Significance
In a breast cellular transformation model, we describe the gene regulatory network that is mediated by joint and direct action of three inflammatory transcription factors on hundreds of target genes in various oncogenic pathways. This inflammatory feedback loop and associated network functions in many types of cancer cells and patient tumors. It forms the basis for a “cancer inflammation index” that defines cancer types by functional criteria, and that may be useful for choosing drugs for personalized cancer treatment. Inflammatory tumor cells, via secreted inflammatory molecules, might help recruit immune cells to the site of the tumor. Thus, detailed analysis of an artificial transformation model uncovers the molecular basis of an inflammatory network relevant for many forms of human cancer.
Keywords: epignetic switch, inflammation, cancer, gene regulatory network
Abstract
Using an inducible, inflammatory model of breast cellular transformation, we describe the transcriptional regulatory network mediated by STAT3, NF-κB, and AP-1 factors on a genomic scale. These proinflammatory regulators form transcriptional complexes that directly regulate the expression of hundreds of genes in oncogenic pathways via a positive feedback loop. This transcriptional feedback loop and associated network functions to various extents in many types of cancer cells and patient tumors, and it is the basis for a cancer inflammation index that defines cancer types by functional criteria. We identify a network of noninflammatory genes whose expression is well correlated with the cancer inflammatory index. Conversely, the cancer inflammation index is negatively correlated with the expression of genes involved in DNA metabolism, and transformation is associated with genome instability. We identify drugs whose efficacy in cell lines is correlated with the cancer inflammation index, suggesting the possibility of using this index for personalized cancer therapy. Inflammatory tumors are preferentially associated with infiltrating immune cells that might be recruited to the site of the tumor via inflammatory molecules produced by the cancer cells.
Inflammation is a hallmark of cancer and plays important regulatory roles during cell transformation, invasion, metastasis, and treatment resistance (1–3). An inflammatory reaction during tumor development occurs in nearly all solid malignancies (4). Preexisting inflammation promotes subsequent cancer development, accounting for 15 ∼ 20% of cancer deaths (3). Inflammation also plays critical roles in immunosuppression (2). For example, STAT3 promotes the expression of PD-L1 and PD-L2 in cancer cells, suppressing immune cell activity (5).
Tumor-associated inflammation results from both an intrinsic pathway, in which mutations in cancer cells activate inflammatory gene expression, and an extrinsic pathway, in which cytokines and chemokines secreted by tumor-associated immune cells create inflammatory microenvironments (3). Oncogenes can trigger a gene expression cascade, resulting in activation or overexpression of proinflammatory transcription factors such as NF-κB, STAT3, and AP-1, with the resulting production of cytokines and chemokines (1, 2, 6). For example, NF-κB signaling pathway is activated upon activation of oncogenes RAS and MYC, through activation of IL-1β (6–8), and the proto-oncoprotein Src kinase can directly phosphorylate and activate STAT3 (9).
In previous work, we have described an inducible model of cellular transformation in which transient activation of v-Src oncoprotein converts a nontransformed breast epithelial cell line into a stably transformed state within 24 h (6, 10). The stably transformed cells form foci and colonies in soft agar, show increased motility and invasion, form mammospheres, and confer tumor formation in mouse xenografts (6, 10). This epigenetic switch between stable nontransformed and transformed states is mediated by an inflammatory positive feedback loop involving NF-κB and STAT3 (6, 11). By integrating motif analysis of DNase hypersensitive regions with transcriptional profiling, we found that >40 transcription factors are important for transformation and identified putative target sites directly bound by these factors (12).
A few transcriptional regulatory circuits involved in this transformation model have been identified, and these are important in some other cancer cell types and human cancers (6, 11, 13, 14). During transformation, STAT3 acts through preexisting nucleosome-depleted regions bound by FOS, and expression of several AP-1 factors is altered in a STAT3-dependent manner (15). However, the connection between STAT3, NF-κB, and AP-1 factors as well as the underlying transcriptional regulatory circuits has not been described on the whole-genome level.
Here, we define the transcriptional network mediated by the combined action of NF-κB, STAT3, and AP-1 factors (JUN, JUNB, and FOS) on a genomic scale in this breast transformation model. In contrast to previous studies (12, 15), this network is defined by genes that are induced during transformation by the binding of NF-κb, STAT3, and AP-1 factors to common target sites either through their cognate motifs or via protein–protein interactions. Based on this common NF-κb, STAT3, and AP-1 network, we develop a cancer inflammation index to define cancer types, both in cell lines and in patients, by functional criteria. As this inflammation index is based on the common regulatory network, it is distinct from, and more specific than, indices based simply on gene expression profiles that arise from multiple regulatory inputs. In addition, we identify many noninflammatory genes whose expression is positively or negatively correlated with the cancer inflammation index, leading to the observation that inflammation is linked functionally to other aspects of cancer as well as genomic instability. Lastly, we show that inflammatory tumor samples preferentially contain contaminating immune and stromal cells from the tumor microenvironment, consistent with the idea that immune cells might be recruited to the site of the tumor via inflammatory molecules produced by the cancer cells.
Results
Colocalization of STAT3, NF-κB, and AP-1 Factors on Target Sites in Vivo.
During transformation, NF-κb, STAT3, and AP-1 (JUN, JUNB, and FOS) levels increase in the nucleus (SI Appendix, Fig. S1A). In accord with previous results using chemical inhibitors and siRNA knockdowns (6), CRISPR-cas9–mediated knockouts of these factors (SI Appendix, Fig. S1B) cause decreased transformation as assayed by growth under conditions of low attachment (16) (SI Appendix, Fig. S1C). Our previous ChIP-seq analysis mapped protein-binding sites to nucleosome-depleted regions before and after (24-h tamoxifen treatment) transformation (15) but did not provide more detailed localization.
