Abstract
Background: Long non-coding RNA homeobox A11 antisense RNA (HOXA11-AS) has been reported to be involved in initiation and development of multiple cancers. However, the detailed biological roles and underlying molecular mechanism of HOXA11-AS remain unclear in retinoblastoma (RB). Herein, the goals of this study were to explore the biological function and the potential mechanism of HOXA11-AS in RB.
Materials and methods: The expression of HOXA11-AS in RB tissues and cell lines was detected using real-time PCR (qRT-PCR). Cell proliferation, cycle arrest and apoptosis were measured using a cell counting kit 8 and flow cytometry. The target miRNAs of HOXA11-AS was predicted by Starbase2.0 software and was confirmed using a dual-luciferase reporter assay.
Result: We found that HOXA11-AS expression was markedly elevated in RB tissues and cell lines compared to normal retina tissues and human retinal epithelial cells, respectively. Functional analysis showed that knockdown of HOXA11-AS in RB cells significantly suppressed cell proliferation, and induced cell cycle arrest at G1/G0 phase and promoted cell apoptosis. We also found that HOXA11-AS could serve as a competing endogenous RNA (ceRNA) that inhibited miR-506-3p expression, which regulated its downstream target NIMA-related kinase 6 (NEK6) in RB. In addition, miR-506-3p inhibitors partially reversed the effect of HOXA11-AS depletion on proliferation, cycle arrest and apoptosis in RB cells.
Conclusion: Taken together, these findings demonstrated that HOXA11-AS could promote RB progression by sponging miR-506-3p, suggesting that HOXA-11-AS might be a potential therapeutic target for RB.
Keywords: retinoblastoma, LncRNA, miR-506-3p, proliferation, apoptosis
Introduction
The retinoblastoma (RB) is the most common intraocular malignant tumor usually occurring during childhood.1 Despite great improvement of diagnostic and surgical techniques, the prognosis of patients with RB is still unsatisfactory.2,3 Therefore, exploring the molecular mechanisms underlying RB tumorigenesis and progression is urgently needed to find the effective diagnostic and therapeutic targets for the treatment of this disease.
Long noncoding RNAs (lncRNAs) are a class of newly identified regulatory RNA member with more than 200 nucleotides in length and without the protein-coding ability.4 Accumulating evidence has suggested that lncRNAs implicated in a wide range of biological processes, including cell proliferation, apoptosis and invasion.5,6 Number of cancer-associated lncRNAs has been identified to be involved in occurrence and development of various cancers.7,8 Besides, several lncRNAs have been reported to be correlated with RB progression functioning as oncogene and tumor suppressor in RB.9,10
Homeobox A11 antisense RNA (HOXA11-AS), a highly conserved lncRNA located in the HOXA gene cluster on chromosome 7p15.2,11 is primordially discovered in a mouse embryonic cDNA library.12 HOXA11-AS has been reported to be involved in tumor progression in several types of cancer.13–15 However, the expression status, detailed biological function and the possible molecular mechanisms of HOXA11-AS involved in RB progression remain unknown. Therefore, the aims of this study were to explore the biological function and underlying molecular mechanism of HOXA11-AS in RB.
Materials and methods
Human tissue samples
A total of 30 RB tissue samples were collected from RB patients treated with surgical resection without preoperative chemotherapy or radiotherapy at the Second Hospital of Jilin University. 10 normal retina tissue samples were obtained from patients with ruptured globes. All samples were extracted from enucleated eyes and immediately stored in liquid nitrogen for further study. All experiments about using human tissues have been conducted in accordance with the World Medical Association Declaration of Helsinki, and that all subjects provided written informed consent. This study was approved by the Clinical Research Ethics Committee of the Second Hospital of Jilin University (Changchun, China).
Cell culture and transfection
The human retinal epithelial cells ARPE-19 and four human RB cell lines Y79, Weri-Rb1, SO-Rb50, and HXO-RB44 were bought from the American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Invitrogen) and 1% penicillin-streptomycin (Invitrogen; Carlsbad, CA, USA) at 37°C in a humidified atmosphere with 5% CO2.
For HOXA11-AS down-regulation, a small interference RNA (siRNA) targeting human HOXA11-AS (si-HOXA11-AS) and its negative control nonsensical sequence (si-NC) were constructed and synthesized by GenePhara (Shanghai, China). The miR-506-3p mimic, the appropriate negative control mimic (miR-NC) and miR-506-3p inhibitor (miR-506-3p in) were purchased from RiboBio (Guangzhou, China). Y79 cells were transiently transfected with one of the aforementioned siRNAs, mimics and inhibitor using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Transfection efficiency was determined in every experiment at 48 hrs post-transfection.
