Abstract
Bivalves, from raw oysters to steamed clams, are popular choices among seafood lovers and once limited to the coastal areas. The rapid growth of the aquaculture industry and improvement in the preservation and transport of seafood have enabled them to be readily available anywhere in the world. Over the years, oysters, mussels, scallops, and clams have been the focus of research for improving the production, managing resources, and investigating basic biological and ecological questions. During this decade, an impressive amount of information using high-throughput genomic, transcriptomic and proteomic technologies has been produced in various classes of the Mollusca group, and it is anticipated that basic and applied research will significantly benefit from this resource. One aspect that is also taking momentum is the use of bivalves as a model system for human health. In this review, we highlight some of the aspects of the biology of bivalves that have direct implications in human health including the shell formation, stem cells and cell differentiation, the ability to fight opportunistic and specific pathogens in the absence of adaptive immunity, as source of alternative drugs, mucosal immunity and, microbiome turnover, toxicology, and cancer research. There is still a long way to go; however, the next time you order a dozen oysters at your favorite raw bar, think about a tasty model organism that will not only please your palate but also help unlock multiple aspects of molluscan biology and improve human health.
Keywords: disseminated neoplasia, HAB, mucosal immunity, microbiome, microplastics, myticalins
Graphical Abstarct
1. Introduction
Bivalves are the second largest phylum of animals after the arthropods with about 200,000 extant species (Runnegar, 1996). Bivalves, characterized by remarkable anatomical and structural dissimilarities between the species, share collective characteristics including a soft, unsegmented body consisting of a muscular foot, a visceral mass, and a mantle (Pechenik, 2000). Marine bivalves populate all latitudes, and they are particularly important in benthic-pelagic coupling filtering large volumes of water, cycling particulate matter and phytoplankton to grow the shell and the soft body fueling higher trophic levels and modifying the community composition (Newell, 2004; Smith et al., 2018a). Bivalves also represent an important source of food and valuable goods around the world, with bivalves making up roughly 20% of aquatic animals production by weight (FAO, 2018). The phylogeny, biology, ecology, and economic importance of bivalve bivalves makes them ideal candidates for investigations targeting critical basic and applied research questions. This is facilitated by their ubiquity and amenability for maintenance in laboratory settings even in laboratories with no direct access to marine water. Bivalves are today used as experimental organisms for research in chronobiology (Perrigault and Tran, 2017), neuroendocrinology (Catapane et al., 1978) bacterial endosymbiosis (Dubilier et al., 2008), innate immunity (Cunningham and Robledo, 2015; Mydlarz et al., 2006; Song et al., 2010), biomineralization (Mount et al., 2004; Takeuchi, 2017), aging (Abele et al., 2009), and various biotechnological applications (Ferreira et al., 2007) as well as for the monitoring of environmental health (Vethaak et al., 2017; Zuykov et al., 2013). The concept of a model organism is applied to one organism that is particularly suited to answer a particular question in biology. These questions can be unique to the clade where the targeted organism belongs (e.g., teka formation in diatom species, apicoplast segregation in the apicomplexan group, quorum sensing) or a shared with a large number of clades (e.g., chromatin packaging, cell division). Most well-establish model organisms (e.g., Drosophila melanogaster, Caenorhabditis elegans, Saccharomyces cerevisiae) are the result of decades of studies and the participation of multiple laboratories; however, the world wide web and a more affordable access the latest technologies are enabling researchers working in less-studied organisms to gain momentum for “non-model model organisms as emerging systems for tackling questions across the whole spectrum of biology (and beyond)” (Russell et al., 2017).
