Significance
Recent studies have shown that humans have adapted to many different environments around the world. However, few studies have centered on Indigenous groups in the Americas. We present a comparative analysis of genetic adaptations in humans across North America using genome-wide scans for signals of natural selection in three populations inhabiting vastly different environments. We find evidence for adaptation to cold and high latitudes in an Alaskan population, whereas infectious disease was a strong selective pressure in the southeastern United States and central Mexico. Because there are few shared signals of selection between populations, these sweeps likely occurred after population differentiation in the Americas. This study fills an important gap in our knowledge of genetic adaptations in humans.
Keywords: population genomics, natural selection, Native Americans, Alaskan Natives, human evolutionary genetics
Abstract
While many studies have highlighted human adaptations to diverse environments worldwide, genomic studies of natural selection in Indigenous populations in the Americas have been absent from this literature until very recently. Since humans first entered the Americas some 20,000 years ago, they have settled in many new environments across the continent. This diversity of environments has placed variable selective pressures on the populations living in each region, but the effects of these pressures have not been extensively studied to date. To help fill this gap, we collected genome-wide data from three Indigenous North American populations from different geographic regions of the continent (Alaska, southeastern United States, and central Mexico). We identified signals of natural selection in each population and compared signals across populations to explore the differences in selective pressures among the three regions sampled. We find evidence of adaptation to cold and high-latitude environments in Alaska, while in the southeastern United States and central Mexico, pathogenic environments seem to have created important selective pressures. This study lays the foundation for additional functional and phenotypic work on possible adaptations to varied environments during the history of population diversification in the Americas.
Since first leaving their ancestral environments in Africa more than 100,000 y ago, humans have spread to nearly every region of the planet. In doing so, different populations have been exposed to many new environments and selective pressures, and they have developed a diversity of adaptations as a result (1). The declining cost of array and sequencing technologies and the improvement of methods for detecting signals of natural selection have allowed researchers to answer questions about selective pressures across a growing number of populations worldwide (2–4).
However, very little is known about the recent evolutionary history of Indigenous populations in North America and the selective pressures that they have experienced. The Indigenous peoples of North America are underrepresented in the population genetics literature as a whole (5) and in studies of selection in particular. Only a handful of genomic studies of natural selection have been conducted in the Americas, and the majority of these have focused on populations in South America (6–9). To our knowledge, only two genomic studies of selection in North American populations have been published. Lindo et al. (10) found evidence of a complex history of selective pressures on the immune gene HLA-DQ1 using exome data from ancient and modern populations in the Canadian Pacific Northwest. Another study with the Greenlandic Inuit found evidence of selection in the FADS genes, which code for fatty acid desaturases that are associated with polyunsaturated fatty acid levels in the blood (11), as well as in the genomic region encompassing TBX15, which plays a role in the differentiation of brown and white adipocytes. The authors suggested that these signals of selection are likely related to adaptation to cold environments.
Here, we present genome-wide scans for natural selection across three populations from different regions of North America. We find evidence of adaptation to cold and high latitudes in an Alaskan Native population from the Arctic and evidence of selection at several genes related to inflammation and immune function in Indigenous populations from the southeastern United States and central Mexico. We find little overlap between putatively selected genes in these three populations, suggesting that local selective pressures in each geographic region have shaped these Indigenous North American populations differently since they settled in distinct regions of the continent.
Results
Data Collection and Genetic Ancestry Estimates.
We collected DNA samples from 150 individuals from three Indigenous populations in North America (Fig. 1 and SI Appendix, Figs. S1 and S2), including 35 Alaskan Iñupiat from the North Slope of Alaska, 47 individuals from the town of Xaltocan in central Mexico, and 68 individuals from several closely related communities in the southeastern United States (populations referred to hereafter as Alaska, central Mexico, and southeastern United States, respectively). In some cases, exact sampling locations and community names are not reported to protect the privacy and anonymity of both the individuals and communities participating in this research. These protections were developed in collaboration with community members and are part of the IRB protocol and informed consent documentation used in this study. We then used the Affymetrix Axiom Human Origins Array to genotype 629,443 genome-wide SNPs for each of these individuals. A total of 563,162 SNPs were included in our analyses after quality control filtering and merging with the 1000 Genomes dataset (12) for comparative analyses.
Fig. 1.
Map of sampling areas. Specific sampling locations, where publicly available, are provided in SI Appendix, Figs. S1 and S2. Blue, Alaska; green, central Mexico; red, southeastern United States.