The ChIP-seq data reveals many target sites bound by STAT3 (89,764 sites), NF-κB (56,539 sites), and AP-1 factors (95,958 sites for JUNB, 152,443 sites for FOS, and 118,245 sites for JUN) (SI Appendix, Figs. S1D and S2A). Less than 17% of factor binding sites are located in promoter regions, and the majority of sites are located in other regulatory regions, including ∼30% that are in active enhancers (SI Appendix, Fig. S2A). As expected from the heteromeric nature of AP-1 factors, binding levels of JUN, JUNB, and FOS are well correlated with each other (Pearson correlation coefficiencies ≥ 0.83), which are nearly comparable to biological replicates (SI Appendix, Fig. S2B). Based on this, we grouped the AP-1 factors together in most subsequent analyses. Also as expected, STAT3 binding sites strongly overlap with FOS binding sites (15) and with the other AP-1 factors (89% of STAT3 binding sites) (SI Appendix, Fig. S3A). In addition, 89% of NF-κB binding sites overlap with AP-1 factors (SI Appendix, Fig. S3A). Thirty-eight percent of STAT3, NF-κB, and AP-1 sites are located in the same cis-regulatory regions (CRRs; SI Appendix, Fig. S3A), which is far more frequent than expected by chance (P < 10−200; χ2 test). Despite the strong colocalization, STAT3, NF-κb, and AP-1 factors can independently bind to specific sites (SI Appendix, Fig. S3A), and the pairwise correlation values are lower than the correlation values between biological replicates (SI Appendix, Fig. S3B).
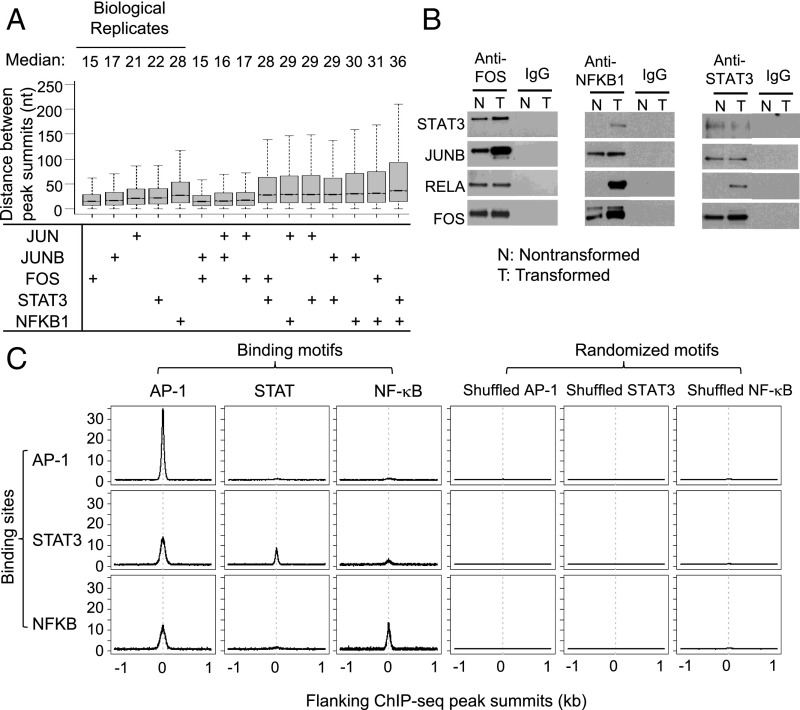
Additional results suggest that STAT3, NF-κB, and AP-1 factors cobind to target sites as a multiprotein complex. Most importantly, the median distance of peak summits for all pairwise combinations of AP-1 factors, STAT3, and NF-κB ranges between 15 and 30 bp, and these values are comparable to those obtained for biological replicates of the relevant individual factors (Fig. 1A). The indistinguishable binding profiles of STAT3, NF-κB, and AP-1 factors on target sites indicate that these proteins all associate (either directly or indirectly) with the same DNA sequence. In this regard, the analytical method used here revealed that the NF-YA and NF-YB subunits of the heteromeric transcription factor NF-Y bind asymmetrically to their target site, with a 15-bp separation between peak binding of the individual subunits (17). Consistent with these common binding profiles, coimmunoprecipitation experiments show that STAT3, NF-κB, and AP-1 factors interact with each other in the nucleus (Fig. 1B).
Fig. 1.
Features of transcription factor binding. (A) Distance between peak summits of indicated factors locating in the same cis-regulatory region. (B) Coimmunoprecipitation experiment showing the interactions between FOS, JUNB, STAT3, and NF-κB in the nucleus of nontransformed (N) and transformed (T) cells. IgG IP was used as the control. (C) Distribution of AP-1, STAT, and NF-κb consensus motif around factor peak summits. As the control, we randomly shuffled AP-1, STAT, and NF-κb consensus motifs and plotted the motif distribution around peak summits.
Sequence Motifs Associated with Binding of STAT3, NF-κb, and AP-1 Factors.
