Significance
An obligatory step in the replication cycle of all retroviruses involves reverse transcription of their RNA genomes into double-stranded DNA for integration into the host cell chromosome. The process of synthesizing double-stranded HIV-1 DNA is carried out by viral reverse transcriptase, which lacks some of the enzymatic activities necessary to accomplish this task. Thus, the process is completed by cellular postreplication DNA repair enzymes. Here, we report that select aspects of the cellular postreplication DNA machinery restrict HIV-1 replication in dividing T cells and that these restrictions are counteracted by HIV-1 Vpr accessory virulence factor. Our studies reveal that specific cellular postreplication DNA repair enzymes can sense and inhibit HIV-1 infection, and hence constitute a class of HIV-1 restriction factors.
Keywords: HIV-1, restriction, postreplication DNA repair, Vpr, HLTF
Abstract
Lentiviruses, including HIV-1, possess the ability to enter the nucleus through nuclear pore complexes and can infect interphase cells, including those actively replicating chromosomal DNA. Viral accessory proteins hijack host cell E3 enzymes to antagonize intrinsic defenses, and thereby provide a more permissive environment for virus replication. The HIV-1 Vpr accessory protein reprograms CRL4DCAF1 E3 to antagonize select postreplication DNA repair enzymes and activates the DNA damage checkpoint in the G2 cell cycle phase. However, little is known about the roles played by these Vpr targets in HIV-1 replication. Here, using a sensitive pairwise replication competition assay, we show that Vpr endows HIV-1 with a strong replication advantage in activated primary CD4+ T cells and established T cell lines. This effect is disabled by a Vpr mutation that abolishes binding to CRL4DCAF1 E3, thereby disrupting Vpr antagonism of helicase-like transcription factor (HLTF) DNA helicase and other DNA repair pathway targets, and by another mutation that prevents induction of the G2 DNA damage checkpoint. Consistent with these findings, we also show that HLTF restricts HIV-1 replication, and that this restriction is antagonized by HIV-1 Vpr. Furthermore, our data imply that HIV-1 Vpr uses additional, yet to be identified mechanisms to facilitate HIV-1 replication in T cells. Overall, we demonstrate that multiple aspects of the cellular DNA repair machinery restrict HIV-1 replication in dividing T cells, the primary target of HIV-1 infection, and describe newly developed approaches to dissect key components.
Replication of retroviruses involves reverse transcription of their RNA genome into double-stranded DNA for integration into the host cell chromosome. Unlike gammaretroviruses, which are generally unable to cross the nuclear membrane, with integration contingent upon nuclear membrane breakdown during mitosis, HIV-1 and other lentiviruses can access the interphase nucleus through the nuclear pore (1–3).
Reverse transcription of the HIV-1 single-stranded RNA genome into double-stranded DNA provirus is initiated by a single tRNALys3 primer (4). Extension of this primer, carried out by a viral polymerase [reverse transcriptase (RT)], results in synthesis of a continuous minus strand provirus DNA. Synthesis of the plus strand of the provirus, on the other hand, is primed by multiple RNA primers (5–7). Hence, the nascent plus strand contains gaps and flaps, displaced branched DNA/RNA structures (7, 8) that cannot be converted into a continuous DNA strand by the enzymatic activities of the viral RT alone. Notably, the noncanonical DNA structures present in HIV-1 reverse transcription intermediates resemble those that are transiently present in the lagging DNA strand during chromosomal DNA replication. The latter are removed by postreplication DNA repair enzymes, which complete lagging strand synthesis during S-phase (9). Thus, postreplication DNA repair proteins are likely candidates for enzymes that process HIV-1 reverse transcription intermediates into fully double-stranded provirus DNA. Little is known, however, about these processes, as they have remained largely unexplored.
Innate immunity and intrinsic immunity employ a broad array of effector mechanisms to sense and inhibit retrovirus infection (10). In response, viruses have evolved countermeasures that neutralize the cellular defenses, and these measures are typically carried out by viral accessory virulence factors (11–13). HIV-1 encodes four such accessory virulence proteins: Vif, Vpu, Vpr, and Nef. These small adaptor proteins nucleate novel protein complexes and use them to subvert key antiviral mechanisms. Vif, Vpu, and Vpr each bind to a specific cellular E3 ubiquitin ligase complex and recruit novel protein substrates possessing anti–HIV-1 activity, thereby directing them for degradation via the ubiquitin/proteasome pathway (12). Whereas Vif and Vpu use their target E3s to antagonize cellular proteins with demonstrated antiviral activity (14–16), the roles of the usurped by HIV-1 Vpr E3 enzyme and its Vpr-recruited substrates in the HIV-1 replication cycle remain poorly understood.
HIV-1 Vpr, a virion-associated protein, reprograms CRL4DCAF1 E3 ubiquitin ligase by binding to its DCAF1 substrate receptor subunit (17–20). Interestingly, known Vpr-recruited substrates of this E3 are key enzymes in select postreplication DNA repair pathways. In particular, the nuclear isoform of uracil-DNA glycosylase (UNG2) initiates uracil-specific base excision repair (21, 22); helicase-like transcription factor (HLTF) DNA helicase processes branched DNA structures and catalyzes replication fork reversal (23–25); MUS81-EME1(-EME2) nuclease also processes branched DNA structures (26–28); and exonuclease 1 (Exo1), a versatile exonuclease possessing FLAP nuclease and ribonuclease H (RNAseH) activity, is involved in homologous recombination-mediated DNA repair processes, as well as in mismatch repair (29, 30). HIV-1 Vpr also targets for degradation the DCAF1-associated methylcytosine dioxygenase TET2 (31). TET2 mediates active DNA demethylation and is involved in damaged DNA repair (32, 33). In view of the above, it is thus not surprising that a subset of Vpr molecules is chromatin-bound and associated with DNA repair foci (34, 35) and that Vpr activates the DNA damage checkpoint, leading to cell cycle arrest in G2 phase (35, 36). The above evidence reveals that HIV-1 Vpr engages in multiple functional interactions with the postreplication DNA repair machinery.
By analogy to other antiviral factors antagonized by viral accessory proteins (11, 13), the above studies proposed a model in which select postreplication DNA repair enzymes restrict HIV-1 replication and are removed from infected cells by HIV-1 Vpr, via CRL4DCAF1 E3, to relieve the restrictions. In support of this model is the finding that UNG2 restricts HIV-1 infection in macrophages by initiating uracil excision in nascent HIV-1 reverse transcripts (37). Furthermore, Exo1 was shown to restrict HIV-1 replication in T cells (29). However, little is known about the roles of HIV-1 Vpr interactions with other aspects of postreplication DNA repair proteins in the HIV-1 replication cycle.
Here, we continued testing the hypothesis that specific aspects of the DNA repair machinery restrict HIV-1 infection in T lymphocytes and that HIV-1 Vpr counteracts these restrictions. Using a sensitive HIV-1 replication fitness assay, we found that HIV-1 Vpr promotes HIV-1 replication by antagonizing the restriction mediated by a DNA repair pathway controlled by HLTF DNA helicase and multifunctional Exo1 nuclease. Overall, our findings reveal that multiple aspects of the cellular DNA repair machinery restrict HIV-1 in dividing T cells.
Results
Vpr Promotes HIV-1 Replication in T Cells.