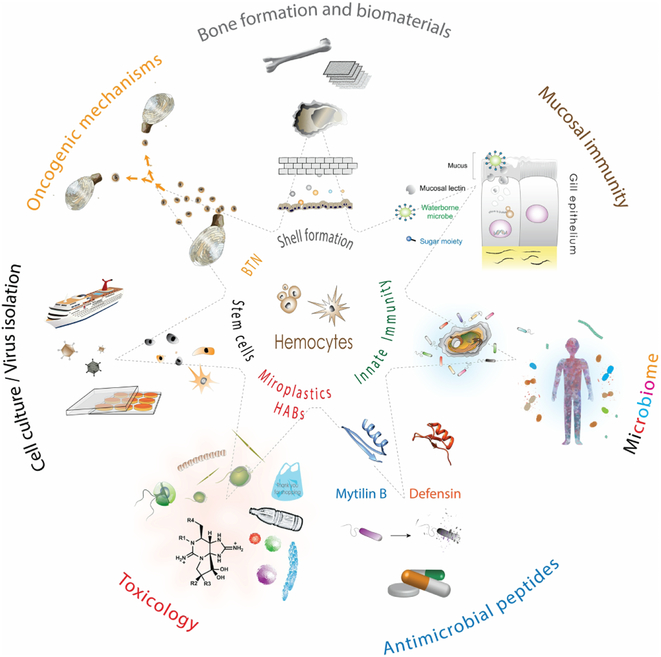
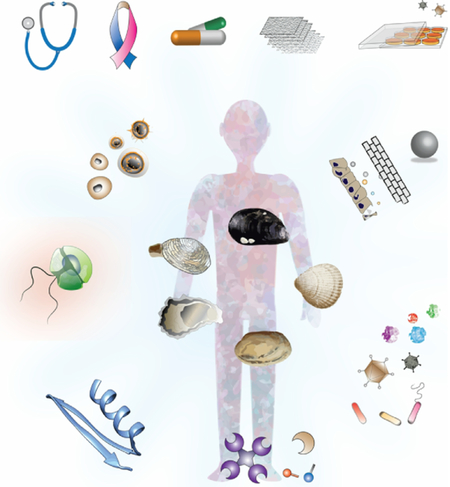
In this review, we focus on aspects of bivalve biology with implications in human health, (Fig. 1). We devoted section 2.1 to the shell formation. Bivalves use the shell to shield the soft body from both predators and environmental stressors, and for physiological homeostasis; in the absence of bones, the shell can be considered an exoskeleton. The biomineralization during shell formation takes place on the external surface of the mantle by specialized epithelial cells; however, there is growing evidence showing that hemocytes are also involved in biomineralization and shell formation. Section 2.2, focuses on innate immunity and what mechanisms the bivalves have for dealing with pathogens, but also with food particle selection. We have broken down this section into mucosal immunity (section 2.2.1), the microbiome (section 2.2.2), and alternatives to antibiotics (section 2.2.3). The components of the mucosal immunity that Lamellibranchiate bivalves uses do allow them to endure not only the myriad of waterborne microbes they are exposed to in the marine environment through their suspension-feeding mechanism but also a wide range of environmental conditions (section 2.2.1). Gills and other pallial organs are continuously encountering waterborne microbes as they enter the pallial cavity. Immune defense factors associated with the mucosal surfaces in the pallial organs combined with the open circulatory system with hemocytes on patrol makes the pallial cavity this first battleground with invading microorganisms. There is mounting evidence on the essential roles of microbiomes (bacteria, archaea, viruses, and microeukaryotes) in the biology, ecology, and evolution of eukaryotic hosts. In addition to waterborne pathogens, the convolution of pallial organs greatly increases the effective surface of these interfaces and enhance their exposure to a rich bacterial community (section 2.2.2). Bivalves can reject some of these microorganisms; some are digested as they go through the digestive tract of the bivalve, while others are retained, colonizing the gut and other organs. With clearly separated growth and reproductive seasons, it is expected that the microbiome of bivalves also changes through the seasons while maintaining a core microbiome. All these features make bivalves an attractive model to study microbiome composition and dynamics. As bivalves lack adaptive immunity, at least as we know it for vertebrates, they have evolved powerful and unique mechanisms and strategies (e.g., antimicrobial peptides) to fight and keep at-large bacterial pathogens and viruses. In section 2.3 we emphasize harmful algal blooms (HABs) and aspects of the concentration and effect on the bivalve, and detoxification mechanisms as well. As the temperature of the planet keeps rising, we are witnessing an increase in the frequency, magnitude, and distribution of harmful algal blooms. With many of these HAB species containing potent biotoxins, it becomes critical to understand better how filter-feeding bivalves concentrate these toxins, vector them to humans and/or bioaccumulate through food chains, and, ultimately, eliminated them. Microplastics (section 2.4) have recently gained attention and notoriety in society and the mass media beyond the reason why they were engineered. The plastics into the environment do not just stay in their original form, they break down, and they are passed from one organism to the next through the trophic web. Section 2.5 focuses on the tissue regeneration and stem cells, an unexplored territory with great interest still lagging in bivalves compared to other marine organisms. Bivalves can repair and regenerate at least shell and mantle, and there is considerable tissue remodeling associated with an early immune response, and it appears there is a degree of conservation between mammalian and at least oyster adult stem cells. Bivalve Transmissible Neoplasia (BTN) (Section 2.6) is a leukemia-like proliferation of cells in the hemolymph of bivalves that has been reported in at least 15 different bivalve species, and it is one of the few cases, and first in a marine organism, of leukemia-like cancers that is horizontally transmitted. Associated with overexpression of the retrotransposon Steamer, the BTN represents a novel model to examine the process of cancer evolution. Finally (Section 3), we review the resources and tools available, from the pioneering efforts in the 1990s for developing systems for delivering genetic material into bivalves to the numerous attempts to establish cell lines. In this section, we also highlight the need for the development of the tools that would make the bivalves an attractive model to attract new students and researchers and at the same time be competitive for funding with other well-established model organisms.
