Significance
More than three-fourths of human germline mutations are paternal in origin and their total number increases with the father’s age at conception. These observations are thought to support the textbook view that germline point mutations stem mostly from DNA replication errors. Analyzing large germline mutation datasets for humans, we find that this understanding cannot explain the observed patterns of new mutations. Instead, we show that the male mutation bias is not driven by spermatogenesis. We further find evidence that a substantial fraction of mutations are not replicative in origin and uncover a potential effect of a mother’s age on the number of mutations that happen early in the development of the embryo.
Keywords: germline mutation, male mutation bias, DNA replication, DNA damage and repair, maternal age effect
Abstract
The textbook view that most germline mutations in mammals arise from replication errors is indirectly supported by the fact that there are both more mutations and more cell divisions in the male than in the female germline. When analyzing large de novo mutation datasets in humans, we find multiple lines of evidence that call that view into question. Notably, despite the drastic increase in the ratio of male to female germ cell divisions after the onset of spermatogenesis, even young fathers contribute three times more mutations than young mothers, and this ratio barely increases with parental age. This surprising finding points to a substantial contribution of damage-induced mutations. Indeed, C-to-G transversions and CpG transitions, which together constitute over one-fourth of all base substitution mutations, show genomic distributions and sex-specific age dependencies indicative of double-strand break repair and methylation-associated damage, respectively. Moreover, we find evidence that maternal age at conception influences the mutation rate both because of the accumulation of damage in oocytes and potentially through an influence on the number of postzygotic mutations in the embryo. These findings reveal underappreciated roles of DNA damage and maternal age in the genesis of human germline mutations.
Despite the fundamental importance of germline mutation as the source of heritable diseases and driver of evolution, its genesis remains poorly understood (1, 2). De novo base substitution mutations could arise from errors made while copying an intact DNA template (i.e., be “replication-driven”), or from damage of the template or free nucleotides that occurred before DNA replication (be “damage-induced”), or an interaction of the two (3). The relative importance of these mutation sources remains unclear but is of inherent interest and carries many implications, including for understanding the erratic behavior of the molecular clock used to date evolutionary events (4–6), for the nature of selection pressures on DNA replication and repair machinery (7, 8), and in humans, for predicting recurrence risks of Mendelian diseases and disease burdens (9, 10).
Since germline mutagenesis in mammals is extremely difficult to study directly, our understanding of this process is based on mutations identified in offspring and their relationship to their parental ages or on phylogenetic comparisons of species with differing life histories. Notably, the textbook view that replication errors are the primary source of human germline mutations (11–14) often invokes the increase in the number of germline mutations with paternal age (11, 13). This increase can arise not only from DNA replication in spermatogonial stem cells, however, but also from other metabolic activities associated with cell division or from unrepaired damage that accrues with the passage of time (15). A further complication is that the rate at which unrepaired DNA lesions are converted into mutations is affected by DNA replication and thus can depend on the cell division rate (16, 17).
Insight into the genesis of germline mutations can also be gained by contrasting male and female mutation patterns, which reflect distinct developmental trajectories and epigenetic dynamics. In mammals, fathers contribute more de novo mutations (DNMs, i.e., changes to the DNA sequence of an individual relative to their parents) to their offspring than do mothers, a phenomenon sometimes termed “male mutation bias.” The male germ cells also undergo more cell divisions in each generation than do those of females, because spermatogonial stem cells are continuously renewed after puberty in males whereas primary oocytes are formed in the fetal stage in females. Given the greater number of cell divisions undergone by male germ cells, the male mutation bias has been widely interpreted as evidence that replication errors are the primary source of point mutations (other than transitions at CpG sites) (11, 12, 18–20). However, already Müller (15) noted that other explanations are possible, as there exist numerous sex differences in germ cell development and gametogenesis other than in the number of cell divisions. To evaluate the evidence that the male mutation bias is driven by replication errors that occur during spermatogenesis, we reanalyzed DNM data from a recent study of over 1,500 parent–offspring trios (21) and contrasted the properties of paternal and maternal mutations. We tested the hypothesis that germline mutations are primarily replication-driven in origin by asking: How well do male and female mutations track the numbers of germ cell divisions? Do the dependencies of the mutation rate on the parent’s sex and age differ by mutation types and, if so, why? Is the higher male contribution to DNMs explained by replication errors in dividing male germ cells?
Results
The Ratio of Paternal to Maternal Mutations Is Already High for Young Parents and Is Stable with Parental Age.
If mutations are replication-driven, the ratio of male to female germline mutations (also known as the strength of male mutation bias, α) should reflect the ratio of the number of germ cell divisions in the two sexes; the two ratios need not be strictly proportional, though, if per-cell division mutation rates vary across developmental stages or between the two sexes (16). While male and female germlines are thought to undergo similar numbers of mitotic cell divisions by the onset of puberty (∼30 to 35 divisions), thereafter the ratio of male to female cell divisions increases rapidly with age, because of frequent mitotic divisions of spermatogonial stem cells (an estimated 23 divisions per year) and the absence of mitosis of female germ cells over the same period (11, 22). Thus, if replication errors are the primary source of germline mutations, α is expected to increase substantially with parental age at conception of the child, although again not necessarily as rapidly as the cell division ratio (16, 23, 24).
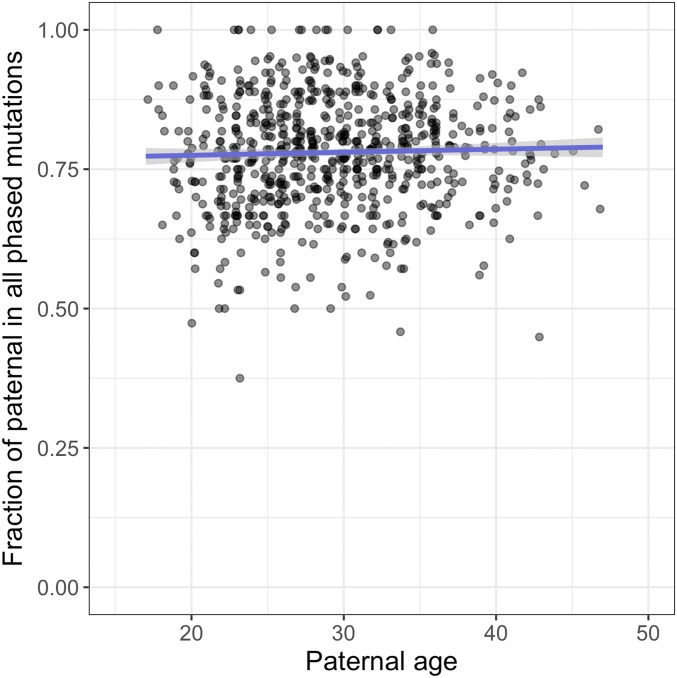
To test this prediction, we analyzed autosomal DNM data from 1,548 Icelandic trios (21) (henceforth, the “deCODE dataset”), initially focusing on phased mutations, that is, the subset of mutations for which the parental origin of the DNM had been determined by either transmission to third-generation individuals or linkage to nearby variants in reads. Given that the phasing rate differs across trios (SI Appendix, Fig. S1), we considered the fraction of paternal mutations in all phased DNMs and compared this fraction against the father’s age (Materials and Methods). We found that, for trios with similar paternal age, GP, and maternal age, GM, the paternal contribution to mutations is strikingly stable with paternal age (i.e., considering 0.9 < GP/GM < 1.1, which is the case for 719 families in the dataset; Fig. 1 and SI Appendix, Table S1). Paternal mutations comprise around 75 to 80% of DNMs (i.e., α = 3 to 4 across paternal ages); moreover, despite the large number of trios, no significant effect of paternal age is detected by regression under various generalized linear models (P > 0.28; see Materials and Methods for details). The stable α remains after excluding C > G mutations, which were previously reported to increase disproportionately rapidly with maternal age (21) (SI Appendix, Fig. S2). Moreover, the same result is seen in an independent DNM dataset containing 816 trios (25, 26), which are also mostly of European ancestry (henceforth the “Inova dataset”; SI Appendix, Fig. S3). The finding of a relatively stable (although not strictly constant) α of 3 to 4 with parental ages calls into question the widespread belief that spermatogenesis drives the male bias in germline mutations (9, 18, 23).
Fig. 1.
The fraction of paternal mutations among phased mutations, as a function of paternal age at conception. Each point represents the data for one child (proband) with similar parental ages (paternal-to-maternal age ratio between 0.9 and 1.1; 719 trios in total with a minimum of 6 and an average of 23.5 phased mutations per trio). The x-axis position, but not the y-axis position, is slightly jittered to show overlapping points. The blue line is the predicted fraction of paternal mutations by binomial regression with logit link, with the shaded area representing the 95% confidence interval (calculated with the “predict” function in R).