RNA isolation and qRT-PCR
Total RNA was extracted from tissue samples and cultured cells using TRIzol® reagent (Invitrogen) following the manufacturer’s instructions. Reverse transcription was performed for quantification using the TaqMan miRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) or a Prime Script First Strand cDNA Synthesis kit (Takara, Dalian, China) according to the manufacturer’s protocol. The cDNAs were amplified using TaqMan miRNA kit (Applied Biosystems) or SYBR Premix Ex Taq (Takara) using the ABI 7900 Fast System (Applied Biosystems). The primers are listed in Table 1. Relative gene expression was calculated according to the 2–ΔΔCt method after normalization to U6 for miR-506-3p or GAPDH for HOXA11-AS or NEK6.
Table 1.
Real-time PCR primers used for mRNA expression analysis
| Target gene | Prime(5ʹ-3ʹ) |
|---|---|
| U6 | F- TCCGATCGTGAAGCGTTC R- GTGCAGGGTCCGAGGT |
| miR-506-3p | F- GCCACCACCATCAGCCATAC R- GCACATTACTCTACTCAGAAGGG |
| HOXA11-AS | F- AGAAATCTGGACCCGAGACG R- CGTCAGCTTACGTCTCCAAA |
| NEK6 | F- TTCCAACAACCTCTGCCACACC R- CACAGTCCTGCCTCGCCTTG |
| GAPDH | F- AAGGTGAAGGTCGGAGTCAA R- -AATGAAGGGGTCATTGATGG |
Abbreviations: F, forward; mRNA, messenger RNA; PCR, polymerase chain reaction; R, reverse.
Cell proliferation assay
Cell proliferation was measured using Cell Counting Kit-8 (CCK8; Beyotime, Beijing) according to the manufacturer’s guide. Briefly, transfected cells in each group were plated into 96-well plates at density of 2,000 cells/well. In indicated time, the CCK8 regent (10 μL) was added to each well for another 4 hrs at 37°C with 5% CO2. The absorbance at the wavelength of 450 nm was finally measured using a microplate reader.
Flow cytometry for cell cycle and apoptosis analysis
Flow cytometry was used for cell cycle arrest and apoptosis analysis. Cells were harvested at 48 hrs post-transfection. For cell cycle arrest, transfected cells were seeded in six-well and were fixed with 75% ethanol overnight at 4°C. Then, cells stained with propidium iodide (PI) supplemented with RNaseA (Sigma) at room temperature in the dark for 15 mins. Cell cycle arrest was determined with a FACScan flow cytometer (BD Biosciences) and analyzed with the ModFit software (BD Biosciences).
The Annexin V/Dead Cell Apoptosis Kit (Invitrogen) was used to determine cell apoptosis according to the manufacturer’s instructions. The cell apoptosis was analyzed using the FACScan flow cytometer equipped with CellQuest Software (BD Biosciences).
Luciferase reporter assays
HOXA11-AS fragment containing the predicated miR-506-3p binding site (wild-type) or mutant putative sequences of the binding sites (mutant-type) were synthesized and subcloned into a pmirGLO Dual-luciferase vector (Promega, USA) to form the reporter vector pmiRGLO-HOXA11-wild type (HOXA11-AS-WT) or pmiRGLO-HOXA11-AS-mutant (HOXA11-AS-MT). Y79 cells were co-transfected with reporter vector HOXA11-AS-WT or HOXA11-MT and miR-506-3p mimic or miR-NC by using Lipofectamine 2000. The relative luciferase activities were determined using a dual-luciferase reporter assay system (Promega) at 48 hrs after transfection according to the manufacturer’s guide.
Statistical analyses
All data are presented as as mean ± SD from at least three separate experiments. Statistical analyses were performed using SPSS 19.0 (SPSS, Chicago) using Student’s t-test and one way ANOVA. Correlations between HOXA11-AS expression and miR-506-3p or NEK6 expression were evaluated by Pearson’s correlation analysis. Significance level was set as P<0.05.
Results
HOXA11-AS expression was upregulated in RB cells and tissues
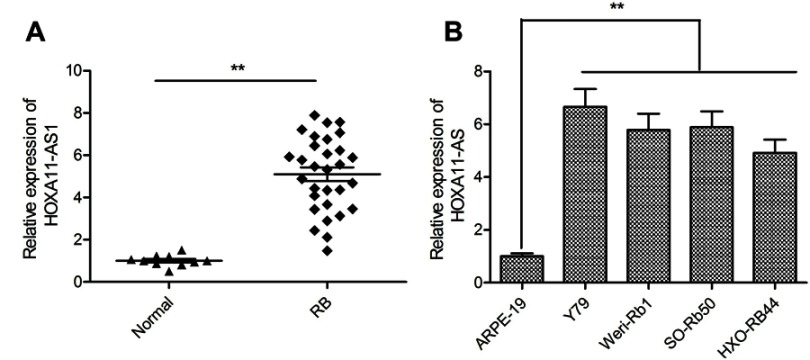
We initially detected the expression of HOXA11-AS in 30 RB tissues and 10 normal retina tissues by qRT-PCR. The results showed that the expression of HOXA11-AS was significantly increased in RB tissues compared to the normal retina tissues (Figure 1A). Subsequently, we analyzed the expression of HOXA11-AS in four human RB cell lines (Y79, Weri-Rb1, SO-Rb50 and HXO-RB44) and human retinal epithelial cells ARPE-19. Consistent with the results in RB tissues, HOXA11-AS expression was also dramatically increased in four RB cell lines as compared to ARPE-19 cells (Figure 1B), especially in Y79 cells. Thus, Y79 cells were chosen for the following experiments.