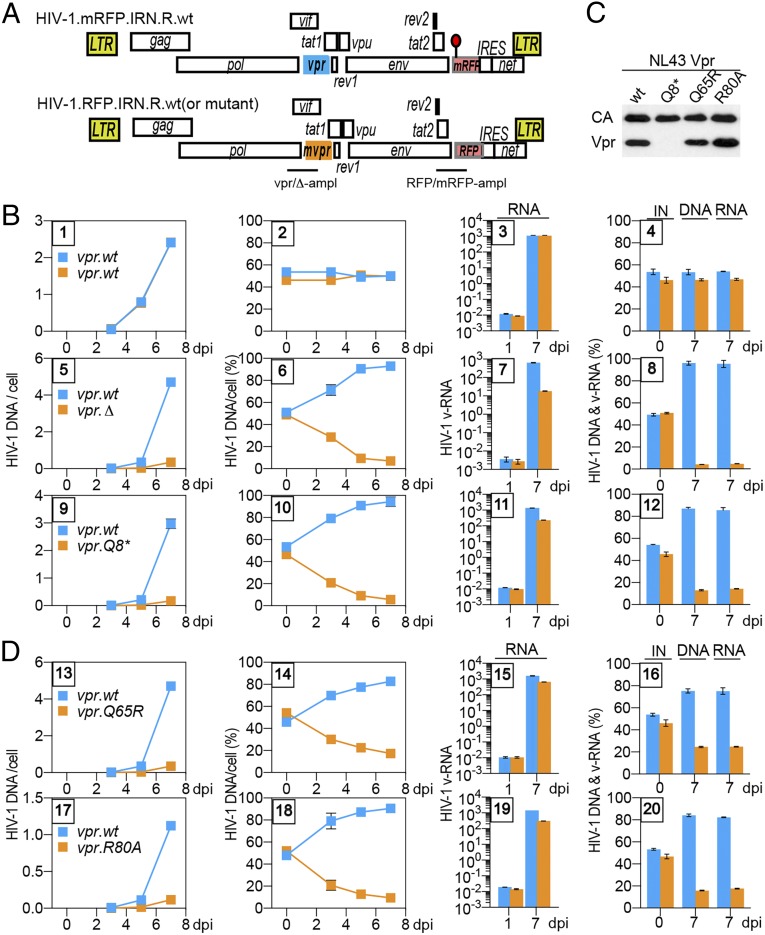
The vpr-dependent differences in HIV-1 replication kinetics in T cells were previously detected using parallel spreading replication assays (29, 38, 39). These differences were not very robust, and hence not widely accepted. Therefore, we implemented a robust, internally controlled pairwise replication competition assay (PRCA) for the purpose of in-depth studies of vpr’s role in HIV-1 replication in T cells. PRCA provides a robust measure of relative HIV fitness as viral variants compete for the same cell population under identical environmental conditions (40, 41). For this assay, we used a pair of HIV-1 NL4-3–derived reporter viruses that were isogenic, except for a cluster of silent mutations in the red fluorescent protein (RFP) marker gene inserted between env and nef ORFs. The wild-type (wt) and mutant RFP (mRFP) sequences provided unique tags for selective detection of each of the two viruses in coinfection experiments by qPCR assay (Fig. 1A). To validate the assay, CEM.SS T cells were infected with a 1:1 mixture of the two viruses, both containing wt NL4-3 vpr gene (vpr.wt), at a low multiplicity of infection (moi). At 3, 5, and 7 d postinfection (dpi), cell-associated HIV-1 DNA and cell-free HIV-1 RNA in cell culture supernatants were quantified by qPCR/qRT-PCR. Of note, only the infected cultures with less than 5% of cells expressing RFP/mRFP marker proteins at 7 dpi were selected for analysis to minimize the likelihood of coinfection by the competing viruses and subsequent generation of recombinants that could lead to suppression of the vpr phenotype. As expected, the viruses replicated at similar rates, with their ratios remaining constant over time (Fig. 1B, panels 1–4). Next, we assessed the relative replication fitness of HIV-1 viruses with the vpr gene disrupted, either by a premature stop codon at Vpr glutamine residue Q8 (vpr.Q8*) or by a frameshift-generating mutation at glycine residue G43 (vpr.Δ). Specifically, CEM.SS T cells were coinfected with mRFP-reporter HIV-1 carrying vpr.wt (HIV-1.mRFP.vpr.wt) and RFP-reporter HIV-1 carrying the vpr.Q8* (HIV-1.RFP.vpr.Q8*) or the vpr.Δ allele (HIV-1.RFP.vpr.Δ). Significantly, both vpr mutations markedly attenuated HIV-1 replication (Fig. 1B, panels 5–12 and SI Appendix, Fig. S1). This effect, seen for both cell-associated DNA and cell-free viral RNA, was already detectable at 3 dpi, and at 7 dpi, the vpr-defective HIV-1 was severely outcompeted by the HIV-1 with wt vpr gene (∼5–10% vpr.Q8*/vpr.Δ compared with ∼90–95% vpr.wt). Importantly, this vpr phenotype was also seen in PRCA with replication-competent HIV-1 carrying vpr.wt or vpr.Δ allele and lacking the RFP/mRFP and internal ribosome entry site (IRES) elements (i.e., devoid of any exogenous sequences) (SI Appendix, Fig. S2), and was seen when the competing vpr.wt and vpr.Q8* viruses harbored red (RFP-IRES) and/or blue (BFP-IRES) reporter cassettes and replication was quantified by flow cytometry (SI Appendix, Fig. S3). Together, the above experiments corroborate, through independent assays, that Vpr promotes HIV-1 replication in T cells. This finding confirms some previous reports (29, 38, 39) and counters a common perception that Vpr facilitates HIV-1 replication only in monocyte-derived macrophages (42–46).
Fig. 1.
Vpr promotes HIV-1 replication in CEM.SS T cells. (A) Schematic representation of HIV-1.mRFP.vpr and HIV-1.RFP.vpr reporter constructs used in the PRCA. ORFs are shown as rectangles. The viruses are isogenic, except for an array of silent mutations in the RFP gene, indicated by a red lollipop, which provides unique primer annealing sites in the wt RFP and its mutant (mRFP) that allow their quantification by qPCR in coinfection assays. In some experiments, mutations were introduced into the vpr gene (mvpr) of the HIV-1.RFP construct. For the mvpr constructs, another set of primers, distinguishing between the vpr.wt and vpr.Δ alleles, was used to quantify viruses carrying those alleles. The locations of the vpr.wt, the deleted vpr.Δ amplicons (vpr-ampl), and the RFP and mRFP amplicons (RFP-ampl) are indicated. (B) PRCA reveals a positive effect of HIV-1 vpr on HIV-1 replication in CEM.SS T cells. CEM.SS T cells were infected with a normalized mixture, at 1:1 ratio, of HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.wt (panels 1–4), HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.Δ (panels 5–8), or HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.Q8* (panels 9–12), at an moi of 0.006–0.02. The infected cultures were sampled 3, 5, and 7 dpi, and cell-associated HIV-1 DNA, as well as cell-free HIV-1 RNA, for each of the competing viruses was quantified by qRT-PCR, using mRFP and RFP amplicons and, in some experiments, also using vpr.wt and vpr.Δ amplicons (SI Appendix, Fig. S1). The number of copies per cell of cell-associated HIV-1 DNA from each of the competing viruses is shown in panels 1, 5, and 9, with the relative percentages shown in panels 2, 6, and 10. Quantification of cell-free viral RNA in culture medium on 1 dpi and 7 dpi is shown in panels 3, 7, and 11. Percentages of the competing viruses in the initial infecting mixture (IN) of cell-associated DNA and cell-free RNA at 7 dpi are shown in panels 4, 8, and 12. The cell-associated HIV-1 DNA data (two leftmost panels) and the virion-associated HIV-1 RNA data (two rightmost panels) are from different experiments. The data are representative of three independent experiments. (C) Virion incorporation of mutant HIV-1 Vpr proteins. Concentrated, normalized HIV-1.mRFP.R viruses with wt or mutated vpr gene were analyzed by immunoblotting with antibodies reacting with p24 capsid or HIV-1 Vpr. (D) Vpr functions mediated via Q65 and R80 account for the positive effect of Vpr on viral fitness. PRCA was performed as described above with the HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.Q65R mixture (panels 13–16) or the HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.R80A mixture (panels 17–20).
The Positive Effect of Vpr on HIV-1 Replication Requires Vpr Glutamine Q65 and Arginine R80.
To assess whether Vpr interaction with CRL4DCAF1 E3 and/or the DNA damage checkpoint has a role in HIV-1 replication, we tested the effects of two Vpr mutations, Q65R and R80A, that disrupt these functions. In particular, Vpr.Q65R binds DCAF1 poorly and is defective for all Vpr functions mediated by the CRL4DCAF1 E3 ligase, including its ability to deplete HLTF, UNG2, Exo1, MUS81, and TET2 (19, 24, 31). The Vpr.R80A variant retains the ability to bind DCAF1 and functions through its associated CRL4 E3 (27, 47). However, neither the Vpr.Q65R variant nor the Vpr.R80A variant arrests cells in G2 phase (19, 48).
PRCA was performed with mixtures of the reference mRFP-reporter HIV-1 vpr.wt and the RFP-reporter HIV-1 vpr.Q65R or vpr.R80A viruses. Of note, both the Vpr.Q65R and Vpr.R80A proteins were well packaged into HIV-1 virions (Fig. 1C). Interestingly, both variants were grossly defective for stimulating HIV-1 replication in CEM.SS T cells, similar to those with vpr.Q8* or vpr.Δ mutation (Fig. 1D). Of note, when competing directly, the vpr.Q65R and vpr.R80A viruses replicated at roughly similar rates, as expected (SI Appendix, Fig. S4).
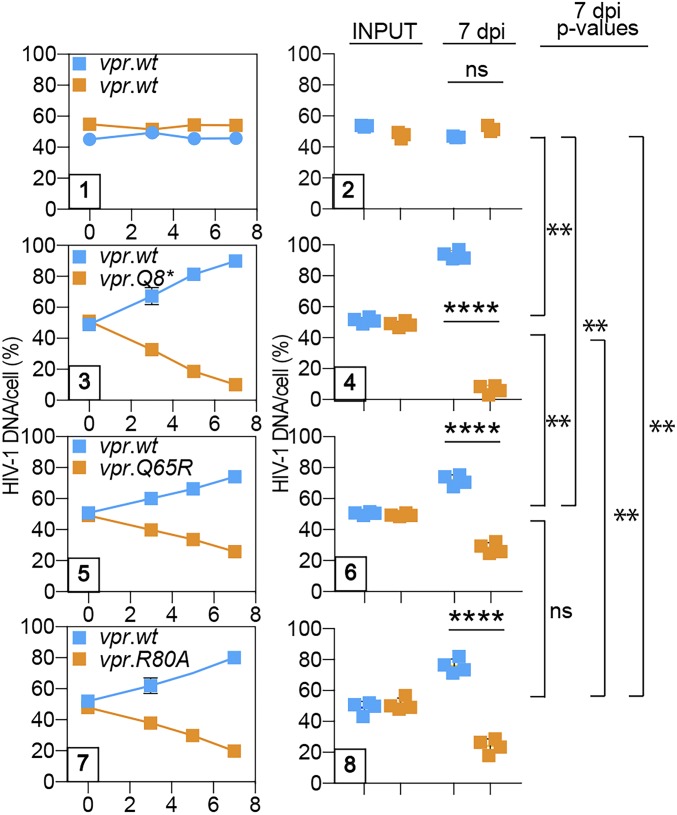
HIV-1 Vpr Q65 and R80 Mediate Enhanced HIV-1 Replication in Activated Primary CD4+ T Cells.
