Significance
Bruton’s tyrosine kinase (Btk) activation on the cell membrane is critical for B cell proliferation and development, and Btk inhibition is a promising treatment for several hematologic cancers and autoimmune diseases. Here, we examine Btk activation using the results of long-timescale molecular dynamics simulations. In our simulations, Btk lipid-binding modules dimerized on the membrane in a single predominant conformation. We observed that the phospholipid PIP3—in addition to its expected role of recruiting Btk to the membrane—allosterically mediated dimer formation and stability by binding at two novel sites. Our results provide strong evidence that PIP3-mediated dimerization of Btk at the cell membrane is a critical step in Btk activation and suggest a potential approach to allosteric Btk inhibitor development.
Keywords: Bruton’s tyrosine kinase, dimerization, allosteric activation, PIP3, enhanced sampling
Abstract
Bruton’s tyrosine kinase (Btk) is critical for B cell proliferation and activation, and the development of Btk inhibitors is a vigorously pursued strategy for the treatment of various B cell malignancies. A detailed mechanistic understanding of Btk activation has, however, been lacking. Here, inspired by a previous suggestion that Btk activation might depend on dimerization of its lipid-binding PH–TH module on the cell membrane, we performed long-timescale molecular dynamics simulations of membrane-bound PH–TH modules and observed that they dimerized into a single predominant conformation. We found that the phospholipid PIP3 stabilized the dimer allosterically by binding at multiple sites, and that the effects of PH–TH mutations on dimer stability were consistent with their known effects on Btk activity. Taken together, our simulation results strongly suggest that PIP3-mediated dimerization of Btk at the cell membrane is a critical step in Btk activation.
Bruton’s tyrosine kinase (Btk), a Tec-family tyrosine kinase, is a peripheral membrane-binding protein present in all blood cells except for T cells and natural killer cells (1, 2). Btk participates in a number of receptor-mediated signaling pathways (2–6). In B cells, where Btk’s biological functions are best understood, Btk activates phospholipase C-γ2, which produces second messengers that are essential for B cell activation and proliferation. The up-regulation of Btk is associated with various B cell malignancies, including chronic lymphocytic leukemia and mantle cell lymphoma. Btk down-regulation causes X-linked agammaglobulinemia (XLA), a severe disease of primary immunodeficiency (7). Btk is also implicated in maintaining a number of autoimmune diseases, such as rheumatoid arthritis, systemic lupus erythematosus, and multiple sclerosis (8, 9). A structural characterization of Btk’s activation mechanism would significantly advance our understanding of Btk’s role in cell signaling and could facilitate the development of Btk-specific inhibitors (10), but such a characterization has thus far remained elusive.
Btk is autoinhibited in the cytoplasm, where it adopts a compact conformation similar to the inactive forms of the kinases c-Src and c-Abl (11–14), in which the SH2 and SH3 domains stabilize an inactive conformation of the kinase domain. In the case of Btk, its lipid-interaction module [the PH–TH module, composed of a pleckstrin homology (PH) domain and a Tec homology (TH) domain] acts in conjunction with the SH2 and SH3 domains to stabilize the inactive conformation of the Btk kinase domain (11, 15). It has been shown that Btk activation is primarily regulated by phosphorylation upon membrane recruitment (recruitment triggers transautophosphorylation or transphosphorylation by membrane-bound Src-family kinases) (16–22). Membrane recruitment occurs by way of interactions between the canonical lipid-binding site of the PH–TH module and PIP3 membrane lipids, but the structural mechanism by which membrane recruitment leads to Btk activation is unknown.
Saraste and Hyvönen (23), after identifying a crystallographic dimer of the PH–TH module (often referred to as the “Saraste dimer”), suggested that dimerization of the PH–TH modules of membrane-bound Btk molecules may promote the transautophosphorylation of their kinase domains, leading to activation. Dimers of the PH–TH module have never been directly detected in solution by biophysical methods, however, making their functional relevance uncertain. Membrane binding might be critical for PH–TH dimerization due to the increased local Btk concentration (24, 25), but it is difficult to obtain atomic structural information about the protein on the membrane by traditional experimental means. Molecular dynamics (MD) simulations have enabled the observation of spontaneous binding events between proteins (26–32), providing an alternative, computational route for obtaining atomic structural information about protein oligomerization on membranes.
In this study, we use long-timescale MD simulations to examine whether and, if so, how the PH–TH module of Btk dimerizes on a membrane. In our simulations, which started from crystal structures of individual PH–TH modules and used no other prior structural information, the PH–TH modules spontaneously dimerized on the membrane into a single predominant conformation closely resembling the Saraste dimer. We observed that PIP3 allosterically stabilized the dimer conformation, binding not only at the canonical PIP3-binding site but at the peripheral IP6-binding site, which has been shown to be important for Btk activation in solution (11). At higher PIP3 concentrations, an additional PIP3 molecule bound at a site between the two PH–TH modules, interacting with both modules simultaneously, and there is evidence that this further stabilized the interface.
Our simulations also provide an explanation for the effects of multiple mutations in the PH–TH module that are known to be associated with Btk dysfunction: In our simulations, mutations that lead to down-regulation of Btk and cause immunodeficiency diseases destabilized the PH–TH dimer, and a mutation that leads to the up-regulation of Btk and causes cell overproliferation stabilized the dimer interface. These observations provide compelling evidence that the Saraste dimer is important for the regulation of Btk. Taking this together with our simulation results showing that PIP3 can allosterically mediate dimer formation, we further propose that PIP3 plays a physiological role as an allosteric activator of Btk by stimulating dimerization of the PH–TH modules and thus promoting transautophosphorylation of the kinase domains.
Results
The PH–TH Module Bound Multiple PIP3 Lipids on the Membrane.
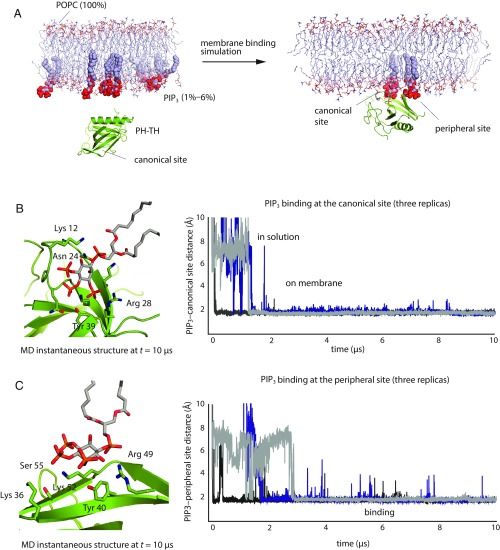
To establish the utility of our MD simulations for studying the behavior of the PH–TH module, we first showed that our simulations could reproduce essential experimentally observed interactions between the PH–TH module and soluble inositol phosphates (SI Appendix, Texts T1 and T2). We then studied the binding of the PH–TH module of Btk onto a membrane containing 94% POPC and 6% PIP3. We started the simulations by placing a PH–TH module in solution, about 10 Å away from the membrane surface, and positioned the module so that the canonical PIP3-binding site did not face the PIP3-containing leaflet of the membrane (Fig. 1A).
Fig. 1.
Spontaneous binding of the PH–TH module to the membrane. (A) Cartoon illustration of a membrane-binding simulation setup (see SI Appendix, Methods for details). (B, Right) Minimum distance between atoms in PIP3 lipids and in the canonical binding site for three independent membrane-binding simulations. (B, Left) A structure taken from one of these simulations (t = 10 μs) showing a PIP3 molecule bound at the canonical site. (C, Right) Minimum distance between atoms in PIP3 lipids and in the peripheral binding site for three independent membrane-binding simulations. (C, Left) A structure taken from one of these simulations (t = 10 μs) showing a PIP3 molecule bound at the peripheral site.
Within 2 μs of simulated time, the PH–TH module was recruited onto the membrane, with a PIP3 molecule bound to the canonical site (Fig. 1B). The binding pose of PIP3 at the canonical site resembles that of IP4 bound to the PH–TH module, as seen in crystal structures (33). Residues Asn-24, Tyr-39, Arg-28, and Lys-12 interacted with phosphate groups on the 2, 3, and 4 positions of the myo-inositol ring. In addition, Ser-21, Ser-14, and Lys-18 were also in close contact with the PIP3 molecule and formed transient interactions with PIP3 in the simulations. The bound PIP3 stayed at the canonical binding site until the end of simulations.
Once the PH–TH module was recruited onto the membrane through PIP3 binding to the canonical site, the peripheral IP6-binding site of the PH–TH module repeatedly bound a second PIP3 lipid (Fig. 1C). Multiple PIP3 binding and unbinding events were observed at the peripheral site in individual trajectories, and the overall occupancy rate of the peripheral site was ∼96%; in further simulations at a lower (1.5%) PIP3 concentration, the peripheral site occupancy was about 80%. The less stable binding of PIP3 in the peripheral site is to be expected, since the peripheral site is relatively flat, whereas the canonical site has a well-defined groove where PIP3 binds. PIP3 assumed multiple binding poses at the peripheral site, with residues Lys-52, Tyr-40, and Arg-49 most frequently coordinating PIP3 binding; these are also the key residues experimentally observed to coordinate IP6 binding (11) (Fig. 1C). Residues Lys-36 and Ser-55, which are outside the area known to bind IP6, also interacted with PIP3 at the peripheral site.
