleneck seen in present-day European populations (51, 59).
A central topic of discussion concerning the megalithic phenomena relates to the character of the communities that erected and used them for funerary rituals (27, 31, 37, 41, 42). The distinction of specific paternal lineages among the megaliths, a greater fraction of males than females in some megaliths, and their kindred relationships suggest that people buried in the megalithic tombs belonged to patrilineal segments of the groups/societies rather than representing a random sample from a larger Neolithic farmer community living in close vicinity. The sex ratio in the Irish megaliths is also in line with this finding. If one of the main functions of the tombs was to contain the remains of the deceased of a patrilineal segment, this would explain the inclusion of more males than females in the tombs. However, the finding that three of the five kinship relationships in these megaliths involved females indicates that female kindred members were not excluded. The observation of paternal continuity across time at the Gotlandic Ansarve megalith and at the Irish megaliths is a strong indication that specific family groups used these stone constructions for burial and other funerary practices. Of course, the patterns that we observe could be unique to the Primrose, Carrowmore, and Ansarve burials, and future studies of other megaliths are needed to provide additional data that can inform us further about social organization in the Neolithic.
Materials and Methods
Archaeological Samples (SI Appendix, sections S1 and S2).
Bones and teeth from human remains representing 27 individuals (Table 1) from seven sites were sampled for ancient DNA analyses; Primrose Grange (Tomb 1) and the Listhogil court cairn at Carrowmore (Ireland), the Lairo and Midhowe chambered tombs in Orkney and the Balintore short cist burial (Scotland), the Ansarve dolmen on the Island of Gotland (Sweden), and the Kolin Rondel site (the Czech Republic). Twelve samples were radiocarbon dated using accelerator mass spectrometry, and datings were available for the other samples.
Sequencing (SI Appendix, section S3).
DNA was extracted from bones and teeth (60, 61), and DNA sequences in the extracts were converted to blunt-ended Illumina libraries. For some individuals, uracil-DNA-glycosylase (UDG)-treated, whole-genome capture-enriched, and/or single-strand libraries were also generated. All samples were prepared in dedicated ancient DNA facilities. The libraries were sequenced on Illumina HiSeq platform 2500 or XTen.
NGS Data Processing and Authentication (SI Appendix, section S4).
Overlapping paired-end reads were trimmed and merged (62), and the fragments were mapped to the human reference genome (63). Fragments with identical start and end positions were considered PCR duplicates and collapsed into consensus sequences. Contamination was estimated based on phylogenetically informative sites on the mitochondrial genome using Contamix (64), on the X chromosome using ANGSD v.0.902 (65), and on the autosomal data using VerifyBamID v.1.1.2 (66). All libraries except the UDG-treated libraries showed signs characteristic of aDNA damage (67).
Uniparental Haplogroups (SI Appendix, sections S6 and S7).
We inferred the most likely haplogroup from mitochondrial consensus sequence from each individual (68, 69). Y chromosomal haplogroups were further assigned by investigating informative single base substitutions obtained from the International Society of Genetic Genealogy (version 11.110 from April 21, 2016; https://isogg.org/). Geographical and temporal distribution of Y chromosomal haplogroups are outlined in SI Appendix, section S11.6.
Population Genetic Analysis (SI Appendix, section S11).
The data from the investigated individuals were merged with various published datasets depending on the nature of the analyses. At each SNP position, a single read (minimum mapping and base quality of 30) was drawn at random to represent a haploid copy from the ancient individual. Transitions were coded as missing data to exclude potential postmortem damage. For each ancient individual, a PCA was conducted together with 203 modern Europeans (70, 71), and ancient individuals were plotted using Procrustes transformation (72). Ancestry components were inferred (73) based on 1,718 modern-day individuals from 179 populations and all ancient individuals (SI Appendix, Table S5). Common modes among the different runs were identified, and clusters were aligned across different values of K using pong (74). f3 and f4 statistics were computed (71) to estimate shared drift between populations.
Kinship Relationship Inferences (SI Appendix, section S10).
Familial relationships were inferred (55) for individuals. Data generated with different library building strategies were handled separately to avoid potential biases.
Data Availability.
Raw sequencing reads produced for this study have been deposited in the European Nucleotide Archive (accession no. PRJEB31045).