To address which factors are primarily responsible for binding site specificity, we performed motif analysis on sequences around peak summits. As expected, 60% of AP-1 factor binding peaks contain an AP-1 motif (SI Appendix, Fig. S3C). Interestingly, 38% of STAT3 binding sites and 30% of NFKB1 sites also have an AP-1 motif, while 24% of STAT3 sites have a STAT motif and 16% NFKB1 sites have a NF-κB motif (SI Appendix, Fig. S3C). Furthermore, the AP-1 motif is well located in STAT3 and NFKB1 peak summits (Fig. 1C). We did not find significant colocalization of AP-1, STAT, and NF-κB motifs in the same CRRs. These results indicate that a significant fraction of STAT3 and NF-κB binding is mediated through the interaction with AP-1 factors. In contrast, only a small minority of AP-1 binding sites contain a STAT or NF-κb motif (Fig. 1C and SI Appendix, Fig. S3C), and STAT and NF-κb motifs show modest enrichment around AP-1 peak summits (Fig. 1C). Thus, binding of AP-1 factors occurs predominantly via interactions with AP-1 motifs, presumably reflecting a direct protein–DNA interaction. In contrast, in addition to directly binding via their motifs, STAT3 and NF-κb can also bind to AP-1 motifs, presumably via protein–protein interactions with AP-1 factors. However, we did not observe significant motif differences in AP-1 sites bound by AP-1 factors alone or those cobound with STAT3 and/or NF-κb.
STAT3, NF-κb, and AP-1 Factors Coregulate Key Genes in Various Oncogenic Pathways.
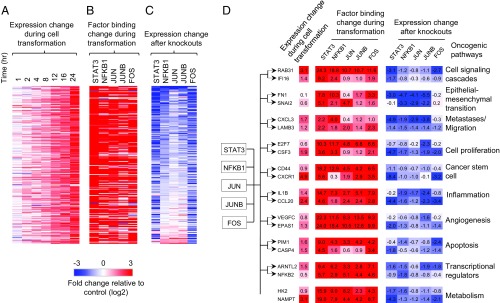
Cobinding of STAT3, NF-κB, and AP-1 factors regulates gene expression and chromatin status. Genes up-regulated during transformation (Fig. 2A) tend to have increased binding of STAT3, NFKB1, JUN, JUNB, and FOS at promoters/enhancers (Fig. 2B). Furthermore, isogenic cell lines lacking individual factors can block the up-regulation of a subset of genes during transformation (Fig. 2C), with the STAT3 knockout showing the most drastic effects. As there are multiple members of the NF-κb and AP-1 families, it is likely that the weaker effect on transcription is due to redundancy among family members. With respect to chromatin structure, regions bound by increasing numbers of these factors tend to have higher accessibility and acetylation levels (SI Appendix, Fig. S4A, B). In addition, differential binding levels of STAT3, NFKB1, and AP-1 factors during transformation are positively correlated with dynamic chromatin accessibility (Pearson correlation coefficient ≥ 0.44) (SI Appendix, Fig. S4C), and open chromatin regions bound by more factors tend to show increased accessibility (SI Appendix, Fig. S4D).
Fig. 2.
Cobinding and common targets of STAT3, NF-κb, and AP-1 factors. (A-C) For each gene that is continuously up-regulated during transformation (A), fold changes of factor binding levels in promoters/enhancers (B), and gene expression changes after factor knockouts at 24 h after transformation (C) are shown. (D) Examples of common target genes of STAT3, NFKB1, and AP-1 factors regulating different oncogenic pathways. The fold changes of expression during transformation, factor binding level changes in promoters/enhancers, and gene expression changes after factor knockout at 24 h after transformation are shown. The degree of up-regulation (red) and down-regulation (blue) is indicated by the color intensity.
We identified 1,461 genes that are common targets of STAT3, NFKB1, JUN, JUNB, and FOS and that show increased binding of at least four factors (>1.5 fold) during transformation and down-regulation upon at least four factor knockouts (Dataset S1). These genes are enriched [Benjamini–Hochberg false discovery rate (FDR) < 0.005] in cancer-related processes (SI Appendix, Fig. S5A), and they include genes involved in cell signaling cascades (e.g., RAB13, IF116, ZAK), inflammatory response (e.g., IL1B, IL1R1, SERPINA1), cell proliferation (e.g., CSF3, E2F7, E2F8) regulation of apoptosis (e.g., PIM1, CARD6, BCL2L1), angiogenesis (e.g., RHOB, CEGFC, EPAS1), and cell migration/metastases (e.g., LAMB3, CXCL3, and PLAU) (Fig. 2D and SI Appendix, Fig. S5B). Cancer stem cells are generated during the transformation process in our model (18), and STAT3, NF-κb, and AP-1 factors activate key genes (e.g., CD44, CXCR1, and ITGA7) mediating cancer stem cell formation. Cancer metabolism is a key component of oncogenesis (19), and important metabolic enzyme genes (e.g., HK2 and NAMPT) are also regulated by these factors (Fig. 2D and SI Appendix, Fig. S5B). Thus, STAT3, NF-κb, and AP-1 are major factors involved in tumor-promoting inflammation, and they bind to and promote the expression of key genes in various oncogenic pathways (SI Appendix, Fig. S5B).
The Inflammatory Positive Feedback Loop That Promotes Transformation Is Extensive.
