Significance
Although constituting the majority of the transcriptional output of the human genome, the functional importance of long noncoding RNAs (lncRNAs) has only recently been recognized. The role of lncRNAs in wound healing is virtually unknown. Our study focused on a skin-specific lncRNA, termed “wound and keratinocyte migration-associated lncRNA 1” (WAKMAR1), which is down-regulated in wound-edge keratinocytes of human chronic nonhealing wounds compared with normal wounds under reepithelialization. We identified WAKMAR1 as being critical for keratinocyte migration and its deficiency as impairing wound reepithelialization. Mechanistically, WAKMAR1 interacts with DNA methyltransferases and interferes with the promoter methylation of the E2F1 gene, which is a key transcription factor controlling a network of migratory genes. This line of evidence demonstrates that lncRNAs play an essential role in human skin wound healing.
Keywords: wound healing, long noncoding RNA, keratinocyte migration
Abstract
An increasing number of studies reveal the importance of long noncoding RNAs (lncRNAs) in gene expression control underlying many physiological and pathological processes. However, their role in skin wound healing remains poorly understood. Our study focused on a skin-specific lncRNA, LOC105372576, whose expression was increased during physiological wound healing. In human nonhealing wounds, however, its level was significantly lower compared with normal wounds under reepithelialization. We characterized LOC105372576 as a nuclear-localized, RNAPII-transcribed, and polyadenylated lncRNA. In keratinocytes, its expression was induced by TGF-β signaling. Knockdown of LOC105372576 and activation of its endogenous transcription, respectively, reduced and increased the motility of keratinocytes and reepithelialization of human ex vivo skin wounds. Therefore, LOC105372576 was termed “wound and keratinocyte migration-associated lncRNA 1” (WAKMAR1). Further study revealed that WAKMAR1 regulated a network of protein-coding genes important for cell migration, most of which were under the control of transcription factor E2F1. Mechanistically, WAKMAR1 enhanced E2F1 expression by interfering with E2F1 promoter methylation through the sequestration of DNA methyltransferases. Collectively, we have identified a lncRNA important for keratinocyte migration, whose deficiency may be involved in the pathogenesis of chronic wounds.
Wound healing is a fundamental and intricate process required to recover the integrity of the skin after injury, which is achieved through a series of well-orchestrated phases: hemostasis, inflammation, proliferation, and remodeling (1). Failure of proceeding through such orderly and timely reparation leads to chronic nonhealing wounds, which often happen in patients with underlying disorders, such as venous insufficiency [venous ulcer (VU)] and diabetes mellitus [diabetic foot ulcer (DFU)] (2). Chronic wounds represent a major, and increasing, health and economic burden to our society. Current treatments for chronic ulcers focus on optimization of controllable healing factors, and efficient targeted approaches are essentially lacking (2). Better understanding of the molecular pathogenesis of chronic wounds is a prerequisite for the development of more precise and effective treatments.
Long noncoding RNAs (lncRNAs) are a large and diverse class of noncoding RNA molecules with a length of more than 200 nt. Although more than 40,000 lncRNA loci have been identified in humans, less than 1% are functionally annotated (3, 4). Study of the noncoding transcriptome reveals new and unanticipated biology, significantly advancing our understanding of normal physiology and disease pathology. Moreover, it has recently been shown that not only the expression but also the function of lncRNAs is more tissue- and cell type-specific than protein-coding genes, underscoring their great potential as precise therapeutic and diagnostic entities (5). Interestingly, emerging studies have revealed important functions of lncRNAs in skin biology [e.g., lncRNAs TINCR and ANCR can regulate epithelial differentiation (6, 7)]. In addition, several lncRNAs have been involved in the pathophysiology of skin diseases, including psoriasis (8, 9), cutaneous squamous cell carcinoma (10), and melanoma (11). However, the role of lncRNAs in skin wound healing remains unexplored.
Our study focused on a previously annotated but uncharacterized lncRNA, LOC105372576, since it was a top down-regulated lncRNA in human DFU compared with foot skin and was specifically expressed in the skin among 27 human tissues. Using both gain- and loss-of-function studies, we demonstrated that LOC105372576 is important for the motility of keratinocytes and reepithelialization of human ex vivo skin wounds; thus, it was termed “wound and keratinocyte migration-associated lncRNA 1” (WAKMAR1). Our further mechanistic studies revealed that WAKMAR1 regulated the expression of transcription factor E2F1, which is upstream of a migratory gene network. WAKMAR1 interacted with multiple DNA methyltransferases (DNMTs) and subsequently affected binding of DNMT1 and DNA methylation at the E2F1 promoter. Collectively, our study identified WAKMAR1 as a promigratory lncRNA in human keratinocytes, and its deficiency may contribute to the pathogenesis of chronic wounds.
Results
WAKMAR1 Is a Nuclear-Localized, RNAPII-Transcribed, and Polyadenylated lncRNA.
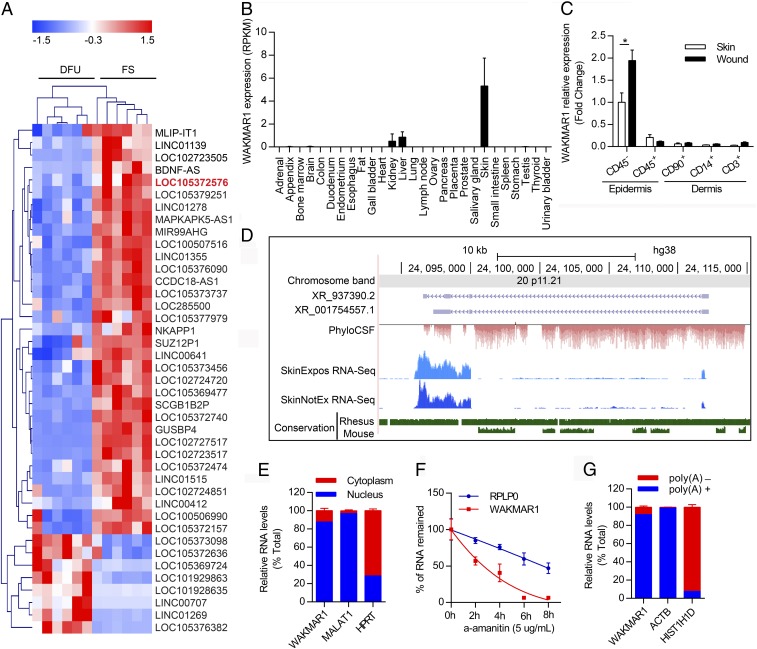
To identify lncRNAs that may be involved in the pathogenesis of human chronic wounds, we examined a recently published microarray dataset comparing human DFU with foot skin, which had not yet been analyzed for lncRNA expression (12). Forty-one lncRNAs were revealed to be significantly differentially expressed (absolute fold change ≥ 3, false discovery rate < 0.01) between DFU and skin, and WAKMAR1 was identified as a top down-regulated lncRNA in DFU (Fig. 1A). By surveying the expression pattern of these 41 differentially lncRNAs in RNA sequencing data of 27 human tissues from 95 individuals (13), we found that WAKMAR1 is predominantly present in the skin, with only scant levels in the kidney and the liver, which may be poised to exert a functional role in the skin (Fig. 1B and SI Appendix, Fig. S1). Moreover, to identify the major cell type(s) expressing WAKMAR1 in human skin and wounds, we isolated epidermal CD45− cells (mainly composed of keratinocytes) and CD45+ cells (leukocytes), dermal CD90+ cells (fibroblasts), CD14+ cells (macrophages), and CD3+ cells (T cells) from the intact skin and day 7 wounds of four healthy donors (SI Appendix, Fig. S2 and Table S1). Among these different cell types, we found that WAKMAR1 was mainly expressed in epidermal keratinocytes and its level was enhanced by wounding (Fig. 1C). Therefore, our study focused on WAKMAR1 in epidermal keratinocytes in wound healing.
