Significance
Although synaptic plasticity is intimately controlled by synthesis of transmitter receptors and regulatory proteins, it remains largely unknown how synthesis and distribution of synaptic proteins could be modulated at the specific synapses. microRNAs (miRNAs) such as miR-134 could adjust synapse development and plasticity due to their regulatory functions for a distinct set of proteins. We developed an imaging method for in situ microRNAs mediated by atomic force microscopy (AFM), which revealed that miR-134s are enriched at nearby dendritic shafts and necks of spines. Notably, the abundance of miR-134s varied among distinct spine types, and their amounts were inversely correlated with maturity and functional states of individual spines. The functional impact of miR-134s would represent a molecular means of miRNAs, dynamically tuning synaptic connectivity and plasticity.
Keywords: atomic force microscopy, force mapping, microRNAs, dendritic spines, structural plasticity
Abstract
Dendritic spines are major loci of excitatory inputs and undergo activity-dependent structural changes that contribute to synaptic plasticity and memory formation. Despite the existence of various classification types of spines, how they arise and which molecular components trigger their structural plasticity remain elusive. microRNAs (miRNAs) have emerged as critical regulators of synapse development and plasticity via their control of gene expression. Brain-specific miR-134s likely regulate the morphological maturation of spines, but their subcellular distributions and functional impacts have rarely been assessed. Here, we exploited atomic force microscopy to visualize in situ miR-134s, which indicated that they are mainly distributed at nearby dendritic shafts and necks of spines. The abundance of miR-134s varied between morphologically and functionally distinct spine types, and their amounts were inversely correlated with their postulated maturation stages. Moreover, spines exhibited reduced contents of miR-134s when selectively stimulated with beads containing brain-derived neurotropic factor (BDNF). Taken together, in situ visualizations of miRNAs provided unprecedented insights into the “inverse synaptic-tagging” roles of miR-134s that are selective to inactive/irrelevant synapses and potentially a molecular means for modifying synaptic connectivity via structural alteration.
Spines, small protrusions emanating from the dendritic shafts of mammalian neurons, are major postsynaptic structures in apposition to axonal terminals of excitatory synapses (1, 2). The strength and modification of synaptic connectivity, which encompasses the ensemble of chemical and electrical connections between neurons, are critical for learning and memory (3). The physiological properties and strength of synapses can be mediated and modified by alterations in spine structure (4), mainly controlled by the polymerization and depolymerization of cytoskeletal components (5). Dendritic spines may be morphologically categorized as filopodia, which protrude from dendrites but lack postsynaptic density (PSD), thin types, which are normally considered to be immature spines, and stubby and mushroom types, which are considered to be mature spines (1, 6). A variety of proteins participate in the regulation of dendritic spine structure, which can be modulated by myriad ligands, such as brain-derived neurotropic factor (BDNF) and neuromodulators released into the synapses (7).
microRNAs (miRNAs) are a diverse class of small noncoding single-stranded RNAs that posttranscriptionally regulate gene expression (8). A distinct set of miRNAs appear to play critical roles in morphological changes of neurons (9, 10). They are also reported to exert various synaptic functions such as synapse formation and maturation (9, 11–13). For instance, activity-dependent miR-134s can regulate dendritic spine morphogenesis by repressing the translation of Limk1 mRNA within dendrites (14–17). However, in situ distribution of miRNAs has not been resolved in synaptic structures, largely due to resolution limits imposed by conventional miRNA detection methods including in situ hybridization (18–20) and fluorophore-labeled probes (14, 15, 18–23).
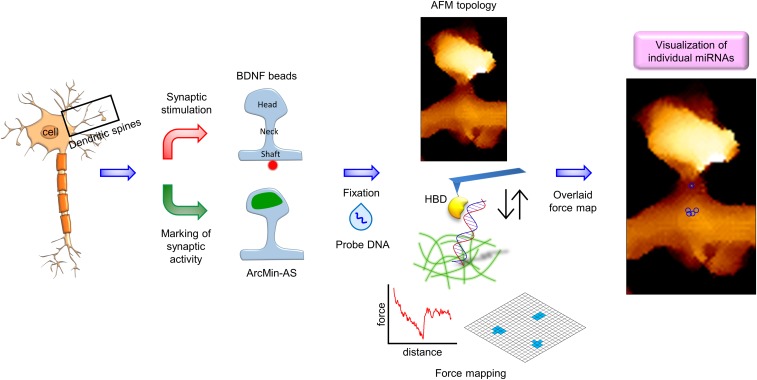
Atomic force microscopy (AFM) enables multiparametric nanoscale imaging for topography and stiffness, even under physiological conditions (24–27). In particular, AFM tips comprise molecular probes that bind to specific targets, thereby generating specific adhesion signals characterized by force-distance curves (28–30). Under ultrahigh lateral resolution, the specific signals produced by AFM may be used to locate individual molecules at nanometer resolution without the need for labeling or amplification. A hybrid binding domain (HBD) binds specifically to RNA/DNA hybrids that do not normally exist in the cytosol (31). We previously took advantage of HBD to detect and quantify miRNAs in single cells (32). Using the HBD with a complementary probe DNA to hybridize with miR-134s as the target, we were able to map individual miR-134s in dendritic shafts and spines at 10-nm resolution (SI Appendix, Fig. S1). Importantly, miR-134 contents were inversely correlated with the postulated maturity of individual spines, which was experimentally substantiated by beads coated with brain-derived neurotropic factor (BDNF) and the expression of a marker for activated-synapse (AS) targeting (Fig. 1).