Figure 1.
HOXA11-AS was up-regulated in RB tissues and cell lines. (A) The relative expression levels of HOXA11-AS was examined in 30 RB tissues and 10 normal retina tissue samples by using qRT-PCR analysis. (B) The relative expression levels of HOXA11-AS was examined in four human RB cell lines (Y79, Weri-Rb1, SO-Rb50 and HXO-RB44) and human retinal epithelial cells ARPE-19 by using qRT-PCR analysis. **P<0.01.
Abbreviation: RB, retinoblastoma.
Knockdown of HOXA11-AS suppressed proliferation and facilitated apoptosis in RB cells
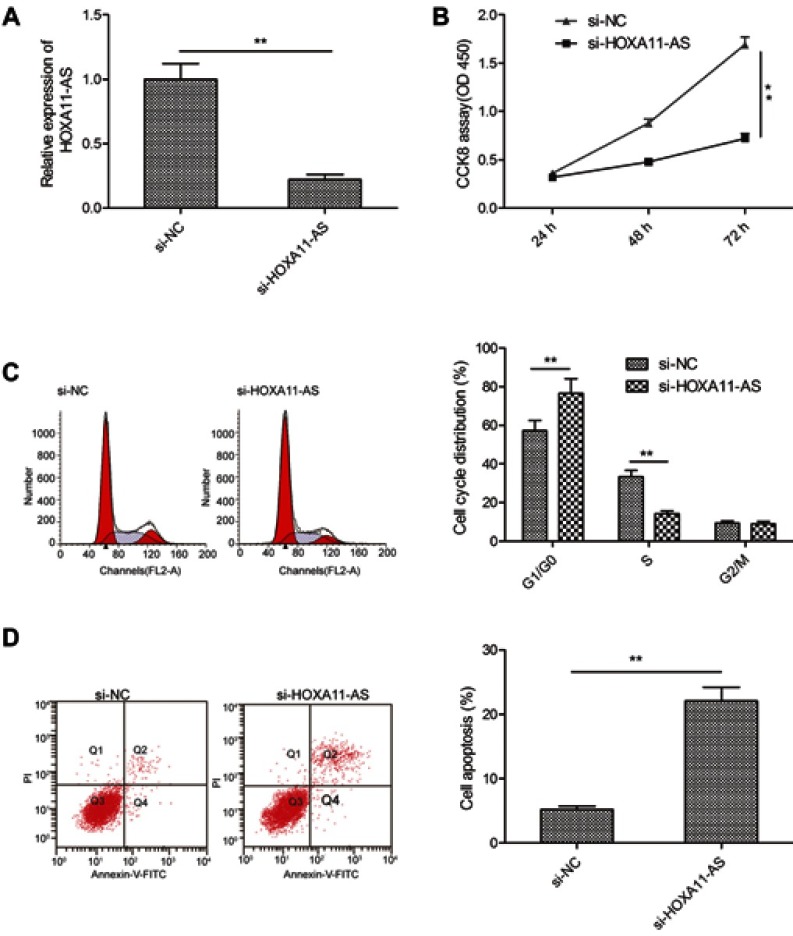
To explore the biological function of HOXA11-AS in RB cells, siRNA against HOXA11-AS (si-HOXA11-AS) were transfected into Y79 cells to decrease the expression of HOXA11-AS (Figure 2A). CCK-8 assay was performed to investigate the effect of HOXA11-AS on cell proliferation. The results demonstrated that downregulation of HOXA11-AS significantly inhibited cell proliferation of Y79 cells in comparison to negative control (si-NC) (Figure 2B). To determine whether the effect of HOXA11-AS on RB cells proliferation reflected cell cycle arrest, cell cycle progression was determined by flow cytometry analysis. The results revealed that knockdown of HOXA11-AS significantly increased cell cycle arrest at G1/G0 phase, and decreased arrest at S phase (Figure 2C). Moreover, we investigate the effect of HOXA11-AS on apoptosis of RB cells. We found that knockdown of HOXA11-AS significantly induced cell apoptosis of Y79 cells (Figure 2D).
Figure 2.
Knockdown of HOXA11-AS suppressed proliferation, and induced apoptosis in RB cells. (A) The relative expression levels of HOXA11-AS was examined in Y79 cells transfected with si-HOXA11-AS or si-NC. Subsequently, (B) cell proliferation, (C) cell cycle distribution and (D) cell apoptosis were determined in Y79 cells transfected with si-HOXA11-AS or si-NC. *P<0.05, **P<0.01.
Abbreviation: RB, retinoblastoma.