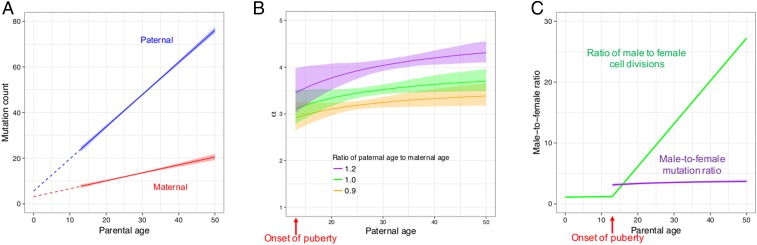
A stable α further implies that the number of maternal mutations increases with the mother’s age almost at the same relative rate as do paternal mutations with father’s age. To obtain more precise estimates of parental age effects, we modeled paternal and maternal age effects jointly, leveraging information from both phased and unphased mutations in the deCODE dataset. Briefly, we modeled the expected number of mutations in a parent as a linear function of her (his) age at conception of the child and assumed that the observed number of maternal (paternal) mutations follows a Poisson distribution. We further modeled the number of maternal (paternal) mutations that were successfully phased as a binomial sample of DNMs (Materials and Methods). We then estimated the sex-specific yearly increases with parental ages by maximum likelihood (SI Appendix, Table S2). Uncertainty in the estimates was evaluated by bootstrap resampling of trios. This analysis replicates previous findings that the mutation counts on the paternal and maternal sets of chromosomes increase with father’s and mother’s ages, respectively (21, 25, 26) (Fig. 2A).
Fig. 2.
Inferred sex and age dependencies of germline mutations (based on a linear model applied to trios with maternal age no greater than 40 y). In all panels, shaded areas and bars represent 95% CIs of the corresponding quantities obtained from bootstrapping. (A) Inferred sex-specific mutation rates as a function of parental ages. Parental ages are measured since birth, that is, birth corresponds to age 0 (throughout the paper). The extrapolated intercepts at age 0 are small but significantly positive for both sexes, implying a weak but significant effect of reproductive age on yearly mutation rates (16). (B) Predicted male-to-female mutation ratio (α) as a function of the ratio of paternal to maternal ages. For reference, the ratio of parental ages is centered around 1.10 in the deCODE DNM dataset (SD = 0.20). (C) Contrast between male-to-female mutation ratio (purple) and the ratio of male to female cell divisions (green), assuming the same paternal and maternal ages. Estimates of the cell division numbers for the two sexes in humans are from Drost and Lee (11).
In addition to a linear model, we considered exponential age effects of either or both sexes. We observed a significant improvement in fit of an exponential maternal age effect over a linear one [∆AIC (Akaike information criterion) = −29.9], consistent with a previous analysis of the Inova dataset that indicated a more rapid increase in the maternal mutation rate at older ages (25). To verify our finding, we divided the 1,548 trios into two groups with maternal age at conception over or under the median age of 27 y and fit a linear model to the two groups separately. As expected from an accelerating increase in the number of maternal mutations with age, the estimated maternal age effect is greater for older mothers than for younger mothers (0.56 vs. 0.24, 95% CI: [0.45,0.66] vs. [0.12,0.38]), whereas the estimates of paternal age effect are similar for the two groups (1.41 vs. 1.40, 95% CI: [1.31, 1.51] vs. [1.29, 1.53]; SI Appendix, Table S5). We further found that the exponential maternal age effect no longer provides a significantly better fit when excluding the 72 trios with maternal age over 40 (SI Appendix, Table S6), suggesting that the linear model is a reasonable approximation for families with maternal age under 40 y. As a sanity check on our estimates, we predicted the paternal mutation fraction for individuals with divergent paternal and maternal ages (GP/GM = 0.9, 1.2, or 1.4); our predictions provide a good fit to the observed patterns for the subset of phased mutations (SI Appendix, Fig. S4).
Next, we used the linear model fitted to trios with maternal ages below 40 y at conception (SI Appendix, Table S6) to examine the male to female mutation ratio. We found that α is already ∼3 (95% CI: [2.8, 3.5]) at the average age of onset of puberty [assumed to be 13 y of age for both sexes (27); Fig. 2 B and C], consistent with our observation of stable fraction of paternal mutations with paternal age (Fig. 1), and indicating that the male germline has accumulated a substantially greater number of DNMs than the female germline by puberty. The same is seen in our reanalysis of the smaller Inova dataset (SI Appendix, Fig. S5). This finding is highly unexpected: Male and female germ cells are thought to experience comparable numbers of divisions by puberty (an estimated 35 vs. 31, respectively) and approximately half of these divisions predate sex determination (11, 22), so males and females should harbor similar numbers of replicative mutations before puberty. Moreover, differences in the mutation spectrum between males and females are subtle (21, 28, 29), suggesting that the sources of most mutations are shared between the two sexes.
How then to explain that α is already high by puberty and persists at roughly the same value throughout adulthood? A possible resolution is that the number of male germ cell divisions from sex determination to puberty has been vastly underestimated. Indeed, a recent study reported that mutations in repetitive microsatellites, which likely result primarily from replication slippage, are already more numerous in teenage fathers compared with teenage mothers (30). A second, nonmutually exclusive explanation is that after sex determination, germ cell divisions are much more mutagenic in males than in females. In principle, these first two possibilities could account for the high α at puberty. However, they would only lead to a stable α throughout adulthood under implausible conditions: Specifically, the numbers of cell divisions and the mutation rates per cell division over developmental stages in both sexes would need to meet specific conditions, such that male-to-female ratio of replication errors before puberty is coincidentally similar to the ratio of yearly increases in mutations in the two sexes after puberty (SI Appendix). Another assumption worth reconsidering is the estimate of the rate of spermatogonial stem cell divisions. If it is slower than has been estimated (24), the ratio of male to female germ cell divisions may be lower than shown in Fig. 2C and more in line with the observed mutation bias. Even so, however, the high α at puberty or its relative stability afterward remain unexplained. Instead, we hypothesize that most germline mutations in both sexes are damage-induced: Under this scenario, both the elevated α by puberty and the stable α after puberty are parsimoniously explained by damage rates that are roughly constant per unit time in both sexes and higher in males (23).
Specific Sources of DNA Damage-Induced Mutations.
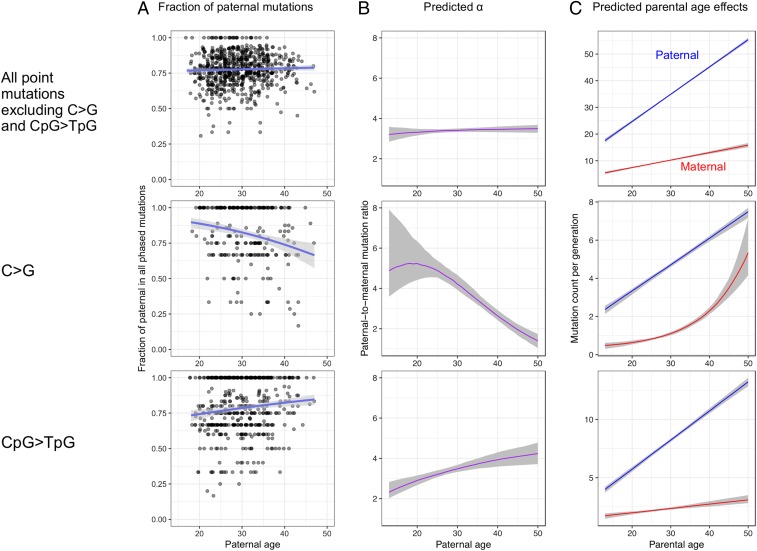
To explore possible mutational mechanisms, following previous studies (28, 31–33), we classified base substitution DNMs into six disjoint and complementary mutation classes based on parental and derived alleles: T > A, T > C, T > G, C > A, C > G, and C > T (each type also includes the corresponding variant on the reverse complement strand). Given the well-characterized hypermutability of methylated CpG sites in primates (34–36), we further divided the C > T transitions into subtypes in non-CpG and CpG contexts (excluding CpG islands which are typically hypomethylated; Materials and Methods). Confirming the original analysis of these data (21), we detected significant paternal and maternal age effects for all seven mutation types, of varying magnitudes (SI Appendix, Fig. S6 and Table S2). While the male bias is stable with parental age for most mutation types (SI Appendix, Fig. S6), C > G transversions and C > T transitions at CpG sites stand out from the general pattern (Fig. 3). In particular, C > G mutations show a decreasing α with age and CpG > TpG mutations an increasing α, evident in both the analysis of phased mutations and in our modeling of all mutations (Fig. 3 A and B). We further found that whereas the linear model is a good fit to the other six mutation types individually, for C > G mutations a model with a linear paternal age effect and an exponential maternal age effect provides a significantly better fit (∆AIC = −18.3; SI Appendix, Table S4). Interestingly, an exponential maternal age effect also provides a significantly better fit for the six non-C > G point mutations combined (ΔAIC < −9; SI Appendix, Table S6), and again the effect is nonsignificant when the 72 trios with maternal age over 40 y are excluded, suggesting that mutation types other than C > G are also increasing at higher rates in mothers of older ages.
