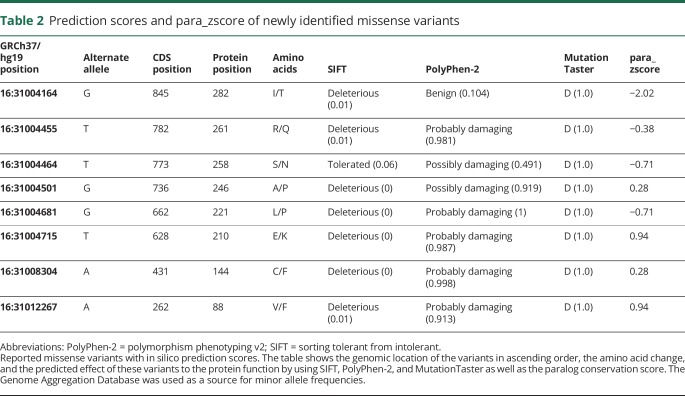
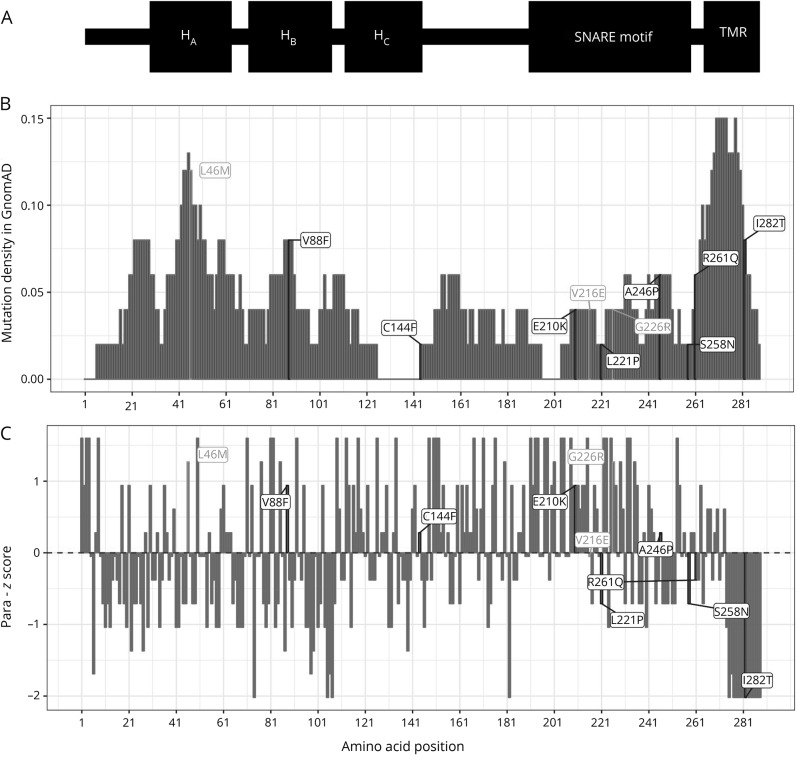
Stefan Wolking
Stefan Wolking, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Patrick May
Patrick May, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Davide Mei
Davide Mei, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Rikke S Møller
Rikke S Møller, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Simona Balestrini
Simona Balestrini, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Katherine L Helbig
Katherine L Helbig, MS
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Cecilia Desmettre Altuzarra
Cecilia Desmettre Altuzarra, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Nicolas Chatron
Nicolas Chatron, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Charu Kaiwar
Charu Kaiwar, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Katharina Stöhr
Katharina Stöhr, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Peter Widdess-Walsh
Peter Widdess-Walsh, MB
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Bryce A Mendelsohn
Bryce A Mendelsohn, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Adam Numis
Adam Numis, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Maria R Cilio
Maria R Cilio, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Wim Van Paesschen
Wim Van Paesschen, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Lene L Svendsen
Lene L Svendsen, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Stephanie Oates
Stephanie Oates, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Elaine Hughes
Elaine Hughes, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Sushma Goyal
Sushma Goyal, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Kathleen Brown
Kathleen Brown, MS
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Margarita Sifuentes Saenz
Margarita Sifuentes Saenz, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Thomas Dorn
Thomas Dorn, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Hiltrud Muhle
Hiltrud Muhle, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Alistair T Pagnamenta
Alistair T Pagnamenta, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Dimitris V Vavoulis
Dimitris V Vavoulis, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Samantha JL Knight
Samantha JL Knight, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Jenny C Taylor
Jenny C Taylor, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Maria Paola Canevini
Maria Paola Canevini, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Francesca Darra
Francesca Darra, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Ralitza H Gavrilova
Ralitza H Gavrilova, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Zöe Powis
Zöe Powis, MS
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Shan Tang
Shan Tang, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Justus Marquetand
Justus Marquetand, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Martin Armstrong
Martin Armstrong, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Duncan McHale
Duncan McHale, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Eric W Klee
Eric W Klee, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Gerhard J Kluger
Gerhard J Kluger, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Daniel H Lowenstein
Daniel H Lowenstein, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Sarah Weckhuysen
Sarah Weckhuysen, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Deb K Pal
Deb K Pal, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Ingo Helbig
Ingo Helbig, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Renzo Guerrini
Renzo Guerrini, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Rhys H Thomas
Rhys H Thomas, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Mark I Rees
Mark I Rees, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Gaetan Lesca
Gaetan Lesca, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Sanjay M Sisodiya
Sanjay M Sisodiya, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Yvonne G Weber
Yvonne G Weber, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Dennis Lal
Dennis Lal, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Carla Marini
Carla Marini, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,
Holger Lerche
Holger Lerche, MD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1,✉,
Julian Schubert
Julian Schubert, PhD