Supplementary Material
Acknowledgments
We thank the teams involved in the excavations; J. Pearson and C. McCullagh of Inverness Museum for access to the Balintore samples; A. R. Munters for processing and curating of NGS data; and K. Dobney and T. Günther for valuable discussions and comments on the manuscript. This project was supported by grants from the Riksbankens Jubileumsfond (to A.G., M.J., and J.S.), the Knut and Alice Wallenberg Foundation (to A.G., M.J., and J.S.), and the Berit Wallenberg Foundation (to M.F.). Sequencing was conducted at the Swedish National Genomics Infrastructure in Uppsala, and computations were performed at Uppsala Multidisciplinary Centre for Advanced Computational Science. Primrose Grange Tomb 1 was excavated under Licenses 98E0168, 98E0169, and 96E0020/ext, and analyses were performed under License 5396 to Export and Alter Archaeological Objects from June 27 to 29, 2012.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Raw sequencing reads produced for this study have been deposited in the European Nucleotide Archive (accession no. PRJEB31045).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818037116/-/DCSupplemental.
References
- 1.Skoglund P, et al. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science. 2012;336:466–469. doi: 10.1126/science.1216304. [DOI] [PubMed] [Google Scholar]
- 2.Skoglund P, et al. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science. 2014;344:747–750. doi: 10.1126/science.1253448. [DOI] [PubMed] [Google Scholar]
- 3.Lazaridis I, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gamba C, et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat Commun. 2014;5:5257. doi: 10.1038/ncomms6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassidy LM, et al. Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome. Proc Natl Acad Sci USA. 2016;113:368–373. doi: 10.1073/pnas.1518445113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Günther T, Jakobsson M. Genes mirror migrations and cultures in prehistoric Europe: A population genomic perspective. Curr Opin Genet Dev. 2016;41:115–123. doi: 10.1016/j.gde.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Omrak A, et al. Genomic evidence establishes Anatolia as the source of the European Neolithic gene pool. Curr Biol. 2016;26:270–275. doi: 10.1016/j.cub.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 8.Kılınç GM, et al. The demographic development of the first farmers in Anatolia. Curr Biol. 2016;26:2659–2666. doi: 10.1016/j.cub.2016.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofmanová Z, et al. Early farmers from across Europe directly descended from Neolithic Aegeans. Proc Natl Acad Sci USA. 2016;113:6886–6891. doi: 10.1073/pnas.1523951113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathieson I, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valdiosera C, et al. Four millennia of Iberian biomolecular prehistory illustrate the impact of prehistoric migrations at the far end of Eurasia. Proc Natl Acad Sci USA. 2018;115:3428–3433. doi: 10.1073/pnas.1717762115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Günther T, et al. Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc Natl Acad Sci USA. 2015;112:11917–11922. doi: 10.1073/pnas.1509851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olalde I, et al. A common genetic origin for early farmers from Mediterranean cardial and central European LBK cultures. Mol Biol Evol. 2015;32:3132–3142. doi: 10.1093/molbev/msv181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradley R. The Prehistory of Britain and Ireland. Cambridge Univ Press; Cambridge, UK: 2007. [Google Scholar]
- 15.Olalde I, et al. The beaker phenomenon and the genomic transformation of northwest Europe. Nature. 2018;555:190–196. doi: 10.1038/nature25738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser M. 2018. People of the Dolmens and Stone Cists: An Archaeogenetic Investigation of Megalithic Graves from the Neolithic Period on Gotland. PhD dissertation (Uppsala University, Uppsala, Sweden)
- 17.Mittnik A, et al. The genetic prehistory of the Baltic Sea region. Nat Commun. 2018;9:442. doi: 10.1038/s41467-018-02825-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joussaume R. Dolmens for the Dead: Megalith-Building Throughout the World. B.T. Batsford; London: 1988. [Google Scholar]
- 19.Whittle AW. Europe in the Neolithic : The Creation of New Worlds. Cambridge Univ Press; Cambridge, UK: 1996. [Google Scholar]
- 20.Sheridan JA. 2003. The chronology of Irish megalithic tombs. Stones and Bones: Formal Disposal of the Dead in Atlantic Europe During the Mesolithic-Neolithic Interface 6000–3000 BC. British Archaeological Reports International Series 1201, eds Burenhult G, Westergaard S (Archaeopress, Oxford, UK), pp 69–73.
- 21.Sheridan JA. 2010. The Neolithization of Britain and Ireland: The “Big Picture.” Landscapes in Transition, Levant Supplementary Series 8, eds Finlayson B, Warren GM (Oxbow, Oxford, UK), pp 89–105.