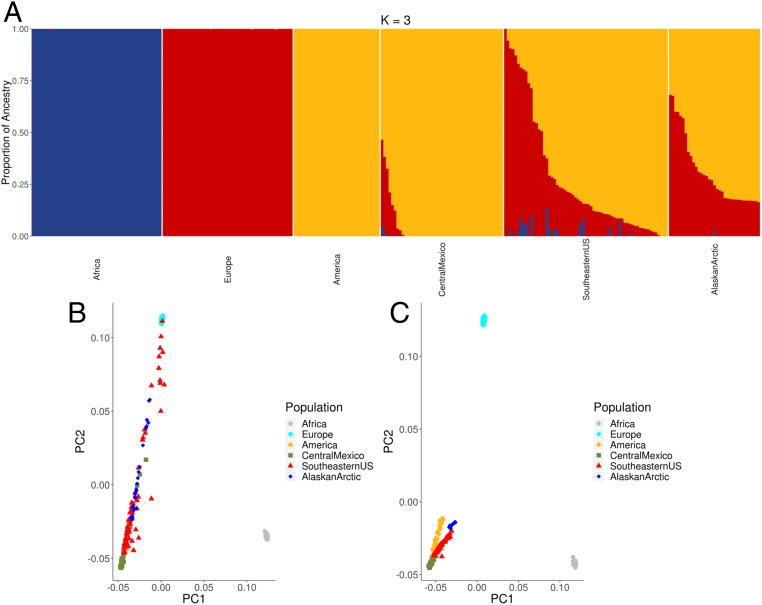
Because many previously genotyped Indigenous populations in the Americas trace a large percentage of genetic ancestry to recent European and African ancestors, which can influence results of genome-wide scans for selection, we first conducted a nonhierarchical clustering analysis of the SNP data implemented in the program ADMIXTURE (13). Fig. 2A shows that many of the Alaska and southeastern US individuals have more European ancestors than the individuals sampled in central Mexico. Local ancestry assignment was then done using RFMIX (14) to assign each chromosomal segment to its most likely ancestral source for each individual in the dataset. To minimize the effects of recent admixture on our selection scans, we masked SNPs from the data for an individual if they were in a section of chromosome inferred to have been inherited from a non-Indigenous ancestor (Fig. 2 B and C).
Fig. 2.
Global ancestry analysis of study populations. (A) ADMIXTURE plot at K = 3, (B) Principal components analysis (PCA) of unmasked genotypes showing principal components 1 (PC1) and 2 (PC2), and (C) PCA of genotypes after study populations were masked.
Genome-Wide Scans for Signals of Natural Selection.
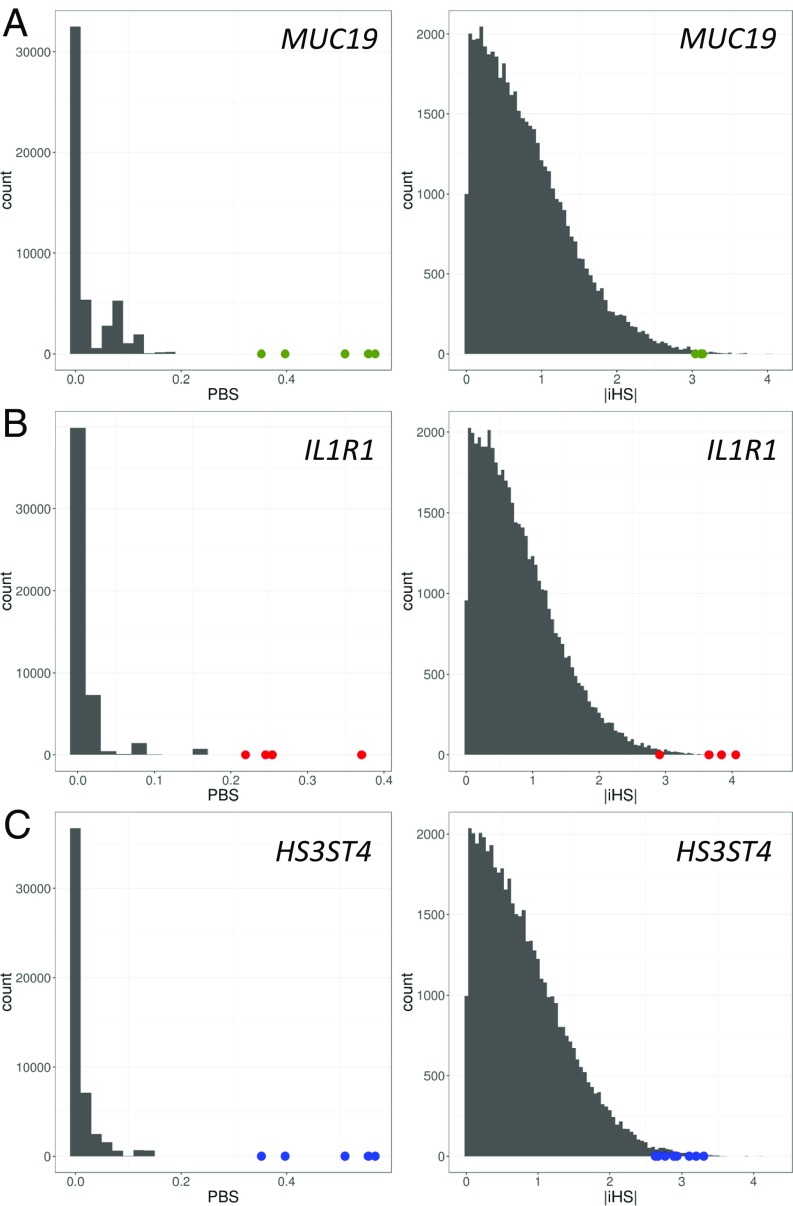
We computed two statistics to identify potential signatures of natural selection in the three study populations. We calculated the population branch statistic for each autosomal SNP in each population using individuals from the 1000 Genomes Peruvian population (PEL) without recent European or African ancestry as an ingroup and the 1000 Genomes Han Chinese population (CHB) as an outgroup. The population branch statistic computes the amount of genetic differentiation at a given locus along a branch leading to a population of interest by comparing transformed pairwise FST values between each pair of three populations (4). A population’s population branch statistic value at a given locus corresponds to the magnitude of allele frequency change relative to its divergence from the other two populations. This approach has proven to be powerful at detecting recent signals of selection (4, 10). We also calculated the integrated haplotype score (iHS), a widely used haplotype-based method of detecting signals of selection, for each autosomal SNP in each of the three study populations. The iHS is a measure of extended homozygosity in the haplotype surrounding a given SNP. Extended stretches of homozygosity relative to the background are a signal of a selective sweep that has not yet reached fixation. P values were calculated for both population branch statistics and iHS using a distribution of each statistic simulated under a demographic model specific to each study population. We