Fig. 1.
The experimental scheme for activity-dependent miRNA detection at single dendritic spines using single-molecule adhesion of HBD. (Left) For nanoscale visualization of individual miRNAs, single spines are stimulated with BDNF-coated beads or functional states of spines are labeled with ArcMin-AS. (Middle) Fluorescence and topology images are obtained to classify dendritic spine types. Force maps of entire spine regions are generated using an HBD-tethering AFM tip. (Right) Spine topology and force maps are overlaid in a final map. As a result, miRNAs appear in a cluster of positive pixels on the adhesion force map.
Results
Visualization of Individual miR-134s in Dendrites and Spines.
To assess the differences between sectioned soma (intracellular) and soma from which the membrane had been removed (soma surface), we first mapped miR-134s at high resolution within sectioned somas of hippocampal neurons. An interaction between the HBD tethered to an AFM probe and the miR-134/DNA hybrid within the sections was observed, with the most probable adhesion force at 23 pN (SI Appendix, Fig. S2A), which revealed an average of 2.4 miR-134s (i.e., observable clusters) per 1.0 μm2. When depolarized with KCl, hippocampal neurons exhibited an increase in the density to 9.0 miR-134s per 1.0 μm2 (SI Appendix, Fig. S2 B–E). As these values corresponded with those obtained at the surface of the soma after membrane removal (32), miR-134s also seem to be homogeneously distributed within the cytosol area, such that data obtained from the intracellular surface are representative of data within the surrounding compartment. While this approach revealed that miR-134s at somas were increased by depolarization (SI Appendix, Fig. S2), application of the same approach for the subcompartments of dendrites was technically challenging because it is difficult to slice the resin-embedded samples along the dendrites due to the morphological characteristics.
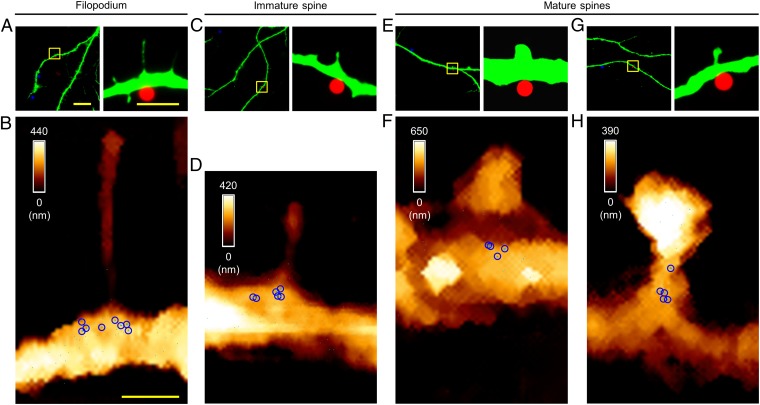
Primary cultured hippocampal neurons were fixed at 14 d in vitro (DIV14). The plasma membranes were removed, and then miR-134-complementary probe DNA was added for hybridization. The neurons were also immunostained to visualize dendrites and spines to guide the placement of an AFM probe tethering the HBD. An AFM morphology map was obtained, which enabled the classification of spine shapes into various types (see SI Appendix, SI Materials and Methods for classification details). Then high-resolution force-based maps were made using the interaction between HBD and the miR-134/DNA duplex, resulting in highly clustered pixels (Fig. 2), as well as the most probable specific adhesion force of 23 pN at dendritic shafts (SI Appendix, Fig. S3 A and B), which are consistent with the measurements from somas. Control experiments, such as mapping without DNA hybridization, after RNase treatment, with noncomplementary DNA, or with RNA hybridization, corroborated the validity of our analytical approaches (SI Appendix, Fig. S3 C–E). Unlike our previous observations in somas (32) (SI Appendix, Fig. S2 B–E), miR-134s within dendrites were highly localized to dendritic shafts below the spines, within an area of 700 (horizontal) × 200 (longitudinal) nm, and occasionally at the necks of the spines, but not within the spine heads (Fig. 2 and SI Appendix, Fig. S4 A–H).