Fig. 1. Bivalves as models for human health.
Hemocytes are involved in multiple aspects from the biology of bivalves with a direct implication on human health. Keystone of the immune defense at the center against a broad variety of pathogens, we can learn how organism lacking adaptive immunity (as defined in vertebrates) deal with viruses, bacteria, and protozoan. Mucosal immunity, maintaining and turnover of the microbiome in an aquatic environment rich in bacteria and as source of antimicrobial peptides in a world where antibiotic resistance is on the rise, are only a few of the aspects where hemocytes and humoral factors are involved. Bivalves can to cope with detoxification during HABs and as a filter feeder can concentration of human viruses responsible for outbreaks and microplastics that are incorporated into the food chain. Shell formation is also an exciting biology aspect that has parallel mechanisms with bone formation. Contagious cancer is clams can bring some light into oncogenic mechanisms, including cancer evolution, metastasis, and the role of transposable elements in oncogenesis. Some drawings modified from IAN (http://ian.umces.edu/).
2. Unique Aspects of Bivalve Biology
2.1. Shell Formation
Bivalves are protected from predators and environmental stressors (desiccation) by shells, a sort of exoskeleton that also provides the means for physiological homeostasis regulation in the soft body. The diversity and complexity of the bivalve shells are astonishing (Aguilera et al., 2017; McDougall and Degnan, 2018) and it is hypothesized to be one of the critical factors for the animal diversity explosion during the Cambrian period (Kocot et al., 2016). The shell formation involves specialized epithelial cells on the dorsal surface of the mantle that secrete the extrapallial fluid constituted of proteins, polysaccharides, glycoproteins, and lipids. This organic membrane (periostracum) is produced from a groove between the external and middle lobes of the mantle. In the eastern oyster, Crassostrea virginica, the formation of the shell starts early in development (14 + days from egg to spat) and an embryonic shell can be seen before the gastrulation is completed (Carriker, 2009). The general biomineralization process has been described although research is still needed to characterize it fully [reviewed by (Furuhashi et al., 2009; Kocot et al., 2016; Marin et al., 2012)]. The diversity of complexity and patterns of the shell makes bivalves an ideal model organism to study the evolution of biomineralization (Kocot et al., 2016). Comparative mantle transcriptome analysis of bivalves and gastropods indicates a lineage- or species-specific genes often including domains with rapid evolution rate and with a tendency to expand, and contract and rearrange in the genome (Aguilera et al., 2017). Genome annotation, and transcriptome and spectroscopy analysis of shell proteins indicate that biogenesis of the calcified shells is more complex than currently understood (Zhang et al., 2012) and it appears that the gene repertoire for shell formation is different in larvae and adults (Zhao et al., 2018). Osteogenic markers are induced in bone marrow-derived human skeletal stem cell growing on polycaprolactone surfaces mimicking Pinctada maxima nacre topographical surfaces (Waddell et al., 2018). Bivalves also provide an excellent model to study the molecular mechanisms involved in symmetry; e.g., oysters having anatomically paired mantle tissues display bilateral symmetry and recent RNA sequencing analyses of the right and left mantle indicates unique gene expression patterns, which might translate in functional differences (Wei et al., 2018a). Transcriptomic studies