then identified the top 1% P values for population branch statistics and iHS from each population (Fig. 3 and SI Appendix, Fig. S6) and cross-referenced them to find SNPs that were significant outliers in both statistics. This approach, which has been used previously (15), should reduce our chances of reporting false positives, as the iHS has been shown to be robust to demographic history that is often a confounding factor in FST-based approaches, such as population branch statistics (16).
Fig. 3.
Top selected gene from each population plotted against the simulated distribution of population branch statistics (PBS) and iHS: (A) MUC19 from the central Mexico population, (B) IL1R1 from the southeastern US population, and (C) HS3ST4 from the Alaskan Arctic population.
We found 153 putatively selected SNPs for the Alaska population, 104 such SNPs for the southeastern US population, and 190 such SNPs for the central Mexico population. Genes with the strongest signals of selection (i.e., those with the largest number of SNPs putatively under selection) for each population are listed in Table 1 along with the average population branch statistics and iHSs for the significant SNPs. A complete list of the putatively selected SNPs for each population is available in Dataset S1.
Table 1.
Genes with strongest signals of selection
| Region | Gene | SNPs | Population branch statistic | P value | |iHS| | P value |
| Central Mexico | MUC19 | 8 | 0.2166 | 1.30E-04 | 3.1143 | 1.70E-03 |
| Alaskan Arctic | HS3ST4 | 9 | 0.4892 | 2.00E-05 | 2.9371 | 4.30E-03 |
| Alaskan Arctic | KCNH1 | 5 | 0.5400 | 2.00E-05 | 3.2764 | 2.00E-03 |
| Alaskan Arctic | OCA2 | 5 | 0.4572 | 2.00E-05 | 3.1052 | 3.20E-03 |
| Southeastern United States | IL1R1 | 5 | 0.2669 | 3.60E-05 | 3.660 | 1.10E-03 |
Functions of the Putatively Selected Genes.
We used GeneCards (17) to gain insight into the functions of the putatively selected genes. The strongest signals of selection in the Alaska population occur within three genes related to heparan sulfate biosynthesis (HS3ST4), adipose tissue production (KCNH1), and melanin production (OCA2). One gene related to immune response (IL1R1) shows a strong signal of selection in the southeastern United States. One gene related to immune response (MUC19) also shows a strong signal of selection in the central Mexico population.
We also conducted a pathway enrichment analysis using the WebGestalt platform (18) to better understand the metabolic pathways involving putatively selected genes and to determine if these genes were overrepresented in any particular pathways. However, after correcting for the false discovery rate, we found no significant enrichment of the putatively selected genes in any metabolic pathways (Dataset S1).