HOXA11-AS acted as a molecular sponge for miR-506-3p in RB cells
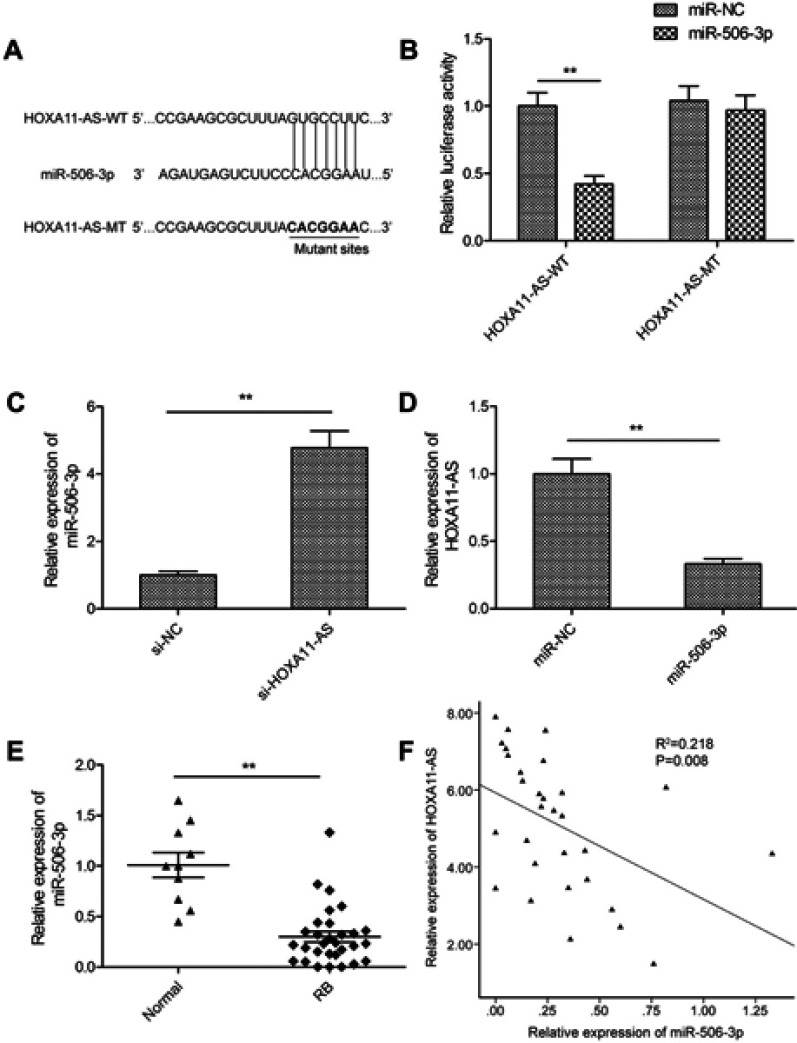
Accumulating evidence suggested that lncRNAs implicated in tumor progression by functioning as competing endogenous RNAs (ceRNAs) to specific miRNAs.16 Here, a bioinformatics prediction program (StarBase 2.0, http://starbase.sysu.edu.cn/) was used to predict the potential binding miRNAs for HOXA11-AS. We observed that miR-506-3p had relevant binding sites in HOXA11-AS (Figure 3A). This was confirmed by a dual-luciferase reporter assay, which revealed that overexpression of miR-506-3p significantly decreased the luciferase activity of the HOXA11-AS-WT, but not of HOXA11-AS-MT (Figure 3B). Furthermore, qRT-PCR showed that knockdown of HOXA11-AS-MT facilitated miR-506-3p expression in Y79 cells (Figure 3C), and that overexpression of miR-506-3p significantly decreased HOXA11-AS expression (Figure 3D). In addition, we also examined the expression of miR-506-3p expression in 30 RB tissues and 10 normal retina tissue samples by qRT-PCR, and found that miR-506-3p expression was downregulated in RB tissues compared to normal tissue samples (Figure 3E). Pearson’s correlation analysis suggested a negative correlation between HOXA11-AS and miR-506-3p expression in RB tissues (Figure 3F).
Figure 3.
HOXA11-AS acted as a molecular sponge for miR-506-3p in RB cells. (A) Schematic representation of the predicted binding sites between miR-506-3p and HOXA11-AS, and the mutagenesis design for the reporter assays. (B) Luciferase activity in Y79 cells co-transfected with miR-506-3p mimics or miR-NC, and luciferase reporters containing HOXA11-AS-WT or HOXA11-AS-MT. (C) The relative expression of miR-506-3p was determined in Y79 cells transfected with si-HOXA11-AS or si-NC. (D) The relative expression levels of HOXA11-AS was determined in Y79 cells transfected with miR-506-3p mimics or miR-NC. (E) The relative expression levels of miR-506-3p were examined in 30 RB tissues and 10 normal retina tissue samples using qRT-PCR analysis. (F) An inverse association between HOXA11-AS and miR-506-3p expression in RB tissues was identified by Pearson’s correlation analysis. **P<0.01.