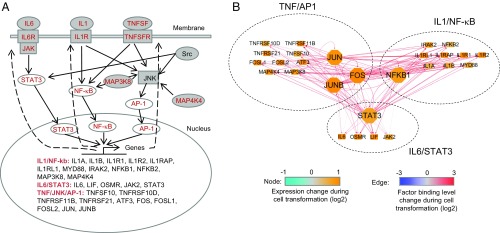
In previous work, we described a few specific regulatory circuits that constitute an inflammatory positive feedback loop required for transformation in this experimental model (6, 11, 15). Here, we show that this positive feedback loop is far more extensive. In particular, during transformation, STAT3, NF-κΒ (NFKB1 and NFKB2), and AP-1 factors (JUN, JUNB, FOS, FOSL1, FOSL2, and ATF3) are transcriptionally up-regulated as are many upstream regulators in the IL6/STAT3, IL1/NF-κB, and TNF/AP-1 signaling pathways, including IL1 (IL1A and IL1B) and IL1 receptors (IL1R1, IL1R2, IL1RAP, and IL1RL1), IL6 (IL6 and LIF) and IL6 receptor, TNF and TNF receptors (TNFRSF10D, TNFRSF11B, and TNFRSF21), and JAK2 and MAP kinases (MAP3K8 and MAP4K4) (Fig. 3A). These genes are all common targets of STAT3, NFKB1, and AP-1 factors, with increased factor binding in promoters/enhancers (Fig. 3B). Thus, the essence of the loop is that STAT3, NFKB1, and AP-1 directly activate upstream regulators that trigger the activation of intracellular signaling cascades, phosphorylate the transcription factors, and promote their nuclear localization and transcriptional activation. In accord with previous studies on IL6 (6), reducing the activation levels of JAK2, JNK, and IL1 receptors via inhibitors or antagonists results in decreased transformation efficiency (SI Appendix, Fig. S6). For subsequent analyses, we define 27 genes directly involved in the IL6/STAT3, IL1/NF-κb, and TNF/AP-1 signaling pathways (Fig. 3A) as the core positive feedback loop that maintains the transformed state.
Fig. 3.
Positive feedback loop among IL1/NF-κb, IL6/STAT3, and TNF/AP-1 pathways. (A) The positive feedback loop of IL1/NF-κb, IL6/STAT3, and TNF/AP-1 pathways. The red colored genes are transcriptionally activated during transformation. (B) The network showing genes in the loop are common targets STAT3, NFκb1, JUN, JUNB, and FOS. The nodes represent genes in the loop. The edges represent binding of transcription factors in promoters/enhancers of target genes.
A Cancer Inflammation Index To Measure the Inflammation Level of a Cancer Cell Line.
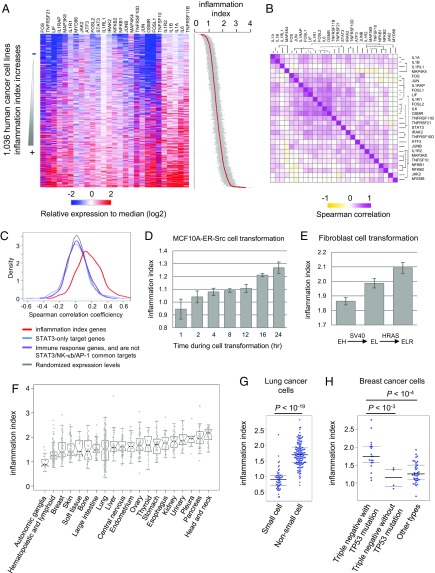
The Cancer Cell Line Encyclopedia (CCLE) database contains gene expression data for 1,036 human cancer cell lines from over 20 developmental lineages (20). The expression levels of 27 genes in the IL6/STAT3, IL1/NF-κB, and JNK/AP-1 pathways across those cancer cell lines (Fig. 4 A and B) are positively correlated beyond chance expectation (median of Spearman’s rank correlation coefficient = 0.17; Wilcoxon rank sum test P value <10−200) (Fig. 4C), consistent with the inflammatory loop being active in different types of cancers. In contrast, genes regulated by STAT3 only and immune response genes do not show significant correlation across cancer cell lines (Fig. 4C). As expected from the extraordinary diversity of cancers, some cancer cell lines have transcriptional profiles much more similar to those the ER-Src model than others.
Fig. 4.
The inflammatory loop is active in different types of cancer cells. (A) Heatmap showing relative expression levels (median normalized values) of genes in the loop among 1,036 cancer cell lines. Samples were sorted by the cancer inflammation index (SEs shown in gray color), which is calculated as the normalized median value of the relative expression levels of the indicated 27 genes in the inflammatory loop. (B) Spearman correlation of expression levels of the gene pairs. (C) Distribution of observed and expected Spearman correlation coefficiencies of expression levels of genes in the loop. The expected correlation values were calculated by the randomly picked genes expressed in the cancer cell lines. We also calculated the Spearman correlation coefficiencies of genes targeted by STAT3 only and immune response genes, which are not targets of STAT3/NF-κB/AP1. (D) Change of cancer inflammation index values during ER-Src cell transformation. Error bars represent SD between two biological replicates. (E) Change of cancer inflammation index values during fibroblast transformation. Error bars represent SD between three biological replicates. (F) Distribution of cancer inflammation index values among cancer cell lines from different developmental lineages. (G) Cancer inflammation indexes in genetic subtypes of lung cancer cell lines. (H) Cancer inflammation indexes in genetic subtypes of breast cancer cell lines.
To measure the inflammation level for each cancer cell line, we developed a scoring system called the cancer inflammation index, which is calculated as the expression values of 27 genes (normalized to the median expression levels in the 1,036 cell lines) in the positive feedback loop (Dataset S2). Importantly, this index is not simply based on a subset of genes that are induced during transformation, but rather genes that are direct targets of the STAT3/NF-κB/AP-1 regulatory network. The inflammatory levels gradually increase during ER-Src transformation (Fig. 4D), and they are highly variable (>fivefold) among different cancer cell lines (Fig. 4A). Similarly, in a fibroblast cell transformation model involving stepwise addition of the SV40 and RAS oncogenes to immortalized fibroblast, the inflammatory levels increase upon transformation (Fig. 4E).
