Abstract
Two North American fungal pathogens caused a coepizootic leading to localized collapse of an outbreak population of the newly invasive planthopper pest, the spotted lanternfly (Lycorma delicatula), in the eastern United States. The pathogens partitioned the habitat, with the majority of L. delicatula on tree trunks killed by Batkoa major, while cadavers of L. delicatula killed by Beauveria bassiana were usually on the ground. The future will show whether these pathogens will be drivers in boom–bust cycles or will result in recurrent low population densities of this new invasive species.
Keywords: coepizootic, pathogen accumulation, invasive species, entomopathogenic fungi, spotted lanternfly
The outcome of incipient biological invasions is difficult to predict (1, 2), and while some species increase and then remain at high densities, others undergo boom–bust cycles (periods of rapid growth followed by precipitous decrease) or remain at lower densities after decreasing in abundance (3, 4). The success of invasive species has been attributed to various causes, including escape from natural enemies present in their native range (the enemy release hypothesis) (5). The causes of population collapse of invasive species are typically unknown, but collapse is often hypothesized as being caused by predators, pathogens, parasites, competitors, or declines in resources (4). Pathogen species can accumulate in invasive species based on the history of the invasion (6, 7), but whether pathogen activity can lead to recovery of native ecosystems due to suppression of invasive species is presently under discussion (2, 8).
The spotted lanternfly, Lycorma delicatula, is native to China, Taiwan, and Vietnam and has invaded South Korea and Japan (9). This univoltine, polyphagous planthopper feeds on 70 or more species of woody plants in 25 families and has caused damage to grape and apple crops (9). In 2014, L. delicatula was discovered in southeastern Pennsylvania where it now occurs in high densities and has subsequently spread to six additional eastern states (10). L. delicatula is only distantly related to hemipteran insects in the region, as there are no native species of Fulgoridae in northeastern North America (11). Few natural enemies have been reported attacking L. delicatula in Pennsylvania (12).
We report a coepizootic caused by two unrelated native fungal entomopathogens attacking an outbreak population of this new invasive insect. We identify and compare the two pathogens and present how they impacted the host population.
Results
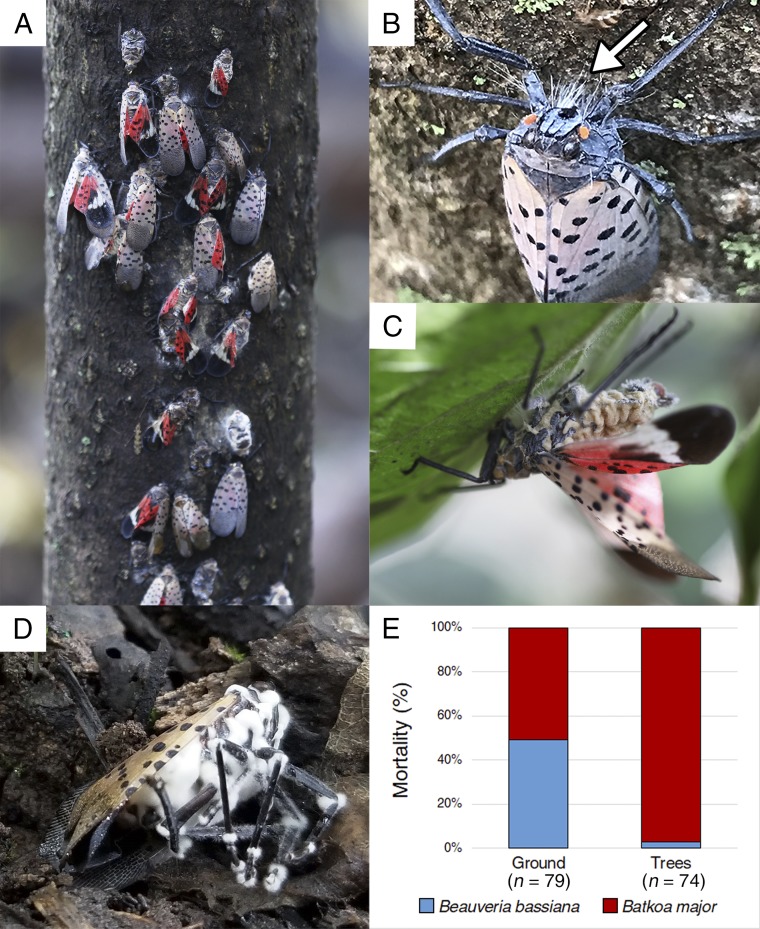
The pathogens, both of which infect and kill insect hosts, are in the Division Zoopagomycota (Order Entomophthorales) and the Division Ascomycota (Order Hypocreales). For the entomophthoralean pathogen, the top-two GenBank references for 18S ribosomal RNA partial sequences were both Batkoa major with 98% identity (GenBank MK483702; February 11, 2019) (Figs. 1 A–C); in addition, morphology matched descriptions (13). For the hypocrealean pathogen, the top-10 search results were all representative of isolates of Beauveria bassiana (GenBank MK574670; April 9, 2019) (Fig. 1D).