Fig. 3.
Distinctive sex and age dependencies for C > G and CpG > TpG DNMs. The shaded areas in all panels represent 95% CIs. See SI Appendix, Fig. S6 for similar plots for other mutation types. The male-to-female mutation ratio at age 17 is significantly lower for CpG > TpG than for other mutation types (discussed in the main text). (A) Fraction of paternal mutations in phased DNMs (similar to Fig. 1). (B) Predicted male-to-female mutation ratio (α). (C) Predicted parental age effects.
Based on their spatial distribution in the genome and their increase with maternal age, maternal C > G mutations were hypothesized to be associated with double-strand breaks in aging oocytes: This mutation type often appears in clusters with strong strand concordance and near de novo copy number variant breakpoints; moreover it is enriched in genomic regions with elevated rates of noncrossover gene conversion, an enrichment that increases rapidly with maternal age (21, 37). C > G mutations are also more frequent in the human pseudoautosomal 1 region, which experiences an obligate crossover in males, than on autosomes and the rest of the X chromosome (29). We additionally found that C > G transversions are significantly more likely than other mutation types to occur on the same chromosome as a de novo deletion (≥5 bp) in the same individual, and conditional on cooccurrence, that the distance to the closest deletion tends to be shorter (SI Appendix, Fig. S7). The same association is not seen for short deletions or insertions (<5 bp), however (SI Appendix, Table S7), which are more likely to arise from replication slippage (38, 39). Together, these observations support imperfect repair of double-strand breaks as an important source of C > G transversions in both sexes (21, 29, 37).
In turn, the high rates of C > T transitions at CpG sites are thought to be due to the spontaneous deamination of methylcytosine (40, 41) that remains unrepaired by the next replication cycle, although recent studies of tumors indicate that they may also result from an interaction between methylation and the DNA replication process (3, 42). In yeast species that are believed to lack DNA methylation, however, the mutation rates at CpG sites are also moderately elevated (43–45), which may suggest that methylation-independent mutational mechanisms are also at play, at least in these species (44).
Since the methylation dynamics of mammalian male and female germlines are relatively well characterized, we can make a priori predictions about when sex differences in C > T transitions should arise in development to examine if their genesis is associated with methylation. Specifically, during embryogenesis, several rounds of global DNA demethylation and remethylation take place to enable the erasure and reestablishment of the epigenetic memory from the parents (46, 47). Because these methylation changes are shared by male and female embryos until sex determination of the embryo, the two sexes should share most methylation-related mutations during early development. Consistent with this prediction, we estimated a lower α for CpG transitions than for other presumably more replication-dependent mutation types at early reproductive ages (e.g., at age 17 y, α = 2.6 [2.2, 3.0] vs. 3.4 [3.2, 3.7] for mutations other than CpG > TpG) (Fig. 3B). After sex determination (around 7 wk postfertilization in humans), the methylation profiles of male and female germ cells diverge: Remethylation takes place early in males, before differentiation of spermatogonia, but very late in females, just shortly before ovulation (46, 47). Therefore, the male germline is markedly more methylated compared with the female germline for the long period from sex determination of the parents to shortly before conception of the child. Accordingly, after puberty, the estimated yearly increase in CpG > TpG mutations is 6.5-fold higher in fathers than in mothers, roughly double what is seen for other mutation types, resulting in a marked increase in α with parental age at CpG > TpG (Fig. 3C). In support of the key role of methylation in CpG transition mutagenesis, the genomic distribution of DNMs in the 1,548 Icelandic trios is strongly associated with the methylation levels in testis, and more weakly with those in ovary (48, 49) (SI Appendix, Fig. S8A). Moreover, the distributions of paternal mutations along the genome show a closer correspondence to the methylation profile of testis than that of ovary, and vice versa for maternal mutations (SI Appendix, Fig. S8 B and C). In summary, the sex and age dependencies of CpG transitions accord with the sex-specific temporal and genomic methylation profiles of the mammalian germ cells, providing further evidence that methylation-related mutagenic processes are the major sources of CpG transitions, and validating our inferences for the one case in which we have independent information about what to expect. Together with C > G mutations, CpG transitions represent approximately one-fourth of germline point mutations accumulated in a parent of age 30 y at conception; both show signatures of DNA damage-induced mutational mechanisms.
The Number of de Novo Mutations Increases Substantially with Maternal Age.
In mammals, primary oocytes are formed and arrested in prophase of meiosis I, before the birth of the future mother, with no further DNA replication occurring until fertilization. On this basis, the maternal age effect detected by recent DNM studies (21, 25, 26) and confirmed here (Fig. 2A) has been interpreted as reflecting the accumulation of DNA lesions or damage-induced mutations in (primary) oocytes during the lengthy meiotic arrest phase (16, 21, 23, 50), exemplified by the rapid increase of maternal C > G mutations. However, other explanations for a maternal age effect are possible (25). For example, such an effect could also arise if oocytes ovulated later in life have undergone more mitoses (51, 52). In this scenario, the substantial increase in maternal DNMs from age 17 y to age 40 y (Fig. 2A) would require oocytes ovulated later in life to go through almost double the number of cell divisions compared with those ovulated early (more, if the per-cell division mutation rate is higher in early cell divisions, discussed below). Moreover, this scenario does not provide an explanation for the stability of the male-to-female ratio with parental ages. Thus, while this phenomenon could hypothetically contribute to the maternal age effect, in practice, it is likely to be a minor effect (see SI Appendix for a more detailed discussion).
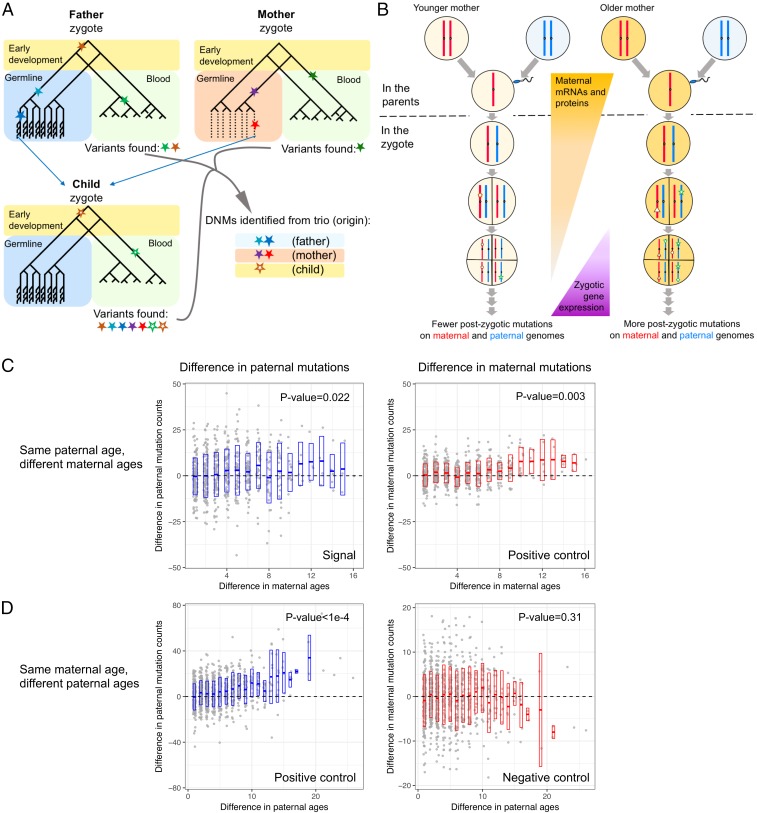
A less appreciated explanation for a maternal age effect on mutation is an effect on postzygotic mutations (25). Although DNMs are usually interpreted as mutations that occur in germ cells of the parents, in fact what are identified as DNMs in trio studies are the genomic differences between the offspring and the parents in the somatic tissues sampled (here, blood). These differences can arise in the parents but also during early development of the child (Fig. 4A). Notably, the first few cell divisions of embryogenesis have been found to be relatively mutagenic, leading to somatic and germline mosaic mutations in cattle (53), mice (54), and to a lesser extent in humans (28, 55–58), as well as to mutations that are discordant between monozygotic twins (21, 59). Increased numbers of point mutations in the first few cell divisions should perhaps be expected, as two key components of base excision repair are missing in spermatozoa, leading lesions accumulated in the last steps of spermatogenesis to be repaired only in the zygote (60). More generally, mammalian zygotes are almost entirely reliant on the protein and transcript reservoirs of the oocyte until the four-cell stage (61–63). Thus, if the replication or repair machinery of the oocytes deteriorates with the mother’s age (64, 65), one consequence could be more mutations in the first few cell divisions of the embryo (Fig. 4B). This scenario predicts that the mother’s age influences not only the number of mutations on the chromosomes inherited from the mother but also from the father (which would be assigned to “paternal mutations”) (Fig. 4B).