Abbreviation: RB, retinoblastoma.
miR-506-3p inhibition restored the si-HOXA11-AS induced effects on cell proliferation, cycle arrest and apoptosis in RB cells
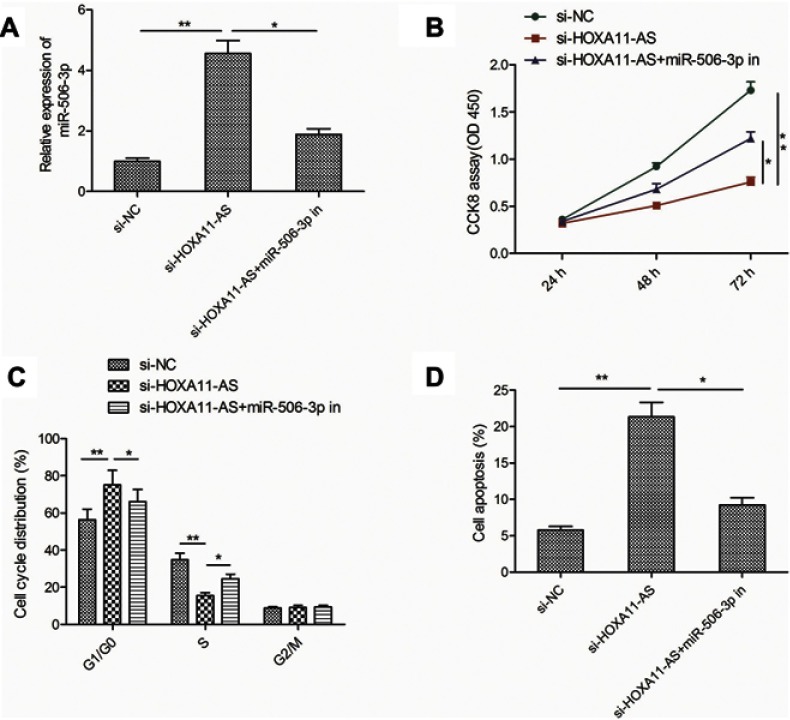
To determine whether HOXA11-AS exerts biological roles in RB cells via regulating miR-506-3p, we carried out the rescue experiments by transfecting si-HOXA11-AS plus miR-506-3p inhibitor into Y79 cells. As shown in Figure 4A, result from qRT-PCR assay revealed that miR-506-3p inhibitor could decrease HOXA11-AS expression in Y79 cells transfected with si-HOXA11-AS (Figure 4A). In addition, CCK8 assay revealed that the inhibitory effect of HOXA11-AS knockdown on RB cell proliferation could be partially restored by miR-506-3p inhibitor (miR-506-3p-in) (Figure 4B). Flow cytometry assay revealed that repression of miR-506-3p largely rescued the effect of cell cycle arrest and apoptosis induced by si-HOXD-AS1 in Y79 cells (Figure 4C and D). These results implied that HOXA11-AS exerted its role in RB by targeting miR-506-3p.
Figure 4.
miR-506-3p inhibition restored the si-HOXA11-AS-induced effects on cell proliferation, cycle arrest and apoptosis in RB cells. (A) The relative expression of miR-506-3p expression was determined in Y79 cells transfected with si-NC or si-HOXA11-AS, with or without the miR-506-3p inhibitor. (B–D) Cell proliferation, cycle arrest and apoptosis were determined in Y79 cells transfected with si-NC or si-HOXA11-AS, with or without the miR-506-3p inhibitor. *P<0.05, **P<0.01.
Abbreviation: RB, retinoblastoma.