also reveal differentially expressed genes in the layers of the mantle (Li et al., 2017a). In the advent of the omics era, numerous studies continue to identify genes involved shell biomineralization using genomics; e.g., gene candidates for the formation of the nacreous layer, a strong, resilient, and iridescent organic-inorganic composite material made by some bivalves as an inner shell layer (Ohmori et al., 2018), formation of prismatic and nacreous layers (Funabara et al., 2014), receptor involved in regulating shell biomineralization (Li et al., 2017b), shell larvae formation (Liu et al., 2017; Liu et al., 2015; Liu et al., 2018), and shell color (Liu et al., 2015). Proteomics studies have also benefited from genomes; e.g., conservation of nacre proteins between bivalve species (Marie et al., 2017), identifying the “basic tool kit” for calcification processes (Arivalagan et al., 2017), and the proteins present in the water-soluble nacre matrix (Oliveira et al., 2012). Transcriptomics and proteomics are also used to ascertain the effect of global warming and ocean acidification on calcifiers (De Wit et al., 2018; Ivanina et al., 2013; Li et al., 2016b; Li et al., 2016c; Timmins-Schiffman et al., 2014).
In bivalves, circulating hemocytes in the hemolymph represent the main component of the internal self-defense system against pathogens which involves both cell-mediated and humoral systems (Allam and Raftos, 2015; Bachére et al., 2015; Vasta et al., 2015). In addition to being the primary immune effector in bivalves against pathogens, the hemocytes are also involved in many other physiological events including nutrient transportation, detoxification, and wound repair (Mount et al., 2004). Histological, structural and fluorescent microscopy, and mass spectrometry of Pinctada fucata indicated the presence of intracellular CaCO3 crystals in circulating granulocytes thought to be released to the extrapallial fluid (Li et al., 2016d). Most recent studies provide evidence for the presence of two functional groups of hemocyte. Comparison of the hemocytes from different body fluids of C. virginica using stained epitopes in conjunction with flow cytometry and functional assays indicate that the cells in the extrapallial space appear to be more actively involved in biomineralization and shell formation compared to the hemolymph (Lau et al., 2017). Transcriptomic profiling of hemocytes showed a marked contrast of gene expression patterns between hemocytes collected from the extrapallial fluid and those samples from the circulatory system in Manila clam Ruditapes philippinarum suggesting functional specialization at the biomineralization site (Allam et al., 2014). Expression analysis of selected biomineralization-related genes in hemocytes and mantle cells from the C. gigas also found a different pattern of expression and distinct phenotypes based on the cell morphology, motility, and adhesion properties (Ivanina et al., 2017). A similar line of evidence in P. fucata is that granulocytes containing calcium-rich vesicles and crystals serve as a calcium pool in the extrapallial cavity while retaining the phagocytic activity (Huang et al., 2018). The same study showed no evidence of the shell matrix proteins in the hemocytes, suggesting that they might not be solely responsible for directing the growth of the shell. The oysters also provide a valuable model to study tissue implanting; the pearl industry produces natural pearls by inserting the mantle graft from a donor together with a nucleus into the gonad of a recipient oyster. Graft recipient is the primary driver of the pearl size while graft donor is responsible for pearl quality traits (Blay et al., 2017).