Fig. 4.
Maternal age effect on mutations that occur on paternally inherited chromosomes. (A) An illustration of mutations occurring during development and gametogenesis. Adapted from ref. 82. Filled stars represent mutations that arise in the parents and hollow stars mutations in the child. The standard trio approach requires allelic balance in the child and no or few reads carrying the alternative allele in the parent, leading to inclusion of some early postzygotic mutations in the child (brown open) and exclusion of a fraction of early mutations in the parents (brown filled). (B) An illustration of a potential maternal age effect on the number of postzygotic mutations. The shade of the oocyte represents its cellular quality, with a darker color indicating a worse condition of the replication or repair machinery. (C) Pairwise comparison conditional on the same paternal age. Each point represents a pair of trios, with the x axis showing the difference in maternal ages and the y axis the difference in paternal mutation counts (Left; older mother – younger mother) or maternal mutation counts (Right; older mother – younger mother); point position is slightly jittered to show overlapping points. P values are evaluated by 10,000 permutations, using Kendall’s rank correlation test statistic (Materials and Methods). (D) Pairwise comparison conditional on the same maternal age, similar to C. The ranges of y axis differ for the plots on the left and right for visualization purposes.
Any maternal age effect on postzygotic mutations is challenging to detect, given the small fraction of postzygotic mutations estimated in humans (66), especially in comparison with the stochasticity in mutation counts across individuals and the noise induced by incomplete phasing of mutations. Further reducing the ability to detect either a pre- or postzygotic effect of maternal age is the high correlation between maternal and paternal ages (which is likely why a maternal age effect was not detected in earlier, smaller studies) (33, 36). As an illustration, if we assume a large increase of 0.3 paternal mutations with each year of maternal age and complete phasing of DNMs, we estimate the power to detect this maternal age effect in 200 trios to be only 45 to 56% (SI Appendix, Fig. S9). Moreover, using trio data alone, DNMs can only be phased based on informative heterozygous variants in the same reads, so only a small fraction (typically 25 to 30%) are phased (21, 25, 26) (SI Appendix, Fig. S1). If we assume no DNM calling errors and a uniform phasing rate of 30% across all trios, the power is reduced to 15 to 20% with 200 trios and is 66 to 73% even with 1,000 trios (SI Appendix, Fig. S9). Moreover, the phasing rate is likely to vary somewhat across families, introducing additional variation in the number of phased mutations and further reducing the power. Finally, these simulations ignore errors in calling DNMs, when in reality error rates are non-zero—especially when a third generation is not available to verify transmission—so the power estimates for the standard trio design are likely too high. Consistent with these considerations, we found that in the deCODE dataset, DNMs from trios with and without third-generation individuals differ in multiple properties, including the dependency of the number of mutations on sex and parental age (SI Appendix). Therefore, we expect that a maternal age effect on postzygotic mutations, if it exists, should only be detectable using data from a sufficiently large number of pedigrees with more than two generations. To our knowledge, the only publicly available dataset that currently satisfies these criteria is the deCODE dataset, which includes 225 three-generation families (21).
To reduce noise due to incomplete phasing and DNM calling errors, we focused on the subset of 199 deCODE probands in which almost all DNMs are phased (>95% phased) by transmission to third-generation individuals. Intriguingly, Poisson regression of the count of paternal mutations on both parental ages revealed a marginally significant effect of maternal age (P = 0.035) and a slight but nonsignificant improvement in the fit compared with a model with paternal age only (∆AIC = −2.4; SI Appendix, Table S8). We verified that such a signal would not arise artifactually from the correlation between maternal and paternal ages and the assignment of parental ages to 1-y bins (P = 0.007; see Materials and Methods for details). We further noted that the maternal age effect becomes more significant when we limited the Poisson regression to 130 probands with >98% DNMs phased (P = 0.0037 in Poisson regression) and remains significant in a negative binomial regression that allows for overdispersion in mutation counts (P = 0.0075; SI Appendix, Table S8). To visualize the effect, we carried out further analyses of DNMs in the 199 probands by comparing all pairs with the same paternal age but different maternal ages. Among 619 such pairs, the child born to the older mother carries more paternal mutations than does the child with the younger mother in 319 cases, fewer in 280 cases, and the same number in 20 cases, and greater differences in the number of paternal mutations are associated with greater differences in maternal ages (Kendall’s rank test τ = 0.09, P = 0.022 by a permutation test; see Materials and Methods for details; Fig. 4C). In contrast, there is no significant effect of paternal age on the number of maternal mutations when matching for the mother’s age (P > 0.31; Fig. 4D and SI Appendix, Table S8), although we caution that the power to detect such an effect is lower because of the smaller numbers of maternal mutations.
The estimated effect of maternal age on maternal mutations is 0.34 mutations per year (SE = 0.04) by Poisson regression (P = 3.4e-13). The estimated maternal age effect on paternal mutations is similar but highly uncertain (0.30, SE = 0.14). Naively, one might expect the maternal age effect on maternal mutations to be stronger, as it includes both prezygotic effects (e.g., damage in the oocyte) and postzygotic effects, whereas the effect on paternal mutations can only be postzygotic. This expectation is implicitly based on the assumption of the same postzygotic effects of maternal age on maternal and paternal genomes, but they need not be similar. Indeed, before fertilization, sperm and oocytes may harbor different levels of DNA damage (e.g., oxidative stress may be higher in male germ cells) (67, 68) and after fertilization but before the first cleavage the two parental genomes experience distinct epigenetic remodeling and are replicated separately in their own pronuclei (69, 70). Thus, the relative contributions of prezygotic and postzygotic effects of maternal age on the maternal genome are not distinguishable without additional data. Regardless, the positive association between maternal age and the number of DNMs on paternal chromosomes supports the hypothesis that a mother’s age at conception affects the postzygotic mutation rate in the developing embryo.
In cattle and humans, the high-frequency mosaic mutations that are likely to have arisen in early embryonic development are enriched for C > A transversions (53, 55, 57), potentially reflecting the accumulation of the oxidative DNA damage 8-hydroxyguanine in oocytes and the last stages of spermatogenesis (67, 68, 71) that remains uncorrected in spermatozoa (60). Hypothesizing that a maternal age effect may be particularly pronounced for these mutations, we focused on C > A mutations in the 199 probands with >95% phasing rates. Although this subset represents only ∼8% of mutations, there is a significant effect of maternal age on paternal mutations (P = 0.02 by Poisson regression), and the point estimate of the maternal age effect on paternal genome (0.095, SE = 0.041) is even stronger than that of paternal age (0.057, SE = 0.033), as well as stronger than the maternal age effect on the maternal genome (0.024, SE = 0.0094; see Materials and Methods for details). Such results are not often obtained in a random subset of mutations of the same size (P = 0.045; Materials and Methods), suggesting paternal C > A mutations are indeed more strongly affected by maternal age than are other DNMs. After excluding C > A mutations, the maternal age effect on paternal point mutations is no longer significant in the 199 probands with >95% DNMs phased (P = 0.13) but remains significant for the subset of 130 probands with phasing rates higher than 98% (P = 0.02). Thus, a mutation type associated with damage in sperm and known to be enriched in early embryogenesis shows a heightened signal of a maternal age effect on paternal mutations, without entirely accounting for the signal.