Fig. 1.
WAKMAR1 is a nuclear-localized, RNAPII-transcribed, and polyadenylated lncRNA. (A) Heat map illustrates the differentially expressed lncRNAs (absolute fold change ≥ 3, false discovery rate < 0.01) in DFU compared with foot skin (FS). LOC105372576/WAKMAR1 is highlighted in red. Z-score transformation was applied for visualization. Data are from a published microarray dataset (GEO accession no. GSE80178; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse80178). (B) WAKMAR1 expression in human tissues. RPKM, reads per kilobase per million reads. (C) qPCR analysis of WAKMAR1 in epidermal CD45− and CD45+ cells and in dermal CD90+, CD14+, and CD3+ cells, which were isolated from the skin and day 7 wounds of healthy donors (n = 4). *P < 0.05, unpaired two-tailed Student’s t test. (D) Genomic snapshot of WAKMAR1 generated in RefSeq (purple), phylogenetic information-based codon substitution frequency (PhyloCSF; red), RNA-sequencing (blue) of human skin with (SkinExpos) or without sun exposure (SkinNotEx), and conservation (green) tracks. (E) qPCR analysis of WAKMAR1, MALAT1, and HPRT in the nucleus or cytoplasm of keratinocytes (n = 3). (F) qPCR analysis of WAKMAR1 and RPLP0 mRNA in keratinocytes treated with α-amanitin (n = 3). (G) qPCR analysis of WAKMAR1, ACTB, and HIS1H1D in Poly(A)+ and Poly(A)− RNA fractions from keratinocytes (n = 3). Data are presented as mean ± SEM (B and C) or mean ± SD (E–G).
WAKMAR1 is annotated in the human genome with two isoforms, XR_937390.2 and XR_001754557.1 [GRCh38/hg38, chromosome 20 (chr20):24,092,278–24,112,234; genome.ucsc.edu] (14) (Fig. 1D). It has no homolog in nonprimate species, including mice (Fig. 1D). Phylogenetic information-based codon substitution frequency analysis suggested that WAKMAR1 had no protein-coding potential, which is in line with the analysis result of the Coding Potential Calculator (15) (SI Appendix, Fig. S3A). Using a cellular fractionation assay, we detected WAKMAR1 mainly in nuclear extracts, which is similar to the well-known nuclear noncoding RNA MALAT1 (16) and distinct from HPRT mRNA, which is mainly present in cytoplasm (17) (Fig. 1E). To identify which RNA polymerase is responsible for WAKMAR1 transcription, we treated human primary keratinocytes with α-amanitin at a concentration (5 μg/mL) that has been shown to specifically inhibit RNA polymerase II (RNAPII) (18). As expected, the expression of RNAPI-transcribed 28S and 18S rRNAs was not affected by α-amanitin treatment (SI Appendix, Fig. S3B). The levels of WAKMAR1 and a known RNAPII-transcribed mRNA, RPLP0, were decreased by α-amanitin, suggesting that WAKMAR1 is also transcribed by RNAPII (Fig. 1F). In addition, we determined a half-life of 3.5 h for WAKMAR1 in keratinocytes. This was further confirmed by treating the cells with actinomycin-D (5 μg/mL), which blocks total cellular transcription (18) (SI Appendix, Fig. S3C). Furthermore, we examined the polyadenylation status of WAKMAR1 by separating Poly(A)+ and Poly(A)− RNA. The qPCR results showed that WAKMAR1 was a polyadenylated transcript similar to ACTB mRNA but, in contrast to HIST1H1D, a known nonpolyadenylated RNA (19) (Fig. 1G). Collectively, we identified WAKMAR1 as a nuclear localized, RNAPII-transcribed, and polyadenylated noncoding transcript in human primary keratinocytes.
WAKMAR1 Expression Is Reduced in Wound-Edge Keratinocytes of Human Chronic Wounds.
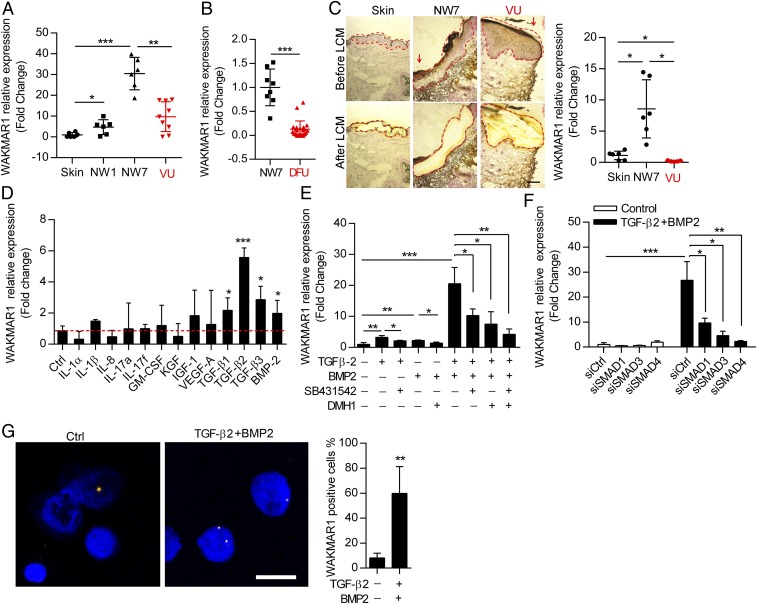
Next, we sought to validate and explore the expression pattern of WAKMAR1 in human normal wounds and in different types of chronic wounds. To this end, we created an excisional normal wound (NW) on the skin of healthy donors and collected wound edges 1 d (NW1) or 7 d (NW7) later, which represented wounds in the inflammatory or proliferative phase of wound repair (1) (SI Appendix, Fig. S2 and Table S1). In addition, we collected chronic nonhealing wound-edge biopsies from patients with VU or DFU (SI Appendix, Fig. S2 and Tables S2 and S3). Expression analysis by qPCR showed that the expression of WAKMAR1 was increased during skin wound healing (Fig. 2A; the intact skin < NW1 < NW7); however, its level was significantly lower in VU and DFU compared with the day 7 wounds from healthy donors (Fig. 2 A and B). Furthermore, we used laser capture microdissection (LCM) to isolate epidermis, which predominantly consists of keratinocytes (20). We could confirm that WAKMAR1 expression was induced in keratinocytes by skin injury (Fig. 2C), as previously shown in Fig. 1C. Interestingly, we found that the WAKMAR1 expression in the wound-edge epidermis of VU was significantly lower compared with the day 7 normal wounds and the skin (Fig. 2C). Based on its increased expression during healing and its loss in chronic wounds, we hypothesize that WAKMAR1 may play an important role in wound repair.