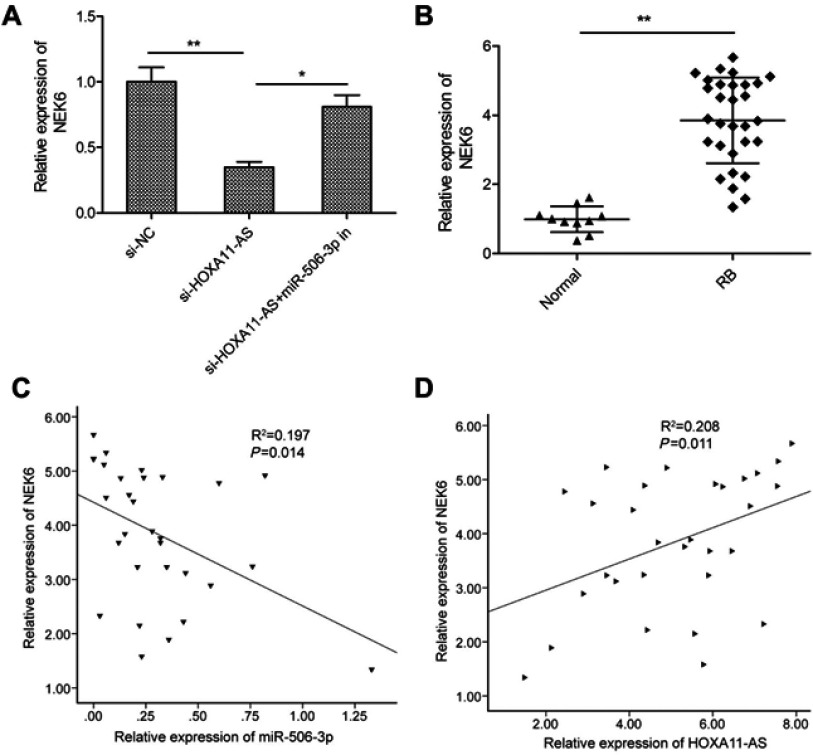
HOXA11-AS regulated the expression of NEK6 through miR-506-3p in RB cells
Growing evidences suggested that lncRNAs exerted its function through acting as molecular sponges or ceRNAs to regulate specific miRNAs and their targets of miRNAs.17 A previous study showed that miR-506-3p inhibited RB progression by targeting mitosis Gene A (NIMA)-related kinase 6 (NEK6).18 Thus, we determine whether HOXA11-AS can regulate NEK6 expression which is mediated by miR-506-3p by a rescue experiment. As shown in Figure 5A, knockdown of HOXA11-AS in Y79 cells significantly decreased the mRNA expression levels of NEK6, while inhibition of miR-506-3p could reverse this trend. In addition, we also examined the expression of NEK6 expression in 30 RB tissue and 10 normal retina tissue samples by qRT-PCR. We found that the NEK6 mRNA expression was upregulated in RB tissues compared to normal tissue samples (Figure 5B). Pearson’s correlation analysis revealed that NEK6 expression was negatively correlated with miR-506-3p expression in RB tissues (Figure 5C), and positively correlated with HOXA11-AS expression in RB tissues (Figure 5D). These data suggest that HOXA11-AS could regulate NEK6 expression through miR-506-3p in RB.
Figure 5.
HOXA11-AS regulated the expression of NEK6 through miR-506-3p in RB cells. (A) The relative expression of NEK6 was measured in Y79 cells transfected with si-NC or si-HOXA11-AS, with or without the miR-506-3p inhibitor. (B) The relative expression levels of NEK6 mRNA were examined in 30 RB tissues and 10 normal retina tissue samples by using qRT-PCR analysis. (C) An inverse association between NEK6 mRNA and miR-506-3p expression in RB tissues was identified by Pearson’s correlation analysis. (D) A positive association between NEK6 mRNA and HOXA11-AS expression in RB tissues was identified by Pearson’s correlation analysis. *P<0.05, **P<0.01.
Abbreviation: RB, retinoblastoma.
Discussion
Recently, number of lncRNAs have been reported to play a significant regulatory role in RB initiation and development by functioning as a tumor suppressor or oncogene.8,9 For example, Hu et al reported that XIST promoted RB progression partially by modulating the miR-124/STAT3 axis.19 Li et al demonstrated that knockdown of LINC00152 significantly inhibited RB cell growth in vitro and in vivo.20 Zhang et al found that H19 inhibited RB progression via counteracting the roles of miR-17–92 cluster.21 Wang et al revealed that DANCR acted as ceRNA for miR-34c and miR-613 to regulate progression and metastasis in RB oncogenesis via targeting MMP-9.22 In this study, we found that HOXA11-AS expression was upregulated in RB tissues and cell lines. Knockdown of HOXA11-AS significantly inhibited RB cell proliferation, and induced cell arrest at G1/G0 phase and cell apoptosis. Mechanic analysis demonstrated that HOXA11-AS contributed to RB progression by modulating miR-506-3p/NEK6 axis.
HOXA11-AS1 (also known as NCRNA00076), a member of the homeobox (HOX) family gene,10 has been reported to be upregulated in multiple types of cancer, including renal cancer,23 hepatocellular carcinoma,24 laryngeal squamous cell carcinoma,25 non-small cell lung cancer,26 glioma,27 breast cancer,28 osteosarcoma29 and gastric cancer,30 suggesting that HOXA11-AS1 played an oncogenic role in these types of cancers. On the contrary, in ovarian cancer31 and colorectal cancer,32 HOXA11-AS1 expression was downregulated and functioned as a tumor suppressor. These inconsistent results suggested that HOXA11-AS1 played different roles in tumor progression depending on details of cancer types. However, until now, no systematic study was performed on the expression and role of HOXA11-AS in RB. Our present data revealed that HOXA11-AS expression was upregulated in RB tissues and cell lines, suggesting that HOXA11-AS might be involved in RB progression. Furthermore, we identified the effects of HOXA11-AS on the biological behaviors of RB, showing that knockdown of HOXA11-AS significantly inhibited RB cell proliferation, and induced cell arrest at G1/G0 phase and cell apoptosis. These results implied that HOXA11-AS functioned as an oncogene in RB.