Discussion
These findings call into question the textbook view that germline mutations arise predominantly from replication errors in germ cells. First, multiple lines of evidence, reported in previous studies and extended here, suggest that CpG transitions and C > G mutation often arise from methylation-associated damage and double-strand break repair, respectively (21, 28, 37). Second, and unexpectedly, even excluding both of these mutation types, roughly threefold more paternal mutations than maternal mutations have occurred in young parents, despite similar numbers of estimated germ cell divisions by that age in both parents (Fig. 2 B and C). Moreover, the male-to-female mutation ratio remains surprisingly stable with parental age, even as the ratio of male-to-female cell divisions increases rapidly (Fig. 2 B and C). The high α of ∼3 in young parents could be explained by a vast underestimation of the number of germ cell divisions in males between birth and puberty (30) or a much higher per-cell division mutation rate during development of spermatogonia in males, but its stability with parental ages cannot. Finally, despite highly variable cell division rates over development, germline mutations appear to accumulate in rough proportion to absolute time in both sexes (Fig. 2A and SI Appendix, Fig. S6). Together, these findings point to a substantial role of DNA damage-induced mutations, raising questions about the relative importance of endogenous versus exogenous mutagens, as well as about why male and female germ cells differ in the balance of DNA damage and repair.
In addition, we identified a tentative signal of a maternal age effect on the number of mutations on the paternal genome, which supports the hypothesis that the age of the mother at conception influences mutagenesis in early embryonic development of her child. Because violations of the assumptions of the regression model may lead to inflated significance and the various tests that we performed were based on the same dataset, our findings need to be replicated in other large, independent datasets. A maternal age effect on the postzygotic mutation rate is plausible, however, given what is known about early mammalian embryogenesis, as well as accumulating evidence for a nonnegligible number of early embryonic mutations among DNMs (28, 53, 54, 66, 72). Given its potential implications, it will be important to investigate further.
Finally, our findings shed light on the divergent conclusions about sex-specific mutation rates reached from phylogenetic analyses versus analyses of DNMs. Pedigree studies in humans and chimpanzees suggest that by typical reproductive ages there is a similar degree of male bias for de novo CpG transitions and other mutation types (21, 73), when in phylogenetic analyses CpG transitions are estimated to have a much weaker male bias (19). Here, we show that the male bias of de novo CpG > TpG mutations is significantly lower than that of other mutations at young reproductive age but higher at older age (Fig. 3). Thus, results from pedigree and phylogenetic studies could be reconciled if humans and chimpanzees long had shorter generation times than at present. In addition, across mammalian species, a longer generation time is associated with a decreased ratio of X to autosome divergence [interpreted as a greater male bias in mutation (6, 18, 74, 75), but see ref. 76 for complications of this method] and a lower substitution rate [interpreted as a lower mutation rate per year (23, 77–79)]. These phylogenetic observations have been widely taken to support a replicative origin of most non-CpG mutations (6, 12, 19, 20, 23, 78, 79). Casting doubt on this interpretation, our analyses of human DNMs show generally weak effects of reproductive age on the male-to-female mutation ratios as well as on yearly mutation rates (SI Appendix, Fig. S6 B and C), an important role for nonreplicative mutations beyond CpG transitions, and a potential maternal age effect on the number of mutations on both maternal and paternal genomes. An alternative explanation for the phylogenetic patterns is that interspecific differences in the male mutation bias and in yearly substitution rates reflect the evolution of the ratio of paternal to maternal ages at reproduction (76) (Fig. 2B) or of rates of DNA damage (e.g., metabolic rates) that covary with life history traits (6, 80).
Materials and Methods
Processing of de Novo Mutation Data.
For each DNM we obtained parental ages at conception of the child (proband) and the position, allele, and parent-of-origin information from the appendix of the publication for one dataset (21) and by personal communication with the authors for the replication dataset (25, 26). We considered a mutation as “phased” if the parental haplotype on which it arose was determined by either informative flanking variant in the read or from transmission to a third generation. See SI Appendix, Table S3 for a comparison of summary statistics of these datasets.
For both datasets, we removed indels and mutations on X chromosome (no Y-linked DNMs were reported), which resulted in 98,858 and 35,793 point mutations (or single nucleotide substitutions) for Jónsson et al. (21) and Goldmann et al. (26), respectively. Each of these mutations was then assigned into one of six mutation types (T > A, T > C, T > G, C > A, C > G, and C > T) based on the original allele present in homozygous state in both parents and the derived allele that is carried by the child in heterozygous state. Complementary combinations (such as C > T and G > A) were combined such that the original allele is always a pyrimidine (C or T). Moreover, each DNM was annotated to be in CpG or non-CpG context based on its two immediate flanking bases extracted from human reference genome. For analyses of C > T mutations at CpG sites, we excluded those present in CpG islands (annotations downloaded from UCSC browser: CpG Islands track), because these sites are thought to be hypomethylated and thus behave differently in terms of mutation rate compared with CpG sites outside CpG islands (CGI) (79). C > T mutations at CpG sites in CGIs were included in analysis of “all point mutations.”
Test for an Effect of Parental Age on the Male Mutation Bias.
We modeled the numbers of paternal and maternal DNMs (denoted by XPi and XMi, where the index i indicates the proband ID) of each trio by two independent Poisson distributions with unknown expected values (denoted as λPi and λMi). Under the assumption that in the same individual the phasing probability (denoted by pi) of each mutation is identical and independent, the numbers of phased paternal and maternal DNMs (denoted by YPi and YMi) also follow independent Poisson distributions with expectations of piλPi and piλMi, based on the thinning property of Poisson process. Moreover, it can be shown that conditional on YPi +YMi, YPi follows a binomial distribution with a success parameter of , which is exactly the expected contribution of paternal mutations in all DNMs that we are interested in. Therefore, the test for a paternal age effect on the male mutation bias becomes a test for an effect of paternal age on the “success rate” ri in a series of binomial samples (YPi, YMi) ∼ binom(YPi +YMi, ri).
We performed binomial regression with phased DNM data of 719 trios with similar parental ages in R using the glm function with option “family=binomial()” and did not detect a significant effect of paternal age with either a logit or an identity link function (P = 0.29 and 0.31, respectively). To take into account the greater dispersion in mutation counts than assumed under Poisson distribution, we tested quasi-binomial models by specifying “family=quasibinomial()” in glm and again found no significant effect with either a logit or an identity link (P = 0.33 and 0.34, respectively). We also tested the effect of the average age of the father and the mother in these 719 trios with the same regression methods and found no significant results. We calculated the predicted fraction of paternal mutations and its 95% confidence interval (shown in Figs. 1 and 3) with the R function “predict.”
Estimation of Sex-Specific Mutation Parameters with a Model-Based Approach.
Similar to Jónsson et al. (21), we modeled the expected number of mutations from a parent as a linear function of her (or his) age at conception of the child, and assumed that the observed maternal (paternal) mutation count follows a Poisson distribution with this expectation. One difference from the Jónsson et al. (21) model is in how we account for the incomplete parental origin information for the unphased DNMs. As described above, we explicitly modeled the phasing process as a binomial sampling of DNMs, assuming identical and independent phasing probabilities of all mutations in the same individual. Therefore, the parental age effects on DNM rates are modeled as the following:
where index i indicates the proband; λMi and λPi are the expected numbers of maternal and paternal mutations; GMi and GPi are ages of the mother and the father at conception, respectively; and β0,M, βM, β0,P, and βP are the mutation parameters that characterize the sex-specific parental age effects and are shared across all probands (note that β0,M and β0,P are the extrapolated intercepts at age zero, which are not biologically meaningful quantities and in particular are not necessarily nonnegative). We assumed linear effects for both sexes in the initial model, but we relaxed this assumption by testing for exponential effects for either or both sexes later (discussed below and SI Appendix, Table S4).
We then modeled the realized numbers of mutations, XMi and XPi, as
The observed phased and unphased mutation counts YMi and YPi are modeled as
where pi is the phasing rate in proband i and YMi, YPi, and YUi represent the numbers of phased maternal, phased paternal, and unphased mutations, respectively. YMi, YPi, and YUi are defined as random variables, and we denote the observed values of these with lowercase notations yMi, yPi, and yUi.
With the parameterization above, the likelihood of the observed data for proband i can be written as
We note that the likelihood function of Jónsson et al. (21) does not include the first term, which is the probability of the observed data given possible partitions of the unphased mutations into paternal and maternal origins (assuming the same phasing rates of maternal and paternal mutations). As an illustration, the set of observations (yMi, yPi, yUi) = (10, 30, 80) is more probable under (xMi, xPi) = (30, 90), where one-third of DNMs were phased for both parental origins, than under (xMi, xPi) = (80, 40), where 75% paternal DNMs were phased but only 12.5% of maternal DNMs.
The likelihood function for proband i can be simplified as (see derivation in SI Appendix):
The first term of the likelihood contains the phasing rate (pi) but is independent of the mutation parameters, whereas the second term is dependent on the mutation parameters but independent of pi. Therefore, the maximum likelihood estimator (MLE) of pi and those of the mutation parameters can be identified by maximizing the first and second terms separately.