Fig. 2.
WAKMAR1 expression is decreased in wound-edge keratinocytes of human chronic wounds and regulated by TGF-β. (A) qPCR analysis of WAKMAR1 in the skin, in day 1 (NW1) and day 7 (NW7) normal wounds from six healthy donors, and in wound edges of nine patients with VU. (B) qPCR analysis of WAKMAR1 in NW7 biopsies from eight healthy donors and wound edges of 29 patients with DFU. (C) qPCR analysis of WAKMAR1 in wound-edge epidermis isolated from the healthy skin and NW7 (n = 6) and VU (n = 5) using LCM. (Scale bar, 200 μm.) Red arrows indicate wound edges. (D) qPCR analysis of WAKMAR1 in keratinocytes treated with wound-related cytokines/growth factors for 24 h (n = 3). Ctrl, control. (E) TGF-β receptor inhibitor SB431542 and/or BMP receptor inhibitor DMH1 was applied 15 min before adding TGF-β2 and/or BMP2 to keratinocytes, and WAKMAR1 was analyzed by qPCR 24 h later (n = 3). (F) qPCR analysis of WAKMAR1 in keratinocytes transfected with SMAD1-, SMAD3-, and SMAD4-specific siRNAs for 24 h and then treated with TGF-β2 and BMP2 for 24 h (n = 3). (G) ISH of WAKMAR1 in keratinocytes treated with TGF-β2 and BMP2 for 24 h. Cell nuclei were costained with DAPI. (Scale bar, 50 μm.) WAKMAR1+ cells were counted. *P < 0.05; **P < 0.01; ***P < 0.001 by Mann–Whitney U test (A–C) and unpaired two-tailed Student’s t test (D–G). Data are presented as mean ± SEM (A–C) or mean ± SD (D–G).
WAKMAR1 Expression Is Induced by TGF-β Signaling in Keratinocytes.
To understand the mechanism(s) regulating WAKMAR1 expression in wound-edge keratinocytes, we treated human primary keratinocytes with a variety of cytokines (IL-1α, IL-1β, IL-8, IL-17a, IL-17f, and GM-CSF) and growth factors (KGF, IGF1, VEGF-A, TGF-β1, TGF-β2, TGF-β3, and BMP-2) that are typically present in the wound environment. The qPCR analysis showed that several members of the TGF-β superfamily (i.e., TGF-β1, TGF-β2, TGF-β3, BMP-2) significantly induced WAKMAR1 expression in keratinocytes (Fig. 2D). As two branches of TGF-β signaling, TGF-β and BMP, engage different receptors and SMAD transcription factors (21), we further explored the contribution of each signaling pathway. Interestingly, we observed a synergistic effect of TGF-β2 and BMP-2 on the induction of WAKMAR1 expression (Fig. 2E). Importantly, inhibition of the TGF-β type I receptor (TGFBR1) with SB431542 and/or blockade of the BMP type I receptors (BMPR1A and BMPR1B) with DMH1 mitigated TGF-β signaling-induced WAKMAR1 expression (22, 23) (Fig. 2E), suggesting that both TGF-β and BMP branches are engaged in WAKMAR1 regulation. In line with this, silencing the expression of SMAD1 or SMAD3, which is specific to the BMP and TGF-β branches, respectively, or SMAD4, which is shared by both branches, with siRNAs (SI Appendix, Fig. S4) significantly decreased the WAKMAR1 expression induced by TGF-β signaling (Fig. 2F). Furthermore, we visualized WAKMAR1 expression in keratinocytes by in situ hybridization (ISH), which confirmed its nuclear localization and its increased expression after the treatment with TGF-β2 and BMP-2 (Fig. 2G and SI Appendix, Fig. S5). Altogether, these findings demonstrate that WAKMAR1 expression is induced by TGF-β signaling in keratinocytes. This is of special interest since the TGF-β signaling pathway has been known to be activated during wound healing, whereas it is deficient in human chronic wounds, such as VU and DFU, which might explain the lack of WAKMAR1 expression in chronic wounds (24, 25).
WAKMAR1 Regulates Keratinocyte Motility and Reepithelialization of Human ex Vivo Wounds.
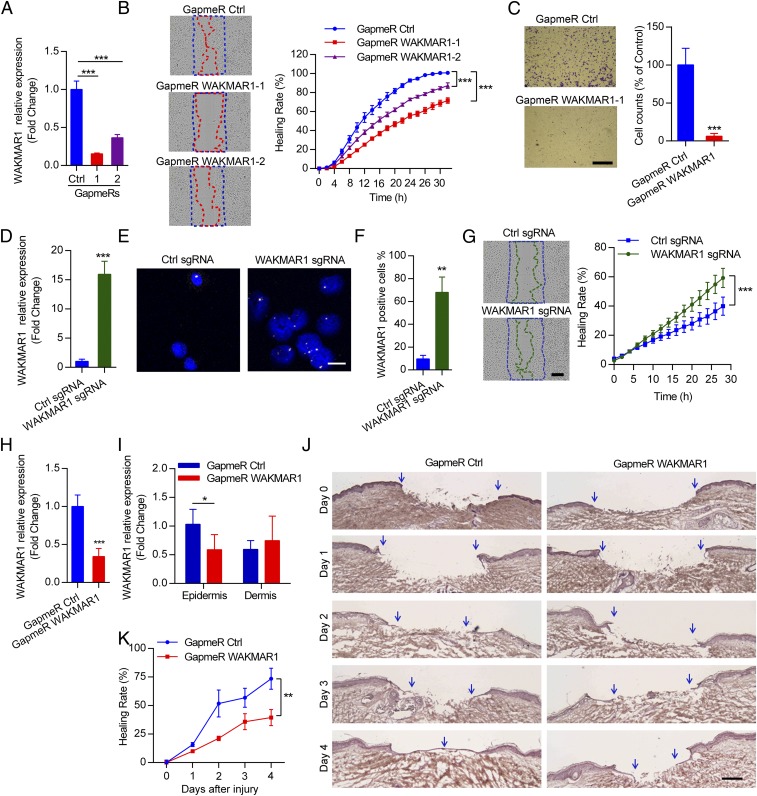
Keratinocyte migration is pivotal for wound reepithelialization, which fails in chronic wounds (26). To study the effect of WAKMAR1 on cell motility, we knocked down WAKMAR1 expression by transfecting human primary keratinocytes with locked nucleic acid-modified antisense oligonucleotides (GapmeRs), which can enter the cell nucleus and catalyze RNase H-dependent degradation of cRNA targets. Two GapmeRs were designed to target different sites of WAKMAR1, and both significantly decreased its expression in keratinocytes, as confirmed by qPCR analysis (Fig. 3A). Using scratch wound assays, we found that knockdown of WAKMAR1 significantly delayed wound closure (Fig. 3B and Movie S1). In line with this, haptotactic transwell migration assay showed that lack of WAKMAR1 significantly reduced the migratory capacity of keratinocytes (Fig. 3C). In addition to knockdown, we activated endogenous WAKMAR1 transcription in human primary keratinocytes using a CRISPR/Cas9 synergistic activation mediator (SAM) system (27). To this end, we designed five single-guide RNAs (sgRNAs) targeting various regions of the WAKMAR1 promoter and found that sgRNA1 significantly increased WAKMAR1 expression, as shown by qPCR and ISH (Fig. 3 D–F and SI Appendix, Figs. S5 and S6). Of note, in ISH analysis, we observed that WAKMAR1 signal presented as a single dot in a nucleus; however, two or more foci were visible after keratinocytes were transfected with CRISPR/Cas9-SAM plasmids (Fig. 3E and SI Appendix, Fig. S5C). Enhanced WAKMAR1 expression significantly accelerated wound closure, as shown in scratch wound assays (Fig. 3G and Movie S2). Moreover, we showed that knockdown or activation of WAKMAR1 expression in keratinocytes did not affect cell proliferation or viability, demonstrating that the effect of WAKMAR1 on keratinocyte migration is not due to altered cell growth or death (SI Appendix, Fig. S7).