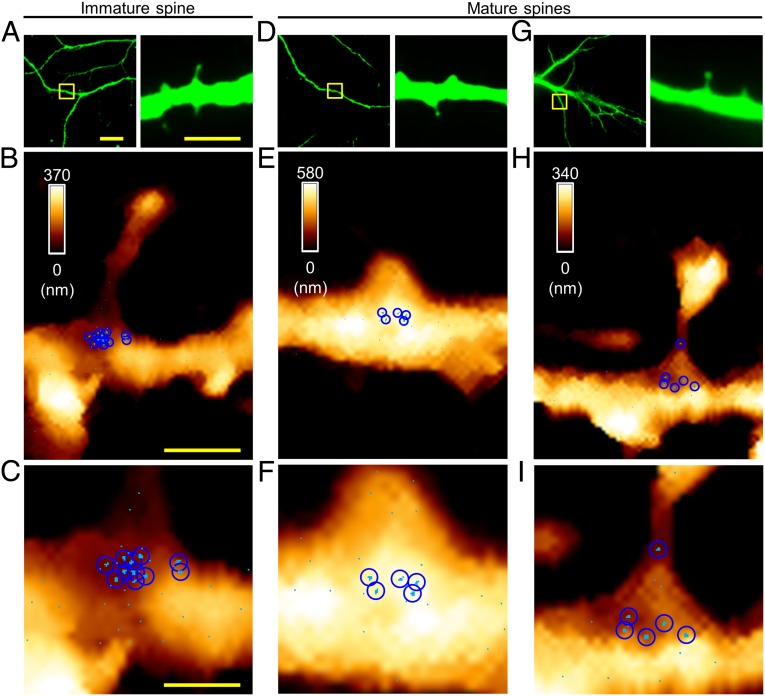
Fig. 2.
Visualization of miR-134s at individual spines. (A) Fluorescence images showing an immature spine (DIV14); the boxed area (Left) is shown at higher magnification (Right); MAP2, green. (B) AFM topographic image (3.0 × 4.0 μm2) of the boxed area in A and the force map showing individual miR-134/DNA hybrids. (C) Magnified force map of the dendritic shaft region in B. (D and G) Fluorescence images showing mature spines; the boxed areas (Left) are shown at higher magnification (Right). (E and H) AFM topographic images and overlaid force maps (3.0 × 4.0 μm2) of the boxed areas in D and G. (F and I) Magnified force maps in E and H; a sky-blue pixel represents a location where specific unbinding events were observed in more than two out of five measurements (pixel size, 10 nm); a blue circle indicates a cluster corresponding to the hydrodynamic radius observed at high resolution. [Scale bars: 20 μm (A, Left), 5.0 μm (A, Right), 1.0 μm (B), 0.50 μm (C). The scale bar for the fluorescence images in D and G is the same as that for the corresponding image in A; the scale bar for the AFM maps in E, F, H, and I is same as that for the corresponding maps in B and C.]
To ascertain the consistency of our measurement of miR-134s for each class of spines, we conducted analysis of each spine, one by one, in as many individual cells as possible. Importantly, the numbers of observed miR-134 clusters varied accordingly for each spine type (stages). For immature (thin) spines, there were 10.40 ± 0.40 (mean ± SEM) miR-134s, whereas for mature (stubby and mushroom) spines, there were only 5.50 ± 0.43 miR-134s (Fig. 2 and SI Appendix, Fig. S4 A–H and Table S1). Indeed, the number of observed miR-134 clusters was negatively correlated with spine head diameter, indicating their functionality (33, 34) (Kendall tau correlation coefficient: −0.76; P < 0.001; SI Appendix, Fig. S4I). Therefore, miR-134s are primarily localized in the dendritic shaft at the base of spines, and appear to be more abundant near immature forms of spines than near mature ones.
miR-134 Contents of Active Spines.
Expression of the immediate early gene Arc is widely used as a cellular marker of neuronal activity. This feature was exploited by a construct, ArcMin-AS, harboring a cassette consisting of mVenus for visualization, a deletion mutant of PSD-95 for accumulation at the postsynaptic sites without PDZ interaction, and the dendritic targeting element (DTE) of Arc mRNA for selective translation at the active dendritic segments (35). We transduced hippocampal neurons with adeno-associated viruses (AAVs) encoding the activity marker (SI Appendix, Fig. S5A), which allowed us to distinguish active spines from inactive ones: mVenus fluorescence was evident at the tips of the active spine heads or dendritic shafts (Fig. 3 A, B, F, and G and SI Appendix, Fig. S6 A, C, E, G, I, K, M, O, and Q). Thus, we were able to report the activation states of sampled spines by simultaneous visualization of both AS-labeled and nonlabeled spines.