The log joint likelihood of all observed data under a set of mutation parameter values can be expressed as
where C is a constant that is independent of the mutation parameters of interest.
We implemented this log likelihood function in R and found the MLEs of the mutation parameters by using function mle2 in the package bbmle with the optimization method BFGS. To avoid being trapped in local maxima, we tested a grid of initial values for the slopes (βP and βM). We performed the estimation for all point mutations altogether as well as for each mutation type separately. We note that the greater overdispersion of the mutation counts than expected under a Poisson distribution is expected to have minimal influence on the MLEs, as application of Poisson regression and negative binomial regression to the same dataset produces nearly identical point estimates of the coefficients, despite differences in estimated SEs.
Confidence Intervals of Male-To-Female Mutation Ratio at Given Parental Ages.
To account for uncertainties in the DNM parameter estimates, we used a bootstrap approach, randomly resampling the probands with replacement 500 times, keeping the same total number of probands in each run. For each replicate, we obtained the MLEs of the DNM parameters as described above, predicted the numbers of paternal and maternal mutations at given ages, and calculated the male-to-female mutation ratio. Thus, each bootstrap replicate provides one point estimate for each of the quantities of interest, and the approximate distribution for each quantity can be obtained by aggregating results from the 500 replicates. The confidence intervals shown in Figs. 2 and 3 represent the ranges between 2.5 and 97.5% quantiles of the empirical distributions, and given that they are estimated by bootstrap, they should be robust to overdispersion of the mutation counts.
Test for Alternative Models for Parental Age Effects.
In addition to the linear model described in the above, we also considered models with exponential parental age effects postpuberty for either or both sexes. Specifically, we modeled the exponential parental age effect as follows:
where P = 13 is the age of onset of puberty assumed for both sexes. We note that results are not sensitive to the choice of the value of P. Under this formulation, models with different values of P are mathematically equivalent to models with the same bP (or bM) value but different cP (or cM) values. Indeed, we confirmed the MLEs for bP and bM are the same for different P values (even for P = 0).
We obtained the MLEs and corresponding log likelihoods of all four models for all point mutations combined and for each mutation type separately and used the AIC to compare the relative fits of different models (a smaller AIC indicates a better fit of the model). We took ΔAIC < −6 as the threshold for evidence for a significantly better fit (∼20-fold more probable). The models with exponential paternal age effect provide worse fits (∆AIC > 0) for all mutation types.
For all DNMs combined, models with exponential effects of maternal age or both parental ages provide significantly better fits but are not significantly different from each other. As verification, we split the 1,548 trios into two groups with maternal age at conception over and under 27 y (the median maternal ages in the dataset), respectively, and fitted both with linear parental age effects (see results in SI Appendix, Table S5).
Among all mutation types considered, C > G transversions are the only type for which the model with exponential maternal age effect provides a significantly better fit by the criterion of ΔAIC < −6 (SI Appendix, Table S4). Therefore, in all analyses for C > G transversions (e.g., calculation of α), we used the estimates from the model with an exponential maternal age effect (and linear paternal age effect) fitted to all 1,548 trios. For all DNMs combined, the model with an exponential maternal age effect also provides a significantly better fit than the linear model. Interestingly, considering all trios, even after C > G transversions (or C > G transversions and CpG transitions) are excluded, an exponential maternal age effect still provides a significantly better fit for other point mutations combined (ΔAIC < −9; SI Appendix, Table S6), suggesting that the signal is not driven by C > G mutations alone. This effect is no longer discernible when trios with maternal age above 40 are excluded (SI Appendix, Table S6).
Processing of Ovary and Testis Methylation Data at CpG Sites.
The methylation data were generated as part of the Roadmap Epigenomics Project (48, 49). We downloaded the methylation data from GEO (https://www.ncbi.nlm.nih.gov/geo/) with accession numbers GSM1010980 for ovary and GSM1127119 for testis (sperm). The methylation levels were measured by bisulfate sequencing of testis spermatozoa primary cells from a male donor (age and descent unknown) and ovary cells from a 30-y-old female donor of European descent, respectively. Methylation levels (measured as percentage methylated) are reported only for CpG sites in the reference human genome: 27,057,581 (94.3%) of the ∼28,700,000 CpG sites (∼57,400,000 bp) have reported methylation levels in ovary and 26,693,016 (93%) have that information for testis. CpG sites with data available were sorted based on their modification levels and grouped into bins of 100,000 sites (i.e., 50,000 CpGs). The average methylation level of each bin was then correlated with the total number of C > T DNMs in the 1,548 Icelandic trios that occurred at the 100,000 sites (an estimate of the average mutation rate of these sites). We note that the methylation profile of ovary cells may be a poor proxy for that of (primary) oocytes, so the correlation between CpG > TpG DNM rate and methylation may be underestimated. In addition, there is likely interindividual variation in methylation profiles, but such variation is typically smaller than intertissue variation (81), so it is expected to reduce the correlation between methylation and mutation rates in both tissues by a small amount.
Detection and Estimation of Maternal Age Effect on Paternal Mutation Rate.
For analyses in this section, we focused on the 199 probands in which almost all DNMs were phased (>95% DNM phased). We first did a Poisson regression (with an identity link) of the count of paternal point mutations on both parental ages and found a marginally significant effect of the maternal age (P = 0.035) and a slight but nonsignificant improvement in the fit compared with a model with paternal age only (∆AIC = −2.4; ∼3.3-fold more probable); P values and AIC were obtained by the glm function in R [with the option “family = poisson(link = “identity”)”]. In contrast, regressing the maternal mutation count on both parental ages does not provide any improvement in the fit compared with a model with maternal age alone (∆AIC = 0.2; SI Appendix, Table S8), although the power to detect an effect of paternal age on maternal mutation, if any, is lower due to the low mutation counts.
Concerned about violation of the equidispersion assumption (i.e., that E[X] = Var[X]) in Poisson regression, we tested for an overdispersion of the mutation counts using the dispersiontest function in the R package AER and found a significant overdispersion in paternal mutation counts even under a model with both parental ages (dispersion factor = 1.36; P = 0.0074). We therefore also tested the maternal age effect on paternal mutations with a negative binomial regression using the glm.nb function with option “link = identity” and significance was somewhat reduced (to P = 0.062; SI Appendix, Table S8). However, when we limited the analysis to a smaller but more stringent dataset, the 130 trios with >98% DNMs phased, the effect of maternal age on paternal mutations was significant at the 5% level (P = 0.0075; SI Appendix, Table S8).
Motivated by these findings, we reestimated the mutation parameters by maximum likelihood under models including a maternal age effect on paternal mutations (i.e., “maternal-on-paternal effect”) of the same size (model 1) or a different size (model 2) than the maternal age effect on maternal mutations. Both models provide slight but insignificant improvements in fit compared with a model without a maternal age effect on paternal mutations (model 0), and the model with the same maternal effect on both maternal and paternal mutations gives the best fit based on AIC (∆AIC = −3.7; MLE of maternal age effect is 0.34 mutations per year; SI Appendix, Table S8).
We also carried out a “pairwise analysis” of the same data conditional on paternal age. Specifically, we compared all pairs of trios with the same paternal age, GP, but different maternal ages, GM, (i.e., a pairwise analysis). Because some pairs include the same probands and are thus not independent, we did a permutation test by swapping the maternal ages within paternal age bin and calculating the adjusted z-score of Kendall’s tau-b statistic. 220 out of 10,000 permutations had statistics equal to or greater than that observed with in real data (corresponding to an empirical one-tailed P value of 0.022). To estimate the effect size of maternal age, we ran weighted linear regression of the difference in paternal counts on the difference in maternal ages for each pair of trios with the same paternal age (with an intercept of zero), with the weight of each data point specified as the inverse of the paternal age, which is approximately proportional to the variance in the observed difference in paternal mutation counts (SI Appendix, Table S8). Because the mutation counts are integers and do not follow a normal distribution, the SEs are inaccurate.
One concern is that parental ages are assigned to integer bins in the Icelandic dataset, and there is potentially a subtle correlation between maternal and paternal ages even within a paternal age bin, in which case variation in paternal age counts caused by small GP variation may be mistakenly ascribed to an effect of GM. To address this concern, we simulated data of 199 trios with similar parental age structure but no maternal age effect on paternal DNMs and asked how frequently analysis of simulated data based on binned parental ages would generate signals comparable to those observed in actual data. To mimic the distributions of maternal and paternal ages and the correlation between them in the actual dataset, we simulated an exact maternal age for each trio by adding a random variable that is uniform on (0,1) to the integer maternal age given in the dataset, and a corresponding exact paternal age taken from 2.70 + 1.076GM + e, where e follows Normal(0, 4.5) (parameters obtained by ordinary linear regression on the binned parental ages in the dataset). We then simulated the paternal DNM count as a Poisson random variable with expectation of either 1.51GP + 6.05 [as estimated by Jónsson et al. (21)] or 1.41GP + 5.56 (estimated by our maximum likelihood model) and ran Poisson regression or pairwise analysis on the mutation counts and integer parts of parental ages, as described above. The simulated data generated either a greater or equal maternal age effect on paternal mutations by Poisson regression or a z-score of Kendall’s tau-b statistic as significant or more significant in only about 3.5% of 10,000 replicates and both in ∼0.7%, suggesting that this scenario is unlikely to lead to the patterns observed (SI Appendix, Table S9).