Although MD simulation studies have shown that several PH domains bind anionic lipids at noncanonical binding sites (34–37), the peripheral site identified in our study has not been previously reported as a lipid-binding site for Btk. In our simulations, we also observed PIP3 repeatedly occupying a third binding site, located on the other side of the IP4-binding loop (SI Appendix, Fig. S1). We have not studied PIP3 binding in this pocket in detail, however, due to its low occupancy rate (∼30%) in our simulations.
Our simulations do not rule out the possibility that other types of anionic lipids, such as PIP2 or PS, might also bind at these regions of the PH–TH module. In this work, we chose to exclusively study PIP3 due to the critical signaling role it is known to play in mediating B cell activation, and its known function as a regulator of Btk activity. The finding that the PH–TH module can simultaneously bind multiple PIP3 lipids on the membrane has, to our knowledge, not been noted in prior studies and has important implications for Btk activation on membranes, as we will show in the results below.
The PH–TH Modules Spontaneously Dimerized on the Membrane with a Single Predominant Interface.
PIP3-bound PH–TH modules diffused freely on membranes in our simulations, and we were thus able to study the encounter process of two membrane-bound PH–TH modules. Here, we show that PH–TH modules can indeed spontaneously dimerize on membranes, with a single dimer interface predominating.
We began by performing conventional MD simulations of two separate, non–membrane-bound PH–TH modules and a piece of membrane containing 6% PIP3 (Fig. 2A). In 20 independent 20-μs trajectories, the individual modules always bound with the membrane, and a variety of PH–TH dimers subsequently formed, in each case within the first few microseconds of simulation time (SI Appendix, Fig. S2). Some dimer interfaces, despite having small interface areas (∼500 Å2) and lacking hydrophobic packing interactions and hydrogen bonds, stayed bound through the end of the 20-μs simulations (SI Appendix, Fig. S2). The fact that such dimer interfaces did not dissociate in the simulations is not unexpected: Observing dissociation of protein–protein complexes is a well-known challenge for conventional MD simulations, since experimentally observed dissociation events often occur on a longer timescale than is accessible to MD simulations (32).
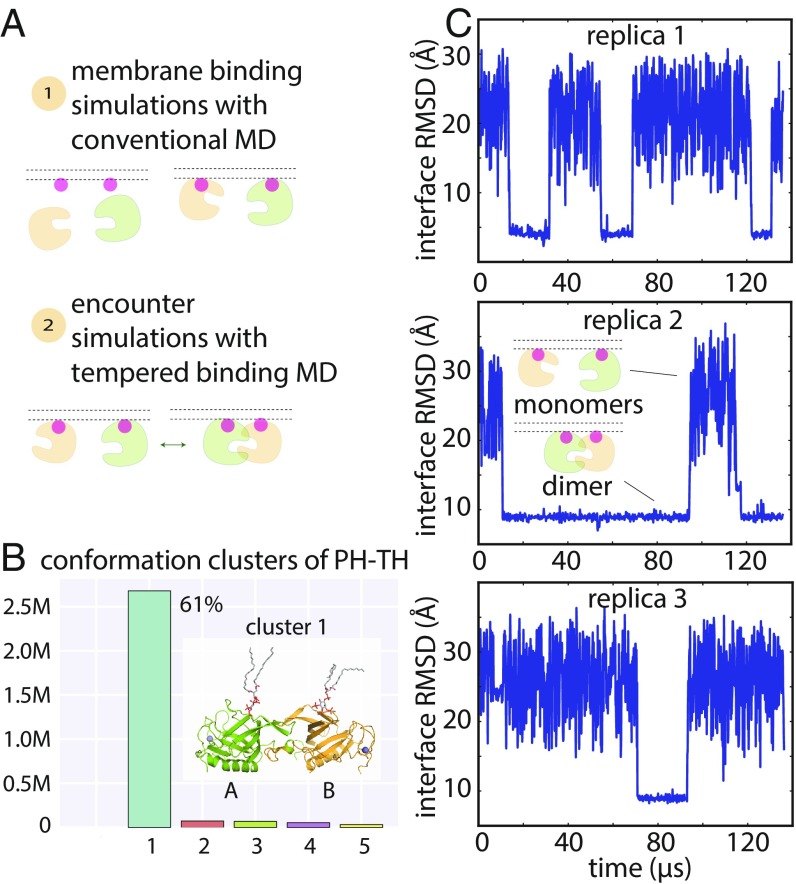
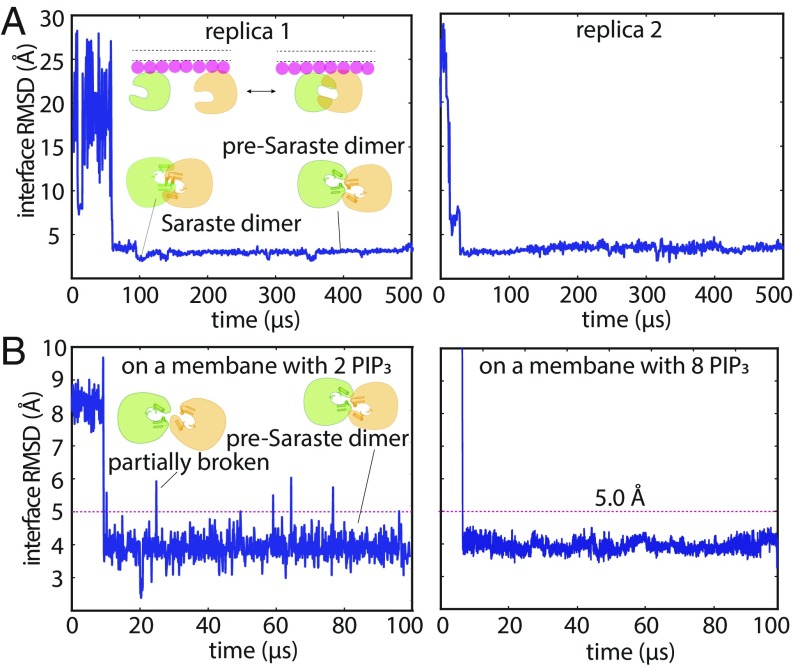
Fig. 2.
Dimerization of the PH–TH module on the membrane. (A) Cartoon illustration of an encounter simulation setup (see SI Appendix, Methods for details). (B) Conformation clusters of PH–TH dimers formed in the simulations (see SI Appendix, Methods for details). A representative structure of the most populated dimer conformation is shown (see Inset). (The PIP3 lipids bound in the canonical binding sites are also shown.) (C) Reversible dimerization of two membrane-bound PH–TH modules on a membrane that contains 1.5% PIP3 and 98.5% POPC in tempered binding simulations. The rmsd at the Saraste interface is calculated for all interface residues (residues 9, 11, 42, 44, 92, and 95) of the two modules with respect to the Saraste dimer interface conformation seen in a crystal structure of the PH–TH module (PDB ID code 1BTK). (This same interface rmsd calculation is used for the rmsd plots in all other figures, unless stated otherwise.)
To sample PH–TH module conformations more extensively than would be possible using conventional MD simulations of reasonably achievable length, we employed an enhanced sampling method called “tempered binding.” (32) This technique intermittently weakens the interactions between the two proteins (and thus all potential PH–TH dimer interfaces), thus increasing the frequency of dimer dissociation and accelerating the exploration of different protein–protein interfaces. The method nonetheless rigorously preserves the property of Boltzmann sampling at each interaction strength, with unscaled portions of the trajectory sampled from the same distribution as a conventional MD simulation with an unmodified Hamiltonian (see SI Appendix, Methods for details).
We performed 24 tempered binding simulations, each based on three replicas, with a total of ∼6 ms of simulated time for all simulations combined. All tempered binding simulations started from the same configuration—in which two PH–TH modules were bound to the membrane but were not in contact with each other—but varied in membrane PIP3 concentration, tempering strength, and protein backbone-correction strength.
In these tempered binding simulations, the two PH–TH modules diffused freely on the membrane, and multiple association/dissociation events between the two modules were observed. We then analyzed all dimer conformations assumed by the PH–TH modules in these simulations: The two modules were in contact in 4 million of the 6 million total frames extracted from the 24 simulations, and we began by clustering the instantaneous structures in these 4 million frames according to their structural similarity (Fig. 2B and SI Appendix, Fig. S3). The largest cluster of these PH–TH dimer structures contained 61% of total dimer structures, while the next-largest cluster contained less than 1%; given the small size of all but the largest, all further structural analysis was based only on the most populated cluster (Fig. 2B). Notably, clustering only those portions of the trajectory in which the protein–protein interactions were untempered led to the same predominant cluster, supporting the relevance of this dimer cluster under untempered conditions.
Dimers in the predominant cluster formed repeatedly in all 24 simulations (across all tempering parameters and PIP3 concentrations). The dissociation time for these dimers varied from simulation to simulation, ranging from at least 15 μs of simulation time to up to 500 μs, which was the length of the longest simulations. We also observed multiple reversible dimerization events for these dimers in individual trajectories under certain tempering conditions (Fig. 2C). Dimer structures in other clusters, on the other hand, were short-lived, and most of them dissociated within a few microseconds of simulation time.
The Predominant PH–TH Dimer Interface Resembles the Saraste Dimer Interface.