The bivalve exoskeleton formation is a fascinating biological process that has also attracted the attention of medical research as a model for bone formation and repair (Furuhashi et al., 2009; Kocot et al., 2016; Marin et al., 2012; Westbroek and Marin, 1998). The water-soluble matrix (WSM) of nacre obtained from the inner shell layer of the oyster P. maxima promotes an increase in alkaline phosphatase in bone marrow stromal cells (Almeida et al., 2001). The MC3T3-E1 pre-osteoblast cell line from mouse can differentiate into osteoblasts and to mineralize in the presence of beta-glycerophosphate and ascorbic acid; WSM also induces differentiation and reduces the mineralization of MC3T3-E1 pre-osteoblast (Rousseau et al., 2003). Nacre injected percutaneously into experimental cavities prepared in the lumbar vertebrae of sheep results in significant activation of bone formation and mineralization (Lamghari et al., 2001). Similar results have been found when nacre is implanted in vivo in bones of mice, rats, and humans (Brion et al., 2015; Lee et al., 2017; Liao et al., 2002). Proteomic studies identified three gigasin-2 isoforms, and a cystatin A2 in WSM of C. gigas nacre that appears to be involved in bone remodeling processes and could be responsible for the biocompatibility shown between bone and nacre grafts (Oliveira et al., 2012). Ostreae Testa, a powder made of oyster shell (Ostrea gigas), enhances the proliferation of primary osteoblasts, differentiation (ALP activity) and bone nodule formation of osteoblast in vitro and has the potential to prevent OVX-induced osteoporosis through osteoblasts activation effects (Han et al., 2007). Shell formation in bivalves is also a model system for studying biomineralization; the BMP-2 signaling pathway plays a recognized role in biomineralization. The pearl oyster, P. fucata SMAD4, an orthologue of the intracellular transmitter of the BMP signaling pathway involved in biomineralization process in mammals, also appears to play a role in the regulation of the biomineralization in pearl oyster (Zhao et al., 2016). Although most of the work focuses on bone formation, oyster shell extracts also have shown lipogenesis inhibition properties (Tran et al., 2015b) and there is an increasing interest of the use of nacre as biomaterial for orthopedic applications (Gerhard et al., 2017; Rodrigues et al., 2017; Shen et al., 2014; Zhang et al., 2017).
Several attempts to generate cell lines from bivalves were based on explants from the mantle edge (Awaji and Suzuki, 1998; Chen and Wang, 1999; Daugavet and Blinova, 2015). The study of the molecular mechanisms of biomineralization of the shell, pearl formation, apoptosis, and terminal differentiation, will benefit from the availability of cell lines (section 2.5).
2.2. Innate Immunology
2.2.1. Mucosal Immunity
Animal mucosal tissues represent the sites of initial interactions between microbes and their hosts (Duncan and Edberg, 1995; Rayner and Wilson, 1997). For this reason, the understanding of how mucosal immune factors interact with and regulate microbes has become a central theme in biological research. Justifiably, most studies on the role of mucosal immune factors in host-microbe interactions are performed on medically-relevant models, and results highlight an intriguing central role of mucosal tissues in animal health, with an elaborate mechanism of feedback controls for the maintenance of mucosal (and host overall) homeostasis. Bivalve bivalves represent an appealing model for the examination of innate effectors involved in mucosal immunity. Their mucosal tissues are readily accessible both for in vivo observation (Lau et al., 2017; Mardones-Toledo et al., 2015; Pales Espinosa et al., 2016; Ward et al., 1991) and sampling (Lau et al., 2017; Pales Espinosa et al., 2016), and recent investigations show the responsiveness of these species to the knockdown of genes involved in microbial interactions in mucosal tissues (Pales Espinosa and Allam, 2018) making the validation of mucosal immune effectors within reach.
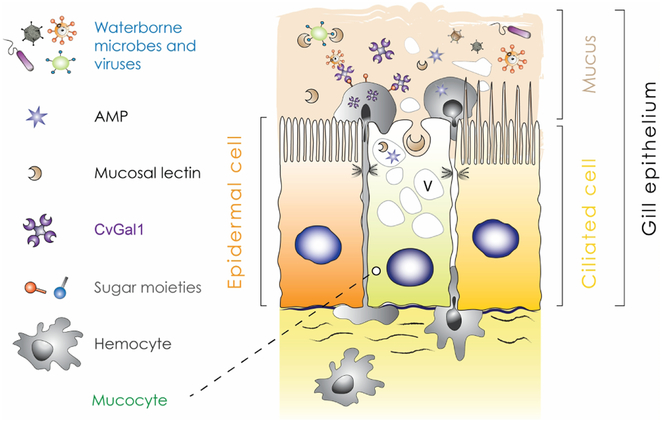
A particular feature of bivalves is their ability to process an extraordinarily large volume of water for nutrient and oxygen extraction. This is performed by the gills that pump and circulate water in the shell (pallial) cavity. In this context, the gills and other pallial organs (e.g., mantle, labial palps) represent the first tissues encountered by waterborne microbes as they enter the pallial cavity. The convolution of pallial organs greatly increases the effective surface of these interfaces and enhance their exposure to waterborne microbes. In fact, bivalve pallial cavity occupies a similar niche as that of the bucconasal and upper respiratory cavities of higher vertebrates (semi-confined compartment, highly regulated fluid circulation, the presence of immune defense factors associated with the mucosal surfaces) and therefore displays similar importance as a portal and barrier to invading microorganisms. It is striking that most fatal infections