- 22.Sheridan JA. Interdigitating pasts: The Irish and Scottish Neolithics. In: Bickle P, Cummings V, Hofmann D, Pollard J, editors. The Neolithic of Europe. Papers in Honour of Alasdair Whittle. Oxbow; Oxford, UK: 2017. pp. 298–313. [Google Scholar]
- 23.Scarre C. The Megalithic Monuments of Britain and Ireland. Thames and Hudson; New York: 2007. [Google Scholar]
- 24.Collard M, Edinborough K, Shennan S, Thomas MG. Radiocarbon evidence indicates that migrants introduced farming to Britain. J Archaeol Sci. 2010;37:866–870. [Google Scholar]
- 25.Darvill T. Megalithic tombs, barrows, and enclosures in forth millennium BC Britain. Giants in the landscape: Monumentality and territories in the European Neolithic. In: Ard V, Pillot L, editors. Proceedings of the XVII UISPP World Congress (1-7 September, Burgos, Spain) Vol 3/Session A25d. Archaeopress; Oxford, UK: 2016. pp. 3–17. [Google Scholar]
- 26.McLaughlin TR, et al. The changing face of Neolithic and Bronze Age Ireland: A big data approach to the settlement and burial records. J World Prehist. 2016;29:117–153. [Google Scholar]
- 27.Schulz Paulsson B. Time and Stone: The Emergence and Development of Megaliths and Megalithic Societies in Europe. Archaeopress; Oxford, UK: 2017. [Google Scholar]
- 28.Cummings V, Midgley M, Scarre C. Chambered tombs and passage graves of Western and Northern Europe. In: Fowler C, Harding J, Hofmann D, editors. The Oxford Handbook of Neolithic Europe. Oxford Univ Press; Oxford, UK: 2015. pp. 813–838. [Google Scholar]
- 29.Midgley MS. The Megaliths of Northern Europe. Routledge; London: 2008. [Google Scholar]
- 30.Midgley M. Monuments and monumentality: The cosmological model of the world of megaliths. Doc Praehist. 2010;XXXVII:55–64. [Google Scholar]
- 31.Midgley MS. Megaliths in North-West Europe. In: Nilsson-Stut L, Tarlow S, editors. The Oxford Handbook of the Archaeology of Death and Burial. Oxford Univ Press; Oxford, UK: 2013. [Google Scholar]
- 32.Fleming A. Tombs for the living. Man (Lond) 1973;8:177. [Google Scholar]
- 33.Sherratt A. The genesis of megaliths: Monumentality, ethnicity and social complexity in Neolithic North-West Europe. World Archaeol. 1990;22:147–167. [Google Scholar]
- 34.Klassen L. Frühes Kupfer im Norden. Untersuchungen zu Chronologie, Herkunft und Bedeutung der Kupferfunde der Nordgruppe der Trichterbecherkultur. Vol 36 Jutland Archaeological Society; Højbjerg, Denmark: 2000. [Google Scholar]
- 35.Müller J. Megaliths and Funnel Beakers: Societies in Change 4100–2700 BC. Drieendertigste Kroon-Voordracht; Amsterdam: 2011. [Google Scholar]
- 36.Lee EJ, et al. Collective burials among agro-pastoral societies in later Neolithic Germany: Perspectives from ancient DNA. J Archaeol Sci. 2014;51:174–180. [Google Scholar]
- 37.Renfrew C. Before Civilisation: The Radiocarbon Revolution and Prehistoric Europe. Penguin; London: 1973. [Google Scholar]
- 38.Sjögren K-G. Kinship, labor, and land in Neolithic southwest Sweden: Social aspects of megalithic graves. J Anthropol Archaeol. 1986;5:229–265. [Google Scholar]
- 39.Sjögren K-G. 2003. Mångfalldige uhrminnes grafar: Megalitgravar och samhälle i Västsverige. GOTARC Series B, Gothenburg Archaeological Theses 27 (Göteborgs Universitet, Göteborg, Sweden)
- 40.Furholt M, Müller J. The earliest monuments in Europe: Architecture and social structures (5000–3000 cal BC) In: Furholt M, Lüth F, Müller J, editors. Early Monuments and Neolithic Societies from the Atlantic to the Baltic. Frühe Monumentalität und soziale Differenzierung; Bonn, Germany: 2011. pp. 15–32. [Google Scholar]
- 41.Renfrew C. Megaliths, territories and populations. In: de Laet SJ, editor. Acculturation and Continuity in Atlantic Europe. De Tempel; Bruges, Belgium: 1976. pp. 198–220. [Google Scholar]
- 42.Renfrew C. The Megalithic Monuments of Western Europe. Thames and Hudson; London: 1981. [Google Scholar]
- 43.Malmer M. 1962. Jungneolitische Studien, Acta Archaeologica Lundensia Series in 8o No 2 (Rudolf Habelt Verlag, Bonn, Germany)
- 44.Malmström H, et al. Ancient DNA reveals lack of continuity between Neolithic hunter-gatherers and contemporary Scandinavians. Curr Biol. 2009;19:1758–1762. doi: 10.1016/j.cub.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 45.Fraser M, et al. New insights on cultural dualism and population structure in the Middle Neolithic Funnel Beaker culture on the island of Gotland. J Archaeol Sci Reports. 2018;17:325–334. [Google Scholar]
- 46.Lazaridis I, et al. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536:419–424. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martiniano R, et al. The population genomics of archaeological transition in west Iberia: Investigation of ancient substructure using imputation and haplotype-based methods. PLoS Genet. 2017;13:e1006852. doi: 10.1371/journal.pgen.1006852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brace S, et al. 2018. Population replacement in early Neolithic Britain. BioRxiv:10.1101/267443. Preprint, posted February 18, 2018. [Google Scholar]