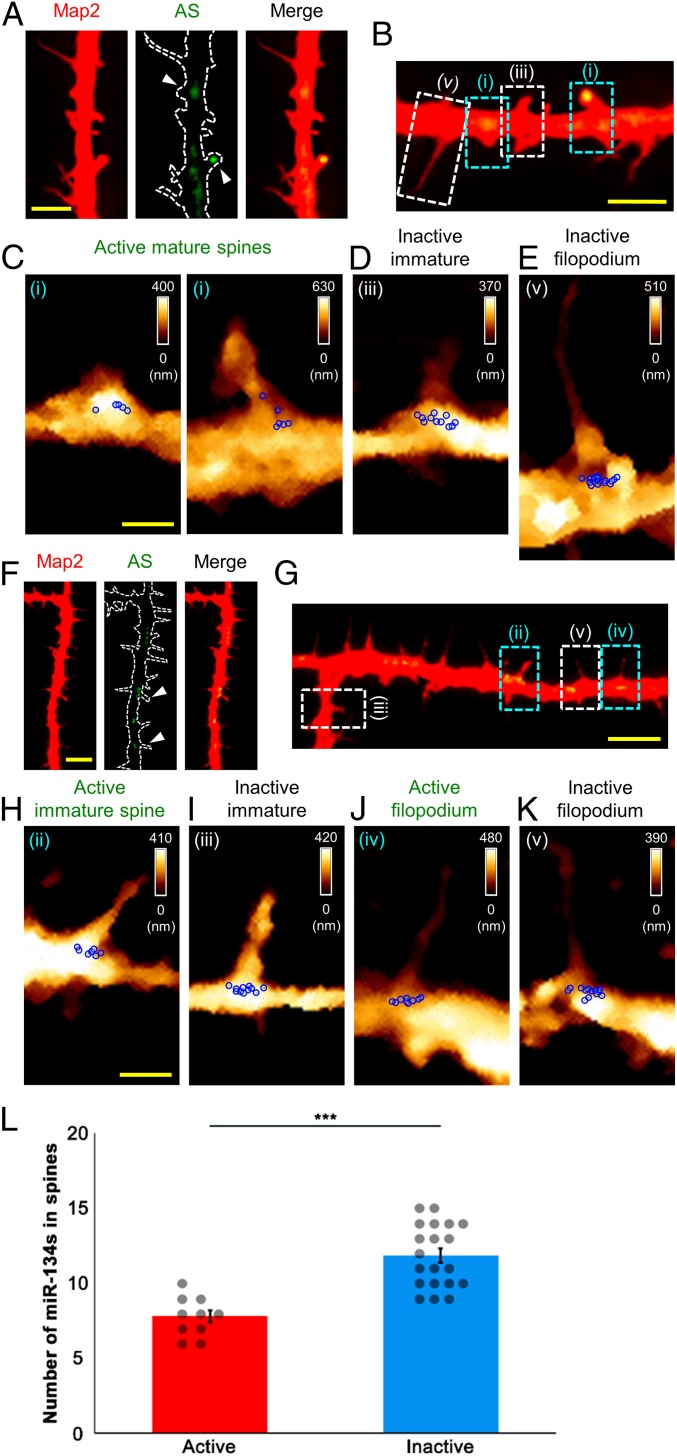
Fig. 3.
Mapping of miR-134s at spontaneously active filopodia and dendritic spines labeled with ArcMin-AS. (A, B, F, and G) Fluorescence images of filopodia and dendritic spines (DIV14); AS, ArcMin-AS. Each arrowhead indicates active filopodium and dendritic spines. (B and G) Active mature (i) and immature spine (ii) and filopodium (iv), and inactive immature spine (iii) and filopodium (v). (C–E and H–K); AFM topographic images and force maps were obtained for the boxed areas in B and G (3.0 × 5.0 μm2 or 3.0 × 5.5 μm2). (L) Numbers of miR-134s for active filopodia and immature spines vs. those for inactive filopodia and immature spines (two tailed t test, ***P < 0.001). [Scale bars: 20 μm (A and F), 5.0 μm (B and G), 1.0 μm (C and H). The scale bar for the AFM maps in D, E, I, J, and K are the same as those for the corresponding maps C or H.]
mVenus-expressing (active) mature spines exhibited 4.70 ± 0.30 miR-134s (Fig. 3C and SI Appendix, Fig. S6 B, D, F, H, J, and L and Table S1). This was similar in value to that of spines displaying the mature morphology (Fig. 2 D–I and SI Appendix, Fig. S4 A–H and Table S1; also see Fig. 5). Intriguingly, we also observed filopodia and immature forms of spines that exhibited mVenus fluorescence, albeit at a much lower frequency than that of mature forms of spines with mVenus signals (total labeling rate of ArcMin-AS in filopodia and dendritic spines analyzed, 22.89%; labeling rate of ArcMin-AS in mature spines, 17.71%; labeling rate of ArcMin-AS in immature spines, 4.08%; labeling rate of ArcMin-AS in filopodia, 1.10%; P < 0.01, one-way ANOVA followed by a Tukey’s post hoc test; SI Appendix, Fig. S5B). At active filopodia and immature spines, respectively, 8.80 ± 0.37 and 6.80 ± 0.37 miR-134s were detected (Fig. 3 H and J and SI Appendix, Fig. S6 H, J, L, N, P, and R and Table S1; also see Fig. 5). By contrast, the filopodia and immature spines devoid of mVenus had, respectively, 13.70 ± 0.30 and 10.00 ± 0.26 miR-134s (Fig. 3 D, E, I, and K and SI Appendix, Fig. S6 B, D, F, H, J, L, N, P, and R and Table S1; also see Fig. 5). Fewer miR-134s were associated with active filopodia and immature spines than with inactive ones (active, 7.80 ± 0.42; inactive, 11.85 ± 0.47; P < 0.001, one-way ANOVA followed by a Tukey’s post hoc test; Fig. 3L). Therefore, miR-134 abundance appears to be better at predicting the functionality of dendritic spines than predicting the morphology of spines. Collectively, miR-134 level is inversely correlated with the activity state of dendritic spines, which prompts us to speculate that miR-134s participate in defining the activity of individual spines and potentially in controlling structural plasticity, akin to their inhibitory effects on spine morphogenesis.