Fig. 3.
WAKMAR1 regulates keratinocyte motility and wound reepithelialization. (A) qPCR analysis of WAKMAR1 in keratinocytes transfected with WAKMAR1-specific GapmeR1, GapmeR2, or control oligos (Ctrl) (n = 3). (B) Scratch wound assay of keratinocytes after WAKMAR1 knockdown (n = 10). (C) Representative photographs of transwell migration assay for keratinocytes with WAKMAR1 knockdown (n = 3). (Scale bar, 1 mm.) The number of cells passing through the membrane was counted. qPCR (D) and ISH (E and F) of WAKMAR1 in keratinocytes transfected with CRISPR/Cas9-SAM plasmids for 48 h are shown. Cell nuclei were costained with DAPI. (Scale bar, 50 μm.) (G) Scratch wound assay of keratinocytes with WAKMAR1 expression activation (n = 10). (Scale bar, 300 μm.) qPCR analyses of WAKMAR1 in full-depth biopsies (H) and in LCM-isolated epidermis and dermis (I) of human ex vivo wounds after topical application of WAKMAR1 GapmeRs for 4 d (n = 6 donors) are shown. (J) Representative photographs of hematoxylin and eosin staining of ex vivo wounds. Blue arrows demarcate the initial wound edges (day 0) and newly formed epidermis (days 1–4). (Scale bar, 200 μm.) (K) Reepithelialization was quantified as healing rate = 100% − percentage of the initial wound size. *P < 0.05; **P < 0.01; ***P < 0.001 by unpaired two-tailed Student’s t test (A, C, D, F, H, and I) or two way-ANOVA (B, G, and K). Data are presented as mean ± SD and are representative of at least two independent experiments.
Next, we examined the effect of WAKMAR1 on wound reepithelialization using a human ex vivo wound model. Briefly, excisional wounds were made on the excised human skin and cultured for reepithelialization in 4–7 d (28). We applied the WAKMAR1-specific GapmeRs mixed with thermos-reversible F-127 hydrogel topically on the wounds immediately following injury and then on every other day for 5 d, which effectively reduced WAKMAR1 levels in human ex vivo wounds (Fig. 3H). Moreover, using LCM to separate the epidermal and dermal compartments of the treated wounds, we found that the reduced WAKMAR1 level by GapmeR treatment mainly occurred in the epidermis (Fig. 3I). Importantly, inhibition of WAKMAR1 significantly delayed reepithelialization of human ex vivo wounds (Fig. 3 J and K and SI Appendix, Fig. S8 and Table S1). Together, our study identified WAKMAR1 as an important positive regulator of keratinocyte motility, and the deficiency of WAKMAR1 may, at least partially, contribute to the failure of reepithelialization in chronic wounds.
WAKMAR1 Regulates a Gene Network Mediating Its Promigratory Function in Keratinocytes.
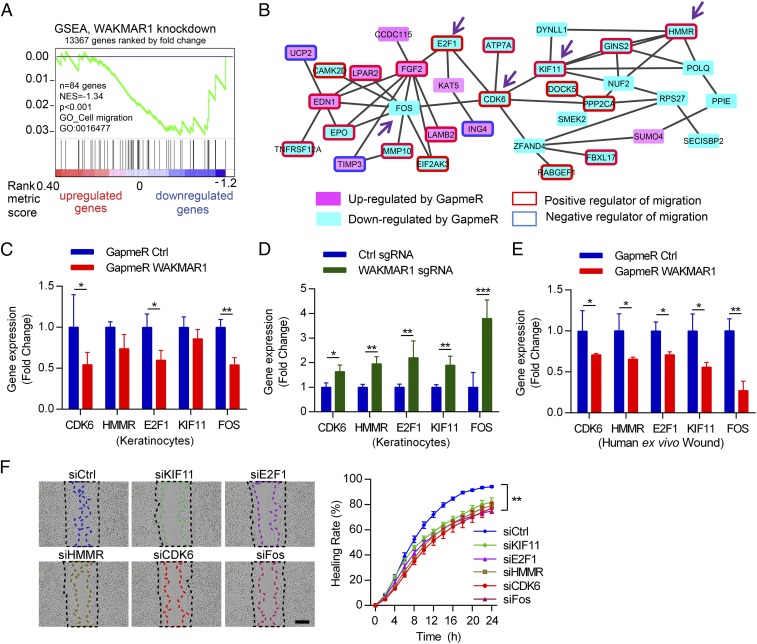
To gain more insight into the mechanisms underlying the biological function of WAKMAR1, we performed a global transcriptomic analysis in human primary keratinocytes upon WAKMAR1 knockdown (29). The microarray analysis identified 119 differentially expressed genes upon the knockdown of WAKMAR1 (absolute fold change ≥ 1.3, P < 0.05), and a large majority (86 of 119) of them were down-regulated (SI Appendix, Fig. S9). Gene set enrichment analysis (GSEA) (30) revealed that a set of cell migration-related genes (Gene Ontology accession GO:0016477) was significantly (P < 0.001) enriched among the genes down-regulated by WAKMAR1 GapmeR (Fig. 4A). Using the STRING network analysis (31), we constructed a functional protein association network with the WAKMAR1-regulated genes (Fig. 4B). Of note, many genes within this network have been previously involved in regulating cell migration. Here, we validated the expression of five hub genes: CDK6, HMMR, E2F1, KIF11, and FOS. They were decreased by WAKMAR1 GapmeR in human primary keratinocytes and ex vivo wound models, whereas activation of WAKMAR1 expression increased their expression (Fig. 4 C–E). Moreover, silencing the expression of these five hub genes with gene-specific siRNAs resulted in significantly slower migration of keratinocytes and mimicked the effect of WAKMAR1 knockdown, suggesting that this gene network may mediate the biological function of WAKMAR1 in keratinocytes (Fig. 4F and Movie S3).