Shared Signals of Selection Between Populations.
We next looked for shared signals of selection among the three study populations by comparing statistically significant results from each analysis. We found no shared signals of selection among all three populations but did identify some putatively selected genes shared between pairs of populations. We found one shared signal of selection at a single SNP within the gene SLIT2 in the Alaska and southeastern US populations. We also found that the southeastern US and central Mexico populations share signals of selection at two SNPs each in the genes MUC19 and CNTN1 (Table 2). We found no shared signals of selection between the Alaska and central Mexico populations. The greater percentage of putatively selected genes shared between the southeastern United States and central Mexico may be due to either similar selective pressures on both populations or the more recent divergence of the southeastern US and central Mexico populations if the selective pressures primarily occurred before they diverged.
Table 2.
SNPs with shared signals of selection between two study populations
| Chromosome | Position | rsid | Gene |
| 12 | 41180074 | rs1596529 | CNTN1 |
| 12 | 41180235 | rs935228 | CNTN1 |
| 12 | 40806911 | rs74565412 | MUC19 |
| 12 | 40958216 | rs7303283 | MUC19 |
| 4 | 20469565 | rs12499199 | SLIT2 |
Discussion
Our results suggest that different selective pressures have been acting on the three study populations sampled from different regions of North America. In the Alaska population, we see evidence for adaptation to both cold and high-latitude environments. The Alaskan Arctic has a tundra climate (Et on the Köppen climate classification) characterized by at least 1 mo with an average temperature >0 °C but no months with an average temperature above 10 °C (19). Being above the Arctic Circle, this region is also subject to low levels of direct sunlight and intermittent periods of complete daylight or darkness. Alaskan Native groups and other Indigenous peoples in the Arctic have developed a number of cultural adaptations in response to this extreme environment. The traditional diet of Alaskan Native peoples in this region relies heavily on both terrestrial (caribou) and marine (seal, whale) mammal resources, as the Arctic environment has low levels of surface vegetation and soil development (20). Traditional Alaskan Native clothing and dwellings are also designed to provide shelter from the extremely cold environment. Our results suggest that genetic adaptations have also arisen in this population.
The three genes that have the strongest evidence of selection in the Alaskan Arctic population could all have adaptive effects in cold, high-latitude environments. The gene HS3ST4 is involved in the production of heparan sulfate, a molecule that affects blood thickness. Previous work has shown that extended exposure to cold temperatures increases blood thickness, which can lead to a number of deleterious effects (21). The gene KCNH1 is involved in the regulation of cell proliferation and differentiation, in particular adipogenic and osteogenic differentiation in bone marrow-derived stem cells (22). Selection on these genes involved in regulating blood thickness and fat storage, respectively, suggests that a cold-resistant phenotype may be under selection in this Alaskan Arctic population. Previous studies of other Arctic populations have similarly found evidence of adaptation to the cold climate, with signals of selection found in genes related to metabolic pathways (11, 23), albeit in different genes than those identified in this study.
OCA2, also under selection in the Alaskan Arctic population, is associated with skin/eye/hair pigmentation in humans, suggesting the possibility of adaptation to high latitudes in this group. Vitamin D is an essential nutrient that is important for skeletal development and the innate immune response among other processes. In humans, the majority of vitamin D synthesis takes place in the skin as a result of the interaction between cholesterol and UVB radiation from sunlight. High-latitude regions, such as Alaska, are exposed to much lower levels of UVB radiation than other parts of the globe, making it difficult for people living in these areas to maintain healthy levels of vitamin D (24). Previous work has shown that variation in the OCA2 gene is correlated with the amount of winter solar radiation (25).
In the southeastern United States, we see the strongest signal of selection on SNPs in and around the gene IL1R1, which codes for a cytokine receptor that plays a key role in the adaptive immune response. Selection on IL1R1 in the southeastern United States makes sense given the colonial history of this region. The colonial period saw the introduction of a variety of diseases into the Americas, including smallpox, measles, influenza, pertussis, cholera, plague, typhus, yellow fever, diphtheria, malaria, and influenza (26). One recent spatial model of the colonial spread of epidemic diseases in North America (27) suggests that such diseases were first introduced during European colonization of coastal areas of the Southeast in the early 16th century, spread slowly toward the Appalachian mountains over the next 140 y, and then, moved very quickly across the interior Southeast. This model is consistent with the history of the region: after the initial Spanish colonization of the coastal Southeast in 1513, five documented expeditions (entradas) were undertaken to map the Southeast before 1545. Interaction between these entradas and Indigenous groups along with the establishment of the Spanish mission system throughout the Southeast likely contributed to the introduction and spread of multiple infectious diseases in this region. Several accounts of disease epidemics in the coastal Southeast are also described in historic records beginning in 1520 and continuing through the early 18th century (28). Groups in the interior Southeast likely avoided the very first epidemics but were affected later (29). While the causal pathogens of many early epidemics remain unknown, accounts of some later epidemics allow the underlying cause to be identified, such as several accounts of a smallpox epidemic in the late 1690s that report its spread from Virginia down into the Carolinas and across the Southeast into Mississippi (30). Altogether, historical documents, ethnohistoric records, Indigenous histories, and archaeological evidence demonstrate that these epidemics in conjunction with other events and practices during the colonial era contributed to a significant population decline and major sociopolitical changes in the region. By the 18th century, for example, many of the Indigenous groups interacting with the Spanish, including those forced into the mission system, had merged, and the ethnogenesis of many of the modern Indigenous southeastern groups was beginning (31). Our results suggest that the repeated epidemics may have also created significant selective pressures, influencing patterns of genetic variation at loci associated with the human immune response in these populations.