affecting bivalves are initiated in pallial organs or hemocytes patrolling the pallial cavity (Allam et al., 2013; Burreson and Ford, 2004; Chagot et al., 1992; Dahl et al., 2010; Ford et al., 2002; Kleeman et al., 2002; Lau et al., 2018; Tasumi and Vasta, 2007; Villalba et al., 2004; Wang et al., 2018b) further stressing the role of pallial mucosa in host-microbe interactions. Therefore, good protection of these soft tissues is required to allow bivalves to maintain microbial homeostasis and health in a microbe-rich environment. The mucus layer, abundantly secreted by pallial organs, represents the first host matrix that interacts with waterborne microbes. Bivalve mucus tridimensional structure follows that of other metazoans (Beninger et al., 1997) with the presence of two distinct layers covering epithelial cells (Ross, 1974) (Fig. 2). The first layer is in direct contact with the cells and is often made of low viscosity mucus while the external layer is typically made of viscous secretions that entrap environmental microbes and is directed by cilia movements. Most prior work investigating bivalve pallial mucus was performed during investigations of the suspension-feeding process (reviewed in (Allam and Pales Espinosa, 2015)). But the pallial mucus layer is also the first host factor encountered by mutualistic (e.g., sulfo-oxidant bacteria that inhabit gill bacteriocytes), commensal, or parasitic microbes. We are only starting to understand the diversity and function of defense factors associated with bivalve mucosal immunity. In addition to representing an efficient physical barrier, bivalve mucus was shown to contain a wide range of molecules involved in host-microbe interactions such as galactose and mannose-binding lectins, C1q-domain containing proteins, and antimicrobial compounds (defensin, lysozyme) (Pales Espinosa et al., 2016). Some of these proteins were shown to be regulated via external stimuli as the levels of mucosal lectins was shown to increase in oysters and mussels exposed to bacterial challenge (Pales Espinosa et al., 2010; Xing et al., 2011).
Fig. 2. Mucosal immunity model.
Mucus covering pallial surfaces of bivalve plays an important role in immunity and homeostasis. Viruses, bacteria, and protozoans, but also particulate matter and microalgae present in seawater directly interact with innate immune effectors present in mucus. Mucosal hemocytes likely play a sentinel role (sensing and signaling) similar to that of dendritic cells in higher vertebrates. Based on (Allam and Pales Espinosa, 2015). Antimicrobial peptide (AMP); Crassostrea virginica Galectin 1 (CvGal1); vacuole (V).
The adhesion of pathogenic microorganisms to mucosal surfaces is the first step of many infections in vertebrates and invertebrates. Due to its carbohydrate- and protein-rich composition, mucus is an excellent environment for microorganism growth (Koren and Rosenberg, 2008; Mark and Peter, 1999; Ritchie, 2006) that can, in turn, produce molecules that enter in mucus composition (Banin et al., 2001) or that alter host mucus (Brun et al., 2000). This result in the establishment of a robust, and often tightly- controlled, population of adapted microbes. Any alteration to this balance (e.g., changes in mucus physicochemical characteristics) can lead to a dramatic shift in microbial dynamics resulting in infection and disease. Previous studies on marine animals showed that mucus secretion could favor the attachment and growth of adapted microbes (Ducklow Hugh and Mitchell, 1979; Ebran et al., 1999; Nagashima et al., 2003; Ritchie, 2006; Vine et al., 2004), and to mediate symbiont recognition (Bulgheresi et al., 2006; Dufour, 2005; Nyholm and McFall-Ngai, 2003; Nyholm et al., 2000; Southward, 1986). Recent investigations showed marked tropism of the alveolate parasite Perkinsus marinus to specific types of mucus from its oyster (C. virginica) host, which induced a significant regulation of the proliferation and virulence of the parasite (Allam et al., 2013; Pales Espinosa et al., 2014; Pales Espinosa et al., 2013). Interestingly, pallial mucus of the non-compatible host C. gigas was strongly inhibitory to P. marinus activities suggesting that host specificity may begin in the mucus (Pales Espinosa et al., 2013). The transformation of Perkinsus olseni zoospores into trophozoites is also influenced by components of R. philippinarum gills and labial palps leading to initial infection primarily within gills and labial palps of the host clam (Wang et al., 2018a). Similarly, a recent study of the gut transcriptome of mussels Mytilus edulis infected with the copepod Mytilicola intestinalis suggested a co-evolution of this host-parasite association as indicated by the predicted mechanistic interactions between both partners at mucosal interfaces (Feis et al., 2018). In this context, bivalves provide a tractable system for the identification of effectors that regulate homeostasis at mucosal interfaces in animals (including higher vertebrates) and how specialized microbes provide benefits to their hosts.