On average, head and neck as well as pancreatic cancer cell lines are most inflammatory, while the autonomic ganglia and blood cancer cell lines are least inflammatory (Fig. 4F). However, cancer cell lines from the same developmental lineage can show high variance in inflammatory levels, which is correlated with their genetic subtypes. For example, cell lines from nonsmall cell lung cancers are more inflammatory than those of small cell lung cancers (Fig. 4G), and triple negative breast cancer cell lines with p53 mutations are more inflammatory than other subtypes (Fig. 4H).
The Inflammatory Loop in Tumors from Cancer Patients.
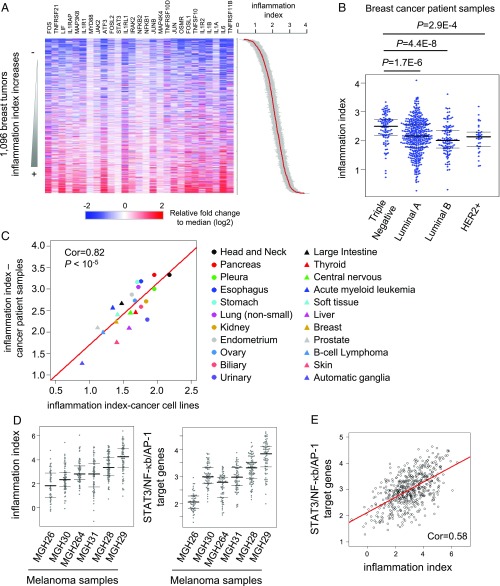
Although cancer cell lines are derived from tumors, long-term propagation of cell lines under artificial conditions raises the possibility that cell line data might be misleading with respect to cancer. We addressed the relevance of the inflammatory loop in human cancers by using RNA-seq data in the Cancer Genome Atlas database (21) to calculate the inflammation index in human tumors (Dataset S3). In accord with the results in cancer cell lines, genes in the IL6/STAT3, IL1/NF-κB, and JNK/AP-1 inflammatory loop are coexpressed in human breast tumors (Fig. 5A). Similarly, triple negative breast tumors are more inflammatory than other types of breast tumors (Fig. 5B). Moreover, the median cancer inflammatory index value of all tumor samples from a given developmental lineage is highly correlated with that of cells lines from the same developmental lineage (Pearson correlation coefficient = 0.82; P value <10−5) (Fig. 5C). As we combined all tumors for a given lineage, it seems unlikely that this analysis is significantly compounded by differences in the tumor microenvironment. These observations indicate that, with respect to inflammation and gene regulation profiles, cancer cell lines are good models for tumors, and the inflammatory loop is relevant for many types of human cancer.
Fig. 5.
The inflammatory levels are variable among cancers with different developmental lineages and genetic subtypes. (A) Heatmap showing the relative expression levels of 27 genes in IL6/STAT3, IL1/NF-κb, and TNF/NF-κb pathways in breast cancer patient samples from the TCGA database. The samples were sorted by the cancer inflammation index (SEs shown in gray color). (B) Inflammatory indexes in genetic subtypes of breast cancer patients. (C) Correlation between inflammatory levels between cancer cell lines and patient tissue samples among different developmental lineages. The median cancer inflammation index value of patient samples from the same developmental lineage indicates its inflammatory level. (D) Inflammatory index and expression levels of of STAT3/NF-κb/AP-1 target genes using RNA-seq data from single cells from melanoma patient samples. Each point represents the expression from a single cell from the indicated tumor. (E) The correlation between inflammatory index and expression levels of target genes of STAT3/NF-κb/AP1 across single cells from melanoma patients.
To avoid potential contributions of noncancer cells in patient samples, we analyzed published gene expression levels in single cancer cells from melanoma patient samples (22). The inflammatory loop and target genes are expressed at variable levels (>10-fold) across single cells in various tumor samples (Fig. 5D). The median inflammatory level in the sample MGH29 is fourfold higher than MGH26 (Fig. 5D). The inflammatory index shows significant correlation with the expression level of STAT3/NF-κB/AP-1 target genes (Pearson correlation coefficiency = 0.58, P value <2.2e-16) (Fig. 5E). These direct measurements of gene expression levels of individual cancer cells from tumors indicates that the inflammatory loop and associated network is used in cancer patients.
Identification of Noninflammatory Genes Whose Expression Is Correlated to the Cancer Inflammation Index.
Previous analyses of the ER-Src model identified oncogenically relevant genes based on differential gene expression upon transformation and/or direct regulation by NF-κB, STAT3, and AP-1 factors. However, it is highly likely that this approach will miss critical genes that are part of the transformation process. As an alternative approach, for every gene, we calculated the Spearman correlation coefficient between its expression level and the inflammation index across cancer cell lines for various developmental lineages. The correlation values are significantly conserved among developmentally distinct cancers, again consistent with a widespread role of the inflammatory loop and it target genes (Fig. 6A).
Fig. 6.