Pathogenic environments seem to have been a major selective force in central Mexico as well, as many of the putatively selected genes in this population are also related to immune system pathways. In Mexico, the spread of European-introduced diseases began shortly after Spanish conquistador Hernán Cortés landed near the present-day city of Veracruz in 1519 and began his military campaign against the Aztec empire (32). In central Mexico, the first documented epidemic was smallpox in 1519–1520 as Cortés marched toward the Aztec capital city of Tenochtitlan, located in present-day Mexico City in central Mexico. This initial epidemic was followed by several subsequent smallpox outbreaks, particularly in the late 16th and early 17th centuries (32). After Tenochtitlan was conquered in 1521, it became known as Mexico City, capital of the Viceroyalty of New Spain, and received a large influx of people from both Europe and Africa (33), no doubt bringing additional pathogens along with them.
This history likely contributed to genomic signatures of selection seen in the central Mexico population in this study. We see the strongest signal of selection on the mucin gene MUC19 in the central Mexico population. Mucin genes are primarily involved in the immune response to parasitic infection (34). Past work has shown that parasite load is strongly correlated with latitude, with populations closer to the tropics having higher levels of parasitic infection (35). Interestingly, the GenomeRNAi database (36) shows that MUC19 is associated with decreased vaccinia virus (VACV) infection. VACV is a close relative of the variola virus, the causal agent of smallpox, and recombinant versions of the VACV were used as a vaccine against smallpox until it was eradicated in the late 1970s (37). However, while these results are suggestive, we cannot be certain that smallpox was the selective agent for this sweep, because there were many more infectious diseases spreading through Mexico at this time (32). Recent work using novel methods to search for ancient pathogen DNA in human ancestral remains has successfully identified some of these unknown pathogens, such as the bacterium Salmonella enterica as a possible causal agent of the 16th century “cocoliztli” epidemic in southern Mexico (38). Future work may help us identify the major pathogens afflicting the people of central Mexico during colonial times.
These results suggest that selective pressures have varied widely across the Americas. The value of investigating selective pressures at a regional level in human populations is becoming increasingly recognized as an important topic of study. In South America, for example, recent work has examined the genetic components of the well-studied adaptation of Andean populations to high altitude (7). In addition, two recent studies have shown evidence of adaptation to an arsenic-rich environment in Andean populations from northwest Argentina (8, 39). Both studies found signals of selection on the gene AS3MT, which is involved in the metabolism of arsenic. Variants identified in these Andean groups allow them to metabolize less of the toxic element. Another recent study of Andean populations farther north in Peru used genomic data collected from both ancient and modern populations to study evolutionary pressures through time (40). The gene showing the strongest signal of selection in this study was MGAM, which is associated with starch digestion. This suggests that the transition to agriculture in this region may have also been a major selective pressure in the past. The only study to our knowledge to look at the history of selective pressures in North America was done with the Tsimshian of British Columbia (10). This study found evidence of selection positive selection at the gene HLA-DQA1 in the Tsimshian population before European colonization and possible evidence of negative selection in that region afterward. These studies conducted with populations thousands of miles apart from each other and in a variety of different ecological environments demonstrate the complex history of human adaptation to the varied environments of the Americas.
Altogether, our analysis of genome-wide signals of selection in three Indigenous populations in North America found evidence for selection on genes related to cold and high-latitude environments in Alaska but selection on genes related to immune function in the southeastern United States and central Mexico. Additional studies may find evidence of other adaptations in different environments on these continents.
Materials and Methods
Ethics and Community Engagement.