The understanding of the role of motile mucosal cells in systemic immunity of higher vertebrates remains relatively limited. Previous work in these species highlighted the role of specific neutrophils and dendritic cells in orchestrating the recognition of, and response to, microbes at mucosal interfaces. Through transepithelial migration, these cells cross epithelial surfaces to monitor environmental microbes (Rescigno et al., 2001), although the molecular cascades involved in their transepithelial migration and signaling remain obscure (Pérez-López et al., 2016). In lower vertebrates and invertebrates, the features and functions of the mobile cellular components of mucosal immunity are, at best, extremely limited (Cima et al., 2006). Bivalves offer an attractive model for such studies. For instance, their mucosal tissues are well irrigated by the blood and hemocytes can cross the basement membrane to wonder at the surface of the epithelial barrier in close association with the mucus layer (Allam and Paillard, 1998; Allam and Pales Espinosa, 2016). Through transepithelial migration, oyster mucosal hemocytes were shown to translocate within hours from pallial surfaces to underlying tissues and the circulatory system (Lau et al., 2018), suggesting these may play a sentinel role similar to that of dendritic cells in vertebrates. Interestingly, mucosal hemocytes in oysters had much higher phagocytic activity than circulating hemocytes (Lau et al., 2017). They also showed epitope signatures (surface carbohydrates and clusters of differentiation) different from those of circulating hemocytes suggesting that they represent a specialized category of hemocytes. Specifically, labelling with the cluster of differentiation 11b (CD11b; specific to integrin alpha M and characteristic of transepithelial leukocytes) and 14 (CD14, a receptor of lipopolysaccharides (LPS) and other pathogen-associated molecular patterns (PAMPs), enriched in sentinel leukocytes) were significantly higher in mucosal hemocytes as compared to circulatory cells underlining a potential sentinel role for these cells. CD14 is a common constituent of vertebrate mucus (Uehara et al., 2003) and may represent a conserved mechanism of innate mucosal immunity in metazoans. The occurrence of two-way movements of hemocytes across mucosal epithelia may facilitate infection by adapted microbes that are capable of surviving phagocytosis, as shown for some pathogens in vertebrates (Nunes-Alves, 2014). With that regard, our preliminary investigations showed the ability of the obligate oyster parasite P. marinus to take advantage of transepithelial migration of mucosal hemocytes to gain access to internal host tissues (Allam and Pales Espinosa, 2016; Lau et al., 2018). Microbial uptake at mucosal surfaces could be further facilitated by lectins associated with hemocytes as shown for CvGal1 (Feng et al., 2015; Tasumi and Vasta, 2007; Vasta et al., 2015). Overall, mucosal hemocytes appear to represent the missing link between the dynamic microbial landscape at mucosal surfaces and the internal immune system. More investigations are needed, however, to unravel if and how these cells monitor the microbial make-up of mucosal tissues before migrating back into tissues to inform the systemic immune system (e.g., the release of cytokines). More specifically, the transepithelial migration of hemocytes may provide a possible mechanism for the transmission of leukemia in bivalves, which was suggested to be acquired from the water column through mucosal interfaces (Metzger et al., 2015; Metzger et al., 2016) (Section 2.6).