As further validation of our PRCA data, we next assessed Vpr’s role in HIV-1 replication in activated primary CD4+ T cells. HIV-1 with the inactivating Vpr Q8* mutation was severely outcompeted (∼19:1 by 7 dpi) by HIV-1 encoding wt Vpr protein (Fig. 2), similar to what we observed in CEM.SS T cells. The effects of the Vpr Q65R and R80A mutations were also strong but less drastic, with the mutant viruses outcompeted by approximately fourfold. The quantitative differences between the effects of the Q8* and Q65R or R80A mutations were statistically significant, suggesting that Vpr acts to promote HIV-1 replication in a more complex manner in activated primary CD4+ T cells than in CEM.SS T cells. These data reveal that HIV-1 Vpr glutamine Q65 and arginine R80 mediate enhanced HIV-1 replication in T cells, thus linking this effect to Vpr interactions with CRL4DCAF1 E3 and its Vpr-recruited substrates (via Q65R), as well as with other aspects of the postreplication DNA repair machinery (via R80A), and, importantly, validate CEM.SS T cells as a model to study the role of these interactions in HIV-1 replication.
Fig. 2.
Positive effect of Vpr on HIV-1 replication in activated primary CD4+ T cells is linked to interaction with CRL4DCAF1 E3 and activation of the DNA damage checkpoint. CD4+ T cells were activated with α-CD3/α-CD28 beads, infected with 1:1 mixture of HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.wt (panels 1–2), HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.Q8* (panels 3–4), HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.Q65R (panels 5–6), or HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.R80A (panels 7–8), at a low moi. The percentage of cell-associated HIV-1 DNA for viruses in each of the competing pairs over time is shown for representative experiments (panels 1, 3, 5, and 7). Percentages of competing viruses in the inocula (INPUT) and of cell-associated DNA at 7 dpi, determined for each virus pair in four biological replicate experiments, are also shown (panels 2, 4, 6, and 8). Each experiment was performed with cells from a different donor. The statistical significance of differences between competing viruses in each pair (t test) within the graphs and among pairs (one-way ANOVA with a post hoc Tukey test) is shown on the right side of the panels. **P < 0.01; ****P < 0.0001. ns, not significant.
HLTF Restricts HIV-1 Replication in T Cells in a Vpr-Dependent Manner.
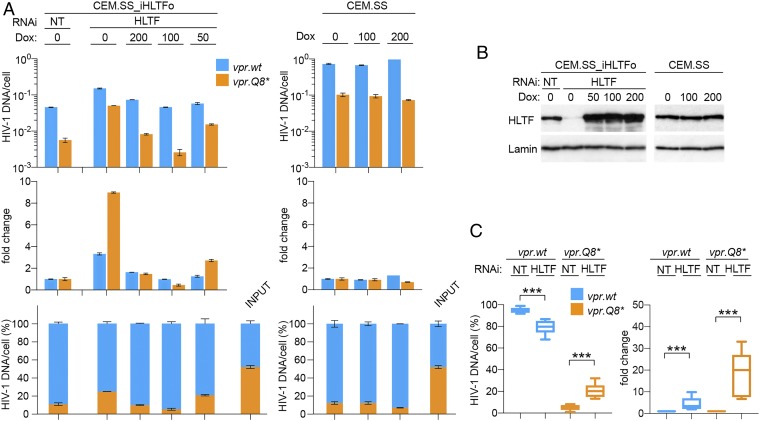
We next focused our attention on the HTLF DNA helicase. HLTF was previously identified as a direct substrate of the CRL4DCAF1 E3 ubiquitin ligase that is reprogrammed by HIV-1 Vpr (24, 25, 49). To test whether HLTF restricts HIV-1 replication, PRCA with a pair of HIV-1 viruses carrying wt or Q8* mutated vpr gene was performed using a CEM.SS T cell population harboring a doxycycline-inducible RNA interference (RNAi)-resistant codon-optimized HLTF transgene (CEM.SS_iHLTFo). The cells were subjected to nontargeting (NT) or endogenous HLTF-targeting RNAi in the absence or presence of doxycycline (Fig. 3B), and PRCA was initiated 3 d later. Replication of HIV-1 with Q8* mutated vpr gene in HLTF-depleted cells was enhanced compared with that in control cells at 7 dpi (Fig. 3A, Top and Middle), indicating that HLTF restricts HIV-1 replication. This effect, consistently observed in multiple independent experiments, was also reflected by an increased percentage of HIV-1 with the defective vpr.Q8* allele in cell-associated viral DNA (Fig. 3 A, Bottom and C). Significantly, replication of HIV-1 with wt vpr gene was also enhanced in HLTF-depleted cells, although to a lesser extent than that of the vpr-deficient HIV-1 (threefold vs. ninefold; Fig. 3A, Middle), indicating that Vpr only partially antagonizes this restriction. Finally, HLTF depletion did not restore the replication of HIV-1 with vpr.Q8* to levels seen for HIV-1 with vpr.wt allele, indicating that Vpr employs additional mechanisms, besides antagonizing HLTF, to promote HIV-1 replication. Similar observations were made in experiments with another T cell line, HPB.ALL T cells, albeit the Vpr antagonism appeared less robust in these cells, indicating that the HTLF restriction phenotype was not limited to CEM.SS T cells (SI Appendix, Fig. S5). Importantly, doxycycline-induced ectopic expression of the RNAi-resistant HLTFo in HLTF-depleted cells suppressed HIV-1 replication and restored the proportions of the viruses with vpr.wt and vpr.Q8* alleles to levels similar to those seen in control CEM.SS T cells subjected to NT RNAi (Fig. 3A). This result excluded the possible involvement of off-target effects of HLTF-directed RNAi and corroborated that the observed virological phenotypes reflect genuine HLTF-mediated restriction of HIV-1 replication.
Fig. 3.
HIV-1 Vpr antagonizes HLTF-mediated restriction of HIV-1 replication in T cells. (A) HLTF restricts HIV-1 replication. CEM.SS T cells harboring doxycycline-inducible codon-optimized HLTF transgene (CEM.SS_iHLTFo) were subjected to NT or targeting endogenous HLTF mRNA (HLTF) RNAi in the absence or presence of the indicated concentrations of doxycycline (ng/mL). (Left) Three days after initiation of RNAi, PRCA was performed with a 1:1 mixture of HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.Q8*. (Right) As a control, PRCA was also performed in parental CEM.SS T cells cultured in the absence or presence of doxycycline. (Upper) Cell-associated DNA of the competing viruses was quantified at 7 dpi. (Middle) Fold-change in HIV-1 DNA copies in cells subjected to HLTF RNAi, normalized to those in cells subjected to NT RNAi. (Lower) Percentages of cell-associated HIV-1 DNA for the competing viruses. (B) HLTF levels in CEM.SS_iHLTFo and parental CEM.SS T cell populations used in experiments shown in A were revealed by immunoblotting. Lamin B1 provided a loading control. (C) HLTF depletion partially restores the HIV-1 replication fitness advantage provided by Vpr. CEM.SS T cells were subjected to NT or HLTF-targeting (HLTF) RNAi and infected with a 1:1 mixture of HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.Q8*. Percentages of cell-associated DNA for the competing viruses at 7 dpi (Left) and fold-change in HIV-1 DNA copies per cell in cells subjected to HLTF-targeting RNAi normalized to those in cells subjected to NT RNAi (Right) are shown. The data shown are from five biological replicate experiments. ***P < 0.001.
HLTF HIRAN Domain Mediates Restriction of HIV-1 Infection.
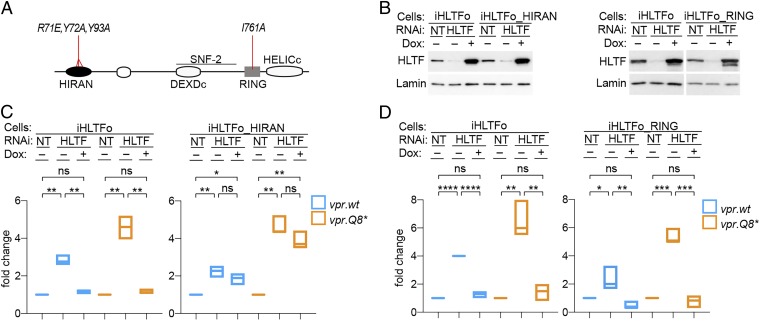
HLTF is a modular multidomain protein (Fig. 4A) controlling distinct postreplication DNA repair pathways. For example, through its RING domain, HLTF acts as an E3 ubiquitin ligase for polyubiquitylation of proliferating cell nuclear antigen (PCNA) and prevents recruitment of error-prone DNA polymerases to PCNA, thereby suppressing this mutagenic repair pathway (50, 51). Distinctively, in a RING domain-independent manner, the HLTF HIRAN domain binds the 3′ end of single-stranded DNA and recruits HLTF to fork-like branched DNA structures, where it acts as an ATP-dependent double-stranded DNA translocase to remodel these structures and displace bound proteins (2, 52, 53). To determine whether RING and/or HIRAN domain-mediated HLTF functions have a role in restricting HIV-1 replication, we took advantage of HLTF variants containing mutations that disrupt HIRAN single-stranded DNA 3′ end binding (K71E/Y72A/Y73A) (53) or disrupt the RING domain surface that mediates binding to the E2 enzyme (I761A) (54).