This project was made possible through active and ongoing collaborations with members of the participating communities. Community authorities and representative bodies were consulted where appropriate, and all individual participants provided written informed consent for the types of analyses conducted in this study. The collection and analyses of samples from all communities were approved by the IRB of the University of Texas at Austin (protocol #2012–05-0105), with additional approval for sample collection and population genetic analyses from the Alaskan Iñupiat provided by the IRB at Northwestern University (protocol #2683–001). In some cases, exact sampling locations and/or community names are not reported to protect the privacy and anonymity of the communities and individuals participating in this research.
DNA Extraction and Genotyping.
We extracted DNA from the saliva samples of 150 individuals from three Indigenous populations in the Americas (Iñupiat from the North Slope of Alaska, n = 35; Xaltocan in central Mexico, n = 47; southeastern United States, n = 68) using the prepIT L2P kit (DNA Genotek) following the manufacturer’s guidelines. Extracts were genotyped using the Affymetrix Human Origins Array. Data are available to researchers who sign a data access agreement with D.A.B. at the University of Connecticut and M.G.H. at Northwestern University.
Data Quality Control and Global Ancestry Analysis.
SNPs not genotyped in the majority of study samples (–geno 0.1) were removed using PLINK v1.9 (41). For ADMIXTURE analysis, we further pruned the SNPs in high linkage disequilibrium (–indep-pairwise 200 25 0.4). A global ancestry analysis was conducted with ADMIXTURE 1.3.0 (13) using the three study populations along with populations from the 1000 Genomes dataset (12) [Yoruba in Ibadan, Nigeria (YRI), Utah residents with northern and western European ancestry (CEU), and unadmixed individuals from Peruvians from Lima, Peru (PEL)] to represent the likely sources of recent admixture in the study populations. All comparative individuals used in this analysis are listed in Dataset S1. Principal component analysis was performed on these samples using the smartpca program in EIGENSOFT (42).
Haplotype Phasing and Genotype Masking.
Sample genotypes were phased using SHAPEIT2 with default parameters and the 1000 Genomes Phase 3 dataset as a reference panel (12, 43). We next used RFMIX (14) to assign each chromosomal segment to its most likely ancestral source for each Indigenous individual in the dataset using 50 YRI individuals, 50 CEU individuals, and 33 unadmixed PEL individuals from the 1000 Genomes Phase 3 dataset to represent the possible ancestral populations for this local ancestry assignment. After ancestry assignment, SNPs in chromosomal segments inferred to have been inherited from an African or European ancestor were masked from the data along with SNPs with low-confidence (<90%) ancestry assignment. SNPs with more than 10% missing data were then removed from the dataset. Using these criteria, the final dataset contained 546,089 SNPs and sample sizes of n = 27 for the Alaskan Arctic (Iñupiat) population, n = 43 for the central Mexico (Xaltocan) population, and n = 47 for the southeastern US population.
Selection Analyses.
The phased masked data were annotated with ancestral allele information using aa_annotate.py (44). iHS values were calculated for each population using hapbin (45). FST values were calculated using the masked dataset for each of the three sampled populations with an ingroup (33 Peruvian individuals without any evidence of recent European or African genetic ancestry selected from the 1000 Genomes Project dataset) (12) and with an outgroup (50 Han Chinese individuals from the 1000 Genomes Project) using vcftools (46). The population branch statistic (4) was then calculated for the three sampled Indigenous populations in North America.
P values were calculated for each population branch statistic and iHS using a distribution of each statistic simulated under a demographic model specific to each study population. The top 1% of P values for population branch statistics and iHSs were identified for each population and then cross-referenced so that additional analysis was done only for SNPs in the top 1% in both tests for selection. This cross-referencing, used previously (15), should reduce our chances of reporting selection results that are actually false positives, since each statistic has different underlying assumptions.
Demographic Models.
Demographic models were constructed for each of the three study populations using fastsimcoal2 (47) to calculate expected distributions of the selection statistics under neutral demographic processes. An overview of the models is shown in SI Appendix, Fig. S11. Briefly, we constructed a model of human demographic history using previously published estimates for the timing of the Out-of-Africa bottleneck and peopling of the Americas (10) using the joint site frequency spectrum of the CHB, unadmixed PEL, and each of our study populations. Arlequin was used to calculate the site frequency spectrum for each population (48). We included the divergence time between the PEL and each study population as an open parameter, as it is likely that our study populations diverged from PEL at different times in the past. We also included the timing and severity of a recent bottleneck as open parameters in the model, as there is archaeological, historical, ethnographic, and genetic evidence for bottlenecks in many, but not all, Indigenous populations in the Americas. Parameters for each of the three population models were estimated by running the model 100 times with 1,000,000 iterations per run. The best likelihood run was then chosen for each population and used to simulate 500,000 sites across 22 chromosomes for a number of individuals equal to that of those used in calculating the joint site frequency spectrum (50 CHB, 33 PEL, 27–47 study populations). This was done 100 times for each of the three models, and population branch statistics and iHSs were then calculated for each of the simulated datasets; 50,000 population branch statistics and iHSs were then randomly selected from the simulated results to form a distribution for comparison with the observed empirical values.