Fig. 1.
Two fungal pathogens causing a coepizootic in an L. delicatula population. (A) L. delicatula during the epizootic: only one of these adults was alive and the remainder had been killed by B. major. A similar degree of mortality was observed on trees throughout the site. (B) Rhizoids from B. major (arrow) attaching a dead adult to a tree. Image courtesy of Kelly Murman (photographer). (C) Adult cadaver with wings and legs extended outward as B. major conidia are released from the abdomen. (D) Adult killed by B. bassiana. (E) Percent L. delicatula killed by either pathogen on the trees or surrounding ground. Sample size is below each bar.
On October 9, 2018, dead L. delicatula adults were abundant throughout the site, on tree-of-heaven (Ailanthus altissima) (Fig. 1A), on neighboring trees and vines, and on the ground. Cadaver locations differed by fungal species (Fisher’s exact test, P = 0.0064; Fig. 1E). Almost all dead L. delicatula adults on tree trunks were killed by B. major (97%) and attached by fungal rhizoids. On the ground was a mix of L. delicatula killed by B. major (51%) or B. bassiana (49%). A few B. bassiana-killed L. delicatula adults were collected from tree trunks (3%) but most were recovered from the ground. Across all collections, 73% of cadavers contained B. major and 27% contained B. bassiana. This coepizootic occurred when females were gravid but before most oviposition, and due to the massive L. delicatula mortality, only 12 egg masses were found.
Discussion
Invasive pests reach high densities because natural enemies are not attacking them (5). We report on the accumulation of two native fungal pathogens causing fatal diseases, leading to the sharp decline of a population of this new invasive pest—an example of biotic resistance (14). Neither pathogen has been previously reported as infecting L. delicatula in North America, despite intensive monitoring of this economically important insect, making these “emerging” pathogens. The probability of increasing disease prevalence generally increases with the age of an invasive infestation (2). The coepizootic described here is unusual because L. delicatula was discovered in North America only a few years before.
The ability of these pathogens to cause a coepizootic could in part be due to density-dependent transmission, host selection plasticity, or evolutionary adaptations (1, 6). B. bassiana principally lives as an arthropod pathogen, with a broad host range and a cosmopolitan distribution, and this pathogen is known to infect L. delicatula in China (15, 16). B. bassiana may also persist in the environment as an endophyte or saprotroph (16). In contrast, B. major is a poorly known obligate pathogen that infects a diversity of insects in the Americas, Europe, and Asia (13). This coepizootic provides an excellent opportunity to compare these pathogens and to study how they responded differentially to a high-density population of this new host. B. major forms rhizoids to hold cadavers in place and then actively ejects relatively short-lived primary and supernumerary infective spores. Conidia produced by entomophthoralean fungi can become airborne and travel longer distances (17). B. bassiana neither forms rhizoids nor actively ejects spores, but cadavers become densely covered with persistent spores that can be spread by direct contact and rainsplash (18). Thus, whether living L. delicatula planthoppers were present on tree trunks, tree bases, or the ground, they were exposed to infective spores produced by one or the other of these fungal pathogens, resulting in this coepizootic.