Fig. 4.
HLTF DNA-binding HIRAN domain mediates restriction of HIV-1 replication. (A) HLTF domain organization and summary of mutations. Schematic representation of the HLTF protein showing the location of the HIRAN, RING, SNF2, and discontinuous DEXDc and HELICc helicase domains. Locations of mutations disrupting DNA binding by the HIRAN domain (R71E, Y72A, Y93A) and E3 ubiquitin ligase activity of the RING finger domain (I761A) are indicated. (B) Endogenous and ectopic HLTF levels in cell lysates were revealed by immunoblotting with α-HLTF antibody. Lamin B1 provided a loading control. Dox, doxycycline. (C) HIRAN domain mediates restriction of HIV-1 replication. Control CEM.SS_iHLTFo (Left) and CEM.SS_iHLTFo_HIRAN (Right) T cells were subjected to NT or HLTF-targeting (HLTF) RNAi in the absence (−) or presence (+) of Dox (100 ng/mL), as indicated. PRCA with a 1:1 mixture of HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.Q8* was initiated 3 d later. Cell-associated DNA for each of the competing viruses was quantified at 7 dpi and normalized to that in cells treated with NT RNAi. The fold-change in HIV-1 DNA copies per cell in cells depleted of endogenous HLTF and ectopically expressing HLTF with wt (Left) or mutant (Right) HIRAN domain is shown. The data shown are from three biological replicate experiments. *P < 0.05; **P < 0.01; ***P < 0.001. ns, not significant. (D) Functional RING domain is dispensable for HLTF-mediated restriction. Experiments with CEM.SS_iHLTFo and CEM.SS_iHLTFo_RING T cells were performed exactly as described for C above.
RNAi-resistant HLTF gene variants harboring HIRAN (HLTFo_HIRAN) or RING domain (HLTFo_RING) mutations were transduced into CEM.SS T cells as doxycycline-inducible transgenes. The resulting cell populations, along with control CEM.SS_iHLTFo T cells, were subjected to NT or HLTF-targeting RNAi, in the absence or presence of doxycycline, leading to efficient depletion of the endogenous HLTF levels by 3 d postinitiation of RNAi (Fig. 4B). Next, the cells were challenged with a 1:1 mixture of HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.Q8*, and their replication was quantified 7 dpi. Depletion of endogenous HLTF was associated with an accelerated replication of both viruses, with the effect being more pronounced for the vpr-defective Q8* HIV-1. Ectopic expression of HLTFo suppressed replication of both viruses to levels similar to those seen in control cells (NT), as expected (Fig. 4 C and D, Left). Notably, expression of HLTFo_HIRAN failed to suppress replication of both viruses in endogenous HLTF-depleted cells, despite expression levels being comparable to those of wt HLTFo (Fig. 4C, Right). In contrast, the RING domain-mutated HLTFo_RING efficiently suppressed HIV-1 replication (Fig. 4D, Right). We conclude that single-stranded DNA 3′ end-binding activity of the HLTF HIRAN domain mediates the restriction of HIV-1 replication, whereas the RING domain E3 ubiquitin ligase activity is likely dispensable.
Exo1 and HLTF Effects Are Largely Codependent.
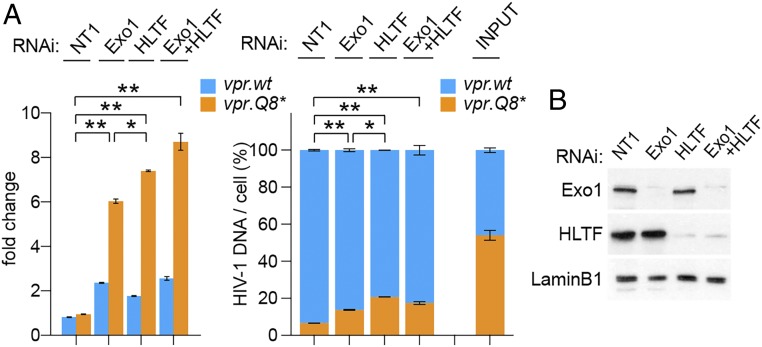
Exo1, a versatile 5′→3′ DNA exonuclease involved in multiple DNA repair pathways, restricts HIV-1 replication in T cells, and this restriction is counteracted by HIV-1 Vpr (29). Epistasis experiments were performed to assess the relation between Exo1- and HLTF-mediated restrictions. Exo1 and HLTF levels were knocked down individually or in combination by RNAi in CEM.SS T cells, and PRCA was performed with a pair of HIV-1 viruses harboring the vpr.wt or vpr.Q8* allele. As shown in Fig. 5, both viruses replicated more rapidly in Exo1-depleted cells, and this effect was more pronounced for the vpr-defective vpr.Q8* HIV-1, similar to what we observed with HLTF-depleted cells. Of note, the latter effect, while not detected previously in parallel replication assays (29), was clearly revealed in the more robust, internally controlled PRCA. Interestingly, virus replication in Exo1- and HLTF-codepleted cells was similar to that seen upon Exo1 or HLTF single depletion. These data suggest that the Exo1- and HLTF-mediated restrictions are codependent, rather than additive, and target the same pool of HIV-1 cDNA.
Fig. 5.
Exo1 and HLTF epistasis studies. (A) CEM.SS T cells were subjected to NT, Exo1-targeting, and/or HLTF-targeting RNAi, as indicated. PRCA with a 1:1 mixture of HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.Q8* was initiated 3 d later, and cell-associated DNA for each of the competing viruses was quantified at 7 dpi. Fold-change (Left) and relative percentages (Right) of cell-associated HIV-1 DNA copies per cell for the competing viruses are shown at 7 dpi. A representative of three experiments is shown. Only P values for HIV-1.RFP.vpr.Q8* lower than 0.05 are shown: *P < 0.05; **P < 0.01. (B) Exo1 and HLTF levels in extracts from cells subjected to RNAi assessed at the time of HIV-1 infection were revealed by immunoblotting. Lamin B1 provided a loading control.
Search for Additional Components to the Positive Effect of Vpr on HIV-1 Replication in T Cells.
The studies described above indicate that HLTF antagonism contributes to the positive effect of Vpr on HIV-1 replication. They also suggest, however, that Vpr employs additional mechanisms, besides antagonizing HLTF, to promote HIV-1 replication. Hence, we were interested in learning whether other previously identified direct targets of HIV-1 Vpr, such as cyclic GMP-AMP synthase (cGAS)-mediated signaling (39) and TET2 (31), contribute significantly to the restriction of HIV-1 replication in CEM.SS T cells. As shown in Fig. 6 A and B, RNAi-mediated depletion of cGAS and TET2 did not significantly alter the competitive advantage provided by the presence of Vpr.
Fig. 6.
Search for additional Vpr-antagonized components that restrict HIV-1 replication in CEM.SS T cells. PRCA with HIV-1.mRFP.vpr.wt and HIV-1.RFP.vpr.Q8* was performed in CEM.SS T cells. Percentages of cell-associated HIV-1 DNA of the competing viruses at 7 dpi are shown. INPUT, percentages of the competing viruses in the initial inoculum. (A) TET2 and cGAS sensing pathway. CEM.SS T cells were subjected to NT RNAi using two different shRNAs (NT1 and NT2) or RNAi targeting cGAS or TET2. The cells were infected with HIV-1 3 d after initiation of RNAi. (B) cGAS and TET2 levels in cell extracts at the time of HIV-1 infection were revealed by immunoblotting. SF2 and lamin B1 provided loading controls. Results representative of two biological replicate experiments are shown. (C) Epigenetic modifiers. CEM.SS T cells were cultured in the presence of SAHA (200 nM), TSA (1 nM), GSK343 (0.7 μM), EX527 (10 μM), SGC0946 (2 μM), C646 (3 μM), and UNC0638 (1 μM), starting 12 h before HIV-1 infection until 7 dpi. Results representative of three biological replicate experiments are shown. (D) HUSH complex. CEM.SS T cells were subjected to NT RNAi (CT1), or shRNAs targeting TASOR. The cells were infected with HIV-1 3 dpi of RNAi. TASOR levels in cell extracts at the time of HIV-1 infection were revealed by immunoblotting. Lamin B1 provided a loading control. Results representative of two biological replicate experiments are shown.
HIV-1 Vpr was reported to modulate epigenetic silencing of HIV-1 proviruses (55, 56). We assessed the potential contribution of such effects to the Vpr-dependent enhancement of HIV-1 replication by interrogating the previously implicated epigenetic silencers or modifiers of HIV-1 provirus expression (57), using a panel of pharmacological inhibitors. In particular, we tested possible roles of (i) histone methyltransferases G9a with UNC0638 inhibitor, DOT1L with SGC0946, and EZH2 with GSK343; (ii) histone deacetylases with suberoylanilide hydroxamic acid (SAHA) and trichostatin A (TSA); (iii) p300 histone acetyltransferase with C646; and (iv) sirtuin 1 (SIRT1) with EX527. Although small effects were detected in some cases, they were not statistically significant (Fig. 6C).