It was well known that lncRNAs exert their biological function by functioning as ceRNAs sponging for miRNAs, thus affecting the expression levels of miRNAs target genes.16,33 To investigate the possible molecular mechanism of HOXA11-AS involved in RB progression, we used starbase 2.0 algorithm to predict miRNAs that targeted HOXA11-AS. Among miRNAs, miR-506 was chosen to act as an object based on its biological roles in RB. miR-506-3p has been reported to be downregulated and functioned as a tumor suppressor in RB.18 Through luciferase reporter assay and qRT-PCR, we confirmed that miR-506-3p was a direct target of HOXA11-AS in RB. We also found that miR-506-3p expression was negatively correlated with HOXA11-AS expression in RB tissues. Importantly, miR-506-3p inhibition restored the si-HOXA11-AS induced effects on cell proliferation, cycle arrest and apoptosis in RB cells. Those data indicated that HOXA11-AS might function as a sponge for miR-506-3p in RB.
LncRNA could function as ceRNAs to regulate their targets of miRNAs.17 NEK6 has been proven to be a target of miR-506-3p in RB.18 Growing evidence showed that NEK6 played crucial roles in tumorigenesis.34,35 Here, we determine whether HOXA11-AS can regulate NEK6 expression. We found that knockdown of HOXA11-AS in Y79 cells significantly decreased the mRNA expression levels of NEK6, while inhibition of miR-506-3p could reverse this trend. In addition, we also found that the mRNA expression levels of NEK6 were upregulated in RB tissues compared to normal tissue samples, which was consistent with the previous result.18 Pearson’s correlation analysis revealed that NEK6 expression was negatively correlated with miR-506-3p expression in RB tissues, and positively correlated with HOXA11-AS expression in RB tissues. These data suggest that HOXA11-AS could regulate NEK6 expression in RB cells through miR-506-3p.
In conclusion, to our knowledge, the present study first provides evidence that HOXA11-AS was upregulated in RB tissues and cell lines, and that HOXA11-AS played an oncogenic role in RB tumorigenesis through sponging miR-506-3p at least partially. These data suggested that HOXA11-AS might serve as a potential therapeutic target for the RB treatment.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Dimaras H, Corson TW. Retinoblastoma, the visible CNS tumor: a review. J Neurosci Res. 2019;97(1):29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabian ID, Onadim Z, Karaa E, et al. The management of retinoblastoma. Oncogene. 2018;37(12):1551–1560. doi: 10.1038/s41388-017-0050-x [DOI] [PubMed] [Google Scholar]
- 4.Schaukowitch K, Kim TK. Emerging epigenetic mechanisms of long non-coding RNAs. Neuroscience. 2014;264:25–38. doi: 10.1016/j.neuroscience.2013.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 6.Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36(1):25–64. doi: 10.1210/er.2014-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Dzakah EE, Shan G. Targetable long non-coding RNAs in cancer treatments. Cancer Lett. 2018;418:119–124. doi: 10.1016/j.canlet.2018.01.042 [DOI] [PubMed] [Google Scholar]
- 8.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J, Yang Y, Fang F, Liu K. MALAT1 modulates the autophagy of retinoblastoma cell through miR-124-mediated stx17 regulation. J Cell Biochem. 2018;119(5):3853–3863. doi: 10.1002/jcb.26464 [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Peng XW. The silencing of long non-coding RNA ANRIL suppresses invasion, and promotes apoptosis of retinoblastoma cells through ATM-E2F1 signaling pathway. Biosci Rep. 2018;38:BSR20180558. doi: 10.1042/BSR20180558 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Yu H, Lindsay J, Feng ZP, et al. Evolution of coding and non-coding genes in HOX clusters of a marsupial. BMC Genomics. 2012;13:251. doi: 10.1186/1471-2164-13-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potter SS, Branford WW. Evolutionary conservation and tissue-specific processing of Hoxa 11 antisense transcripts. Mamm Genome. 1998;9(10):799–806. doi: 10.1007/s003359900870 [DOI] [PubMed] [Google Scholar]
- 13.Xue JY, Huang C, Wang W, Li HB, Sun M, Xie M. HOXA11-AS: a novel regulator in human cancer proliferation and metastasis. Onco Targets Ther. 2018;11:4387–4393. doi: 10.2147/OTT.S166961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu S, Jiang X, Su Z, Cui Z, Fu W, Tai S. The role of the long non-coding RNA HOXA11-AS in promoting proliferation and metastasis of malignant tumors. Cell Biol Int. 2018;42:1596–1601. doi: 10.1002/cbin.v42.12 [DOI] [PubMed] [Google Scholar]