Gene Annotation and Pathway Enrichment Analysis.
We used the Ensembl Variant Effect Predictor (49) to assign gene names to SNPs in the top 1% of population branch statistic and iHS P values. To identify any metabolic pathways that are overrepresented in the putatively selected genes in each of the three study populations, we performed a path enrichment analysis using the WebGestalt platform (18), with the parameters hsapiens > overrepresentation enrichment Analysis > geneontology > Biological process and hsapiens > overrepresentation enrichment Analysis > Pathway > KEGG.
Supplementary Material
Acknowledgments
We thank the communities involved in this research, the participants who provided samples for analysis, and David Reich for helping to process the Affymetrix Human Origins Array data, including merging the data collected in this study with the previously published Human Origins Array data. A.W.R. was supported by an NSF Graduate Research Fellowship. This work was supported by grants from the Norman Hackerman Advanced Research Program of the Texas Higher Education Coordinating Board, NSF Awards SMA-1408876 and BCS-1412501, and Wenner–Gren Foundation for Anthropological Research Grant 8773.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819467116/-/DCSupplemental.
References
- 1.Fan S, Hansen MEB, Lo Y, Tishkoff SA. Going global by adapting local: A review of recent human adaptation. Science. 2016;354:54–59. doi: 10.1126/science.aaf5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perry GH, et al. Adaptive, convergent origins of the pygmy phenotype in African rainforest hunter-gatherers. Proc Natl Acad Sci USA. 2014;111:E3596–E3603. doi: 10.1073/pnas.1402875111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karlsson EK, et al. Natural selection in a bangladeshi population from the cholera-endemic ganges river delta. Sci Transl Med. 2013;5:192ra86. doi: 10.1126/scitranslmed.3006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolnick DA, Raff JA, Springs LC, Reynolds AW, Miró-Herrans AT. Native American genomics and population histories. Annu Rev Anthropol. 2016;45:319–340. [Google Scholar]
- 6.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bigham A, et al. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6:e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlebusch CM, et al. Human adaptation to arsenic-rich environments. Mol Biol Evol. 2015;32:1544–1555. doi: 10.1093/molbev/msv046. [DOI] [PubMed] [Google Scholar]
- 9.Mychaleckyj JC, et al. Genome-wide analysis in Brazilians reveals highly differentiated Native American genome regions. Mol Biol Evol. 2017;34:559–574. doi: 10.1093/molbev/msw249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindo J, et al. A time transect of exomes from a Native American population before and after European contact. Nat Commun. 2016;7:13175. doi: 10.1038/ncomms13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fumagalli M, et al. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science. 2015;349:1343–1347. doi: 10.1126/science.aab2319. [DOI] [PubMed] [Google Scholar]
- 12.Auton A, et al. 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: A discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet. 2013;93:278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergey CM, et al. Polygenic adaptation and convergent evolution on growth and cardiac genetic pathways in African and Asian rainforest hunter-gatherers. Proc Natl Acad Sci USA. 2018;115:E11256–E11263. doi: 10.1073/pnas.1812135115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stelzer G, et al. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1–1.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45:W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peel MC, Finlayson BL, McMahon TA. Updated world map of the Köppen-Geiger climate classification. Hydrol Earth Syst Sci Discuss. 2007;4:439–473. [Google Scholar]
- 20.Freisen TM, Mason OK. Archaeology of the North American arctic. In: Freisen TM, Mason OK, editors. The Oxford Handbook of the Prehistoric Arctic. Oxford Univ Press; New York: 2016. pp. 1–16. [Google Scholar]
- 21.De Lorenzo F, Kadziola Z, Mukherjee M, Saba N, Kakkar VV. Haemodynamic responses and changes of haemostatic risk factors in cold-adapted humans. QJM. 1999;92:509–513. doi: 10.1093/qjmed/92.9.509. [DOI] [PubMed] [Google Scholar]
- 22.Zhang YY, et al. BKCa and hEag1 channels regulate cell proliferation and differentiation in human bone marrow-derived mesenchymal stem cells. J Cell Physiol. 2014;229:202–212. doi: 10.1002/jcp.24435. [DOI] [PubMed] [Google Scholar]