Finally, using our sensitive PRCA, we reexamined the possible role of the human silencing hub (HUSH) repressor complex by RNAi-mediated depletion of its TASOR subunit. The HUSH complex silences a wide range of retroposons and retroviruses, including HIV-1, and is antagonized by HIV-2 Vpx and related Vpx and Vpr proteins found in lentiviruses infecting nonhuman primates (58–60). We found that TASOR depletion stimulated the replication of HIV-1 containing wt vpr gene by approximately twofold over a 7-d PRCA, but in a Vpr-independent manner (Fig. 6D). Of note, HIV-1 Vpr did not appear to detectably deplete any of the subunits of the HUSH complex, in contrast to simian immunodeficiency virus (SIV) mac239 Vpx (SI Appendix, Fig. S6), in agreement with previous reports (58, 61). Together, these data suggest that antagonism of the HUSH complex, or the epigenetic modifiers tested, by Vpr does not account for the Vpr-elicited HIV-1 replication phenotype in our experimental system. Additional studies are needed to identify relevant downstream targets of HIV-1 Vpr in T cells.
Discussion
HIV-1 Vpr has been thought to facilitate HIV-1 replication mainly in nondividing macrophages, and evidence for its role in cycling T lymphocytes has been sparse. Here, using pairwise replication competition assays, we demonstrate that Vpr potently stimulates HIV-1 replication in both CEM.SS and HPB.ALL T cell lines as well as in activated primary CD4+ T cells, the main cellular target of HIV-1 infection. Whereas previously used parallel replication assays can reveal vpr-dependent differences in HIV-1 replication rate, to be robust, the assays must be performed at a low moi and parallel cultures have to be treated in exactly the same way, with the experimental end point well before target cells become depleted by viral infection, since Vpr’s ability to arrest cell growth could differentially affect target cell availability. As such, noncompetitive replication assays are more error-prone and less well suited to reveal the positive effect of vpr on HIV-1 fitness compared with internally controlled PRCAs in which viral variants compete for the same cell population under identical environmental conditions. The latter, however, appear prone to generation of recombinant viruses, which could result in quantitative data variation.
The positive effect of HIV-1 Vpr is linked to its interactions with specific aspects of the cellular postreplication DNA repair machinery. We provide evidence that this effect is mediated, in part, by Vpr antagonism of the HLTF and Exo1 nuclease, the HIV-1 Vpr-recruited CRL4DCAF1 E3 substrate proteins (24, 29), and an as yet to be identified mechanism linked to the DNA damage checkpoint. Overall, our findings provide direct evidence for the existence of novel restrictions on HIV-1 replication conferred by postreplication DNA repair enzymes in dividing T cells, and their counteraction by HIV-1 Vpr.
Our data clearly show that HLTF restricts HIV-1 replication. The requirement for a functional HLTF HIRAN domain suggests a likely underlying mechanism. HIRAN, a single-stranded DNA 3′ end-binding domain, cooperates with HLTF translocase activity to unwind the leading DNA strand at stalled replication forks, thereby providing a template for error-free DNA synthesis and bypassing the lesion on the parental strand (53, 62). During replication fork remodeling, HLTF can displace a wide range of proteins bound to DNA (63). Significantly, branched DNA structures resembling the gapped-fork model of HLTF substrates are generated during plus strand DNA displacement synthesis by RT at early steps of retrovirus infection. The central DNA flap is an example of such a structure and results from displacement DNA synthesis through the RNAseH-resistant central polypurine track, which primes plus strand DNA at the center of the HIV-1 genome (7, 8). Similar structures may also be generated during LTR synthesis and in other regions of the HIV-1 genome (6, 64, 65). Notably, previous findings revealed that reversed fork structures, such as those generated by HLTF, can be substrates for nucleolytic degradation by Exo1 (66, 67), providing a possible explanation for the codependence of HLTF- and Exo1-mediated restrictions of HIV-1 infection, as observed here in epistasis studies. Thus, overall, a model in which sensing and processing of fork-like branched DNA structures by HLTF and Exo1 interfere with ordered progression of plus strand synthesis could explain our finding.
Interestingly, HLTF was recently identified to be a restriction factor for human cytomegalovirus (HCMV), which replicates and is maintained in the nucleus of the infected cells as a chromatinized episome (68). Significantly, the early gene product UL145 of HCMV antagonizes HLTF-mediated restriction, also via a CRL4 E3 enzyme (68). Thus, HLTF DNA helicase appears to have rather broad antiviral activity, restricting both DNA viruses and HIV-1, which replicates through a double-stranded DNA intermediate.
Our finding that both the Vpr glutamine Q65 and arginine R80 are required for the stimulation of HIV-1 replication in T cells links this effect to Vpr’s ability to activate the DNA damage checkpoint controlled by the ATR kinase (19, 36, 69). This checkpoint is canonically activated by sensing single-stranded DNA regions and junctions between single-stranded DNA and double-stranded DNA (70). Whereas the HIV-1 Vpr antagonisms with Exo1, HLTF, and Mus81 are not sufficient for checkpoint activation (24, 29) and the underpinning biochemical mechanism(s) remains to be elucidated, the dual requirement for glutamine Q65, which mediates Vpr subversion of the CRL4DCAF1 E3 complex, and for arginine R80 suggests a model in which R80 mediates recruitment of a novel CRL4DCAF1 E3 substrate protein(s) that restricts HIV-1 infection. The depletion of this protein(s), while relieving restriction, also leads to checkpoint activation and cell cycle arrest in G2 phase. Eventual identification of the HIV-1 Vpr target impinging on the ATR-mediated checkpoint will allow this model to be tested. Of note, HIV-1 Vpr arginine R80 probably mediates an additional function(s) that does not involve CRL4DCAF1 E3, as the consequences of mutating this residue are more severe than those resulting from disrupting Vpr binding with DCAF1 by glutamine Q65R mutation.
It is tempting to speculate that the ability of HIV-1 reverse transcription complex (RTC)/preintegration complex (PIC) to enter the nucleus when the postreplication DNA repair machinery is active in S phase sets the stage for restriction by DNA repair enzymes. Regarding the timing, one possible scenario is that repair processing of the HIV-1 reverse transcripts takes place before integration, and thereby interferes with ordered progression of plus strand DNA synthesis. This scenario is supported by our finding that vpr stimulates the levels of HIV-1 reverse transcription intermediates by roughly 20–30% in single infection cycle PRCA. As shown in SI Appendix, Fig. S7, this effect is also observed when PRCA is carried out in the presence of the integrase inhibitor raltegravir, and hence reflects vpr action before integration. Notably, this preintegration component appears to be a major contributor to the vpr effect on HIV-1 replication in CEM.SS T cells. Interestingly, HLTF was recently shown to restrict replication of HCMV, which persists in infected cells in the form of unintegrated episomes (68). Alternatively, the restriction could result from an encounter between an active replication fork and a not fully double-stranded proviral DNA newly integrated into an active replicon. In support of the latter scenario, viral DNA in HIV-1 PIC was shown to be competent for integration despite containing gapped/nicked DNA (6, 65). While we favor the former model, as the cellular postreplication DNA repair machinery has evolved to process DNA at chromosomal loci, rather than in the context of unintegrated episomes, a contribution from a postintegration component to HLTF-mediated HIV-1 restriction cannot be excluded based on existing data.
Evidence from this study and previous reports supports the existence of multiple components to HIV-1 Vpr function. One set constitutes antagonisms with several postreplication DNA repair proteins, mediated by the reprogrammed by Vpr CRL4DCAF1 E3 ubiquitin ligase. In particular, in T cells, Vpr uses this E3 to counteract the HLTF-mediated, Exo1-mediated (29), and likely additional restrictions of HIV-1 replication. Distinctively, in monocyte-derived macrophages and/or monocytic cell lines, Vpr uses CRL4DCAF1 to antagonize HIV-1 restriction by base excision repair enzyme UNG2 and TET2 methylcytosine dioxygenase (31, 37). In primary T cells, Vpr likely exerts additional effects that are not mediated by CRL4DCAF1 E3. This is supported by our observation that disruption of Vpr binding to DCAF1, by Q65R mutation, only partially suppressed Vpr function in these cells. Candidate mediators are the previously reported Vpr targets in RNA splicing, protein translation, and other cellular machineries (71–73). The above evidence reveals flexible use of multiple Vpr functions, including the reprogrammed CRL4DCAF1 E3, in a cell type-dependent manner to promote HIV-1 replication. The experimental system described here can be used to scrutinize contributions of individual components to virological effects of Vpr in distinct HIV-1 target cell types.
In sum, this and other recent studies have begun to reveal complex restrictions placed on HIV-1 replication by postreplication DNA repair enzymes, and their counteraction by HIV-1 Vpr (27, 29, 37). HIV-1 Vpr antagonism of select aspects of postreplication DNA repair provides evidence that certain modes of repair processing of proviral DNA can interfere with HIV-1 replication. Further studies on HIV-1 replication restrictions placed by DNA repair enzymes should reveal the full impact and detailed antiviral mechanisms exploited by this so far poorly characterized class of HIV-1 restriction factors.