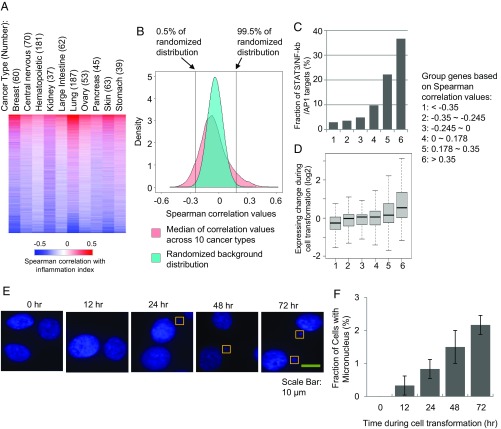
Genes and pathways with expression levels correlated with inflammatory loop. (A) Heatmap showing the Spearman correlation coefficiencies between gene expression levels and inflammatory index across 10 types of cancers. (B) Distribution of the median correlation values across 10 cancer types. For the background distribution, we randomized the correlation values in each cancer type and calculated the median values of the randomized values. (C) Fractions of genes are direct targets of STAT3/NF-κb/AP1. Genes were grouped based on Spearman correlation values with the cancer inflammation index as in B. (D) Expression change of the gene groups during transformation. (E) Fluorescent nuclear imaging of micronuclei reveals increased genome instability. (F) Fraction of cells showing micronucleus formation during transformation.
Genes showing higher positive correlation with the cancer inflammation index are more likely to be direct transcriptional targets of STAT3/NF-κB/AP1 and tend to be up-regulated during transformation (Fig. 6 B–D). We identified 1,303 genes showing significant positive correlation of expression with the cancer inflammation index (median Spearman’s rank correlation coefficient across cancer cell types >0.18, false discovery rate <0.005) (Fig. 6B). Twenty-eight percent of these genes are common and direct targets of STAT3, NF-κB, and AP-1 factors (SI Appendix, Fig. S5B). Aside from genes involved in the inflammatory response, these genes are enriched in biological pathways such as angiogenesis, cell proliferation, apoptosis, intracellular signaling cascade, and cell migration (Fig. 7 and SI Appendix). These pathways, which are defined by the inflammatory index and not transformation per se, are in excellent accord with the pathways accord with pathways activated during ER-Src transformation (SI Appendix, Fig. S5B), thereby providing independent evidence that the inflammatory loop and oncogenic pathways uncovered in the ER-Src cell transformation model are relevant for different types of human cancer.
Fig. 7.
Correlation between inflammation index, cancer progression index, and tumor purity. (A) Correlation between tumor purity, levels of infiltrating immune cells (CD8+/CD4+ T cells and macrophages), and cancer inflammation index of breast cancer patient samples. (B) Randomly picked breast tumor samples with different inflammation levels and similar purity were examined for gene expression levels in the indicated oncogenic pathways that are direct targets of STAT3/NF-κb/AP-1 and also show significant positive correlation of expression with the cancer inflammation index in Fig. 6B. Genes associated with immune/inflammatory response were removed from the analyses. (C) Randomly picked tumor samples with different inflammation levels and similar purity were examined for expression levels of target genes of STAT3/NF-κb/AP1 in lung cancers, head and neck cancers, glioblastomas, and melanoma. (D) Correlation between inflammation index and cancer progression index. (E) Correlation between tumor purity and cancer progression index.
A Correlation Between Inflammation and Genome Instability in Cancer Cell Lines.
One-thousand three hundred sixty-nine genes show negative correlation between expression levels and the cancer inflammation index, and these are enriched in pathways including DNA metabolic process, DNA replication, DNA repair, and cell cycle (Benjamin FDR < 10−13). This observation suggests that oncogenic-associated inflammation is inversely related to genome stability, and indeed the genes showing most negative correlation (e.g., MSH2, FANCF, BRCA1) are regulators of genome instability. Interestingly, the genes related to genome instability are not transcriptionally regulated during ER-Src cell transformation.
To examine the link between inflammation and genome instability, we performed micronucleus staining of cells during ER-Src transformation. Indeed, more cells contain micronuclei as transformation processes (Fig. 6 E and F), with 2.2% cells containing micronuclei after 72 h after induction of transformation compared with virtually no cells before transformation. Thus, the inflammation-mediated process of transformation is associated with decreased genome stability.
Inflammatory Tumor Samples Contain Increased Levels of Noncancer Cells.
As shown above, the inflammation indices of cancer cell lines for a particular cancer type are highly correlated with the corresponding tumor samples (Fig. 5C). However, unlike samples from cancer cell lines, tumor samples also contain immune and stromal cells from the tumor microenvironment in addition to the cancer cells. Such tumor impurity complicates the analysis of gene expression profiles of tumor cells, particularly as immune cells are also inflammatory. We therefore took several approaches to disentangle the inflammatory nature of cancer cells from other cells in a tumor.
First, for numerous tumor samples we analyzed the relationship between the cancer inflammation index, tumor purity (fraction of cancer cells in a sample), and infiltrating immune cells, which has been estimated from cancer-specific genetic mutations or cell type-specific gene signatures (23, 24). In general, the inflammation level increases as the tumor purity decreases and the fraction of infiltrating immune cells increase (Fig. 7A and SI Appendix, Fig. S8 A–G). However, triple negative breast tumors have higher inflammatory levels than those of other genetic subtypes, although all of these forms of breast cancer have similar purity levels (SI Appendix, Fig. S8H). The higher inflammatory level of triple negative breast tumors is consistent with the results in the corresponding cancer cell lines (Fig. 5B), indicating that it reflects the intrinsic properties of the cancer cells and not the degree of tumor purity.