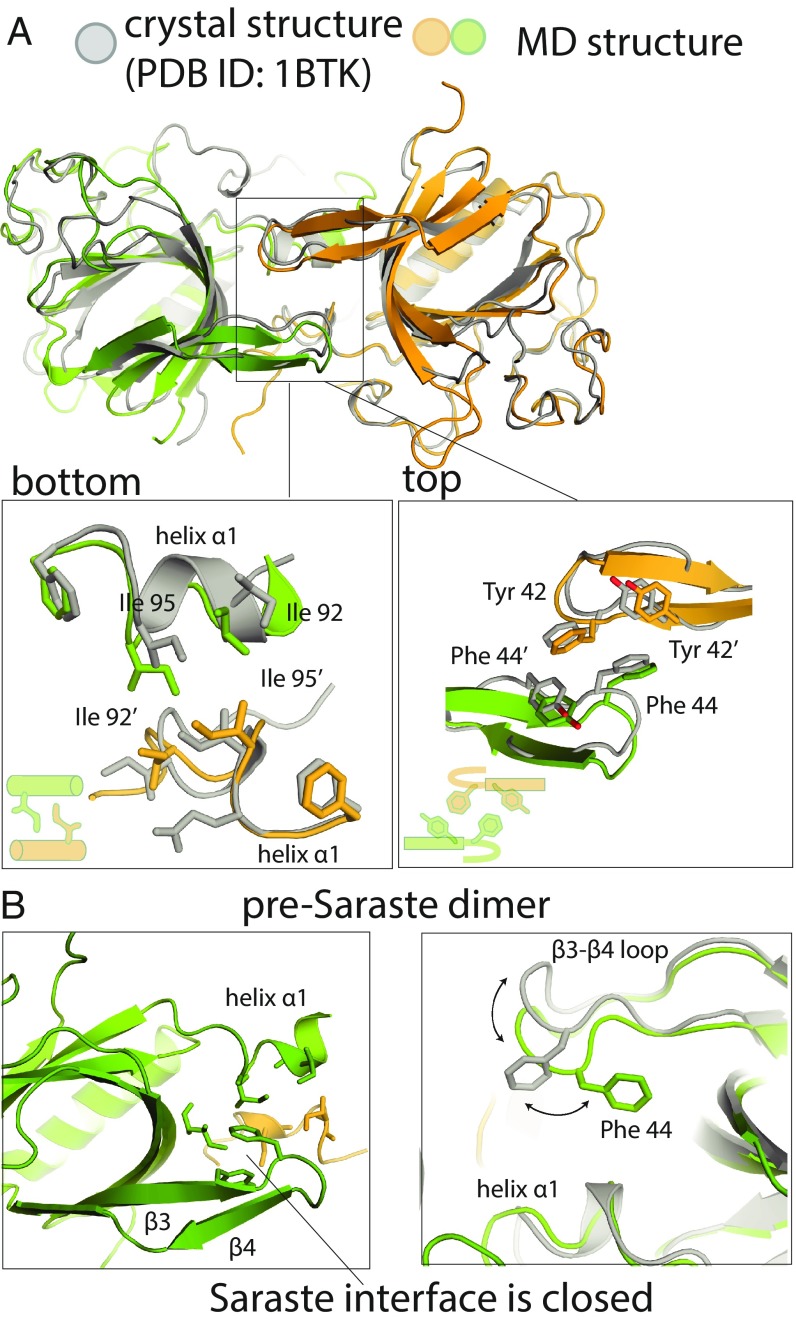
The most populated PH–TH dimer interface in our simulations resembles the Saraste dimer observed in crystal structures (Fig. 2B): The root-mean-square deviations (rmsds) of the PH–TH dimer interfaces in the largest cluster range from 2.5 to 4.5 Å compared with the Saraste dimer. The Saraste-like dimer interfaces that formed in our simulations had two distinct conformations (Fig. 3), with the primary structural difference between them occurring at the β3–β4 hairpin (Fig. 3B). In the conformation closest to the Saraste crystal structure, with an rmsd at the dimer interface of only ∼2.5 Å (Fig. 3A), the interface is tightly packed. In the other conformation, the dimer interface is loosely packed, and the interface regions of the individual PH–TH modules are partially closed (Fig. 3B): The Phe-44 of each module adopts an alternative rotamer conformation (to that seen in crystal structures) and remains packed against its own module’s Ile-92 and Ile-95 residues, as is also the case in simulations of a single PH–TH module. The backbone of the β3–β4 hairpin (residues 42–50) moves ∼3.2 Å toward the center of the interface. The rmsd of the second conformation at the dimer interface is ∼4.0 Å compared with the Saraste dimer crystal structure.
Fig. 3.
Structural analysis of Saraste dimers formed on membranes. (A) Structural comparison of Saraste dimers formed in the encounter simulations and in the Saraste dimer crystal structure (PDB ID code 1BTK). (B) An instantaneous structure of the pre-Saraste interface formed in the encounter simulations.
This loosely packed conformation of the Saraste interface has not been observed in crystal structures of the PH–TH dimer. Nevertheless, we find this conformation very informative: It provides evidence for a structural mechanism by which the tightly packed Saraste dimer interface can form while the interface regions of the individual PH–TH modules remain partially closed (SI Appendix, Text T1). In our simulations of an individual PH–TH module, residues at the Saraste interface region rarely adopted the open conformation, which is the conformation compatible with directly forming the tightly packed Saraste interface with another module. We thus did not expect the direct formation of the Saraste dimer interface to occur frequently in our simulations, since it would require the spontaneous opening of the interface regions of both PH–TH modules simultaneously during the encounter process. Indeed, none of the tightly packed Saraste dimer interfaces we observed in our tempered binding simulations formed through the direct encounter of the two modules with open Saraste interface regions: All Saraste dimer interfaces formed by way of conformational transitions through the loosely packed interface. We thus call the loosely packed interface the “pre-Saraste” interface. Furthermore, in one of our conventional MD simulations of two initially separated PH–TH modules on a membrane, we also observed the PH–TH modules spontaneously form the pre-Saraste dimer, strongly suggesting that the pre-Saraste dimer is an obligatory intermediate state between the unbound modules and the tightly packed Saraste dimer.
The Stability of the Saraste Interface Was Sensitive to Mutations on the PH–TH Module.
The observation in our simulations that the PH–TH modules spontaneously dimerized with a single predominant interface on the membrane, where Btk performs its biological functions, suggests that this Saraste dimer interface may be biologically relevant. Additionally, in solution the transient dimerization of PH–TH modules at the Saraste interface in the presence of IP6 can activate Btk molecules by promoting transautophosphorylation of their kinase domains, and mutations that destabilize the Saraste interface slow Btk activation in solution (11). Together, these findings suggest that the stability of the Saraste dimer may regulate activation of membrane-bound Btk. To test this hypothesis, we used MD simulations to study two mutations on the PH–TH module that are known to affect Btk activity, but for which a structural explanation is lacking. In our simulations, we examined whether and, if so, how these mutations influence the stability of the Saraste dimer on a membrane.
Initially, we explored using tempered binding simulations to study the thermodynamic equilibrium between a Saraste dimer and the unbound modules, but the results were inconclusive due to the small number of association and dissociation events that could be observed on our simulation timescale. Instead, we proceeded by qualitatively analyzing the potential structural consequences for Saraste dimer stability when different mutations were introduced. In this set of simulations, we used the tightly packed conformation of the Saraste dimer on a membrane as a starting structure and performed tempered binding simulations until the dimer dissociated (Fig. 4A).
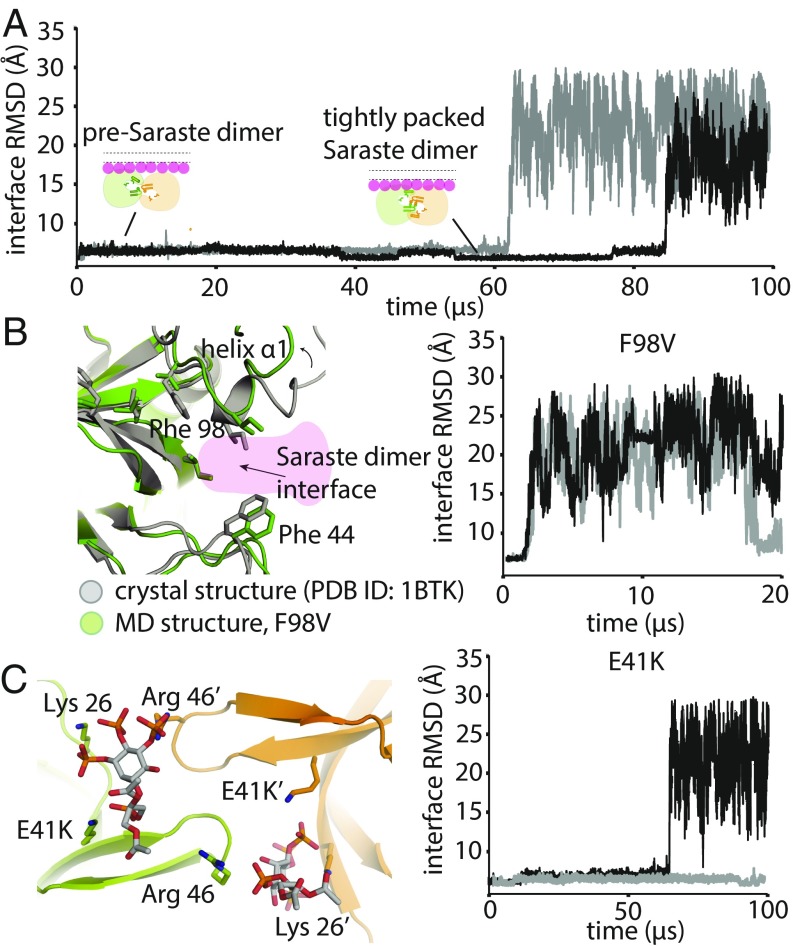
Fig. 4.