Materials and Methods
HIV-1 Reporter Viruses.
HIV-1.RFP.vpr.wt and its derivatives are replication-competent HIV-1 NL4-3 viruses with a tagRFP-IRES cassette inserted between the second and third nef codons. Translation of Nef protein is directed by the IRES element. HIV-1.mRFP.vpr.wt is isogenic to HIV-1.RFP.vpr.wt, except for a variant tagRFP gene (mRFP) containing an array of synonymous changes: (RFP) GCT ACC AGC TTC ATG TAC GGC AGC AGA ACC > (mRFP) GCC ACA TCT TTT ATG TAT GGG TCT CGC ACA. The vpr mutations are vpr.Q8* (CAA > TAA), vpr.Δ (TAGGACAACA125 > Δ), vpr.Q65R (CAA > AGA), and vpr.R80A (AGA > GCC). HIV-1.BFP reporter viruses contain tagBFP (74) in place of the RFP/mRFP coding sequence. HIV-1 vpr.wt and vpr.Δ lacking any exogenous sequences (e.g., lacking RFP/mRFP-IRES cassettes) were constructed by replacing the BamH1-XhoI fragment comprising 3′-segment_of_env–RFP/mRFP–IRES-5′_segment_of_nef with BamHI-XhoI fragment derived from the HIV-1 NL43 molecular clone, thereby restoring the native NL43 sequence. All mutations and constructs were verified by DNA sequencing. Virus particles were produced, concentrated, and normalized by quantitative Gag immunoblotting, as previously described (75, 76). For PRCA coinfection assays, the competing virus pairs were additionally normalized by qRT-PCR quantification of virion-associated HIV-1 RNA.
Retroviral Expression Vectors.
HLTFo cDNA was obtained from GenScript. HLTF mutants were generated using a Quikchange Lightning Kit (Agilent). HLTFo and its variants were subcloned into pEasiLv-puro lentiviral vector (pEasiLv-puro_HLTFo). The pEasiLv-puro was constructed by replacing the SpeI-SalI fragment comprising E2-Crimson gene in pEasiLv (77) (kindly provided by Mike Malim, King’s College London, London) with puromycin N-acetyl-transferase gene, using a NEBuilder HiFi DNA Assembly Cloning Kit (New England Biolabs). Viral particles were produced from HEK293 T cells as previously described (24, 75).
PRCA in T Cell Lines.
Pairs of HIV-1 viruses to be tested in PRCA were normalized by qPCR quantification of reverse-transcribed viron-associated HIV-1 RNA, mixed at an ∼1:1 ratio, quantified again by qPCR, and then mixed again at a corrected 1:1 ratio. In some experiments, viruses were normalized based on their infectivity to CEM.SS T cells in single-cycle infections. The virus mixtures were incubated with 1 μL of Turbo DNase (catalog no. AM2238; Ambion) in 50 μL of RPMI supplemented with 1× Turbo DNase reaction buffer at 37 °C for 30 min before infection. CEM.SS or HPB.ALL T cells (1 × 105 cells) were infected with the virus mixture at a combined moi between 0.006 and 0.02, in a dose–response experiment, in 1 mL of cell culture medium in wells of a 24-well plate. The cultures were counted, and cell density was adjusted to 0.5 × 105 cells per milliliter on 3 dpi and 5 dpi. At 7 dpi, the percentage of RFP+ cells was determined by flow cytometry, and the infected cultures with fewer than 5% RFP+ cells were then selected for quantification of the competing viruses.
PRCA in Primary CD4+ T Cells.
CD4+ T cells were purified from human peripheral blood mononuclear cells (PBMCs), as described previously (24). Then, 1 × 106 cells per milliliter in six-well plates were activated with Dynabeads Human T-Activator CD3/CD28 (Invitrogen) in the presence of IL-2 (30 U⋅mL−1) in RPMI 1640 medium supplemented with 10% FBS, penicillin, and streptomycin. Two days postactivation, the cells were seeded at 1 × 105 cells, in 100 μL of complete RPMI medium, in 96-well round-bottom plates; the cells were then infected with 1:1 mixtures of the viruses to be tested, at an moi of 0.006–0.02, by spinoculation at 2,500 × g for 90 min at 22 °C; subsequently, cells were plated in 0.5 mL of complete RPMI medium supplemented with 30 U⋅mL−1 IL-2 in 24-well plates. The infected cultures were counted on 1 dpi and 3 dpi, and RFP+ cells were determined by flow cytometry. Supernatants from cultures with fewer than 5% of RFP+ cells (200 μL and 40 μL) were used to spinoculate a fresh batch of CD4+ T cells that had been activated 2 d earlier, as described above. The competing viruses were quantified 3, 5, and 7 d after the initial HIV-1 infection. Each of the experiments shown in Fig. 2 was performed with PBMCs from a different donor.
Quantification of Cell-Associated HIV-1 DNA by Real-Time PCR.
Cell cultures were collected into 1.5-mL maximum recovery tubes (Axygen), and cells were pelleted by centrifugation at ∼200 × g for 5 min. Two hundred fifty microliters of the supernatant was collected for isolation of cell-free viral RNA. Total DNA was isolated from cell pellets with a DNeasy Blood & Tissue Kit (Qiagen) and quantified using a Qubit dsDNA HS Assay Kit, and DNA was diluted to 10 ng/μL with tRNA solution (10 ng/μL). Twenty-microliter qPCR reactions contained 10 μL of Power SYBR Green PCR Master Mix (Applied Biosystems), 1 μL of each primer solution (5 μM), 3 μL of H2O, and 5 μL of total DNA at 10 ng/μL, or 5 μL of 1:50 diluted reverse-transcribed cell-free RNA. Three technical replicates were analyzed for each DNA sample, with primers specific for the RFP and mRFP, or vpr.wt and vpr.Δ, amplicons (listed in SI Appendix, Table S1). qPCR was performed with a denaturation step at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s, primer annealing at 60 °C for 15 s, and primer extension at 72 °C for 20 s, on an ABI StepOnePlus Real-Time PCR system. Data were collected during the elongation step. Standards for quantification of the RFP and mRFP amplicons, ranging from 10−3 to 101 HIV-1 copies per cell, were prepared by serially 10-fold–diluting the appropriate HIV-1 NL4-3 provirus plasmid DNA (∼14 kb) into CEM.SS T cell genomic DNA. For each experiment, a standard curve of the amplicon measured was run in duplicate.
Quantification of HIV-1 RNA.
Total viral RNA was isolated from 250 μL of cell culture supernatant using TRIzol LS (Ambion; Life Technologies) and resuspended in 35 μL of RNase-free water, and reverse transcription was performed using 10.5 μL of RNA solution, 1 μL of 100 ng/μL random hexamers (New England Biolabs), and 0.5 μL of SuperScript II Reverse Transcriptase (Invitrogen). The reverse transcription products were diluted 1:50 with tRNA (10 μg/mL) for qPCR quantification of the RFP/mRFP amplicons using primers listed in SI Appendix, Table S1.
Cell Lines and Gene Transfer.
HEK293T cells were maintained in DMEM supplemented with 10% FBS, 2 mM l-glutamine, and penicillin/streptomycin in 5% CO2 at 37 °C. CEM.SS and HPB.ALL T cells (78, 79) and their derivatives (obtained through the NIH AIDS Reagent Program) were maintained in RPMI 1640 medium supplemented as above. Cell lines harboring the puromycin N-acetyl-transferase resistance marker were maintained in the presence of puromycin (2 μg/mL). CEM.SS_iHLTFo T cells were constructed by transduction of CEM.SS and T cells with pEasiLv-puro_HLTFo lentiviral vector. HLTFo expression was induced with doxycycline (100 ng/mL; Sigma–Aldrich), unless indicated otherwise.
Drug Treatments.
CEM.SS T cells were cultured in the presence of the following drugs starting ∼12 h before HIV-1 infection and continuing through the duration of PRCA: prostratin (0.1 μM), JQ1 (0.1 μM), GDC-0941 (1 μM), rapamycin (2.5 μM), SAHA (200 nM), GSK343 (2 μM), EX527 (10 μM), SGC0946 (2 μM), C646 (3 μM), UNC0638 (1 μM), and TSA (3 nM). The chosen concentrations were the highest that resulted in >80% cell viability. The drugs were replenished at the time of each subculturing.
RNAi.
Stable RNAi was performed with the following: retroviral MSCV(EGFP)-miR30E vector expressing NT or gene-specific enhanced mir30E shRNAs (80); pSRG vector expressing NT1 or NT2, cGAS-targeting, or TASOR-targeting shRNAs; and pGIPZ-lentiviral vectors expressing NT or TET2-targeting shRNA (31). The mir30E-based shRNAs targeted the following sequences: NT: TAAGGCTATGAAGAGATAC, HLTF.31: CAGATGTACACTTGAATTTTA, HLTF.33: AACTATAATGCTTCTGTCCTA, Exo1.33: GACGACAAGCCAATCTTCTTA (29), and Exo1.34: CAGATGTAGCACGTAATTCAA (29). The pSRG-based shRNAs targeted the following: NT1: TAAGGCTATGAAGAGATAC, NT2: CTCCCGTGAATTGGAATCC, cGAS.2: GATGCTGTCAAAGTTTAGGAA, cGAS.3: CAACTACGACTAAAGCCATTT (81), and TASOR: GAGGAAGCTTGAGGATCTA (59). The cells were transduced with retroviruses expressing individual NT mir30E-based shRNAs and shRNA at an moi of 3:1 or a mixture of mir30E shRNAs (HLTF) or shRNAs (cGAS) at a combined moi of 3:1. In some experiments, the positively transduced GFP+ cells were isolated by cell sorting on a FACS Aria and expanded for 3 to 4 d. The cells were used for PRCA at 3 dpi of RNAi, or at 6–7 dpi of RNAi if enriched by cell sorting.