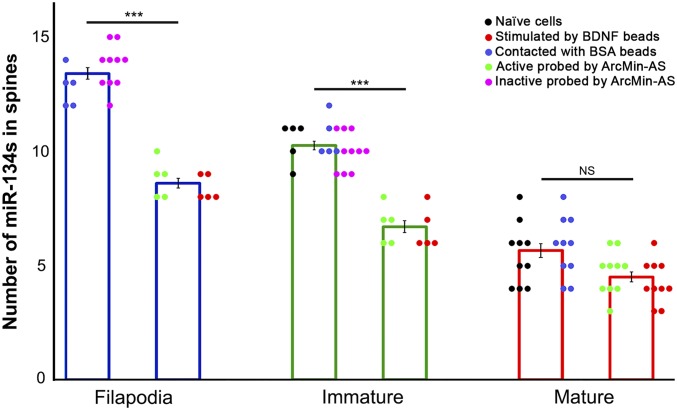
Fig. 5.
Numbers of miR-134 clusters at filopodia and various types of dendritic spines; miR-134s associated with filopodia and various spine types were quantified under each condition (n ≥ 5 spines); naïve cells refer to untreated spines; error bars indicate the SEM (***P < 0.001 via one-way ANOVA followed by a Tukey’s post hoc test; NS, not significant).
miR-134 Contents of Locally Activated Spines.
BDNF-mediated stimulation of TrkB induces synaptic activation of the subject synapses and subsequently increases intracellular Ca2+ levels, leading to structural changes in dendritic spines (7, 36). For direct comparison of the contents of miR-134s at filopodia and spines between BDNF and bovine serum albumin (BSA) groups, we applied beads covalently coated with BDNF and beads coated with BSA to cultured hippocampal neurons (DIV14), and then incubated them for 60 min. BDNF-dependent stimulation was verified by (i) measuring the expression of the photoinducible genetically encoded calcium indicator CaMPARI (37) followed by 405-nm laser illumination (SI Appendix, Fig. S7 A and B) and (ii) confirming an increase in spine head diameters from 0.78 ± 0.07 to 1.40 ± 0.20 μm after BDNF apposition (P < 0.05, one-way ANOVA followed by a Tukey’s post hoc test; SI Appendix, Fig. S6C) as well as altered spine population (SI Appendix, Fig. S7 D and E). As expected, treatment with BSA-coated beads produced no significant change in spine head diameter (0.82 ± 0.08 μm).
In agreement with our results using ArcMin-AS (Fig. 3 H and J and SI Appendix, Fig. S6 H, J, L, N, P, and R), BDNF-stimulated filopodia and immature forms of spines were associated with 8.40 ± 0.24 and 6.60 ± 0.40 miR-134s, respectively (Figs. 4 A–D and 5 and SI Appendix, Fig. S8 and Table S1), which was significantly different from mean numbers of miR-134s associated with nonstimulated filopodia and immature spines (P < 0.001, one-way ANOVA followed by a Tukey’s post hoc test; Figs. 2B, 3 I and K, and 5 and SI Appendix, Fig. S6 B, D, F, H, J, L, N, P, and R and Table S1). Interestingly, BDNF-stimulated mature spines were associated with 4.30 ± 0.30 miR-134s (Figs. 4 E–H and 5 and SI Appendix, Fig. S8 and Table S1), which was comparable to the number of miR-134s associated with nonstimulated mature spines (Fig. 5). No effect of treatment with BSA-coated beads was observed with miR-134 counts of 12.80 ± 0.37 for filopodia, 10.60 ± 0.40 for immature spines, and 5.80 ± 0.42 for mature spines (SI Appendix, Fig. S9 and Table S1). These outcomes were similar to those of the initial visualization studies (Fig. 5). As an additional control experiment, we treated hippocampal neurons with ANA-12, a selective and noncompetitive antagonist of TrkB, before application of BDNF-coated beads. ANA-12 blocked BDNF-mediated changes in miR-134 contents at filopodia and at each dendritic spine type (SI Appendix, Fig. S10). These data indicate that, consistent with what was observed in active filopodia and immature spines, synapse stimulation by BDNF also reduced the number of associated miR-134s at spines that had the immature morphology, but not at spines that had the mature morphology. Thus, synaptic activity may substantially control miR-134 contents at immature (determined by morphology) or inactive (determined by the activity marker) spines, but not at mature or active spines. Since fully mature spines already contain a reduced amount of miR-134s, they are most likely to be indifferent to additional increases in synaptic and neural functionality.