Stability analysis of Saraste dimer variants on a membrane. (A) Representative tempered binding simulation trajectories showing dissociation of the tightly packed Saraste conformation in two individual trajectories. The tempered binding simulations started from a Saraste dimer on a membrane containing 6% PIP3 and 94% POPC. (B) Stability analysis of the F98V mutant, starting from the Saraste dimer conformation, on a membrane. An instantaneous structure from a simulation (t = 20 μs) shows the local conformational rearrangement that occurred at the Saraste interface before the dimer dissociated. Representative tempered binding simulation trajectories show the dissociation of the F98V dimer in two individual trajectories. (C) Stability analysis of the E41K mutant, starting from the Saraste dimer conformation, on a membrane. An instantaneous structure from a simulation (t = 100 μs) shows PIP3 bound to residue Lys-41 at the bridging site before the dimer dissociated. Representative tempered binding simulation trajectories show the dissociation of the E41K dimer in two individual trajectories.
We first studied the loss-of-function mutant F98V, which is located in the PH domain and has been identified in patients with the immunodeficiency disease XLA (38). The molecular basis for the loss of function produced by F98V is not clear: Phe-98 is involved neither in the hydrophobic-core packing interactions of the PH–TH module nor in directly mediating PIP3-binding interactions, suggesting that the loss-of-function effect of F98V is unlikely to be related to misfolding of the individual PH–TH module or to interference with its membrane recruitment. One effect we did observe in our simulations, however, was that substituting Phe with the less bulky Val loosened the packing between residues Leu-11 and Ile-9 (Fig. 4B). Ile-9 is part of the Saraste interface, suggesting that the loosened packing at the end of the α1 helix, allosterically produced by F98V, could affect the stability of the Saraste interface. Consistent with this notion, in our simulations, we observed that the F98V dimer was not stable in the tightly packed Saraste interface conformation, quickly relaxed into the pre-Saraste conformation, and then dissociated into separate PH–TH modules (Fig. 4B and SI Appendix, Fig. S4). In addition to F98V, we observed that the Saraste interface was less stable in another mutant known to cause XLA, F25S (SI Appendix, Fig. S4).
We next studied the effects of the mutation E41K, a gain-of-function mutation in the PH–TH module of Btk (39). Glu-41 is located on the β3–β4 hairpin, near the Saraste interface, and its mutation to lysine constitutively activates Btk and causes cell overproliferation (39). Previous studies have shown that the PIP2-mediated membrane localization of the E41K mutant is more significant than that of the wild-type protein in resting cells (which have no PIP3 lipids), pointing to an important role for PIP2 binding in Btk activation in the absence of stimulation (40–43). Notably, the E41K mutant is also found to generate markedly enhanced BCR-induced Ca2+ signal in hematopoietic cells under stimulation conditions, leading to PIP3 production (44, 45). This signaling enhancement effect of E41K in activated cells is likely not solely the result of augmented PIP2-mediated membrane localization, however, since both the mutant and the wild-type protein are known to be recruited to the membranes through the well-established PIP3 binding mechanism. The structural basis of the stronger membrane localization of the E41K mutant in activated cells, and the interactions between E41K dimers and membranes after the mutant is recruited onto the membrane, are not well understood. In our simulations, we observed Lys-41 repeatedly bind a third PIP3 lipid at the top of the β3–β4 hairpin, confirming the notion, based on previous crystallographic studies, that the E41K mutant can bind an additional PIP3 molecule (33). Although the stability of the E41K dimer was qualitatively comparable to that of the wild-type dimer in our simulations, the PIP3 that bound at the top of the β3–β4 hairpin interacted simultaneously with both modules in the Saraste dimer (leading us to term this binding location the “bridging site”), revealing a structural mechanism that could further strengthen Saraste-interface stability (Fig. 4C). We thus speculate that the activating effect of E41K could be the result of additional PIP3 molecules stabilizing the Saraste interface once the dimer has formed on the membrane, which could further promote the transautophosphorylation of Btk kinase domains.
Finally, we note that, as expected, Saraste dimers with mutations at the dimer interface (I9R/L29R, Y42R/F44R) either barely stayed in the tightly packed Saraste interface conformation or dissociated immediately in our simulations (SI Appendix, Fig. S4). These mutants have been previously shown to impair Btk activation by IP6 in solution (11), and our simulation results strongly suggest that they are also likely to impede Btk activation on membranes.
The qualitative correlation between the stability of the mutated Saraste dimers and the functional consequences of these mutations suggests that the Saraste interface plays an important role in regulating Btk activation.
The Stability of the Saraste Dimer Interface Was also Sensitive to the Amount of PIP3 in the Membrane.
In B cells, down-regulation of Btk activity causes immunodeficiency diseases, and up-regulation of Btk activity is correlated with autoimmune diseases and various blood cancers, suggesting that the activation of Btk must be tightly controlled to maintain normal B cell functions. The observation that Btk function is very sensitive to the stability of the Saraste dimer interface suggests that the stability of the Saraste dimer itself might be tightly controlled in cells. We now describe a set of simulation observations that collectively suggest that PIP3, the anchor of Btk on membranes, is a potential allosteric regulator for Btk that may control the stability of the Saraste interface.
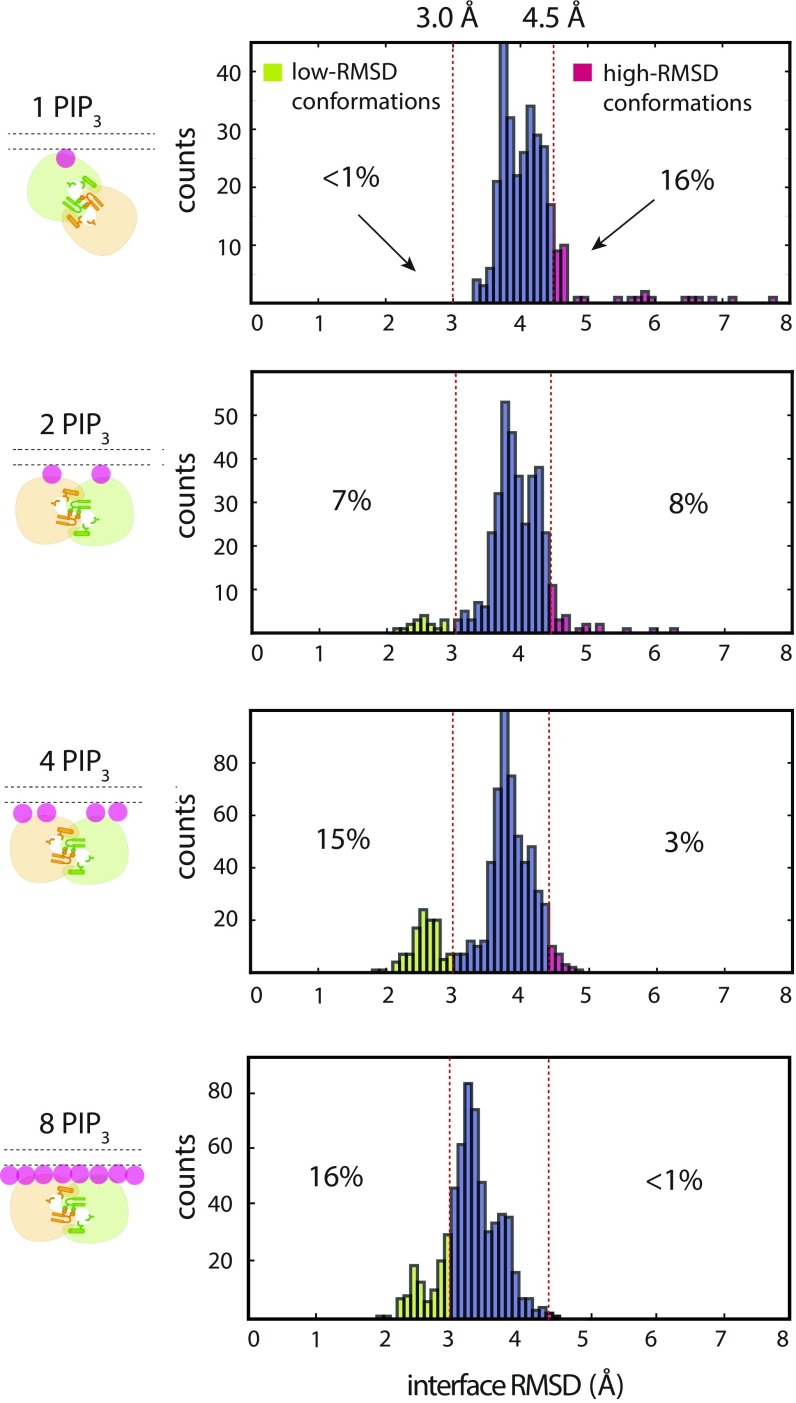
The first insight came from analyzing the tempered binding simulations discussed above, in which we found that Saraste dimers that formed on membranes with 6% PIP3 lipid content did not dissociate as frequently as those that formed on membranes with 1.5% PIP3 (as seen in Figs. 5A and 2C, respectively). Although we were unable to quantitatively compare the dissociation constants of the Saraste dimer at 1.5% and 6.0% PIP3 membrane lipid concentrations due to the small number of association and dissociation events, we found that fluctuations at the pre-Saraste interface, which preceded dissociation in our simulations, were more pronounced in simulations with 1.5% membrane PIP3 content than in those with 6% PIP3 (Fig. 5B). Dimer conformations with rmsds greater than 4.5 Å from the Saraste dimer crystal structure at the interface were repeatedly visited in the 1.5% PIP3 condition (Fig. 5B). These conformations resembled the partially broken Saraste dimer conformations seen in our solution-based conventional simulations (SI Appendix, Text T3 and Fig. S5). The large, relatively frequent conformational fluctuations at the dimer interface that repeatedly visit a structure on the dissociation pathway suggest that the dimers were weaker in the 1.5% PIP3 condition. In simulations with 6% membrane PIP3 content, the rmsd from the crystal structure at the Saraste interface consistently stayed below 4.5 Å. Overall, our observations provide strong evidence that the Saraste dimer was more stable in the 6.0% PIP3 lipid condition than in the 1.5% PIP3 lipid condition.