Flow Cytometry and Cell Sorting.
When the percentage of GFP-expressing CEM.SS T cells subjected to RNAi was <85%, the GFP+ cells were purified by cell sorting on a FACSAria (3 × 105 GFP+ cells) and then expanded for 4 d before the initiation of PRCA. To visualize proliferation rates, CEM.SS T cells (2 × 105) were suspended in PBS (200 μL), reacted with Celltracer Far Red (1.5 μL; Thermo Fisher Scientific) at 37 °C for 20 min, washed with RPMI 1640 for 5 min, pelleted by centrifugation, resuspended in 1 mL of complete RPMI 1640 medium, and cultured for 5 d. The cell fluorescence was characterized 10 min after staining and then 2 d and 5 d later with a BD LSR Fortessa flow cytometer and analyzed using FlowJo software.
Antibodies and Immunoblotting.
Whole-cell extracts were separated by SDS/PAGE, transferred to a PVDF membrane, and immunoblotted with appropriate primary antibodies, and immune complexes were revealed with HRP-conjugated antibodies specific for the Fc fragment of mouse or rabbit IgG (Jackson ImmunoResearch Laboratories) and enhanced chemiluminescence, or with fluorescent secondary antibodies (KPL) and an Odyssey Infrared Imager (Licor), as previously described (18). The following primary antibodies were used: α-HLTF (A300-230A; Bethyl Laboratories, Inc.), α-α-tubulin (sc-5286; Santa Cruz Biotechnology), α-TFIID (sc-204; Santa Cruz Biotechnology), α-lamin B1 (ab16048; Abcam), α-Flag (F1804; Sigma-Aldrich), α-FAM208A (HPA006735; Sigma–Aldrich), α-PPHLN1 (HPA038902; Sigma–Aldrich), α-DDB1 (37-6200; Invitrogen), α-SF2 (provided by A. Krainer, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), and α-HA (12CA5) (82), α-c-myc (9E10), and α-HIV-1 CA (183-H12-5C) (all produced in-house) (24). The α-HIV-1 Vpr antibody and 183-H12-5C hybridoma were obtained from Jeffrey Kopp and Bruce Chesebro, respectively, AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH, Germantown, MD (83).
Statistical Analyses.
Statistical significance of the data was analyzed using one-way ANOVA with a post hoc Tukey test or t test, and graphs were prepared using GraphPad Prism 7 for Mac OS X (GraphPad Software). P values lower than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
The following reagents were obtained through the AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH: CEM.SS cells from Dr. Peter L. Nara, α-HIV-1 Vpr antibody from Dr. Jeffrey Kopp, and α-HIV-1 p24 hybridoma (183-H12-5C) from Dr. Bruce Chesebro. The pEasiLv vector was kindly provided by Dr. Mike Malim. We thank Juan Qian and Margaret Pinkevitch for excellent technical assistance. We thank Drs. Linda VanAelst and Teresa Brosenitsch for critical reading of the manuscript and editorial help, Dr. Angela Gronenborn for support, and Dr. Tomek Swigut for help with statistical analyses. This work was supported by NIH Grants GM123973 and AI100673 and a P50GM082251 subcontract (to J.S.). The Flow Cytometry Facility at Case Western Reserve University is supported by Center for AIDS Research Center Grant P30 AI036219.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818401116/-/DCSupplemental.
References
- 1.Roe T, Reynolds TC, Yu G, Brown PO. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achar YJ, Balogh D, Haracska L. Coordinated protein and DNA remodeling by human HLTF on stalled replication fork. Proc Natl Acad Sci USA. 2011;108:14073–14078. doi: 10.1073/pnas.1101951108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matreyek KA, Engelman A. Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes. Viruses. 2013;5:2483–2511. doi: 10.3390/v5102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu WS, Hughes SH. HIV-1 reverse transcription. Cold Spring Harb Perspect Med. 2012;2:a006882. doi: 10.1101/cshperspect.a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klarmann GJ, Yu H, Chen X, Dougherty JP, Preston BD. Discontinuous plus-strand DNA synthesis in human immunodeficiency virus type 1-infected cells and in a partially reconstituted cell-free system. J Virol. 1997;71:9259–9269. doi: 10.1128/jvi.71.12.9259-9269.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller MD, Wang B, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes containing discontinuous plus strands are competent to integrate in vitro. J Virol. 1995;69:3938–3944. doi: 10.1128/jvi.69.6.3938-3944.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charneau P, et al. HIV-1 reverse transcription. A termination step at the center of the genome. J Mol Biol. 1994;241:651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 8.Charneau P, Clavel F. A single-stranded gap in human immunodeficiency virus unintegrated linear DNA defined by a central copy of the polypurine tract. J Virol. 1991;65:2415–2421. doi: 10.1128/jvi.65.5.2415-2421.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgers PMJ, Kunkel TA. Eukaryotic DNA replication fork. Annu Rev Biochem. 2017;86:417–438. doi: 10.1146/annurev-biochem-061516-044709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sáez-Cirión A, Manel N. Immune responses to retroviruses. Annu Rev Immunol. 2018;36:193–220. doi: 10.1146/annurev-immunol-051116-052155. [DOI] [PubMed] [Google Scholar]
- 11.Nan Y, Wu C, Zhang YJ. Interplay between Janus kinase/signal transducer and activator of transcription signaling activated by type I interferons and viral antagonism. Front Immunol. 2017;8:1758. doi: 10.3389/fimmu.2017.01758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauter D, Kirchhoff F. Multilayered and versatile inhibition of cellular antiviral factors by HIV and SIV accessory proteins. Cytokine Growth Factor Rev. 2018;40:3–12. doi: 10.1016/j.cytogfr.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Viswanathan K, Früh K, DeFilippis V. Viral hijacking of the host ubiquitin system to evade interferon responses. Curr Opin Microbiol. 2010;13:517–523. doi: 10.1016/j.mib.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desimmie BA, et al. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J Mol Biol. 2014;426:1220–1245. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain P, et al. Large-scale arrayed analysis of protein degradation reveals cellular targets for HIV-1 Vpu. Cell Rep. 2018;22:2493–2503. doi: 10.1016/j.celrep.2018.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margottin F, et al. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 17.Belzile JP, et al. HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4AVPRBP E3 ubiquitin ligase. PLoS Pathog. 2007;3:e85. doi: 10.1371/journal.ppat.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hrecka K, et al. Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc Natl Acad Sci USA. 2007;104:11778–11783. doi: 10.1073/pnas.0702102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Rouzic E, et al. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle. 2007;6:182–188. doi: 10.4161/cc.6.2.3732. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S, Feng Y, Narayan O, Zhao LJ. Cytoplasmic retention of HIV-1 regulatory protein Vpr by protein-protein interaction with a novel human cytoplasmic protein VprBP. Gene. 2001;263:131–140. doi: 10.1016/s0378-1119(00)00583-7. [DOI] [PubMed] [Google Scholar]
- 21.Bouhamdan M, et al. Human immunodeficiency virus type 1 Vpr protein binds to the uracil DNA glycosylase DNA repair enzyme. J Virol. 1996;70:697–704. doi: 10.1128/jvi.70.2.697-704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krokan HE, Bjørås M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blastyák A, Hajdú I, Unk I, Haracska L. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol Cell Biol. 2010;30:684–693. doi: 10.1128/MCB.00863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hrecka K, et al. HIV-1 and HIV-2 exhibit divergent interactions with HLTF and UNG2 DNA repair proteins. Proc Natl Acad Sci USA. 2016;113:E3921–E3930. doi: 10.1073/pnas.1605023113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lahouassa H, et al. HIV-1 Vpr degrades the HLTF DNA translocase in T cells and macrophages. Proc Natl Acad Sci USA. 2016;113:5311–5316. doi: 10.1073/pnas.1600485113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- 27.Laguette N, et al. Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell. 2014;156:134–145. doi: 10.1016/j.cell.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 28.Matos J, West SC. Holliday junction resolution: Regulation in space and time. DNA Repair (Amst) 2014;19:176–181. doi: 10.1016/j.dnarep.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan J, et al. HIV-1 Vpr reprograms CLR4DCAF1 E3 ubiquitin ligase to antagonize exonuclease 1-mediated restriction of HIV-1 infection. MBio. 2018;9:e01732-18. doi: 10.1128/mBio.01732-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keijzers G, Liu D, Rasmussen LJ. Exonuclease 1 and its versatile roles in DNA repair. Crit Rev Biochem Mol Biol. 2016;51:440–451. doi: 10.1080/10409238.2016.1215407. [DOI] [PubMed] [Google Scholar]
- 31.Lv L, et al. Vpr targets TET2 for degradation by CRL4VprBP E3 ligase to sustain IL-6 expression and enhance HIV-1 replication. Mol Cell. 2018;70:961–970.e5. doi: 10.1016/j.molcel.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kafer GR, et al. 5-Hydroxymethylcytosine marks sites of DNA damage and promotes genome stability. Cell Rep. 2016;14:1283–1292. doi: 10.1016/j.celrep.2016.01.035. [DOI] [PubMed] [Google Scholar]