Fig. 4.
WAKMAR1 regulates a gene network mediating its promigratory function in keratinocytes. Microarray analysis was performed in human keratinocytes with WAKMAR1 knockdown (n = 3). (A) GSEA evaluated enrichment for the cell migration-related genes in the microarray data. NES, normalized enrichment score. (B) Functional protein association network was identified by STRING APP in Cytoscape software among the genes regulated by WAKMAR1 (absolute fold change ≥ 1.3, P < 0.05). Genes up- or down-regulated by WAKMAR1 GapmeR are colored in pink or cyan, respectively. Genes previously reported to promote or inhibit cell migration are highlighted with red or blue frames, respectively. The expression of CDK6, HMMR, E2F1, KIF11, and FOS was analyzed by qPCR in keratinocytes transfected with WAKMAR1 GapmeRs (C), or with CRISPR/Cas9-SAM plasmids (D), and in human ex vivo wounds treated with WAKMAR1 GapmeRs (E). (F) Scratch wound assay of keratinocytes transfected with siRNAs specific to KIF11, E2F1, HMMR, CDK6, or FOS (n = 8). (Scale bar, 300 μm.) *P < 0.05; **P < 0.01; ***P < 0.001 by unpaired two-tailed Student’s t test (C–E) or two way-ANOVA (F). Data are presented as mean ± SD and are representative of at least two independent experiments.
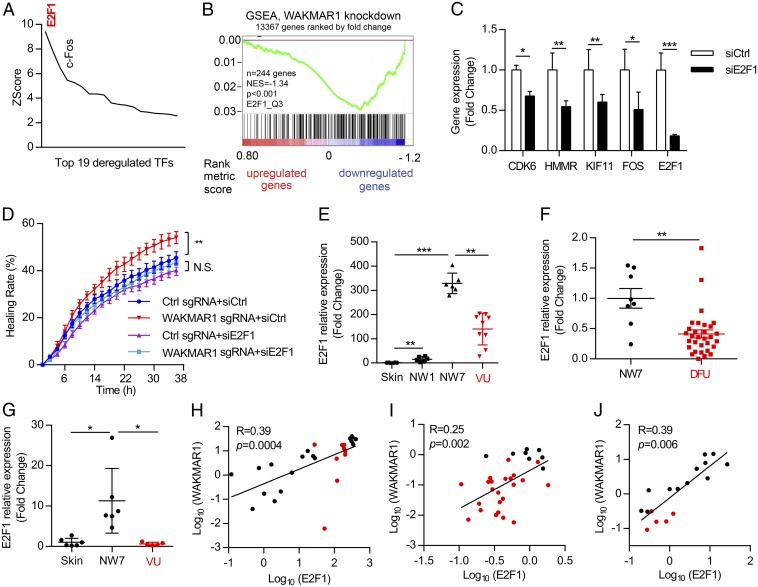
To determine if the WAKMAR1-regulated genes were coordinately controlled by common transcription factors, we performed MetaCore analysis, which identified E2F1 as the top WAKMAR1-regulated transcription factor with its targets overrepresented in the differentially expressed genes (Fig. 5A). In line with this, GSEA revealed that the E2F1-regulated genes [gene set E2F1_Q3 from Molecular Signatures Database v6.1 (32)] were significantly (P < 0.001) enriched among the genes down-regulated by WAKMAR1 GapmeR (Fig. 5B). Silencing of E2F1 expression also reduced the levels of the aforementioned CDK6, HMMR, KIF11, and FOS (Fig. 5C), indicating that E2F1 acts as an upstream regulator within the WAKMAR1-regulated gene network. We further addressed this functionally, and we found that silencing of E2F1 completely abolished the promigratory effect of WAKMAR1, suggesting that E2F1 is a key protein-coding gene mediating the biological function of WAKMAR1 in keratinocytes (Fig. 5D and SI Appendix, Fig. S10). Importantly, we showed that the expression of both WAKMAR1 and E2F1 was reduced in human chronic nonhealing wounds (i.e., VU, DFU) compared with normal day 7 wounds under the healing process (Fig. 5 E–G), and there was a significant positive correlation between their levels (Fig. 5 H–J), supporting that WAKMAR1 regulates E2F1 expression in vivo.
Fig. 5.
E2F1 acts as an upstream regulator in the WAKMAR1-regulated gene network. (A) MetaCore analysis identified transcription factors (TFs) with overrepresented binding sites among WAKMAR1-regulated genes. Only TFs with changed expression after WAKMAR1 knockdown are shown. (B) GSEA evaluated the enrichment of E2F1-target genes in the WAKMAR1-regulated genes. NES, normalized enrichment score. (C) qPCR analysis of CDK6, HMMR, KIF11, FOS, and E2F1 in keratinocytes transfected with E2F1 siRNA for 24 h (n = 3). (D) Scratch wound assay of keratinocytes after WAKMAR1 knockdown and/or E2F1 silencing (n = 8). qPCR analyses of E2F1 in the skin in day 1 (NW1) and day 7 (NW7) normal wounds from six healthy donors and in the wound edges of nine patients with VU (E and H) in NW7 (n = 8) vs. DFU (n = 29) (F and I) and in wound-edge epidermis isolated from healthy skin and NW7 (n = 6) and VU (n = 5) with LCM (G and J) are shown. (H–J) Expression correlation of WAKMAR1 with E2F1 in human skin, normal wounds (black dots), and chronic wounds (red dots). *P < 0.05; **P < 0.01; ***P < 0.001 by unpaired two-tailed Student’s t test (C), two way-ANOVA (D), Mann–Whitney U test (E–G), or Pearson’s correlation test (H–J). N.S., not significant. Data are presented as mean ± SD (C) or mean ± SEM (D–G).
WAKMAR1 Activates E2F1 Expression by Inhibiting Methylation of E2F1 Promoter.