Fig. 4.
Mapping of miR-134s at filopodia and dendritic spines stimulated with BDNF-coated beads. (A) Fluorescence images of a filopodium (DIV14); the boxed area (Left) is shown at higher magnification (Right), where a BDNF-coated bead (in red) is seen in contact with the dendrite (MAP2, green). (B) AFM topographic image of the boxed area in A (3.0 × 5.0 μm2) overlaid with the force map for the stimulated filopodium. (C) Fluorescence images of an immature spine. (D) AFM topographic image of the boxed area in C (3.0 × 4.0 μm2) overlaid with the force map for the stimulated immature spine. (E and G) Fluorescence images of mature spines. (F and H) AFM topographic images of the boxed areas in E and G (3.0 × 5.0 μm2) overlaid with the force maps for the stimulated mature spines; BSA-coated beads are shown in blue, to the left in A, C, E, and G. [Scale bars: 20 μm (A, Left), 5.0 μm (A, Right), 1.0 μm (B). The scale bar for the fluorescence images in C, E, and G is same as that for the corresponding image in A, and the scale bar for the AFM maps in D, F, and H is same as that for the corresponding map in B.]
Discussion
We employed AFM and further improved adhesion force mapping methods for in situ visualization of miRNAs along the neurites of hippocampal neurons, which enabled us to directly analyze miR-134s at individual postsynaptic areas. Indeed, this nanoscale imaging revealed that miR-134s are enriched in the necks of spines and beneath the dendritic shafts but not within spine heads. Importantly, miR-134 abundance varied among the morphologically categorized spine types, such that the number of miR-134s was lower near spines with high-functionality states, as determined by the expression of ArcMin-AS or after stimulation with BDNF (Fig. 5).
Synaptic plasticity is a cellular mechanism underlying learning and memory. In many cases, long-lasting memory relies on long-term synaptic plasticity accompanied by dynamic remodeling of pre- and postsynaptic structures, which allows for the recruitment of glutamate receptors and their incorporation into the PSD (38). Synaptic plasticity also requires de novo synthesis of proteins that interact with the PSD and the cytoskeleton (39–41). miRNAs can intimately regulate protein synthesis by sequence-selective binding to the 3′-untranslated regions of their target mRNAs. For example, miR-134s may modulate dendritic spine sizes and potentially their activity by curbing the synthesis of Limk1 (14). Our observation that miR-134s were more abundant at immature (inactive) spines than at mature (active) spines would lend support to of the notion that miR-134s have roles in regulating the functional states of individual spines and filopodia.
The stimulation of select synapses induces the local translation of mRNAs encoding indispensable proteins for synaptic modification (42). Although the regulatory mechanisms have not been fully clarified, they likely involve polysomes consisting of an mRNA and multiple ribosomes within the dendritic shaft beneath spines. Therefore, miRNAs should be situated in these synaptodendritic compartments to act as local regulators of the translation of those mRNAs. The detection of miR-134s at the base of dendritic spines is consistent with this scenario. Furthermore, it was previously shown that mature forms of miRNAs are localized to stimulated synapses (23) and that miR-134s negatively regulate the size of dendritic spines (14). Despite the apparent likelihood, there have been no reports examining whether or not each miRNA resides distinctly in synapse structures or that miR-134 abundance can predict or control the functional state of individual dendritic spines. In this study, AFM-aided nanoscale imaging and a fluorescence marker of synaptic activity provided in situ evidence that miR-134 abundance is inversely correlated with the activity of individual spines.
The synaptic tagging, or capture, theory explains how particular synapses produce targets for subsequent trafficking of plasticity-related proteins (PRPs) (41, 43) to achieve input-specific synaptic and structural plasticity. For instance, PKMzeta (44), transported mRNAs (45) and Ca2+ microdomains, and short-term plasticity itself (46) have been proposed to act as synaptic tags that regulate the local synthesis of synaptic proteins. Conversely, trafficking of Arc into spine heads has been proposed as a tagging mechanism in ambient inactive synapses for the maintenance of the synaptic balance of potentiated neurons and the enhancement of signal/noise values for the onset of synaptic plasticity (47). Given the activity-dependent expression of miRNAs (23) and their extended stability in the cytoplasm (48), our results support the possibility that miR-134s might also serve as inverse tags for the marking of inactive (irrelevant) synapses and the prevention of nonspecific potentiation. Thus, it is possible to speculate that synaptic modification can arise and persist only at spines associated with reduced levels of miR-134s.