Epizootics observed in insect populations usually include host species principally being attacked by a dominant pathogen species [e.g., western tent caterpillars (Malacosoma californicum pluviale) by the nucleopolyhedrovirus McplNPV (19) and manuka beetles (Pyronota spp.) by Rickettsiella pyronotae (20)]. Therefore, this coepizootic is uncommon, in that significant levels of infection were caused by two unrelated entomopathogenic fungi within the same host population. We hypothesize that due to the coepizootic, increased densities of these pathogens persisting in the environment would impact native communities of insects due to spillover events. However, the increased pathogen reservoir would also provide inocula for future infection of this invasive herbivore (7, 8).
Materials and Methods
On October 9, 2018, an epizootic caused by two fungal pathogens was observed in a high-density L. delicatula population (Fig. 1A) in a wooded area (690 m2, 161 trees, and 43 shrubs) alongside an apple orchard at Angora Fruit Farm (40°21′30.6′′N, 75°53′00.4′′W), near Reading, Pennsylvania. Abundant A. altissima trees were used as a basis for sampling (72% of the trees in these woods were A. altissima). Two species of entomopathogenic fungi that cause fatal diseases were identified and isolated from L. delicatula cadavers that contained fungal cells internally or externally.
For isolation of the entomophthoralean pathogen, primary conidia were obtained via the “ascending conidia” showering method, and hyphal growth occurred in cell culture media (21). Beauveria was isolated using selective media (22).
Fungal Identification.
To identify both pathogens, sequencing with subsequent comparison with the National Center for Biotechnology Information nucleotide database was used as well as morphological evaluation (13, 16). For the entomophthoralean species, primers NS1/NS4 (23) amplified a region of the 18S ribosomal DNA (rDNA), and primers LROR/LR5 (24, 25) amplified 28S rDNA. For both primers, two phase autoextension protocols were used. For the hypocrealean isolate, nucleotide sequences were determined for the B locus nuclear intergenic region using primers B22Udg/B3.3R following PCR conditions described by Rehner et al. (16).
Describing the Epizootic.
To compare the locations of cadavers by fungal pathogen, ∼180 L. delicatula cadavers were collected from three randomly chosen canopy-height A. altissima trees. For each tree, 30 cadavers were collected from the lower 2 m of the tree trunk and 30 cadavers were collected from the ground around the tree base.
Acknowledgments
We thank Heather Leach and David Harris for field assistance and Saskya van Nouhuys, Dylan Parry, Andrew Liebhold, and Patrick Tobin for manuscript assistance. This work was supported by funding from the US Department of Agriculture (USDA) Animal and Plant Health Inspection Service (Grant AP18PPQS&T00C080) and the USDA Forest Service (Grant 18-CA-11420004-058).
Footnotes
The authors declare no conflict of interest.
References
- 1.Young HS, Parker IM, Gilbert GS, Sofia Guerra A, Nunn CL. Introduced species, disease ecology, and biodiversity—Disease relationships. Trends Ecol Evol. 2017;32:41–54. doi: 10.1016/j.tree.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Flory SJ, Alba C, Clay K, Holt RD, Goss EM. Emerging pathogens can suppress invaders and promote native species recovery. Biol Invasions. 2018;20:5–8. [Google Scholar]
- 3.Simberloff D, Gibbons L. Now you see them, now you don’t—Population crashes of established introduced species. Biol Invasions. 2004;6:161–172. [Google Scholar]
- 4.Strayer DL, et al. Boom-bust dynamics in biological invasions: Towards an improved application of the concept. Ecol Lett. 2017;20:1337–1350. doi: 10.1111/ele.12822. [DOI] [PubMed] [Google Scholar]
- 5.Keane RM, Crawley MJ. Exotic plant invasions and the enemy release hypothesis. Trends Ecol Evol. 2002;17:164–170. [Google Scholar]
- 6.Mitchell CE, Blumenthal D, Jarošík V, Puckett EE, Pyšek P. Controls on pathogen species richness in plants’ introduced and native ranges: roles of residence time, range size and host traits. Ecol Lett. 2010;13:1525–1535. doi: 10.1111/j.1461-0248.2010.01543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flory SJ, Clay K. Pathogen accumulation and long-term dynamics of plant invasions. J Ecol. 2013;101:607–613. [Google Scholar]
- 8.Policelli N, et al. Pathogen accumulation cannot undo the impact of invasive species. Biol Invasions. 2018;20:1–4. [Google Scholar]
- 9.Liu H, Mottern J. An old remedy for a new problem? Identification of Ooencyrtus kuvanae (Hymenoptera: Encyrtidae), an egg parasitoid of Lycorma delicatula (Hemiptera: Fulgoridae) in North America. J Insect Sci. 2017;17:1–6. doi: 10.1093/jisesa/iew114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.New York State Integrated Pest Management Program 2019 Spotted lanternfly. Available at https://nysipm.cornell.edu/environment/invasive-species-exotic-pests/spotted-lanternfly/. Accessed February 5, 2019.