Second, we selected breast patient samples with similar levels of tumor purity but different levels of inflammation (divided into four bins) and analyzed expression of noninflammatory genes whose expression is strongly correlated with the inflammatory index (Fig. 7B). Expression of these transformation-related, but noninflammatory, genes, which are direct targets of STAT3/NF-κb/AP1 and are involved in angiogenesis, apoptosis, and cell migration, increases as the cancer inflammation index increases. Similar results are observed in all other types of cancer we examined such as melanoma, lung, head and neck, and glioblastomas (Fig. 7C and SI Appendix, Fig. S9). These results indicate that the inflammatory loop and associated network are active in various types of cancers, although we can’t exclude the formal possibility that nonmalignant cells in the various samples might be in different states.
Third, we created a “progression index” that is strongly correlated with the inflammatory index (Fig. 7D), but is based on 98 genes that regulate “migration/metastasis,” “apoptosis,” and “angiogenesis,” but are not currently annotated as inflammatory or part of the immune response. This progression index should reflect the regulatory circuits involved in cellular transformation and tumor formation, but not normal immune cells. Indeed, across the large set of tumor samples, the score of noninflammatory index inversely correlated with tumor purity (Fig. 7E). All of these observations indicate that tumors containing inflammatory cancer cells preferentially contain noncancer cells. Thus, tumor-associated inflammation level is positively correlated with the complexity of microenvironment and the presence of immune cells.
Identifying Drugs Whose Efficacy Is Correlated with the Cancer Inflammation Index.
The Genomics of Drug Sensitivity in Cancer measured the drug efficacy of 267 compounds in 1,001 cancer cell lines (25). Based on the RNA expression levels, we calculated the inflammation index of the cell lines. Then for each compound, we calculated the Spearman correlation coefficiency between inflammation index and the drug potency indicated by the half maximal inhibitory concentration (IC50) values across the cell lines (SI Appendix, Fig. S10 and Dataset S4). Interestingly, a few compounds showed significantly better effect in inhibiting the growth of inflammatory cells (P value <10−5), such as inhibitors targeting MEK1/MEK2 (Trametinib and Refametinib), HSP90 (Tanespimycin), and HER2/EGFR (Lapatinib). Also, inflammatory cells require higher doses of HDAC inhibitors and bromodomain family inhibitors to achieve the growth inhibition effect. These observations suggest the potential use of the cancer inflammation index for personalized cancer treatment.
Discussion
NF-κB, STAT3, and AP-1 Factors via Their Common Targets Mediate the Positive Feedback Loop Controlling Inflammatory Cancers.
In the ER-Src cellular transformation model, a transient inflammatory stimulus mediates an epigenetic switch from a stable nontransformed cell to a stable transformed cell (6). Epigenetic switches are the basis of multicellular development, and they occur by activating a positive feedback loop that maintains the altered state. In the ER-Src model, Src activates the inflammatory transcription factors STAT3 and NF-κB that form the basis of the inflammatory feedback loop that is required for maintenance of the transformed state (6, 11, 12, 15).
Here, we show that AP-1 factors play a critical role in the inflammatory feedback loop. AP-1 factors are not only important for transformation, but they form complexes with STAT3 and/or NF-κB that bind target sites. Specifically, these factors coimmunoprecipitate, and their binding profiles are coincident at many target sites. Most, and perhaps all, of the sites where cobinding occurs contain AP-1 motifs, suggesting that the AP-1 factors directly interact with DNA, whereas STAT3 and NF-κB are often recruited via interactions with the AP-1 factors. At some sites, STAT3 and NF-κB bind via their own motifs in the absence of AP-1 factors. At present, it is unclear whether AP-1, NF-κB, and STAT3 can form a ternary complex at individual sites or if NF-κB and STAT3 form independent complexes with AP-1 factors at these sites.
In addition to their roles in inflammation per se, STAT3, NF-κB, and AP-1 work together to regulate key genes in oncogenic pathways such as angiogenesis, apoptosis, cell migration, and epithelial to mesenchymal transition. Expression of many genes in these pathways is induced upon transformation in a manner that is linked to increased binding of all these factors. Moreover, expression of many common genes is reduced upon knockouts of individual factors, indicating that all of these factors contribute to expression of these genes. Our identification of a common STAT3/NF-κB/AP-1 network is distinct from, but not inconsistent with, previous observations that STAT3 and NF-κB have different (i.e., nonoverlapping) binding sites and gene expression effects during the transformation process (15).
The positive feedback loop elucidated here is considerably more complex than previously described, and it is maintained in two distinct ways. The key transcription factors directly bind and activate the expression of each other, thereby reinforcing the core transcriptional state. This mutual coregulation of key transcription factors is similar to what occurs in muscle development (MyoD and related factors), embryonic stem cells (Nanog, Oct4), and presumably all epigenetic states. In addition, these transcription factors directly regulate nearly all upstream components in the IL6/STAT3, IL1/NF-κΒ, and TNF/AP-1 signaling pathways, thereby maintaining the activity of the individual factors and hence the gene regulatory pattern. Thus, this extensive positive feedback loop represents a coherent regulatory system in which numerous interconnected components stably maintain a common, yet complex, transcriptional program.
The Inflammatory Gene Signature as an Approach to Type Human Cancers.
Historically, cancer types were classified by their developmental origin as well as by crude cellular phenotypes. More recently, cancers have been classified by the genetic mutations that drive the oncogenic state. Such classification, together with drugs targeted to specific mutations, has been the primary basis for the idea of personalized medicine approaches to cancer treatment. Here, we develop a functional approach to classify human cancers that utilizes gene expression signatures, specifically a cancer inflammation index based on the STAT3/NF-κB/AP-1 network. Importantly, this index is based on an integrated regulatory network and hence is significantly different from indices based solely on transcriptional profiles that arise from multiple regulatory inputs.