Fig. 5.
Fluctuations at the pre-Saraste dimer interface. (A) Representative tempered binding simulation trajectories of two PH–TH modules on a membrane containing eight PIP3 lipids. (B) Fluctuations at the pre-Saraste dimer interface that formed on membranes with two and eight PIP3 lipids (1.5% and 6% membrane PIP3 content, respectively).
PIP3 Binding at the Canonical and Peripheral Sites of the PH–TH Modules Reduced Fluctuations at the Saraste Dimer Interface.
Some polyphosphoinositides, such as PIP2 and PIP3, are known to spontaneously form clusters under certain conditions (46). One possible explanation for the apparently rare dissociation of the Saraste dimer in the 6% PIP3 condition is that PIP3 clusters spatially constrained the PH–TH dimer on the membrane, leading to infrequent dimer dissociation. We examined the localization pattern of PIP3 in our simulations to investigate this possibility, but found that no stable PIP3 clusters formed in any of our simulations. Indeed, the PIP3 molecules were quite isolated from each other, as a result of binding to distinct sites on the PH–TH module. We found that this binding of PIP3 at both the canonical site and the peripheral site of PH–TH modules had profound effects on the stability of the Saraste interface, suggesting that PIP3 plays a role in regulating the PH–TH dimer allosterically.
To study more rigorously whether both the canonical and peripheral sites are involved in the stabilizing effect of PIP3 binding described in the previous section, we performed another set of tempered binding simulations, varying only the number of PIP3 lipids in the membrane (from one to eight), and analyzed the fluctuations at the dimer interface after the formation of the pre-Saraste interface. We first observed that the Saraste dimer interface was not stable in solution, and that PIP3 binding significantly reduced fluctuations at the Saraste interface (SI Appendix, Fig. S5A). PIP3’s stabilizing effect on the Saraste interface became evident when comparing the interface conformations when one or both of the two canonical sites in the Saraste dimer had bound PIP3 ligands (Fig. 6 and SI Appendix, Fig. S5A): In simulations with only one PIP3 molecule in the membrane, one PH–TH module in the Saraste dimer was attached to the membrane at the canonical site, while the other module remained in solution. We observed that conformations with large rmsds (4.5 Å or more) from the Saraste crystal structure at the dimer interface appeared frequently in simulations with only one PIP3 molecule (Fig. 6). When two PIP3 lipids were present, occupying both canonical binding sites in the Saraste dimer, the population of conformations with large rmsds at the Saraste dimer interface shrank from 16 to 8%, and the population of conformations with small rmsds (3.0 Å and below) at the Saraste dimer interface increased to 7% (Fig. 6).
Fig. 6.
Sensitivity of fluctuations at the pre-Saraste dimer interface to membrane PIP3 concentration. Fluctuations at the pre-Saraste dimer interface are shown, measured under various membrane PIP3 concentrations. The histograms of the rmsd of the interface residues are calculated from 100-μs simulation trajectories initiated from Saraste dimers that originally formed in tempered binding simulations; only data from the portion of the trajectories where the interactions were unscaled are used.
We then observed that with four PIP3 lipids in the membrane, the stability of the Saraste dimer improved further (Fig. 6). In this set of simulations, PIP3 lipids typically occupied both the canonical and peripheral sites in the Saraste dimer: The occupancy rate at each peripheral binding site was ∼80%. In these simulations, the population of conformations with large rmsds at the dimer interface fell further, to 3%. The population of conformations with small rmsds at the Saraste dimer interface increased from 7 to 15%, consistent with the notion that binding of PIP3 at the peripheral site further stabilizes Saraste interface.
In the presence of membranes with eight PIP3 molecules (6% membrane PIP3 content), the canonical and peripheral sites of the Saraste dimer were almost continuously occupied in our simulations. We observed a further reduction in the population of conformations with large rmsds, from 3 to <1%. In addition, we found that the peak rmsd distribution was shifted from 3.7 Å to 3.2 Å, suggesting that the PIP3 occupying the peripheral site of each PH–TH module stabilizes the Saraste interface.
Further evidence for the importance of the PH–TH module’s peripheral binding site in stabilizing the Saraste interface is seen in simulations we performed to reveal the thermodynamic equilibrium between the tightly packed Saraste conformation and the more loosely packed pre-Saraste conformation (SI Appendix, Fig. S6A; see SI Appendix, Methods for details). In simulations with 6% PIP3 content, the PH–TH dimer reversibly converted between the pre-Saraste conformation (45%) and the tightly packed Saraste conformation (55%) before dissociating (SI Appendix, Fig. S6B). The free energy difference of the transition between the two Saraste conformations was 0.05 ± 0.2 kcal⋅mol−1. In the presence of a membrane with 1.5% PIP3 content, we found that the fraction of the tightly packed Saraste dimer fell to 40%, and the free energy difference between the two Saraste conformations decreased to −0.4 ± 0.15 kcal⋅mol−1, consistent with the notion that the tightly packed Saraste dimer conformation is less stable in the presence of a membrane with 1.5% PIP3 content.
PIP3 Binding at the Canonical Site and the Peripheral Site Rigidified the Saraste Dimer Interface in an Individual PH–TH Module.
Our simulations point to important roles for the canonical site and the peripheral site in controlling Saraste dimer formation, but the precise mechanisms by which PIP3 binding strengthens Saraste dimer stability are not clear. After the binding of PIP3 at the two sites, we observed no obvious structural change in the PH–TH module that might lead to a more stable dimer conformation. We thus investigated the possibility that the binding of two PIP3 molecules could restrict the range of conformations that the Saraste interface region can adopt.
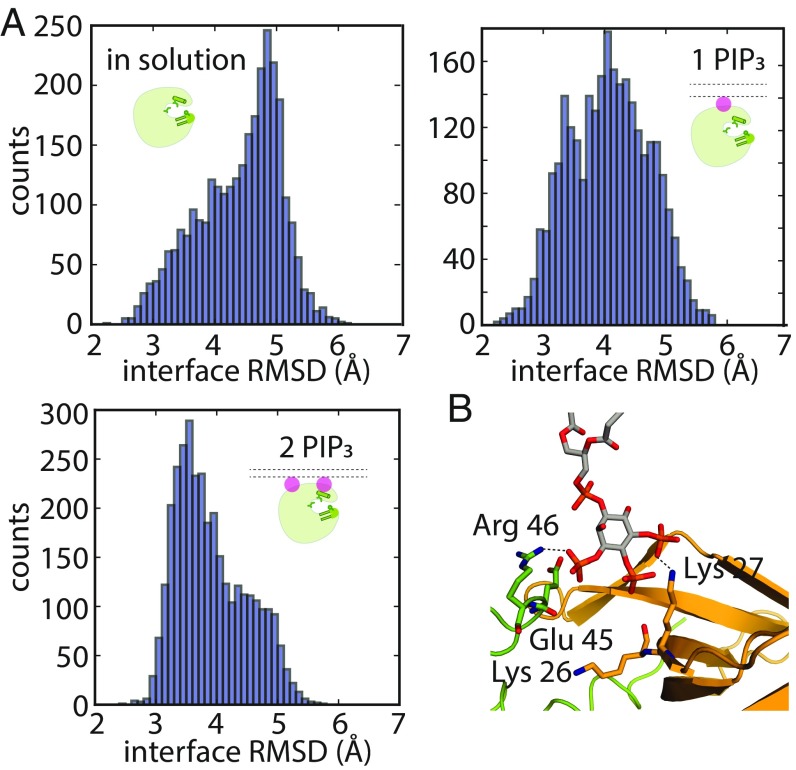
To investigate this possibility, we studied the conformational dynamics of an individual PH–TH module under various conditions: In solution-based simulations, the rmsd from the Saraste crystal structure at the dimer interface ranged from 2.5 to 6.5 Å, with a peak distribution at ∼4.8 Å (Fig. 7A). When the PH–TH module was recruited onto a membrane with only one PIP3 lipid occupying the canonical binding site, the fluctuations at the Saraste dimer interface decreased, with the distribution peak shifting from 4.8 to 4.2 Å rmsd (Fig. 7A). In the presence of two PIP3 lipids, the peripheral site was also occupied, and the peak distribution decreased further, to 3.5 Å (Fig. 7A). These observations indicate that the conformational dynamics of the PH–TH module are more restrained when both the canonical site and the peripheral site are occupied by PIP3.