In summary, we provide in situ evidence showing that miR-134s are localized at nearby dendritic shafts and necks of spines and are differentially enriched at spines depending on their activity. The miR-134–mediated inverse synaptic tagging mechanism proposed here could represent an effective means of preserving synaptic weight differences between active and inactive synapses and enhancing the fidelity of synaptic modification when neuronal ensembles participate in computation of conditioned and unconditioned stimuli (SI Appendix, Fig. S11). Future studies directed toward the molecular and cellular mechanisms by which miRNAs determine input-specific synaptic plasticity will advance our understanding of the information processing at single synapses and the connectivity changes within entire neuronal circuits for memory formation and storage.
Materials and Methods
All animal care and use were in accordance with the institutional guidelines of Pohang University of Science and Technology. Neuronal cell cultures and AFM imaging were performed as previously described (32). Statistical analysis was performed using IBM SPSS Version 21.0. A detailed description of materials and methods is provided in the SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Wei Yang (NIH) for the kind donation of the HBD construct. We also thank Dr. Min Jeong Kye (Universität zu Köln) for her critical reading of the manuscript. J.W.P. acknowledges National Research Foundation of Korea (NRF) grants funded by the Korean government (Ministry of Science and ICT) (Grants 2017R1A2B3008478 and 2015M3C7A1030964). J.-H.K. acknowledges grants from the NRF (Grants 2018R1A3B1052079, 2018M3C7A1024152, and 2017R1A2B2004122). H.K. acknowledges CREST (Grant JPMJCR1652) from the Japan Science and Technology Agency (JST) and the World Premier International Research Center Initiative (WPI) from the Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819374116/-/DCSupplemental.
References
- 1.Hering H, Sheng M. Dendritic spines: Structure, dynamics and regulation. Nat Rev Neurosci. 2001;2:880–888. doi: 10.1038/35104061. [DOI] [PubMed] [Google Scholar]
- 2.Yuste R. Dendritic spines and distributed circuits. Neuron. 2011;71:772–781. doi: 10.1016/j.neuron.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogerson T, et al. Synaptic tagging during memory allocation. Nat Rev Neurosci. 2014;15:157–169. doi: 10.1038/nrn3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araya T, et al. CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc Natl Acad Sci USA. 2014;111:2029–2034. doi: 10.1073/pnas.1319953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch M, Hayashi Y. Structural plasticity of dendritic spines. Curr Opin Neurobiol. 2012;22:383–388. doi: 10.1016/j.conb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maiti P, Manna J, Ilavazhagan G, Rossignol J, Dunbar GL. Molecular regulation of dendritic spine dynamics and their potential impact on synaptic plasticity and neurological diseases. Neurosci Biobehav Rev. 2015;59:208–237. doi: 10.1016/j.neubiorev.2015.09.020. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka J, et al. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science. 2008;319:1683–1687. doi: 10.1126/science.1152864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 9.Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- 10.Edbauer D, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y-W, Ruiz CR, Eyler EC, Lin K, Meffert MK. Dual regulation of miRNA biogenesis generates target specificity in neurotrophin-induced protein synthesis. Cell. 2012;148:933–946. doi: 10.1016/j.cell.2012.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajgor D, Sanderson TM, Amici M, Collingridge GL, Hanley JG. NMDAR-dependent Argonaute 2 phosphorylation regulates miRNA activity and dendritic spine plasticity. EMBO J. 2018;37:e97943. doi: 10.15252/embj.201797943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 15.Fiore R, et al. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiore R, et al. MiR-134-dependent regulation of Pumilio-2 is necessary for homeostatic synaptic depression. EMBO J. 2014;33:2231–2246. doi: 10.15252/embj.201487921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chai S, Cambronne XA, Eichhorn SW, Goodman RH. MicroRNA-134 activity in somatostatin interneurons regulates H-Ras localization by repressing the palmitoylation enzyme, DHHC9. Proc Natl Acad Sci USA. 2013;110:17898–17903. doi: 10.1073/pnas.1317528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong H, et al. MicroRNA: Function, detection, and bioanalysis. Chem Rev. 2013;113:6207–6233. doi: 10.1021/cr300362f. [DOI] [PubMed] [Google Scholar]
- 19.Urbanek MO, Nawrocka AU, Krzyzosiak WJ. Small RNA detection by in situ hybridization methods. Int J Mol Sci. 2015;16:13259–13286. doi: 10.3390/ijms160613259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng Y, Dong L, Zhang J, Zhao Y, Li Z. Recent advances in microRNA detection. Analyst (Lond) 2018;143:1758–1774. doi: 10.1039/C7AN02001E. [DOI] [PubMed] [Google Scholar]