Overall, expression levels of the 27 key genes that make up the cancer inflammation index are strongly correlated with each other. Not only does this observation provide further support for the coherence of the feedback loop, but it suggests that the loop and associated network functions to various degrees in different cancer cell lines and tumors. Transformed cell lines have a higher cancer inflammation index than observed with nontransformed cells, consistent with a general role of inflammation in cancer. However, there is a wide range of inflammation scores among various cancers that occurs in developmentally unrelated cancers. Using data from the Cancer Genome Atlas, we did not observe a significant correlation between the cancer inflammation index and any particular mutation or the total mutation number. Importantly, the inflammation indices of cell lines and tumors with similar developmental origins are strongly related, suggesting its relevance for human disease. We identified drugs whose efficacy correlates with the cancer inflammation index, suggesting that determining the inflammation index of tumors from individual cancer patients might be useful for choosing drugs for personalized therapy.
Inflammatory Cancers May Preferentially Recruit Immune Cells to the Tumor Site.
Patient tumor samples contain cancer cells and immune (and other noncancer) cells, all of which contribute to the transcriptional profile. Interestingly, over a large number of tumor samples, there is an inverse relationship between the inflammatory index and the estimated degree of sample purity. In principle, this relationship could merely reflect the fact that immune cells also express many inflammatory genes, which could significantly contribute to the observed cancer inflammation index of the sample.
We attempted to distinguish the contributions of cancer and immune cells to the transcriptional profile by analyzing many noninflammatory genes (either in specific pathways or as an overall index) whose expression is very strongly correlated with the inflammatory index and hence to the STAT3, NF-κB, AP-1 regulatory network. These observations suggest a dynamic interplay between cancer cells and immune cells that is linked to the cancer inflammation index of the cancer cells in the sample. We propose that cytokines and chemokines secreted by the cancer cells attract immune cells to their vicinity of the tumor, thereby resulting in a less pure tumor sample. In addition, these cytokines and chemokines secreted by the cancer cells also help activate the inflammatory gene expression program in immune cells. Thus, we propose that the mutual and perhaps synergistic functional interactions between cancer cells and immune cells can create an inflammatory microenvironment in a manner that depends on the inflammatory properties of the cancer cells.
Materials and Methods
Detailed information on cell lines, generation of CRISPR knockouts, Western blotting, micronucleus assays, ChIP-seq, DNA-seq, and RNA-seq to determine chromatin states and map transcription factor binding sties, analyzing transcriptional profiles in cancer cell lines (CCLE) and cancer patients (TCGA database), calculation of the cancer inflammation and cancer progression indices, and gene ontology analyses are described in SI Appendix, Materials and Methods. Sequencing data have been deposited in the National Cancer for Biotechnology Information Gene Expression Omnibus with accession numbers GSE115597, GSE115598, and GSE115599.
Supplementary Material
Acknowledgments
This work was supported by the Searle Leadership Fund in the Life Sciences from Northwestern University (to Z.J.), National Cancer Institute K99 CA 207865 (to Z.J.), and National Institutes of Health Research Grant CA 107486 (to K.S.). A.R. is a Howard Hughes Investigator.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE115597, GSE115598, and GSE115599).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821068116/-/DCSupplemental.
References
- 1.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 4.de Martel C, Franceschi S. Infections and cancer: established associations and new hypotheses. Crit Rev Oncol Hematol. 2009;70:183–194. doi: 10.1016/j.critrevonc.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest. 2015;125:3384–3391. doi: 10.1172/JCI80011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerra C, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Shchors K, et al. The Myc-dependent angiogenic switch in tumors is mediated by interleukin 1beta. Genes Dev. 2006;20:2527–2538. doi: 10.1101/gad.1455706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turkson J, et al. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirsch HA, et al. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell. 2010;17:348–361. doi: 10.1016/j.ccr.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji Z, et al. Genome-scale identification of transcription factors that mediate an inflammatory network during breast cellular transformation. Nat Commun. 2018;9:2068. doi: 10.1038/s41467-018-04406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iliopoulos D, Rotem A, Struhl K. Inhibition of miR-193a expression by Max and RXRα activates K-Ras and PLAU to mediate distinct aspects of cellular transformation. Cancer Res. 2011;71:5144–5153. doi: 10.1158/0008-5472.CAN-11-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polytarchou C, Iliopoulos D, Struhl K. An integrated transcriptional regulatory circuit that reinforces the breast cancer stem cell state. Proc Natl Acad Sci USA. 2012;109:14470–14475. doi: 10.1073/pnas.1212811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleming JD, et al. STAT3 acts through pre-existing nucleosome-depleted regions bound by FOS during an epigenetic switch linking inflammation to cancer. Epigenetics Chromatin. 2015;8:7. doi: 10.1186/1756-8935-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rotem A, et al. Alternative to the soft-agar assay that permits high-throughput drug and genetic screens for cellular transformation. Proc Natl Acad Sci USA. 2015;112:5708–5713. doi: 10.1073/pnas.1505979112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleming JD, et al. NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res. 2013;23:1195–1209. doi: 10.1101/gr.148080.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci USA. 2011;108:1397–1402. doi: 10.1073/pnas.1018898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinstein JN, et al. Cancer Genome Atlas Research Network The Cancer Genome Atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iorio F, et al. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166:740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.