Fig. 7.
Speculation on the mechanisms by which binding multiple PIP3 molecules stabilizes the Saraste dimer on the membrane. (A) Fluctuations at the Saraste dimer interface region in an individual PH–TH module in conventional MD simulations in solution and on the membrane. (B) An instantaneous structure from a tempered binding simulation showing that a PIP3 lipid can simultaneously interact with both modules in the Saraste dimer at the bridging site, which formed near the dimer interface.
At High Concentrations, PIP3 Bound at a Bridging Site, Where It Interacted with both PH–TH Modules in a Saraste Dimer.
In the presence of membranes with 6% PIP3 content, the canonical and peripheral sites of the Saraste dimer were mostly occupied in our simulations. Notably, at this concentration, we found that the area between the tip region of the β3–β4 hairpin of one PH–TH module and the edge of the canonical binding site of the other module also bound a PIP3 molecule (Fig. 7B). In an earlier section, we suggested that PIP3 binding at this bridging site might explain the activating effect of the E41K mutant; in the presence of a membrane with 6% PIP3 content, the bridging site was transiently occupied by a PIP3 lipid even in the absence of this mutation. In one representative structure extracted from our simulations, the phosphates at positions 1 and 4 of the inositol ring of PIP3 interacted simultaneously with the Arg-46 residue of one module and Lys-27 of the other, which we label modules A and B, respectively (Fig. 7B). The hydroxyl group at position 2 also occasionally formed a hydrogen bond with Glu-45 of module A. The average occupancy rate of the bridging site was low, at ∼40% when the membrane had 6% PIP3 content, suggesting that PIP3 binding at this site is of low affinity. We speculate that the bridging site might contribute to Saraste dimer stability only in the presence of a large number of PIP3 lipids.
The two mechanisms we have described—stabilization by PIP3 of the individual PH–TH modules in the Saraste dimer-compatible conformation and, at high concentrations, the interaction of PIP3 with both modules simultaneously to stabilize the dimer once it has formed—together support the notion that PIP3 can allosterically stabilize the Saraste dimer. Our simulations also showed that the phosphate group at the position 3′ of the inositol ring contributes to the binding of PIP3 at both the bridging site and the peripheral site (Figs. 1C and 4C), supporting the idea that PIP3 is a stronger binder at these alternative sites than its precursor molecule PIP2. (Consistent with this structural observation, the binding of PIP2 in the alternative sites is less stable than that of PIP3 in our simulations; SI Appendix, Text T6.) Because the concentration of PIP2 in the inner leaflet of the plasma membrane is generally thought to be much higher than that of PIP3, however, it is possible that PIP2 may also bind at these alternative sites.
The B cell plasma membrane is composed of many lipids—such as cholesterols, PE, PS, and sphingolipids—in addition to the PIP3 and PC lipids studied in this work. These differences in membrane composition mean that our results concerning binding at different PIP3 concentrations should be viewed in a qualitative manner. Little is known about the actual local concentration of PIP3 near B cell receptors, where Btk primarily functions. Although the overall concentration of PIP3 is found to be <1% in B cells, it is also known that PIP3 can form mobile clusters or be found in small lipid domains. PIP2 has been found in protein microdomains with a local concentration as high as 82% (47). PIP3 has also been found in protein microdomains, with a local concentration up to 16% (48). In our simulations, the concentration of PIP3 was 1–6%, falling within the range of potential PIP3 concentration in cells. It is thus reasonable to expect that both mechanisms observed in our simulations may occur in nature.
Discussion
A Structural Model of Btk Activation on Membranes.
Based on our atomic-level MD simulations, we have presented two important findings, which together provide a potential mechanism for Btk activation (SI Appendix, Fig. S7). First, we observed that Btk PH–TH modules spontaneously dimerized on membranes into a single predominant conformation, one which resembles the Saraste dimer. This finding provides a structural explanation for the effects of mutations in the PH–TH module known to cause severe Btk dysfunction, as these mutations affected the stability of the dimer.
The other major finding we have presented in this work is that PIP3 can allosterically stabilize the Saraste dimer interface, and may thus play a regulatory role in Btk activation. In prior models of Btk regulation, PIP3 has been considered a membrane anchor for the kinase, but no further regulatory role for PIP3 has previously been proposed. Our simulations provide evidence that PIP3 controls Saraste dimer stability, which may in turn have a profound impact on Btk activation: In order for Btk to be activated by transautophosphorylation, the two kinases need to come close together and form a transient enzyme–substrate complex. At a single-molecule level, the lifetime of the enzyme–substrate complex ought to be longer than the time required for transferring the phosphate from ATP to the substrate tyrosine. Increasing the lifetime of the Saraste dimer would thus make the transautophosphorylation reaction more efficient. [Although for this work we have not studied in detail whether PIP3 binding affects the on rate of PH–TH dimer formation, which would also promote Btk activation, there is some evidence from our simulations that tends to support this proposition (SI Appendix, Texts T4 and T5).]
PH domains, which act as membrane-binding modules and protein–protein interaction modules, are frequently found in the human genome (49, 50). Dimerization of membrane-bound PH domains has received little attention in the past, but PH domain dimerization may play an important role in enzyme regulation. Recently, the PH domain of phosphoinositide-dependent kinase-1 (PDK1), a serine/threonine kinase that is important for cell differentiation and proliferation pathways, was shown to dimerize on a supported bilayer system (51). The structural basis for this dimerization is not clear, but it has been speculated that the PDK1 dimer represents an inhibitory state of the kinase that is not capable of substrate binding (52). This notion, together with the activating role of the PH–TH module of Btk proposed in this paper, leads us to speculate that PH domain dimerization might be a common regulatory mechanism of PH domain-containing enzymes that function on membranes.
Some receptor tyrosine kinases, such as epidermal growth factor receptor (EGFR), self-associate into high-order oligomers as part of the activation process (53, 54). We have not studied whether dimers of the PH–TH module might form high-order oligomers on membranes, but we note that such high-order oligomers, if they exist, could further strengthen the stability of the Saraste dimer interface and contribute to the activation of Btk.
The Saraste Dimer Interface and the Peripheral Binding Site in Btk Are Less Conserved in Early Stages of B Cell Evolution.
Neither the Saraste dimer interface nor the peripheral binding site in Btk is conserved in the PH–TH modules of Tec and Itk, the other two members of the Tec family of kinases. This raises the question of when the PIP3-dependent dimerization of the PH–TH module of Btk originated.
We examined the conservation of those residues that constitute the Saraste dimer interface and the peripheral binding site, comparing the sequence of human Btk with that of other mammals (105 species), birds (47 species), and fish (41 species). We found that these residues are for the most part well conserved in mammals and birds, and much less extensively conserved in fish (SI Appendix, Fig. S8). In contrast, PI3K (the enzyme responsible for generating PIP3) and residues in the canonical binding site of Btk are conserved in all eukaryotes. This analysis suggests that membrane binding is a more ancient function than allosteric binding for PIP3 in its interactions with the PH domain. The origins of the adaptive immune system, in particular B cells, are thought to be linked to the emergence of jawed fish, such as Chondrichthyes and Teleost fish (55, 56). B cells in fish have antigen-specific IgM responses similar to those in mammals, but the time required for fish B cells to generate a significant antigen-specific response is generally much longer than that required in mammals (56), suggesting that the signal transduction pathways that control the antigen-specific response, in which Btk plays an important regulatory role, are much less sensitive in fish than in mammals. One possibility is that the utilization of a PIP3-dependent dimerization mechanism for Btk activation in mammals is a consequence of natural selection for high sensitivity of the B cell response.
Alternative Strategies for Developing Btk Inhibitors That Are Selective and Can Inhibit the Ibrutinib-Resistant C481S Mutant.
A number of ATP-competitive Btk inhibitors have been identified in recent years (57, 58). The most clinically tested of these is ibrutinib (59), which has been approved by the Food and Drug Administration for the treatment of mantle cell lymphoma (in 2013), chronic lymphocytic leukemia (in 2014), and a particular form of non-Hodgkin lymphoma (in 2015). Despite ibrutinib’s high efficacy in the treatment of multiple B cell cancers, ibrutinib-resistant mutations—in particular, C481S—have emerged in a substantial fraction of all chronic lymphocytic leukemia patients treated with ibrutinib (60). In addition, ibrutinib has off-target effects on EGFR, ITK, and Tec-family kinases, which may result in various adverse effects in some patients.
The emerging resistance to and off-target side effects of ibrutinib have led to the active development of second-generation and more specific Btk inhibitors. Our finding that PIP3-dependent dimerization may be a feature of Btk activation suggests a potential alternative approach to the development of such inhibitors in which small molecules disrupt the dimerization of PH–TH modules. Such molecules could potentially bind either at the Saraste dimer interface or in other regions on the PH–TH module that could allosterically impair Saraste dimer formation. Inhibitors targeting the PH–TH module should act on both wild-type Btk and the C481S mutant, and should not interfere with functions of other kinases, thus addressing both of these ibrutinib-related shortcomings at the same time.
Methods
Detailed methods can be found in SI Appendix. A brief summary is provided here.
General Simulation Details.