1From the University of Tübingen (S. Wolking, J.M., Y.G.W., H.L., J.S.), Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, Tübingen, Germany; Luxembourg Centre for Systems Biomedicine (P.M.), University of Luxembourg, Esch-sur-Alzette; Pediatric Neurology and Neurogenetics Unit and Laboratories (D.M., R.G., C.M.), Children's Hospital Anna Meyer, University of Florence, Italy; Danish Epilepsy Centre (R.S.M.), Dianalund; Institute for Regional Health Services (R.S.M.), University of Southern Denmark, Odense; Department of Clinical and Experimental Epilepsy (S.B.), UCL Institute of Neurology and Epilepsy Society, UK, London; Division of Neurology (K.L.H., I.H.), Children's Hospital of Philadelphia, PA; Department of Pediatric Neurology (C.D.A.), Centre de Compétences Maladies Rares, CHU Besançon; Service de Génétique (N.C.), Hospices Civils des Lyon, Bron; GENDEV Team (N.C.), Neurosciences Research Center of Lyon, Bron, France; Neuropediatric Clinic and Clinic for Neurorehabilitation (K.S.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Beaumont Hospital (P.W.-W.), Dublin, Ireland; Department of Pediatrics, Division of Medical Genetics, Institute of Human Genetics (B.A.M.), Departments of Neurology and Pediatrics (A.N.), and Departments of Neurology and Pediatrics, and Institute of Human Genetics (M.R.C.), University of California, San Francisco; Department of Neurology (W.V.P.), University Hospitals Leuven, Belgium; Department of Pediatrics (L.L.S.), Hvidovre Hospital, Denmark; King's College Hospital (S.O., E.H., S.G., D.K.P.), London; Evelina London Children's Hospital (S.O., E.H., S.G.), London, UK; Section of Genetics (K.B., M.S.S.), Department of Pediatrics, University of Colorado and Children's Hospital Colorado, Aurora; Clinique Bernoise Montana (T.D.), Crans-Montana, Switzerland; Department of Neuropediatrics (H.M.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; National Institute for Health Research Oxford Biomedical Research Centre, Wellcome Centre for Human Genetics (A.T.P., S.J.L.K., J.C.T.) and Department of Oncology (D.V.V.), University of Oxford, UK; Epilepsy Center (M.P.C.), Health Sciences Department, San Paolo Hospital, University of Milan; Child Neuropsychiatry (F.D.), Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, Italy; Departments of Neurology and Clinical Genomics (R.H.G.) and Health Sciences Research and Clinical Genomics (E.W.K., C.K.), Mayo Clinic, Rochester, MN; Ambry Genetics (Z.P.), Aliso Viejo, CA; Department of Clinical Neuroscience (S.T.), King's College London; New Medicines (M.A., D.M.), UCB Pharma, Slough, UK; Neuropediatric Clinic and Clinic for Neurorehabilitation (G.J.K.), Epilepsy Center for Children and Adolescents, Schoen Klinik Vogtareuth, Germany; Research Institute for Rehabilitation, Transition and Palliation (G.J.K.), PMU Salzburg, Austria; Department of Neurology (D.H.L.), University of California, San Francisco; Neurogenetics Group (S. Weckhuysen), Center for Molecular Neurology, VIB, Antwerp; Laboratory of Neurogenetics (S. Weckhuysen), Institute Born-Bunge, University of Antwerp; Department of Neurology (S. Weckhuysen), Antwerp University Hospital, Antwerp, Belgium; Department of Basic & Clinical Neuroscience, Institute of Psychiatry, Psychology & Neuroscience (D.K.P.), MRC Centre for Neurodevelopmental Disorders (D.K.P.), King's College London, UK; Evelina London Children's Hospital (D.K.P.), London, UK; Department of Neuropediatrics (I.H.), University Medical Center Schleswig-Holstein, Christian-Albrechts University, Kiel, Germany; Institute of Neuroscience (R.H.T.), Henry Wellcome Building, Newcastle University; Neurology Research Group (M.I.R.), Institute of Life Science, Swansea University Medical School, Swansea, UK; Service de Génétique (G.L.), Hospices Civils des Lyon, Bron; GENDEV Team (G.L.), Neurosciences Research Center of Lyon, Bron, France; NIHR University College London Hospitals Biomedical Research Centre (S.M.S.), UCL Institute of Neurology, London, UK; Cologne Center for Genomics (D.L.), University of Cologne, Germany; Stanley Center for Psychiatric Research (D.L.) and Program in Medical and Population Genetics (D.L.), Broad Institute of MIT and Harvard, Cambridge; Psychiatric and Neurodevelopmental Genetics Unit (D.L.), Massachusetts General Hospital and Harvard Medical School, Boston.
1