The highly localized expression pattern of WAKMAR1 in the keratinocyte nucleus, as shown by ISH (Figs. 2G and 3E and SI Appendix, Fig. S5 B and C), suggests that it may act in a cis-acting manner. To this end, we assessed whether WAKMAR1 regulates the expression of genes at the local genomic region neighboring the WAKMAR1 locus. In the microarray data from WAKMAR1 knockdown keratinocytes, we found E2F1 as the top down-regulated gene among all of the genes encoded at chr20 (Fig. 6A). Since E2F1 is also an upstream regulator of the gene network mediating WAKMAR1’s biological function in keratinocytes, we next investigated how WAKMAR1 modulated E2F1 expression.
Fig. 6.
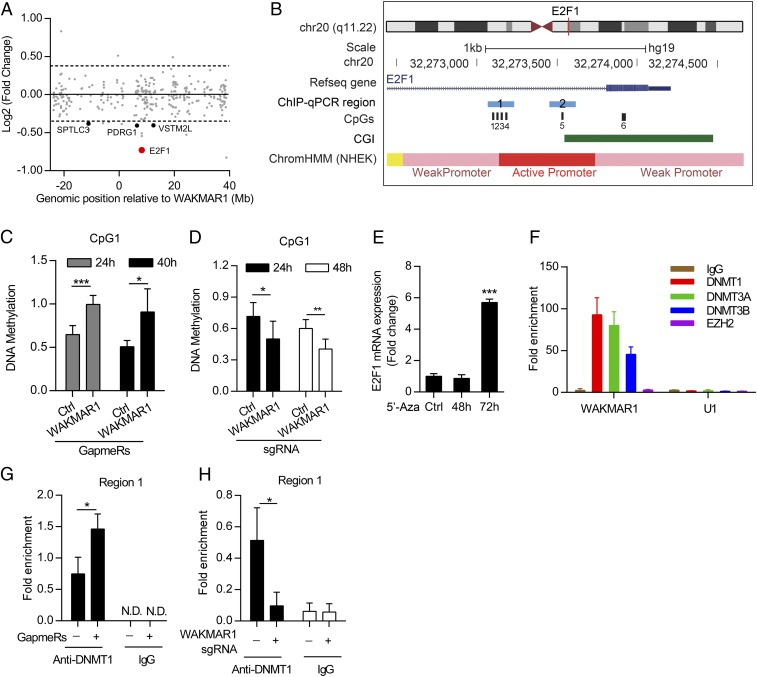
WAKMAR1 activates E2F1 expression by suppressing E2F1 promoter methylation. (A) Chr20 gene expression in keratinocytes with WAKMAR1 knockdown. The gene start distance from the WAKMAR1 transcriptional start site is shown on the x axis. The log-twofold change of expression levels between control and WAKMAR1 knockdown is shown on the y axis. Genes with absolute fold change ≥ 1.3 and P < 0.05 are highlighted. (B) Genomic snapshot of E2F1 promoter: CpG sites (black bars) analyzed by MSRE-qPCR (CpG1, CpG5, and CpG6) and bisulfite pyrosequencing (CpG1, CpG2, CpG3, and CpG4) and regions analyzed by ChIP-qPCR (blue bars) are highlighted. CGI, CpG island; ChromHMM, chromatin state segmentation by hidden Markov model from ENCODE/Broad; NHEK, normal human epidermal keratinocytes. MSRE-qPCR analyses of DNA methylation at CpG1 in keratinocytes transfected with WAKMAR1 GapmeR for 24 h and 40 h (n = 6) (C) or CRISPR/Cas9-SAM plasmids for 24 h and 48 h (n = 6) (D) are shown. Ctrl, control. (E) qPCR of E2F1 in keratinocytes treated with 5′-Aza-2′-deoxycytidine (5′-Aza) (n = 3). (F) qPCR analysis of WAKMAR1 and U1 small nuclear RNA immunoprecipitated from keratinocytes with DNMT1, DNMT3A, DNMT3B, and EZH2 antibodies or IgG (n = 3). ChIP-qPCR of E2F1 promoter region 1 was performed in keratinocytes transfected with WAKMAR1 GapmeRs (G) or CRISPR/Cas9-SAM plasmids (H) and immunoprecipitated using DNMT1 antibody or IgG (n = 3). *P < 0.05; ***P < 0.001 by unpaired two-tailed Student’s t test (C–E, G, and H). N.D., not detected. Data are presented as mean ± SD.
As most nuclear lncRNAs associate with chromatin and regulate gene expression through epigenetic mechanisms (33), we analyzed DNA methylation of the predicted active promoter of E2F1 in human primary keratinocytes (GRCh37/hg19, chr20:32,273,140–32,273,739) (Fig. 6B). Using methyl-sensitive restriction enzyme (MSRE) qPCR, we showed that knockdown of WAKMAR1 increased the degree of methylation at CpG1 and CpG5 in this region, whereas up-regulation of WAKMAR1 expression decreased it (Fig. 6 B–D and SI Appendix, Fig. S11 A and B). The change of CpG1 methylation by WAKMAR1 was further confirmed by using bisulfite-pyrosequencing of a 100-bp region encompassing CpG1, with a tendency for a similar change at three adjacent CpGs (CpG2–CpG4; SI Appendix, Fig. S11 E–G). Of note, no difference in DNA methylation could be detected upstream of this region (CpG6; SI Appendix, Fig. S11 C and D). These data suggest that DNA methylation may mediate the effect of WAKMAR1 on E2F1 expression, which was supported by significantly increased E2F1 mRNA levels in keratinocytes following treatment with demethylating agent 5′-Aza-2′-deoxycytidine (Fig. 6E).
This result prompted us to speculate that WAKMAR1 may physically interact with DNMTs, key enzymes in establishing and maintaining genomic methylation patterns (34). Supporting this idea, DNMTs, but not the cytoplasmic protein HPRT, were predicted to interact with WAKMAR1 by a web-based algorithm, LncPro (35) (SI Appendix, Fig. S12). To test this hypothesis, we performed RNA immunoprecipitation (RIP) in human primary keratinocytes using antibodies against DNMT1, DNMT3A, DNMT3B, and EZH2 (a histone-lysine N-methyltransferase enzyme) (Fig. 6F). Notably, antibodies against all three DNMTs retrieved significant amounts of WAKMAR1. However, EZH2 did not show any interaction with WAKMAR1. As another negative control, U1 small nuclear RNA was not retrieved by any of these antibodies, indicating a specific interaction between WAKMAR1 and DNMTs (Fig. 6F). Furthermore, chromatin immunoprecipitation (ChIP) coupled to detection by real-time qPCR (ChIP-qPCR) analysis revealed that knockdown of WAKMAR1 increased, whereas activation of WAKMAR1 decreased, the binding of DNMT1, but not DNMT3A and DNMT3B, to the two aforementioned CpG1 and CpG5 regions at the E2F1 promoter (Fig. 6 B, G, and H and SI Appendix, Fig. S13), which explained the changed degree of methylation in these regions (Fig. 6 C and D and SI Appendix, Fig. S11). Together, our data suggested a model in which WAKMAR1 RNA sequesters DNMTs, interfering with methylation of the E2F1 promoter and thus enhancing its transcription (SI Appendix, Fig. S14).