All production simulations were run on Anton (61), a specialized machine for MD simulations, using the Amber ff99SB-ILDN force field for proteins (62–64), the CHARMM TIP3P model for water (65), and the CHARMM36 force field for POPC lipids (66). The initial parameters of soluble inositol phosphates and PIP3 lipids were generated with the generalized Amber force field (67), with the atomic charges refit by restrained electrostatic potential fitting. The system was neutralized and salted with NaCl, with a final concentration of 0.15 M. The system was equilibrated in the NPT ensemble for 100 ns using gDesmond (68) on a commodity GPU cluster. Production runs were subsequently performed in the NVT ensemble from the final frame of the NPT relaxation simulation (69) by coupling the system to a variant (70) of the Nosé–Hoover thermostat (71) at 310 K with a relaxation time of 1 ps. A RESPA (72) integrator was used, with an inner time step of 2.5 fs. The long-range electrostatic forces were calculated in k-space using a grid-based method with Gaussian spreading to the grid every 7.5 fs (73).
Tempered Binding Simulations.
To enhance the sampling in our PH–TH module encounter simulations, we used an approach called “tempered binding,” (32) in which the system’s Hamiltonian (energy function) is tempered as opposed to its temperature, as is done in conventional tempering methods. Tempered binding dynamically scales various atomic interactions during an MD simulation by a factor, λ, that is updated among a ladder of discrete values, λi. The atomic interactions are unscaled at the lowest rung of the ladder (rung 0). The structures sampled at rung 0 are sampled from the same Boltzmann distribution as a conventional MD simulation. Our tempered binding simulations linearly scaled the near electrostatic interactions between the positively charged atoms on one protein and the negatively charged atoms on the other protein, and vice versa (interactions within a protein, and between protein and water, were not scaled). We varied the maximum tempering strength (i.e., the scale factor reached at the highest rung; the scale factor at the lowest rung is 1) from 0.991 to 0.997, and found that the efficiency of dimer formation was not sensitive to the maximum tempering strength in this range.
Clustering of Dimers Formed on Membranes.
All frames from all replicas of 24 runs (for a total of 6.55 ms of simulation time) were extracted. Protein structures from these frames were clustered using hierarchical density-based spatial clustering of applications with noise (74). In total, there were 5,571,555 such structures. Frames in which there was no dimer structure were removed. The filtered dataset contained n = 4,379,672 structures. A total of five clusters were identified, with 1,132,602 (26%) of the points analyzed categorized as noise. The primary cluster contained 2,679,781 points (61%). The structures for each cluster’s best approximate representation vector were identified and visualized.
Calculation of Interface Rmsd.
The rmsd values at the Saraste interface were calculated using all of the nonhydrogen atoms in residues at the dimer interface (Ile-9, Leu-29, Tyr-42, Phe-44, Ile-92, Ile-95). The positions of these residues in the crystal structure of the PH–TH dimer [Protein Data Bank (PDB) ID code 1BTK] were used as the reference.
Equilibrium Simulations Between the Saraste Conformation and the Pre-Saraste Conformation.
The population fractions of the tightly packed Saraste conformation and of the pre-Saraste conformation were calculated as follows: First, the rmsd values for all frames at rung 0 were calculated with respect to the crystal structure, as in the other analyses. Only frames with rmsd <7 Å were considered. We classified frames with rmsd <3.1 Å as being in the tightly packed conformation, and frames with rmsd >3.1 Å (and <7 Å) as being in the pre-Saraste conformation. The percentage of the population for each conformation was then calculated as the fraction of the total frames associated with that conformation. Simulation trajectories with more than 10 transition events between the Saraste and pre-Saraste conformations were used for the free energy calculation. The final free energy value reported was averaged from five (for the 8-PIP3 condition) and three (for the 2-PIP3 condition) independent simulations. The total numbers of transition events for the 8-PIP3 and 2-PIP3 conditions were 86 and 81, respectively.
Bioinformatics Analysis.
The Btk sequences used for evolution analysis were compiled using a series of protein–protein BLAST searches in the National Center for Biotechnology Information nonredundant protein database, using the human Btk PH–TH module (residues 1–172) as the reference (75, 76). The initial search generated 772 sequences with a minimal sequence identity of 48%. Btk sequences of mammals, birds, and fish were then extracted from the initial search results based on taxonomy IDs. The sequence pool was then filtered based on the following rules: (i) duplicate entries were removed; (ii) sequences labeled as “partial” or “synthetic” were removed; (iii) sequences annotated as “BMX kinase” were removed. The final number of sequences used for multiple-sequence alignment was 334. The multiple-sequence alignment of the 334 sequences was performed in Clustal Omega (77, 78) with default settings, and sequence logos were generated using the online tool WebLogo3 (79, 80). The final figures were manually adjusted from the WebLogo3 result to show only the residues at the sites of interest.
Supplementary Material
Acknowledgments
We thank Konstantin Yatsenko, Thomas Weinreich, Cristian Predescu, and Tamas Szalay for helpful discussions and help with tempered binding simulations, Je-Luen Li for help with parameterization of lipids, Yibing Shan and Michael Eastwood for a critical reading of the manuscript, and Berkman Frank and Jessica McGillen for editorial assistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819301116/-/DCSupplemental.
References
- 1.Lindvall JM, et al. Bruton’s tyrosine kinase: Cell biology, sequence conservation, mutation spectrum, siRNA modifications, and expression profiling. Immunol Rev. 2005;203:200–215. doi: 10.1111/j.0105-2896.2005.00225.x. [DOI] [PubMed] [Google Scholar]
- 2.Tsukada S, Simon MI, Witte ON, Katz A. Binding of beta gamma subunits of heterotrimeric G proteins to the PH domain of Bruton tyrosine kinase. Proc Natl Acad Sci USA. 1994;91:11256–11260. doi: 10.1073/pnas.91.23.11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dal Porto JM, et al. B cell antigen receptor signaling 101. Mol Immunol. 2004;41:599–613. doi: 10.1016/j.molimm.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Jefferies CA, et al. Bruton’s tyrosine kinase is a Toll/interleukin-1 receptor domain-binding protein that participates in nuclear factor kappaB activation by Toll-like receptor 4. J Biol Chem. 2003;278:26258–26264. doi: 10.1074/jbc.M301484200. [DOI] [PubMed] [Google Scholar]
- 5.Kawakami Y, et al. Tyrosine phosphorylation and activation of Bruton tyrosine kinase upon Fc epsilon RI cross-linking. Mol Cell Biol. 1994;14:5108–5113. doi: 10.1128/mcb.14.8.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bence K, Ma W, Kozasa T, Huang XY. Direct stimulation of Bruton’s tyrosine kinase by Gq-protein alpha-subunit. Nature. 1997;389:296–299. doi: 10.1038/38520. [DOI] [PubMed] [Google Scholar]
- 7.Tsukada S, et al. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. Cell. 1993;72:279–290. doi: 10.1016/0092-8674(93)90667-f. [DOI] [PubMed] [Google Scholar]
- 8.Di Paolo JA, et al. Specific Btk inhibition suppresses B cell- and myeloid cell-mediated arthritis. Nat Chem Biol. 2011;7:41–50. doi: 10.1038/nchembio.481. [DOI] [PubMed] [Google Scholar]
- 9.Honigberg LA, et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci USA. 2010;107:13075–13080. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrd JC, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, et al. Autoinhibition of Bruton’s tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate. eLife. 2015;4:e06074. doi: 10.7554/eLife.06074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagar B, et al. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 13.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 14.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 15.Joseph RE, Wales TE, Fulton DB, Engen JR, Andreotti AH. Achieving a graded immune response: BTK adopts a range of active/inactive conformations dictated by multiple interdomain contacts. Structure. 2017;25:1481–1494.e4. doi: 10.1016/j.str.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park H, et al. Regulation of Btk function by a major autophosphorylation site within the SH3 domain. Immunity. 1996;4:515–525. doi: 10.1016/s1074-7613(00)80417-3. [DOI] [PubMed] [Google Scholar]
- 17.Rawlings DJ, et al. Activation of BTK by a phosphorylation mechanism initiated by SRC family kinases. Science. 1996;271:822–825. doi: 10.1126/science.271.5250.822. [DOI] [PubMed] [Google Scholar]
- 18.Cheng G, Ye ZS, Baltimore D. Binding of Bruton’s tyrosine kinase to Fyn, Lyn, or Hck through a Src homology 3 domain-mediated interaction. Proc Natl Acad Sci USA. 1994;91:8152–8155. doi: 10.1073/pnas.91.17.8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Afar DE, et al. Regulation of Btk by Src family tyrosine kinases. Mol Cell Biol. 1996;16:3465–3471. doi: 10.1128/mcb.16.7.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whyburn LR, et al. Reduced dosage of Bruton’s tyrosine kinase uncouples B cell hyperresponsiveness from autoimmunity in lyn−/− mice. J Immunol. 2003;171:1850–1858. doi: 10.4049/jimmunol.171.4.1850. [DOI] [PubMed] [Google Scholar]
- 21.Wahl MI, et al. Phosphorylation of two regulatory tyrosine residues in the activation of Bruton’s tyrosine kinase via alternative receptors. Proc Natl Acad Sci USA. 1997;94:11526–11533. doi: 10.1073/pnas.94.21.11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawakami Y, et al. Redundant and opposing functions of two tyrosine kinases, Btk and Lyn, in mast cell activation. J Immunol. 2000;165:1210–1219. doi: 10.4049/jimmunol.165.3.1210. [DOI] [PubMed] [Google Scholar]