- 49.Rowley-Conwy P. Westward ho! Curr Anthropol. 2011;52:S431–S451. [Google Scholar]
- 50.Rivollat M, et al. When the waves of European neolithization met: First paleogenetic evidence from early farmers in the southern Paris basin. PLoS One. 2015;10:e0125521. doi: 10.1371/journal.pone.0125521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karmin M, et al. A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res. 2015;25:459–466. doi: 10.1101/gr.186684.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haak W, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lipson M, et al. Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature. 2017;551:368–372. doi: 10.1038/nature24476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goldberg A, Günther T, Rosenberg NA, Jakobsson M. Ancient X chromosomes reveal contrasting sex bias in Neolithic and Bronze Age Eurasian migrations. Proc Natl Acad Sci USA. 2017;114:2657–2662. doi: 10.1073/pnas.1616392114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monroy Kuhn JM, Jakobsson M, Günther T. Estimating genetic kin relationships in prehistoric populations. PLoS One. 2018;13:e0195491. doi: 10.1371/journal.pone.0195491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones ER, et al. The Neolithic transition in the Baltic was not driven by admixture with early European farmers. Curr Biol. 2017;27:576–582. doi: 10.1016/j.cub.2016.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Günther T, et al. Population genomics of Mesolithic Scandinavia: Investigating early postglacial migration routes and high-latitude adaptation. PLoS Biol. 2018;16:e2003703. doi: 10.1371/journal.pbio.2003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mathieson I, et al. The genomic history of southeastern Europe. Nature. 2018;555:197–203. doi: 10.1038/nature25778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng TC, Aw AJ, Feldman MW. Cultural hitchhiking and competition between patrilineal kin groups explain the post-Neolithic Y-chromosome bottleneck. Nat Commun. 2018;9:2077. doi: 10.1038/s41467-018-04375-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang DY, Eng B, Waye JS, Dudar JC, Saunders SR. Technical note: Improved DNA extraction from ancient bones using silica-based spin columns. Am J Phys Anthropol. 1998;105:539–543. doi: 10.1002/(SICI)1096-8644(199804)105:4<539::AID-AJPA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 61.Dabney J, et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci USA. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kircher M. Analysis of high-throughput ancient DNA sequencing data. In: Shapiro B, Hofreiter M, editors. Ancient DNA, Methods in Molecular Biology. Humana Press; Totowa, NJ: 2012. pp. 197–228. [DOI] [PubMed] [Google Scholar]
- 63.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu Q, et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr Biol. 2013;23:553–559. doi: 10.1016/j.cub.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Korneliussen TSTS, Albrechtsen A, Nielsen R. ANGSD: Analysis of next-generation sequencing data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jun G, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet. 2012;91:839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ginolhac A, Rasmussen M, Gilbert MTP, Willerslev E, Orlando L. mapDamage: testing for damage patterns in ancient DNA sequences. Bioinformatics. 2011;27:2153–2155. doi: 10.1093/bioinformatics/btr347. [DOI] [PubMed] [Google Scholar]
- 68.van Oven M. PhyloTree Build 17: Growing the human mitochondrial DNA tree. Forensic Sci International Genet Suppl Ser. 2015;5:e392–e394. [Google Scholar]
- 69.Weissensteiner H, et al. HaploGrep 2: Mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016;44:W58–W63. doi: 10.1093/nar/gkw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patterson N, et al. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang C, et al. Comparing spatial maps of human population-genetic variation using Procrustes analysis. Stat Appl Genet Mol Biol. 2010;9:13. doi: 10.2202/1544-6115.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Behr AA, Liu KZ, Liu-Fang G, Nakka P, Ramachandran S. pong: Fast analysis and visualization of latent clusters in population genetic data. Bioinformatics. 2016;32:2817–2823. doi: 10.1093/bioinformatics/btw327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing reads produced for this study have been deposited in the European Nucleotide Archive (accession no. PRJEB31045).