- 23.Cardona A, et al. Genome-wide analysis of cold adaptation in indigenous Siberian populations. PLoS One. 2014;9:e98076. doi: 10.1371/journal.pone.0098076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jablonski NG, Chaplin G. Colloquium paper: Human skin pigmentation as an adaptation to UV radiation. Proc Natl Acad Sci USA. 2010;107:8962–8968. doi: 10.1073/pnas.0914628107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hancock AM, et al. Adaptations to climate-mediated selective pressures in humans. PLoS Genet. 2011;7:e1001375. doi: 10.1371/journal.pgen.1001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ubelaker DH. Population size, contact to nadir. In: Ubelaker DH, editor. Handbook of North American Indians, Volume 3: Environment, Origins, and Population: Environment, Origins, and Population. Smithsonian Institution Press; Washington, DC: 2006. pp. 694–791. [Google Scholar]
- 27.Jones EE. A spatiotemporal analysis of old world diseases in North America, A.D. 1519-1807. Am Antiq. 2014;3:487–506. [Google Scholar]
- 28.Stojanowski CM. Biocultural Histories in La Florida: A Bioarchaeological Perspective. Univ of Alabama Press; Tuscaloosa, AL: 2005. [Google Scholar]
- 29.Ramenofsky AF. Loss of innocence: Explanations of differential persistence in the sixteenth-century Southeast. In: Thomas DH, editor. Columbian Consequences: The Spanish Borderlands in Pan-American Perspective. Vol 2. Smithsonian Institution Press; Washington, DC: 1990. pp. 31–48. [Google Scholar]
- 30.Kelton P. Shattered and infected: Epidemics and the origins of the Yamasee War, 1696–1715. In: Ethridge R, Shuck-Hall SM, editors. Mapping the Mississippian Shatter Zone: The Colonial Indian Slave Trade and Regional Instability in the American South. Univ of Nebraska Press; Lincoln, NE: 2009. pp. 312–332. [Google Scholar]
- 31.Galloway P. Choctaw Genesis, 1500-1700. Univ of Nebraska Press; Lincoln, NE: 1995. [Google Scholar]
- 32.Acuna-Soto R, Stahle DW, Cleaveland MK, Therrell MD. Megadrought and megadeath in 16th century Mexico. Emerg Infect Dis. 2002;8:360–362. doi: 10.3201/eid0804.010175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Restall M, Schwaller R. The gods return: Conquest and conquest society (1502- 1610) In: Beezley WH, editor. A Companion to Mexican History and Culture. Wiley-Blackwell; Malden, MA: 2011. pp. 195–208. [Google Scholar]
- 34.Hicks SJ, Theodoropoulos G, Carrington SD, Corfield AP. The role of mucins in host-parasite interactions. Part I-protozoan parasites. Parasitol Today. 2000;16:476–481, erratum (2001) 17:135. doi: 10.1016/s0169-4758(00)01773-7. [DOI] [PubMed] [Google Scholar]
- 35.Guernier V, Hochberg ME, Guégan J-F. Ecology drives the worldwide distribution of human diseases. PLoS Biol. 2004;2:e141. doi: 10.1371/journal.pbio.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt EE, et al. GenomeRNAi: A database for cell-based and in vivo RNAi phenotypes, 2013 update. Nucleic Acids Res. 2013;41:D1021–D1026. doi: 10.1093/nar/gks1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobs BL, et al. Vaccinia virus vaccines: Past, present and future. Antiviral Res. 2009;84:1–13. doi: 10.1016/j.antiviral.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vågene AJ, et al. Salmonella enterica genomes from victims of a major sixteenth-century epidemic in Mexico. Nat Ecol Evol. 2018;2:520–528. doi: 10.1038/s41559-017-0446-6. [DOI] [PubMed] [Google Scholar]
- 39.Eichstaedt CA, et al. Positive selection of AS3MT to arsenic water in Andean populations. Mutat Res. 2015;780:97–102. doi: 10.1016/j.mrfmmm.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindo J, et al. The genetic prehistory of the Andean highlands 7000 years BP though European contact. Sci Adv. 2018;4:eaau4921. doi: 10.1126/sciadv.aau4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 44.Cadzow M, et al. A bioinformatics workflow for detecting signatures of selection in genomic data. Front Genet. 2014;5:293. doi: 10.3389/fgene.2014.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maclean CA, Chue Hong NP, Prendergast JG. hapbin: An Efficient Program for performing haplotype-based scans for positive selection in large genomic datasets. Mol Biol Evol. 2015;32:3027–3029. doi: 10.1093/molbev/msv172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Danecek P, et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M. Robust demographic inference from genomic and SNP data. PLoS Genet. 2013;9:e1003905. doi: 10.1371/journal.pgen.1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 49.McLaren W, et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.