- 23.Hyvönen M, Saraste M. Structure of the PH domain and Btk motif from Bruton’s tyrosine kinase: Molecular explanations for X-linked agammaglobulinaemia. EMBO J. 1997;16:3396–3404. doi: 10.1093/emboj/16.12.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grasberger B, Minton AP, DeLisi C, Metzger H. Interaction between proteins localized in membranes. Proc Natl Acad Sci USA. 1986;83:6258–6262. doi: 10.1073/pnas.83.17.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groves JT, Kuriyan J. Molecular mechanisms in signal transduction at the membrane. Nat Struct Mol Biol. 2010;17:659–665. doi: 10.1038/nsmb.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shan Y, et al. Molecular basis for pseudokinase-dependent autoinhibition of JAK2 tyrosine kinase. Nat Struct Mol Biol. 2014;21:579–584. doi: 10.1038/nsmb.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmad M, Gu W, Helms V. Mechanism of fast peptide recognition by SH3 domains. Angew Chem Int Ed Engl. 2008;47:7626–7630. doi: 10.1002/anie.200801856. [DOI] [PubMed] [Google Scholar]
- 28.Ahmad M, Gu W, Geyer T, Helms V. Adhesive water networks facilitate binding of protein interfaces. Nat Commun. 2011;2:261. doi: 10.1038/ncomms1258. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt AG, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci USA. 2013;110:264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plattner N, Doerr S, De Fabritiis G, Noé F. Complete protein-protein association kinetics in atomic detail revealed by molecular dynamics simulations and Markov modelling. Nat Chem. 2017;9:1005–1011. doi: 10.1038/nchem.2785. [DOI] [PubMed] [Google Scholar]
- 31.Blöchliger N, Xu M, Caflisch A. Peptide binding to a PDZ domain by electrostatic steering via nonnative salt bridges. Biophys J. 2015;108:2362–2370. doi: 10.1016/j.bpj.2015.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan AC, et al. Atomic-level characterization of protein-protein association. Proc Natl Acad Sci USA. 2019;116:4244–4292. doi: 10.1073/pnas.1815431116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baraldi E, et al. Structure of the PH domain from Bruton’s tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate. Structure. 1999;7:449–460. doi: 10.1016/s0969-2126(99)80057-4. [DOI] [PubMed] [Google Scholar]
- 34.Lai CL, et al. Molecular mechanism of membrane binding of the GRP1 PH domain. J Mol Biol. 2013;425:3073–3090. doi: 10.1016/j.jmb.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto E, Kalli AC, Yasuoka K, Sansom MSP. Interactions of pleckstrin homology domains with membranes: Adding back the bilayer via high-throughput molecular dynamics. Structure. 2016;24:1421–1431. doi: 10.1016/j.str.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buyan A, Kalli AC, Sansom MSP. Multiscale simulations suggest a mechanism for the association of the Dok7 PH domain with PIP-containing membranes. PLoS Comput Biol. 2016;12:e1005028. doi: 10.1371/journal.pcbi.1005028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan KC, Lu L, Sun F, Fan J. Molecular details of the PH domain of ACAP1BAR-PH protein binding to PIP-containing membrane. J Phys Chem B. 2017;121:3586–3596. doi: 10.1021/acs.jpcb.6b09563. [DOI] [PubMed] [Google Scholar]
- 38.Kanegane H, et al. Clinical and mutational characteristics of X-linked agammaglobulinemia and its carrier identified by flow cytometric assessment combined with genetic analysis. J Allergy Clin Immunol. 2001;108:1012–1020. doi: 10.1067/mai.2001.120133. [DOI] [PubMed] [Google Scholar]
- 39.Li T, et al. Activation of Bruton’s tyrosine kinase (BTK) by a point mutation in its pleckstrin homology (PH) domain. Immunity. 1995;2:451–460. doi: 10.1016/1074-7613(95)90026-8. [DOI] [PubMed] [Google Scholar]
- 40.Várnai P, Rother KI, Balla T. Phosphatidylinositol 3-kinase-dependent membrane association of the Bruton’s tyrosine kinase pleckstrin homology domain visualized in single living cells. J Biol Chem. 1999;274:10983–10989. doi: 10.1074/jbc.274.16.10983. [DOI] [PubMed] [Google Scholar]
- 41.Pilling C, Landgraf KE, Falke JJ. The GRP1 PH domain, like the AKT1 PH domain, possesses a sentry glutamate residue essential for specific targeting to plasma membrane PI(3,4,5)P3. Biochemistry. 2011;50:9845–9856. doi: 10.1021/bi2011306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohamed AJ, et al. Bruton’s tyrosine kinase (Btk): Function, regulation, and transformation with special emphasis on the PH domain. Immunol Rev. 2009;228:58–73. doi: 10.1111/j.1600-065X.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 43.Li T, et al. Constitutive membrane association potentiates activation of Bruton tyrosine kinase. Oncogene. 1997;15:1375–1383. doi: 10.1038/sj.onc.1201308. [DOI] [PubMed] [Google Scholar]
- 44.Fluckiger AC, et al. Btk/Tec kinases regulate sustained increases in intracellular Ca2+ following B-cell receptor activation. EMBO J. 1998;17:1973–1985. doi: 10.1093/emboj/17.7.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang SW, et al. PKCbeta modulates antigen receptor signaling via regulation of Btk membrane localization. EMBO J. 2001;20:5692–5702. doi: 10.1093/emboj/20.20.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levental I, Cebers A, Janmey PA. Combined electrostatics and hydrogen bonding determine intermolecular interactions between polyphosphoinositides. J Am Chem Soc. 2008;130:9025–9030. doi: 10.1021/ja800948c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Bogaart G, et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature. 2011;479:552–555. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J, Richards DA. Segregation of PIP2 and PIP3 into distinct nanoscale regions within the plasma membrane. Biol Open. 2012;1:857–862. doi: 10.1242/bio.20122071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lemmon MA. Pleckstrin homology (PH) domains and phosphoinositides. Biochem Soc Symp. 2007;74:81–93. doi: 10.1042/BSS0740081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 51.Ziemba BP, Pilling C, Calleja V, Larijani B, Falke JJ. The PH domain of phosphoinositide-dependent kinase-1 exhibits a novel, phospho-regulated monomer-dimer equilibrium with important implications for kinase domain activation: Single-molecule and ensemble studies. Biochemistry. 2013;52:4820–4829. doi: 10.1021/bi400488f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Masters TA, et al. Regulation of 3-phosphoinositide-dependent protein kinase 1 activity by homodimerization in live cells. Sci Signal. 2010;3:ra78. doi: 10.1126/scisignal.2000738. [DOI] [PubMed] [Google Scholar]
- 53.Huang Y, et al. Molecular basis for multimerization in the activation of the epidermal growth factor receptor. eLife. 2016;5:e14107. doi: 10.7554/eLife.14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Needham SR, et al. EGFR oligomerization organizes kinase-active dimers into competent signalling platforms. Nat Commun. 2016;7:13307. doi: 10.1038/ncomms13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: Genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sunyer JO. Evolutionary and functional relationships of B cells from fish and mammals: Insights into their novel roles in phagocytosis and presentation of particulate antigen. Infect Disord Drug Targets. 2012;12:200–212. doi: 10.2174/187152612800564419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akinleye A, Chen Y, Mukhi N, Song Y, Liu D. Ibrutinib and novel BTK inhibitors in clinical development. J Hematol Oncol. 2013;6:59. doi: 10.1186/1756-8722-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burger JA. Bruton’s tyrosine kinase (BTK) inhibitors in clinical trials. Curr Hematol Malig Rep. 2014;9:44–49. doi: 10.1007/s11899-013-0188-8. [DOI] [PubMed] [Google Scholar]
- 59.Pan Z, et al. Discovery of selective irreversible inhibitors for Bruton’s tyrosine kinase. ChemMedChem. 2007;2:58–61. doi: 10.1002/cmdc.200600221. [DOI] [PubMed] [Google Scholar]
- 60.Woyach JA, et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370:2286–2294. doi: 10.1056/NEJMoa1400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shaw DE, et al. Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis (SC14) IEEE; New York: 2014. Raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer. [Google Scholar]
- 62.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 65.MacKerell AD, Jr, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 66.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 68.Bowers KJ, et al. Proceedings of the 2006 ACM/IEEE conference on Supercomputing. ACM; New York: 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters. [Google Scholar]
- 69.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 70.Lippert RA, et al. Accurate and efficient integration for molecular dynamics simulations at constant temperature and pressure. J Chem Phys. 2013;139:164106. doi: 10.1063/1.4825247. [DOI] [PubMed] [Google Scholar]
- 71.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys Rev A Gen Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 72.Tuckerman M, Berne BJ, Martyna GJ. Reversible multiple time scale molecular dynamics. J Chem Phys. 1992;97:1990–2001. [Google Scholar]
- 73.Shan Y, Klepeis JL, Eastwood MP, Dror RO, Shaw DE. Gaussian split Ewald: A fast Ewald mesh method for molecular simulation. J Chem Phys. 2005;122:54101. doi: 10.1063/1.1839571. [DOI] [PubMed] [Google Scholar]
- 74.Campello RJGB, Moulavi D, Sander J. 2013. Density-based clustering based on hierarchical density estimates. Advances in Knowledge Discovery and Data Mining, PAKDD 2013 (Springer, Berlin), pp 160–172.
- 75.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 76.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goujon M, et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.