Supplementary Material
Acknowledgments
We thank Michel Georges for useful discussions in person and in his BioRxiv preprint on postzygotic mutations in cattle; Ipsita Agarwal, Mike Eisen, Arbel Harpak, Jonathan Pritchard, Zev Williams, and members of the M.P. and Pritchard laboratories for helpful discussions; and Graham Coop’s laboratory, Chuck Langley, Wendy Wong, Vladimir Seplyarskiy, and anonymous reviewers for comments on an earlier version of the manuscript. This work was supported by a Burroughs Wellcome Fund Career Award at the Scientific Interface (to P.M.); NIH Grants R01GM122975 (to M.P.) and R01GM059290, R01GM104390, and R35GM118335 (to L.B.J.); NIH National Human Genome Research Institute Grants R01HG006693 and R01HG009141 (to A.R.Q. and B.S.P.); National Institute of General Medical Sciences Grant R01GM124355 (to A.R.Q. and B.S.P.); National Cancer Institute Grant U24CA209999 (to A.R.Q. and B.S.P.); and NIH Training Grant T32GM007464 (to T.A.S.). Z.G. is partially supported by NIH Grants U01HG009431 and R01HG008140 (awarded to J. Pritchard).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901259116/-/DCSupplemental.
References
- 1.Huttley GA, Jakobsen IB, Wilson SR, Easteal S. How important is DNA replication for mutagenesis? Mol Biol Evol. 2000;17:929–937. doi: 10.1093/oxfordjournals.molbev.a026373. [DOI] [PubMed] [Google Scholar]
- 2.Kumar S, Subramanian S. Mutation rates in mammalian genomes. Proc Natl Acad Sci USA. 2002;99:803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poulos RC, Olivier J, Wong JWH. The interaction between cytosine methylation and processes of DNA replication and repair shape the mutational landscape of cancer genomes. Nucleic Acids Res. 2017;45:7786–7795. doi: 10.1093/nar/gkx463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu CI, Li WH. Evidence for higher rates of nucleotide substitution in rodents than in man. Proc Natl Acad Sci USA. 1985;82:1741–1745. doi: 10.1073/pnas.82.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang DG, Green P. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci USA. 2004;101:13994–14001. doi: 10.1073/pnas.0404142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson Sayres MA, Venditti C, Pagel M, Makova KD. Do variations in substitution rates and male mutation bias correlate with life-history traits? A study of 32 mammalian genomes. Evolution. 2011;65:2800–2815. doi: 10.1111/j.1558-5646.2011.01337.x. [DOI] [PubMed] [Google Scholar]
- 7.Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch M, et al. Genetic drift, selection and the evolution of the mutation rate. Nat Rev Genet. 2016;17:704–714. doi: 10.1038/nrg.2016.104. [DOI] [PubMed] [Google Scholar]
- 9.Crow JF. The high spontaneous mutation rate: Is it a health risk? Proc Natl Acad Sci USA. 1997;94:8380–8386. doi: 10.1073/pnas.94.16.8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Acuna-Hidalgo R, Veltman JA, Hoischen A. New insights into the generation and role of de novo mutations in health and disease. Genome Biol. 2016;17:241. doi: 10.1186/s13059-016-1110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drost JB, Lee WR. Biological basis of germline mutation: Comparisons of spontaneous germline mutation rates among drosophila, mouse, and human. Environ Mol Mutagen. 1995;64:48–64. doi: 10.1002/em.2850250609. [DOI] [PubMed] [Google Scholar]
- 12.Li W-H, Ellsworth DL, Krushkal J, Chang BHJ, Hewett-Emmett D. Rates of nucleotide substitution in primates and rodents and the generation-time effect hypothesis. Mol Phylogenet Evol. 1996;5:182–187. doi: 10.1006/mpev.1996.0012. [DOI] [PubMed] [Google Scholar]
- 13.Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 14.Strachan T, Read A. Human Molecular Genetics. 5th Ed Garland; New York: 2018. [Google Scholar]
- 15.Müller H. The nature of genetic effects produced by irradiation. In: Hollaender A, editor. Radiation Biology: Ionizing Radiations. McGraw-Hill; New York: 1954. pp. 351–473. [Google Scholar]
- 16.Gao Z, Wyman MJ, Sella G, Przeworski M. Interpreting the dependence of mutation rates on age and time. PLoS Biol. 2016;14:e1002355. doi: 10.1371/journal.pbio.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seplyarskiy VB, et al. Error-prone bypass of DNA lesions during lagging-strand replication is a common source of germline and cancer mutations. Nat Genet. 2019;51:36–41. doi: 10.1038/s41588-018-0285-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makova KD, Li WH. Strong male-driven evolution of DNA sequences in humans and apes. Nature. 2002;416:624–626. doi: 10.1038/416624a. [DOI] [PubMed] [Google Scholar]
- 19.Taylor J, Tyekucheva S, Zody M, Chiaromonte F, Makova KD. Strong and weak male mutation bias at different sites in the primate genomes: Insights from the human-chimpanzee comparison. Mol Biol Evol. 2006;23:565–573. doi: 10.1093/molbev/msj060. [DOI] [PubMed] [Google Scholar]
- 20.Thomas GWC, et al. Reproductive longevity predicts mutation rates in primates. Curr Biol. 2018;28:3193–3197.e5. doi: 10.1016/j.cub.2018.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jónsson H, et al. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature. 2017;549:519–522. doi: 10.1038/nature24018. [DOI] [PubMed] [Google Scholar]
- 22.Vogel F, Rathenberg R. Spontaneous mutation in man. Adv Hum Genet. 1975;5:223–318. doi: 10.1007/978-1-4615-9068-2_4. [DOI] [PubMed] [Google Scholar]
- 23.Ségurel L, Wyman MJ, Przeworski M. Determinants of mutation rate variation in the human germline. Annu Rev Genomics Hum Genet. 2014;15:47–70. doi: 10.1146/annurev-genom-031714-125740. [DOI] [PubMed] [Google Scholar]
- 24.Scally A. Mutation rates and the evolution of germline structure. Philos Trans R Soc B Biol Sci. 2016;371:20150137. doi: 10.1098/rstb.2015.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong WSW, et al. New observations on maternal age effect on germline de novo mutations. Nat Commun. 2016;7:10486. doi: 10.1038/ncomms10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldmann JM, et al. Parent-of-origin-specific signatures of de novo mutations. Nat Genet. 2016;48:935–939. doi: 10.1038/ng.3597. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen CT, et al. Onset of the release of spermatozoa (spermarche) in boys in relation to age, testicular growth, pubic hair, and height. J Clin Endocrinol Metab. 1986;62:532–535. doi: 10.1210/jcem-62-3-532. [DOI] [PubMed] [Google Scholar]
- 28.Rahbari R, et al. UK10K Consortium Timing, rates and spectra of human germline mutation. Nat Genet. 2016;48:126–133. doi: 10.1038/ng.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agarwal I, Przeworski M. January 15, 2019. Signatures of replication, recombination and sex in the spectrum of rare variants on the human X chromosome and autosomes. bioRxiv:10.1101/519421.