- 15.Lu CW, Zhou DD, Xie T, et al. HOXA11 antisense long noncoding RNA (HOXA11-AS): a promising lncRNA in human cancers. Cancer Med. 2018;7(8):3792–3799. doi: 10.1002/cam4.1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu L, Chen Z, Xing Y. MiR-506-3p-3p inhibits cell proliferation, induces cell cycle arrest and apoptosis in retinoblastoma by directly targeting NEK6. Cell Biol Int. 2018. doi: 10.1002/cbin.11041 [DOI] [PubMed] [Google Scholar]
- 19.Hu C, Liu S, Han M, Wang Y, Xu C. Knockdown of lncRNA XIST inhibits retinoblastoma progression by modulating the miR-124/STAT3 axis. Biomed Pharmacother. 2018;107:547–554. doi: 10.1016/j.biopha.2018.08.020 [DOI] [PubMed] [Google Scholar]
- 20.Li S, Wen D, Che S, et al. Knockdown of long noncoding RNA 00152 (LINC00152) inhibits human retinoblastoma progression. Onco Targets Ther. 2018;11:3215–3223. doi: 10.2147/OTT.S160428 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Zhang A, Shang W, Nie Q, Li T, Li S. Long non-coding RNA H19 suppresses retinoblastoma progression via counteracting miR-17-92 cluster. J Cell Biochem. 2018;119(4):3497–3509. doi: 10.1002/jcb.26521 [DOI] [PubMed] [Google Scholar]
- 22.Wang JX, Yang Y, Li K. Long noncoding RNA DANCR aggravates retinoblastoma through miR-34c and miR-613 by targeting MMP-9. J Cell Physiol. 2018;233(10):6986–6995. doi: 10.1002/jcp.26621 [DOI] [PubMed] [Google Scholar]
- 23.Yang FQ, Zhang JQ, Jin JJ, et al. HOXA11-AS promotes the growth and invasion of renal cancer by sponging miR-146b-5p to upregulate MMP16 expression. J Cell Physiol. 2018;233(12):9611–9619. doi: 10.1002/jcp.26864 [DOI] [PubMed] [Google Scholar]
- 24.Zhan M, He K, Xiao J, et al. LncRNA HOXA11-AS promotes hepatocellular carcinoma progression by repressing miR-214-3p. J Cell Mol Med. 2018;22:3758–3767. doi: 10.1111/jcmm.2018.22.issue-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu L, Jin M, Yang L, et al. Expression of long non-coding RNA HOXA11-AS is correlated with progression of laryngeal squamous cell carcinoma. Am J Transl Res. 2018;10(2):573–580. [PMC free article] [PubMed] [Google Scholar]
- 26.Yu W, Peng W, Jiang H, Sha H, Li J. LncRNA HOXA11-AS promotes proliferation and invasion by targeting miR-124 in human non-small cell lung cancer cells. Tumour Biol. 2017;39(10):1010428317721440. doi: 10.1177/1010428317721440 [DOI] [PubMed] [Google Scholar]
- 27.Xu C, He T, Li Z, Liu H, Ding B. Regulation of HOXA11-AS/miR-214-3p/EZH2 axis on the growth, migration and invasion of glioma cells. Biomed Pharmacother. 2017;95:1504–1513. doi: 10.1016/j.biopha.2017.08.097 [DOI] [PubMed] [Google Scholar]
- 28.Li W, Jia G, Qu Y, Du Q, Liu B, Liu B. Long non-coding RNA (LncRNA) HOXA11-AS promotes breast cancer invasion and metastasis by regulating epithelial-mesenchymal transition. Med sci monit. 2017;23:3393–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui M, Wang J, Li Q, Zhang J, Jia J, Zhan X. Long non-coding RNA HOXA11-AS functions as a competing endogenous RNA to regulate ROCK1 expression by sponging miR-124-3p in osteosarcoma. Biomed Pharmacother. 2017;92:437–444. doi: 10.1016/j.biopha.2017.05.081 [DOI] [PubMed] [Google Scholar]
- 30.Liu Z, Chen Z, Fan R, et al. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Mol Cancer. 2017;16(1):82. doi: 10.1186/s12943-017-0651-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards EJ, Permuth-Wey J, Li Y, et al. A functional variant in HOXA11-AS, a novel long non-coding RNA, inhibits the oncogenic phenotype of epithelial ovarian cancer. Oncotarget. 2015;6(33):34745–34757. doi: 10.18632/oncotarget.5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li T, Xu C, Cai B, et al. Expression and clinicopathological significance of the lncRNA HOXA11-AS in colorectal cancer. Oncol Lett. 2016;12(5):4155–4160. doi: 10.3892/ol.2016.5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 34.De Donato M, Righino B, Filippetti F, et al. Identification and antitumor activity of a novel inhibitor of the NIMA-related kinase NEK6. Sci Rep. 2018;8(1):16047. doi: 10.1038/s41598-018-34471-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Z, Ni X, Xia L, Shao Z. Overexpression of NIMA-related kinase 6 (NEK6) contributes to malignant growth and dismal prognosis in human breast cancer. Pathol Res Pract. 2018;214(10):1648–1654. doi: 10.1016/j.prp.2018.07.030 [DOI] [PubMed] [Google Scholar]