- 11.Bartlett CR. 2018 North American Fulgoridae: Lanternflies or fulgorid planthoppers. Available at canr.udel.edu/planthoppers/north-america/north-american-fulgoridae/ Accessed January 31, 2019.
- 12.Barringer LE, Smyers E. Predation of the spotted lanternfly, Lycorma delicatula (White) (Hemiptera: Fulgoridae) by two native Hemiptera. Entomol News. 2016;126:71–73. [Google Scholar]
- 13.Balazy S. 1993. Entomophthorales. Flora of Poland Fungi (Mycota) (Instytut Botaniki, Kraków, Poland), Vol XXIV.
- 14.Gilbert G, Parker I. Invasion and the regulation of plant populations by pathogens. In: Cadotte MC, Mcmahon SM, Fukami T, editors. Conceptual Ecology and Invasion Biology: Reciprocal Approaches to Nature. Springer; Berlin: 2006. pp. 289–305. [Google Scholar]
- 15.Li MY, Lin HF, Li SG, Xu AM, Feng MF. Efficiency of entomopathogenic fungi in the control of eggs of the brown planthopper Nilaparvata lugens Stål (Homopera: Delphacidae) Afr J Microbiol Res. 2013;6:7162–7167. [Google Scholar]
- 16.Rehner SA, et al. Phylogeny and systematics of the anamorphic, entomopathogenic genus Beauveria. Mycologia. 2011;103:1055–1073. doi: 10.3852/10-302. [DOI] [PubMed] [Google Scholar]
- 17.Bittner TD, Hajek AE, Liebhold AM, Thistle H. Modification of a pollen trap design to capture airborne conidia of Entomophaga maimaiga and detection by quantitative PCR. Appl Environ Microbiol. 2017;83:1–11. doi: 10.1128/AEM.00724-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyling NV, Pell JK, Eilenberg J. Dispersal of Beauveria bassiana by the activity of nettle insects. J Invertebr Pathol. 2006;93:121–126. doi: 10.1016/j.jip.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 19.Myers JH, Cory JS. Ecology and evolution of pathogens in natural populations of Lepidoptera. Evol Appl. 2015;9:231–247. doi: 10.1111/eva.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshall SDG, Townsend RJ, Kleespies RG, van Koten C, Jackson TA. An epizootic of Rickettsiella infection emerges from an invasive scarab pest outbreak following land use change in New Zealand. Ann Clin Cytol Pathol. 2017;3:1058–1060. [Google Scholar]
- 21.Hajek AE, Papierok B, Eilenberg J. Methods for study of the Entomophthorales. In: Lacey L, editor. Manual of Techniques in Invertebrate Pathology. 2nd Ed. Academic Press; Amsterdam: 2012. pp. 285–316. [Google Scholar]
- 22.Goettel GM, Inglis GD. Fungi: Hyphomycetes. In: Lacey L, editor. Manual of Techniques in Insect Pathology. Academic Press; San Diego: 1997. pp. 213–250. [Google Scholar]
- 23.Gargas A, Taylor JW. Polymerase chain reaction (PCR) primers for amplifying and sequencing nuclear 18s rDNA from lichenized fungi. Mycologia. 1992;84:589–592. [Google Scholar]
- 24.Rehner SA, Samuels GJ. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res. 1994;98:625–634. [Google Scholar]
- 25.Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]