Discussion
Our study identified WAKMAR1 as a critical promigratory lncRNA in human wound-edge keratinocytes. It acts through activation of E2F1 expression, a key transcription factor upstream of a migratory gene network, by sequestering DNMTs and interfering with methylation of the E2F1 promoter. This finding underscores the importance of epigenetic regulation in skin wound healing. Epigenetic mechanisms, including covalent DNA and histone modifications, as well as chromatin remodeling, have been shown to be essential for homeostatic skin maintenance; however, their role in the wound healing response remains largely unexplored (36). Recent studies unravel lncRNAs as major players in epigenetic regulation by interacting with histone modifiers or chromatin remodelers, while their role in regulating DNA methylation is less understood (37). As DNMTs lack sequence specificity, several mechanisms have been involved in determining the DNA regions modified by DNMTs, such as interaction of DNMTs with histone modifiers or transcription factors that can recognize specific genomic loci (38–41). Interestingly, several lncRNAs [e.g., ecCEBP (42), Dali (43), Dum (44), Dacor1 (45), LincRNA-p21 (44)] have been recently discovered to interact physically with DNMTs and regulate sequence-specific DNA methylation. Our finding identified WAKMAR1 as a DNMT-associated lncRNA in humans. Similar to ecCEBP and Dali, WAKMAR1 sequesters DNMTs and inhibits promoter methylation of its target gene. As DNMT1 has higher binding affinity with structured RNA than its DNA substrate, lncRNA ecCEBP has been demonstrated to act as a shield halting DNMT1 (42). Here, we speculate a similar mechanism; that is, WAKMAR1 may compete for binding to DNMTs with either E2F1 promoter or protein cofactors (e.g., transcription factors) that are important in loading and orienting DNMTs on DNA.
Here, we show that WAKMAR1 regulates methylation of the E2F1 promoter, which is 9 Mb away from WAKMAR1 locus. This evidence sets WAKMAR1 apart from ecCEBP that is regulating its own parental locus (42). A low degree of complementarity between the WAKMAR1 sequence and E2F1 promoter indicates that direct RNA/DNA interaction is unlikely. We hypothesize that chromatin looping, which brings transcriptional regulatory elements and promoters together (46), may enable WAKMAR1-mediated regulation of E2F1 over such a long distance. In line with this, we noticed that E2F1 expression was decreased after silencing of RAD21 and NIBPL (SI Appendix, Fig. S15), which are critical factors for chromatin occupancies of the cohesion complex, facilitating enhancer–promoter looping (47–49). Further study to determine if disruption of the long-range chromatin interaction may impair WAKMAR1-mediated activation of E2F1 expression is warranted.
E2F1 is an essential transcription factor in skin wound healing, as its deficiency delays wound repair in mice (50). In line with previous findings (50), we show that lack of E2F1 in keratinocytes impairs cell migration, which reproduces the effects of WAKMAR1 knockdown. Moreover, silencing of E2F1 completely abolished the promigratory effect of WAKMAR1, strongly supporting that the biological function of WAKMAR1 is mediated through E2F1. Interestingly, increased E2F1 expression has been found in migrating keratinocytes at the wound edge compared with the resting epidermis of human skin (50). Here, we observed increased E2F1 expression during wound healing, whereas its level in human chronic wounds was lower compared with the normal wounds under reepithelialization. These findings suggest that well-controlled E2F1 expression may be critical in wound repair. In addition, we found that WAKMAR1 expression was induced by TGF-β, which is an indispensable signaling pathway driving wound healing (51). Keratinocytes lacking E2F1 exhibited a blunted chemotactic response to TGF-β1 (50), which prompted us to speculate that WAKMAR1-mediated regulation of E2F1 may serve as a key link underlying the keratinocyte chemotactic response to TGF-β.
As WAKMAR1 is a recently evolved, nonconserved human lncRNA, our study provides human-specific mechanistic insights into skin wound healing. Intriguingly, the vast majority of human lncRNAs are not conserved in nonprimate species (52), while little is known about their clinical and functional relevance. Investigation of these recently evolved genomic regulatory features may open new opportunities in translational medicine. However, it is challenging to perform in vivo studies due to the lack of homologous mechanisms in rodents. Here, we applied a human ex vivo wound model and demonstrated the critical role of WAKMAR1 in wound reepithelialization in a complex and in vivo-like setting. Further studies using human-relevant in vivo models, such as human skin transplant in mouse models (53), and nonhuman primate studies will be helpful to fully understand the in vivo function of WAKMAR1 and other nonconserved human lncRNAs.
Collectively, we identified a lncRNA WAKMAR1 critical for human keratinocyte migration, and its deficiency impaired reepithelialization of human wounds. This line of evidence shows that lncRNAs play an essential functional role in human skin wound healing. Moreover, our findings underscore the importance of lncRNAs in regulating locus-specific DNA methylation, which may offer the possibility to correct aberrant DNA methylation at specific genomic loci. Further exploration of the epigenetic regulatory network in human skin wound healing will open exciting opportunities for developing more effective wound therapy.
Materials and Methods
Written informed consent was obtained from all donors for the collection and use of clinical samples. The study was approved by the Stockholm Regional Ethics Committee and the Ethics Committee of the Second Hospital of Dalian Medical University. The study was conducted according to the Declaration of Helsinki’s principles.
Detailed materials and methods used for human wound sample collection; cell culture; RNA extraction and qPCR; magnetic cell separation; ISH; CRISPR-SAM; analysis of cell motility, proliferation, and viability; human ex vivo wound modeling; gene expression microarray; LCM; cell fractionation; polyadenylation analysis; MSRE-qPCR; bisulfite pyrosequencing; RIP assay; and ChIP assay are provided in SI Appendix, Supplemental Experimental Procedures. Oligos and reagents used in this study are listed in SI Appendix, Table S4.
The microarray data of human primary keratinocytes with WAKMAR1 knockdown have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (accession no. GSE114908) (29). Published microarray data of comparison of human DFU with foot skin are available in the GEO (accession no. GSE80178; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse80178).
Supplementary Material
Acknowledgments
We thank the patients and healthy donors participating in this study. We thank Francesco Marabita, Helena Griehsel, Maria Lundqvist, Samudyata, and Gonçalo Castelo-Branco for technical support and constructive discussion of the manuscript, and we thank Desiree Wiegleb Edström, Peter Berg, Fredrik Correa, Martin Gumulka, and Mahsa Tayefi for clinical sample collection. We thank the Bioinformatics and Expression Analysis (BEA) core facility at Novum, which is supported by the board of research at the Karolinska Institute and the research committee at the Karolinska University Hospital. This work was supported by the Swedish Research Council (Vetenskapsradet, Dnr. 2015-06246 and 2016-02051), Ragnar Söderbergs Foundation (M31/15), National Natural Science Foundation of China (Grant 81611130075), Hedlunds Foundation, Welander and Finsens Foundation (Hudfonden), Åke Wibergs Foundation, Jeanssons Foundation, Swedish Psoriasis Foundation, Ming Wai Lau Centre for Reparative Medicine, Tore Nilsons Foundation, Lars Hiertas Foundation, and Karolinska Institute. L.K. was supported by a fellowship from the Margaretha af Ugglas Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences and metadata reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE114908).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814097116/-/DCSupplemental.