- 21.Siegel G, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kye MJ, et al. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA. 2007;13:1224–1234. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambandan S, et al. Activity-dependent spatially localized miRNA maturation in neuronal dendrites. Science. 2017;355:634–637. doi: 10.1126/science.aaf8995. [DOI] [PubMed] [Google Scholar]
- 24.Stroh CM, et al. Simultaneous topography and recognition imaging using force microscopy. Biophys J. 2004;87:1981–1990. doi: 10.1529/biophysj.104.043331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Husale S, Persson HH, Sahin O. DNA nanomechanics allows direct digital detection of complementary DNA and microRNA targets. Nature. 2009;462:1075–1078. doi: 10.1038/nature08626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kodera N, Yamamoto D, Ishikawa R, Ando T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 2010;468:72–76. doi: 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
- 27.Dufrêne YF, et al. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat Nanotechnol. 2017;12:295–307. doi: 10.1038/nnano.2017.45. [DOI] [PubMed] [Google Scholar]
- 28.Yu H, Siewny MG, Edwards DT, Sanders AW, Perkins TT. Hidden dynamics in the unfolding of individual bacteriorhodopsin proteins. Science. 2017;355:945–950. doi: 10.1126/science.aah7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milles LF, Schulten K, Gaub HE, Bernardi RC. Molecular mechanism of extreme mechanostability in a pathogen adhesin. Science. 2018;359:1527–1533. doi: 10.1126/science.aar2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinterdorfer P, Dufrêne YF. Detection and localization of single molecular recognition events using atomic force microscopy. Nat Methods. 2006;3:347–355. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- 31.Nowotny M, et al. Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD. EMBO J. 2008;27:1172–1181. doi: 10.1038/emboj.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koo H, et al. Visualization and quantification of microRNA in a single cell using atomic force microscopy. J Am Chem Soc. 2016;138:11664–11671. doi: 10.1021/jacs.6b05048. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Wang XB, Frerking M, Zhou Q. Spine expansion and stabilization associated with long-term potentiation. J Neurosci. 2008;28:5740–5751. doi: 10.1523/JNEUROSCI.3998-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Q, Homma KJ, Poo MM. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44:749–757. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 35.Hayashi-Takagi A, et al. Labelling and optical erasure of synaptic memory traces in the motor cortex. Nature. 2015;525:333–338. doi: 10.1038/nature15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lang SB, Stein V, Bonhoeffer T, Lohmann C. Endogenous brain-derived neurotrophic factor triggers fast calcium transients at synapses in developing dendrites. J Neurosci. 2007;27:1097–1105. doi: 10.1523/JNEUROSCI.3590-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fosque BF, et al. Neural circuits. Labeling of active neural circuits in vivo with designed calcium integrators. Science. 2015;347:755–760. doi: 10.1126/science.1260922. [DOI] [PubMed] [Google Scholar]
- 38.Bard L, Groc L. Glutamate receptor dynamics and protein interaction: Lessons from the NMDA receptor. Mol Cell Neurosci. 2011;48:298–307. doi: 10.1016/j.mcn.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 40.Spence EF, Soderling SH. Actin out: Regulation of the synaptic cytoskeleton. J Biol Chem. 2015;290:28613–28622. doi: 10.1074/jbc.R115.655118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frey U, Morris RG. Synaptic tagging and long-term potentiation. Nature. 1997;385:533–536. doi: 10.1038/385533a0. [DOI] [PubMed] [Google Scholar]
- 42.Dieterich DC, et al. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Redondo RL, Morris RG. Making memories last: The synaptic tagging and capture hypothesis. Nat Rev Neurosci. 2011;12:17–30. doi: 10.1038/nrn2963. [DOI] [PubMed] [Google Scholar]
- 44.Sajikumar S, Korte M. Metaplasticity governs compartmentalization of synaptic tagging and capture through brain-derived neurotrophic factor (BDNF) and protein kinase Mzeta (PKMzeta) Proc Natl Acad Sci USA. 2011;108:2551–2556. doi: 10.1073/pnas.1016849108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tübing F, et al. Dendritically localized transcripts are sorted into distinct ribonucleoprotein particles that display fast directional motility along dendrites of hippocampal neurons. J Neurosci. 2010;30:4160–4170. doi: 10.1523/JNEUROSCI.3537-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Redondo RL, et al. Synaptic tagging and capture: Differential role of distinct calcium/calmodulin kinases in protein synthesis-dependent long-term potentiation. J Neurosci. 2010;30:4981–4989. doi: 10.1523/JNEUROSCI.3140-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okuno H, et al. Inverse synaptic tagging of inactive synapses via dynamic interaction of Arc/Arg3.1 with CaMKIIβ. Cell. 2012;149:886–898. doi: 10.1016/j.cell.2012.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinjo ER, et al. A possible new mechanism for the control of miRNA expression in neurons. Exp Neurol. 2013;248:546–558. doi: 10.1016/j.expneurol.2013.07.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.