- 33.Pastor WA, Aravind L, Rao A. TETonic shift: Biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belzile JP, Abrahamyan LG, Gérard FC, Rougeau N, Cohen EA. Formation of mobile chromatin-associated nuclear foci containing HIV-1 Vpr and VPRBP is critical for the induction of G2 cell cycle arrest. PLoS Pathog. 2010;6:e1001080. doi: 10.1371/journal.ppat.1001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai M, Zimmerman ES, Planelles V, Chen J. Activation of the ATR pathway by human immunodeficiency virus type 1 Vpr involves its direct binding to chromatin in vivo. J Virol. 2005;79:15443–15451. doi: 10.1128/JVI.79.24.15443-15451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roshal M, Kim B, Zhu Y, Nghiem P, Planelles V. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J Biol Chem. 2003;278:25879–25886. doi: 10.1074/jbc.M303948200. [DOI] [PubMed] [Google Scholar]
- 37.Hansen EC, et al. Diverse fates of uracilated HIV-1 DNA during infection of myeloid lineage cells. eLife. 2016;5:e18447. doi: 10.7554/eLife.18447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goh WC, et al. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: A mechanism for selection of Vpr in vivo. Nat Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 39.Vermeire J, et al. HIV triggers a cGAS-dependent, Vpu- and Vpr-regulated type I interferon response in CD4+ T cells. Cell Rep. 2016;17:413–424. doi: 10.1016/j.celrep.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 40.Manocheewa S, et al. Pairwise growth competition assay for determining the replication fitness of human immunodeficiency viruses. J Vis Exp. 2015:e52610. doi: 10.3791/52610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quiñones-Mateu ME, et al. A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J Virol. 2000;74:9222–9233. doi: 10.1128/jvi.74.19.9222-9233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen R, Le Rouzic E, Kearney JA, Mansky LM, Benichou S. Vpr-mediated incorporation of UNG2 into HIV-1 particles is required to modulate the virus mutation rate and for replication in macrophages. J Biol Chem. 2004;279:28419–28425. doi: 10.1074/jbc.M403875200. [DOI] [PubMed] [Google Scholar]
- 43.Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 44.Mashiba M, Collins DR, Terry VH, Collins KL. Vpr overcomes macrophage-specific restriction of HIV-1 Env expression and virion production. Cell Host Microbe. 2014;16:722–735. doi: 10.1016/j.chom.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subbramanian RA, et al. Human immunodeficiency virus type 1 Vpr is a positive regulator of viral transcription and infectivity in primary human macrophages. J Exp Med. 1998;187:1103–1111. doi: 10.1084/jem.187.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vodicka MA, Koepp DM, Silver PA, Emerman M. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 1998;12:175–185. doi: 10.1101/gad.12.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou X, DeLucia M, Ahn J. SLX4-SLX1 protein-independent down-regulation of MUS81-EME1 protein by HIV-1 viral protein R (Vpr) J Biol Chem. 2016;291:16936–16947. doi: 10.1074/jbc.M116.721183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zimmerman ES, et al. Human immunodeficiency virus type 1 Vpr induces DNA replication stress in vitro and in vivo. J Virol. 2006;80:10407–10418. doi: 10.1128/JVI.01212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou X, et al. HIV-1 Vpr protein directly loads helicase-like transcription factor (HLTF) onto the CRL4-DCAF1 E3 ubiquitin ligase. J Biol Chem. 2017;292:21117–21127. doi: 10.1074/jbc.M117.798801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unk I, Hajdú I, Blastyák A, Haracska L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst) 2010;9:257–267. doi: 10.1016/j.dnarep.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 51.Unk I, et al. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc Natl Acad Sci USA. 2008;105:3768–3773. doi: 10.1073/pnas.0800563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hishiki A, et al. Structure of a novel DNA-binding domain of helicase-like transcription factor (HLTF) and its functional implication in DNA damage tolerance. J Biol Chem. 2015;290:13215–13223. doi: 10.1074/jbc.M115.643643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kile AC, et al. HLTF’s ancient HIRAN domain binds 3′ DNA ends to drive replication fork reversal. Mol Cell. 2015;58:1090–1100. doi: 10.1016/j.molcel.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 55.Shimura M, et al. Epigenetic displacement of HP1 from heterochromatin by HIV-1 Vpr causes premature sister chromatid separation. J Cell Biol. 2011;194:721–735. doi: 10.1083/jcb.201010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romani B, et al. HIV-1 Vpr reactivates latent HIV-1 provirus by inducing depletion of class I HDACs on chromatin. Sci Rep. 2016;6:31924. doi: 10.1038/srep31924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mbonye U, Karn J. The molecular basis for human immunodeficiency virus latency. Annu Rev Virol. 2017;4:261–285. doi: 10.1146/annurev-virology-101416-041646. [DOI] [PubMed] [Google Scholar]
- 58.Chougui G, et al. HIV-2/SIV viral protein X counteracts HUSH repressor complex. Nat Microbiol. 2018;3:891–897. doi: 10.1038/s41564-018-0179-6. [DOI] [PubMed] [Google Scholar]
- 59.Tchasovnikarova IA, et al. GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science. 2015;348:1481–1485. doi: 10.1126/science.aaa7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu Y, Wang GZ, Cingöz O, Goff SP. NP220 mediates silencing of unintegrated retroviral DNA. Nature. 2018;564:278–282. doi: 10.1038/s41586-018-0750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yurkovetskiy L, et al. Primate immunodeficiency virus proteins Vpx and Vpr counteract transcriptional repression of proviruses by the HUSH complex. Nat Microbiol. 2018;3:1354–1361. doi: 10.1038/s41564-018-0256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chavez DA, Greer BH, Eichman BF. The HIRAN domain of helicase-like transcription factor positions the DNA translocase motor to drive efficient DNA fork regression. J Biol Chem. 2018;293:8484–8494. doi: 10.1074/jbc.RA118.002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Achar YJ, et al. Human HLTF mediates postreplication repair by its HIRAN domain-dependent replication fork remodelling. Nucleic Acids Res. 2015;43:10277–10291. doi: 10.1093/nar/gkv896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fuentes GM, Rodríguez-Rodríguez L, Palaniappan C, Fay PJ, Bambara RA. Strand displacement synthesis of the long terminal repeats by HIV reverse transcriptase. J Biol Chem. 1996;271:1966–1971. doi: 10.1074/jbc.271.4.1966. [DOI] [PubMed] [Google Scholar]
- 65.Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: Studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kolinjivadi AM, et al. Smarcal1-Mediated fork reversal triggers mre11-dependent degradation of nascent DNA in the absence of Brca2 and stable Rad51 nucleofilaments. Mol Cell. 2017;67:867–881.e7. doi: 10.1016/j.molcel.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lemaçon D, et al. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat Commun. 2017;8:860. doi: 10.1038/s41467-017-01180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nightingale K, et al. High-definition analysis of host protein stability during human cytomegalovirus infection reveals antiviral factors and viral evasion mechanisms. Cell Host Microbe. 2018;24:447–460.e11. doi: 10.1016/j.chom.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andersen JL, Le Rouzic E, Planelles V. HIV-1 Vpr: Mechanisms of G2 arrest and apoptosis. Exp Mol Pathol. 2008;85:2–10. doi: 10.1016/j.yexmp.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maréchal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guenzel CA, Hérate C, Benichou S. HIV-1 Vpr-a still “enigmatic multitasker”. Front Microbiol. 2014;5:127. doi: 10.3389/fmicb.2014.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharma A, Yilmaz A, Marsh K, Cochrane A, Boris-Lawrie K. Thriving under stress: Selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 2012;8:e1002612. doi: 10.1371/journal.ppat.1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang X, Aida Y. HIV-1 Vpr: A novel role in regulating RNA splicing. Curr HIV Res. 2009;7:163–168. doi: 10.2174/157016209787581517. [DOI] [PubMed] [Google Scholar]
- 74.Subach OM, et al. Conversion of red fluorescent protein into a bright blue probe. Chem Biol. 2008;15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hrecka K, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Srivastava S, et al. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. 2008;4:e1000059. doi: 10.1371/journal.ppat.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goujon C, et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Foley GE, et al. CONTINUOUS CULTURE OF HUMAN LYMPHOBLASTS FROM PERIPHERAL BLOOD OF A CHILD WITH ACUTE LEUKEMIA. Cancer. 1965;18:522–529. doi: 10.1002/1097-0142(196504)18:4<522::aid-cncr2820180418>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 79.Nara PL, et al. Simple, rapid, quantitative, syncytium-forming microassay for the detection of human immunodeficiency virus neutralizing antibody. AIDS Res Hum Retroviruses. 1987;3:283–302. doi: 10.1089/aid.1987.3.283. [DOI] [PubMed] [Google Scholar]
- 80.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 81.Lahaye X, et al. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity. 2013;39:1132–1142. doi: 10.1016/j.immuni.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 82.Wilson IA, et al. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 83.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: Definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.