References
- 1.Reinke JM, Sorg H. Wound repair and regeneration. Eur Surg Res. 2012;49:35–43. doi: 10.1159/000339613. [DOI] [PubMed] [Google Scholar]
- 2.Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: Mechanisms, signaling, and translation. Sci Transl Med. 2014;6:265sr6. doi: 10.1126/scitranslmed.3009337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volders PJ, et al. An update on LNCipedia: A database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:4363–4364. doi: 10.1093/nar/gkv295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu SJ, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355:aah7111. doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kretz M, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonkoly E, et al. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J Biol Chem. 2005;280:24159–24167. doi: 10.1074/jbc.M501704200. [DOI] [PubMed] [Google Scholar]
- 9.Széll M, Danis J, Bata-Csörgő Z, Kemény L. PRINS, a primate-specific long non-coding RNA, plays a role in the keratinocyte stress response and psoriasis pathogenesis. Pflugers Arch. 2016;468:935–943. doi: 10.1007/s00424-016-1803-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponzio G, et al. A new long noncoding RNA (lncRNA) is induced in cutaneous squamous cell carcinoma and down-regulates several anticancer and cell differentiation genes in mouse. J Biol Chem. 2017;292:12483–12495. doi: 10.1074/jbc.M117.776260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hulstaert E, Brochez L, Volders PJ, Vandesompele J, Mestdagh P. Long non-coding RNAs in cutaneous melanoma: Clinical perspectives. Oncotarget. 2017;8:43470–43480. doi: 10.18632/oncotarget.16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramirez HA, et al. Staphylococcus aureus triggers induction of miR-15B-5P to diminish DNA repair and deregulate inflammatory response in diabetic foot ulcers. J Invest Dermatol. 2018;138:1187–1196. doi: 10.1016/j.jid.2017.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fagerberg L, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong L, et al. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doose G, et al. ICGC MMML-Seq Consortium MINCR is a MYC-induced lncRNA able to modulate MYC’s transcriptional network in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 2015;112:E5261–E5270. doi: 10.1073/pnas.1505753112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 19.Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolarsick PAJ, Kolarsick MA, Goodwin C. Anatomy and physiology of the skin. J Dermatol Nurses Assoc. 2011;3:203–213. [Google Scholar]
- 21.Massagué J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 22.Hao J, et al. In vivo structure-activity relationship study of dorsomorphin analogues identifies selective VEGF and BMP inhibitors. ACS Chem Biol. 2010;5:245–253. doi: 10.1021/cb9002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inman GJ, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 24.Pastar I, et al. Attenuation of the transforming growth factor beta-signaling pathway in chronic venous ulcers. Mol Med. 2010;16:92–101. doi: 10.2119/molmed.2009.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jude EB, Blakytny R, Bulmer J, Boulton AJM, Ferguson MWJ. Transforming growth factor-beta 1, 2, 3 and receptor type I and II in diabetic foot ulcers. Diabet Med. 2002;19:440–447. doi: 10.1046/j.1464-5491.2002.00692.x. [DOI] [PubMed] [Google Scholar]
- 26.Pastar I, et al. Epithelialization in wound healing: A comprehensive review. Adv Wound Care (New Rochelle) 2014;3:445–464. doi: 10.1089/wound.2013.0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joung J, et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548:343–346. doi: 10.1038/nature23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stojadinovic O, Tomic-Canic M. Human ex vivo wound healing model. Methods Mol Biol. 2013;1037:255–264. doi: 10.1007/978-1-62703-505-7_14. [DOI] [PubMed] [Google Scholar]
- 29.Li D. 2018 A global transcriptome analysis of human epidermal keratinocytes upon inhibition of lncRNA WAKMAR1. Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse114908. Deposited May 25, 2018.
- 30.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Mering C, et al. STRING 7–Recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberzon A, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun Q, Hao Q, Prasanth KV. Nuclear long noncoding RNAs: Key regulators of gene expression. Trends Genet. 2017;34:142–157. doi: 10.1016/j.tig.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- 35.Lu Q, et al. Computational prediction of associations between long non-coding RNAs and proteins. BMC Genomics. 2013;14:651. doi: 10.1186/1471-2164-14-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis CJ, Mardaryev AN, Sharov AA, Fessing MY, Botchkarev VA. The epigenetic regulation of wound healing. Adv Wound Care (New Rochelle) 2014;3:468–475. doi: 10.1089/wound.2014.0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, Sun H, Wang H. Long noncoding RNAs in DNA methylation: New players stepping into the old game. Cell Biosci. 2016;6:45. doi: 10.1186/s13578-016-0109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viré E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 39.Li H, et al. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem. 2006;281:19489–19500. doi: 10.1074/jbc.M513249200. [DOI] [PubMed] [Google Scholar]
- 40.Hogart A, et al. NIH Intramural Sequencing Center Genome-wide DNA methylation profiles in hematopoietic stem and progenitor cells reveal overrepresentation of ETS transcription factor binding sites. Genome Res. 2012;22:1407–1418. doi: 10.1101/gr.132878.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feldmann A, et al. Transcription factor occupancy can mediate active turnover of DNA methylation at regulatory regions. PLoS Genet. 2013;9:e1003994. doi: 10.1371/journal.pgen.1003994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Ruscio A, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chalei V, et al. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. eLife. 2014;3:e04530. doi: 10.7554/eLife.04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L, et al. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015;25:335–350. doi: 10.1038/cr.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merry CR, et al. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum Mol Genet. 2015;24:6240–6253. doi: 10.1093/hmg/ddv343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taberlay PC, et al. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell. 2011;147:1283–1294. doi: 10.1016/j.cell.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips-Cremins JE, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–535, and erratum (2011) 472:247. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D’Souza SJA, et al. E2F-1 is essential for normal epidermal wound repair. J Biol Chem. 2002;277:10626–10632. doi: 10.1074/jbc.M111956200. [DOI] [PubMed] [Google Scholar]
- 51.Penn JW, Grobbelaar AO, Rolfe KJ. The role of the TGF-β family in wound healing, burns and scarring: A review. Int J Burns Trauma. 2012;2:18–28. [PMC free article] [PubMed] [Google Scholar]
- 52.Hezroni H, et al. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015;11:1110–1122. doi: 10.1016/j.celrep.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong VW, Sorkin M, Glotzbach JP, Longaker MT, Gurtner GC. Surgical approaches to create murine models of human wound healing. J Biomed Biotechnol. 2011;2011:969618. doi: 10.1155/2011/969618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.