- 30.Forster P, et al. Elevated germline mutation rate in teenage fathers. Proc R Soc B Biol Sci. 2015;282:1–7. doi: 10.1098/rspb.2014.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat. 2003;21:12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- 32.Hodgkinson A, Ladoukakis E, Eyre-Walker A. Cryptic variation in the human mutation rate. PLoS Biol. 2009;7:e1000027. doi: 10.1371/journal.pbio.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Francioli LC, et al. Genome of the Netherlands Consortium Genome-wide patterns and properties of de novo mutations in humans. Nat Genet. 2015;47:822–826. doi: 10.1038/ng.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elango N, Kim SH, Vigoda E, Yi SV. Mutations of different molecular origins exhibit contrasting patterns of regional substitution rate variation. PLOS Comput Biol. 2008;4:e1000015. doi: 10.1371/journal.pcbi.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldmann JM, et al. Germline de novo mutation clusters arise during oocyte aging in genomic regions with high double-strand-break incidence. Nat Genet. 2018;50:487–492. doi: 10.1038/s41588-018-0071-6. [DOI] [PubMed] [Google Scholar]
- 38.Montgomery SB, et al. The origin, evolution, and functional impact of short insertion–deletion variants identified in 179 human genomes. Genome Res. 2013;23:749–761. doi: 10.1101/gr.148718.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kloosterman WP, et al. Genome of Netherlands Consortium Characteristics of de novo structural changes in the human genome. Genome Res. 2015;25:792–801. doi: 10.1101/gr.185041.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- 41.Fryxell KJ, Zuckerkandl E. Cytosine deamination plays a primary role in the evolution of mammalian isochores. Mol Biol Evol. 2000;17:1371–1383. doi: 10.1093/oxfordjournals.molbev.a026420. [DOI] [PubMed] [Google Scholar]
- 42.Tomkova M, Mcclellan M, Kriaucionis S, Schuster-böckler B. DNA replication and associated repair pathways are involved in the mutagenesis of methylated cytosine. DNA Repair. 2018;62:1–7. doi: 10.1016/j.dnarep.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 43.Zhu YO, Siegal ML, Hall DW, Petrov DA. Precise estimates of mutation rate and spectrum in yeast. Proc Natl Acad Sci USA. 2014;111:E2310–E2318. doi: 10.1073/pnas.1323011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Behringer MG, Hall DW. Genome-wide estimates of mutation rates and spectrum in Schizosaccharomyces pombe indicate CpG sites are highly mutagenic despite the absence of DNA methylation. G3 (Bethesda) 2015;6:149–160. doi: 10.1534/g3.115.022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharp NP, Sandell L, James CG, Otto SP. The genome-wide rate and spectrum of spontaneous mutations differ between haploid and diploid yeast. Proc Natl Acad Sci USA. 2018;115:E5046–E5055. doi: 10.1073/pnas.1801040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293 doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi H, et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res. 2013;23:616–627. doi: 10.1101/gr.148023.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gascard P, et al. Epigenetic and transcriptional determinants of the human breast. Nat Commun. 2015;6:6351. doi: 10.1038/ncomms7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goriely A. Decoding germline de novo point mutations. Nat Genet. 2016;48:823–824. doi: 10.1038/ng.3629. [DOI] [PubMed] [Google Scholar]
- 51.Polani PE, Crolla JA. A test of the production line hypothesis of mammalian oogenesis. Hum Genet. 1991;88:64–70. doi: 10.1007/BF00204931. [DOI] [PubMed] [Google Scholar]
- 52.Fulton N, Silva SJM, Bayne RAL, Anderson RA. Germ cell proliferation and apoptosis in the developing human ovary. J Clin Endocrinol Metab. 2005;90:4664–4670. doi: 10.1210/jc.2005-0219. [DOI] [PubMed] [Google Scholar]
- 53.Harland C, et al. October 9, 2016 Frequency of mosaicism points towards mutation-prone early cleavage cell divisions. bioRxiv:10.1101/079863. [Google Scholar]
- 54.Lindsay SJ, Rahbari R, Kaplanis J, Keane TM, Hurles ME. May 23, 2018 Striking differences in patterns of germline mutation between mice and humans. bioRxiv:10.1101/082297. [Google Scholar]
- 55.Huang AY, et al. Postzygotic single-nucleotide mosaicisms in whole-genome sequences of clinically unremarkable individuals. Cell Res. 2014;24:1311–1327. doi: 10.1038/cr.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Acuna-Hidalgo R, et al. Post-zygotic point mutations are an underrecognized source of de novo genomic variation. Am J Hum Genet. 2015;97:67–74. doi: 10.1016/j.ajhg.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ju YS, et al. Somatic mutations reveal asymmetric cellular dynamics in the early human embryo. Nature. 2017;543:714–718. doi: 10.1038/nature21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lim ET, et al. Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat Neurosci. 2017;20:1217–1224. doi: 10.1038/nn.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dal GM, et al. Early postzygotic mutations contribute to de novo variation in a healthy monozygotic twin pair. J Med Genet. 2014;51:455–459. doi: 10.1136/jmedgenet-2013-102197. [DOI] [PubMed] [Google Scholar]
- 60.Smith TB, et al. The presence of a truncated base excision repair pathway in human spermatozoa that is mediated by OGG1. J Cell Sci. 2013;126:1488–1497. doi: 10.1242/jcs.121657. [DOI] [PubMed] [Google Scholar]
- 61.Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332:459–461. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- 62.Dobson AT, et al. The unique transcriptome through day 3 of human preimplantation development. Hum Mol Genet. 2004;13:1461–1470. doi: 10.1093/hmg/ddh157. [DOI] [PubMed] [Google Scholar]
- 63.Zhang P, et al. Transcriptome profiling of human pre-implantation development. PLoS One. 2009;4:e7844. doi: 10.1371/journal.pone.0007844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Titus S, et al. Impairment of BRCA1-related DNA double-strand break repair leads to ovarian aging in mice and humans. Sci Transl Med. 2013;5:172ra21. doi: 10.1126/scitranslmed.3004925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei H, et al. Age-specific gene expression profiles of Rhesus monkey Ovaries detected by microarray analysis. BioMed Res Int. 2015;2015:625192. doi: 10.1155/2015/625192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jónsson H, et al. Multiple transmissions of de novo mutations in families. Nat Genet. 2018;50:1674–1680. doi: 10.1038/s41588-018-0259-9. [DOI] [PubMed] [Google Scholar]
- 67.De Iuliis GN, et al. DNA damage in human spermatozoa is highly correlated with the efficiency of chromatin remodeling and the formation of 8-hydroxy-2′-deoxyguanosine, a marker of oxidative stress. Biol Reprod. 2009;81:517–524. doi: 10.1095/biolreprod.109.076836. [DOI] [PubMed] [Google Scholar]
- 68.Lim J, Luderer U. Oxidative damage increases and antioxidant gene expression decreases with aging in the mouse ovary. Biol Reprod. 2011;84:775–782. doi: 10.1095/biolreprod.110.088583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferreira J, Carmo-Fonseca M. Genome replication in early mouse embryos follows a defined temporal and spatial order. J Cell Sci. 1997;110:889–897. doi: 10.1242/jcs.110.7.889. [DOI] [PubMed] [Google Scholar]
- 70.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 71.Ohno M, et al. 8-oxoguanine causes spontaneous de novo germline mutations in mice. Sci Rep. 2014;4:4689. doi: 10.1038/srep04689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sasani TA, et al. 2019. Large, three-generation CEPH families reveal post-zygotic mosaicism and variability in germline mutation accumulation. bioRxiv:10.1101/552117. Preprint, posted February 17, 2019.
- 73.Venn O, et al. Strong male bias drives germline mutation in chimpanzees. Science. 2014;344:1272–1275. doi: 10.1126/science.344.6189.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chang BH, Shimmin LC, Shyue SK, Hewett-Emmett D, Li WH. Weak male-driven molecular evolution in rodents. Proc Natl Acad Sci USA. 1994;91:827–831. doi: 10.1073/pnas.91.2.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang BHJ, Hewett-Emmett D, Li WH. Male-to-female ratios of mutation rate in higher primates estimated from intron sequences. Zool Stud. 1996;35:36–48. [Google Scholar]
- 76.Amster G, Sella G. Life history effects on the molecular clock of autosomes and sex chromosomes. Proc Natl Acad Sci USA. 2016;113:1588–1593. doi: 10.1073/pnas.1515798113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li W-H, Tanimura M. The molecular clock runs more slowly in man than in apes and monkeys. Nature. 1987;326:93–96. doi: 10.1038/326093a0. [DOI] [PubMed] [Google Scholar]
- 78.Kim SH, Elango N, Warden C, Vigoda E, Yi SV. Heterogeneous genomic molecular clocks in primates. PLoS Genet. 2006;2:e163. doi: 10.1371/journal.pgen.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moorjani P, Amorim CEG, Arndt PF, Przeworski M. Variation in the molecular clock of primates. Proc Natl Acad Sci USA. 2016;113:10607–10612. doi: 10.1073/pnas.1600374113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin AP, Palumbi SR. Body size, metabolic rate, generation time, and the molecular clock. Proc Natl Acad Sci USA. 1993;90:4087–4091. doi: 10.1073/pnas.90.9.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schultz MD, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523:212–216. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moorjani P, Gao Z, Przeworski M. Human germline mutation and the erratic evolutionary clock. PLoS Biol. 2016;14:e2000744. doi: 10.1371/journal.pbio.2000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.