Finally, bivalves offer a tractable system for understanding the role of mucosal microbial communities in animal health given the interplay between mutualistic, commensal and pathogenic microbes at mucosal interfaces (Section 2.2.2). This is supported by a growing body of evidence highlighting the role mucosal microbiomes in regulating host resistance to infection either directly microbe-microbe interactions (Barr et al., 2013; Mack et al., 1999; Tuomola et al., 1999) or indirectly via immune stimulation and maturation (Russell, 2015). How mucosa responds to probionts and how changes in mucus physicochemical characteristics (either caused by disease, by other beneficial/harmful microbes or by natural cycles) affect microbial homeostasis at mucosal surfaces are among the many questions that still need to be answered, and that could lead to better disease management strategies.
2.2.2. The Microbiome of Bivalves
The recent surge of microbiome studies in humans and other species have highlighted the essential roles of microbiomes (bacteria, archaea, and viruses) in the biology, ecology, and evolution of eukaryotic hosts (Parfrey et al., 2018; Petersen and Osvatic, 2018; Russell et al., 2014). For example, the human-associated microbiomes have been extensively studied within many contexts, especially by the Human Microbiome Project (Turnbaugh et al., 2007). Despite their economic and ecological importance, little is known about the diversity and function of bivalve-associated microbiomes. A recent extension of ‘omics in bivalves includes the identification of host-associated microbial populations using 16S rRNA sequencing and functional inference. These high-throughput technologies can be used to explore and better understand a range of diverse subjects in biology and ecology, including the basic mechanisms driving selective associations between microbes and hosts (from mutualism to parasitism), and the roles of microbial-bivalve interactions in bivalve biology and health, ecosystem services, resilience to environmental changes, and human health and food safety. Bivalve microbiomes can provide a model for how human-microbe interactions may be studied, particularly within the context of host diseases and environmental change.
As described in previous sections, bivalves are exposed to a rich bacterial community while filter feeding on phytoplankton (Pierce and Ward, 2018). Many of these microorganisms are rejected during the complex process of particle selection; some are digested as they go through the organism, while others are able to colonize the gut and other organs, such as the gills. It is expected that the microbiome of bivalves changes through the seasons due to shifts in phytoplankton communities (Carrier and Reitzel, 2017). Consistent with these features of bivalve biology, the microbiomes of bivalves are proven to be highly dynamic and to respond rapidly to environmental changes. For example, adult oysters have shown differences in microbiome composition according to tissue types, seasons, environmental conditions, and geographical locations (Chauhan et al., 2014; King et al., 2012; Lokmer et al., 2016; Pierce, 2016; Roterman et al., 2015). Moreover, differences in the microbiome of mussels and oysters collected from the same location appear to reflect differences in the biology of these two species, e.g. while the functional diversity of the microbiome of oysters significantly decreased in the winter, the functional diversity in mussels in the winter remained similar to that of other seasons. These differences are consistent with the fact that mussels continue to filter feed during the winter (when temperatures are below 10°C), while oysters do not (Pierce, 2016).
Despite the high variability in microbial communities associated with bivalves derived from variability in their environments, studies of the eastern and Pacific oyster (C. virginica and C. gigas) microbiomes have suggested the potential existence of a core microbiome in the oyster that is significantly enriched as compared to the surrounding water, often dominated by Proteobacteria, Cyanobacteria, and Firmicutes (King et al., 2012; Lokmer et al., 2016). Similarly, a study comparing the microbiomes in Crassostrea hongkongensis and the surrounding waters has shown that microbiome composition of the surrounding waters was more significantly affected by water salinity while microbiome composition of the oysters was more significantly affected by changes in water temperature (Wang et al., 2016a). Microbiome studies of C. gigas larvae in hatcheries and postlarvae grow-out cultivation has found that even though the microbiome in the hatchery/rearing water changes throughout the year, this background microbiome has limited effect on the larval microbiome (Asmani et al., 2016; Powell et al., 2013; Trabal Fernández et al., 2014). The microbiome of gills in blue mussels collected in an estuary in the Northeast US was found to be highly dominated by an Endozoicomonas similar to symbionts present in another marine host (oysters and tunicates) (Schill et al., 2017). Additionally, a comparative study of two lucinid bivalves has revealed that their endosymbiotic populations are species dependent and reestablishes in the host after perturbation under starvation stress (Elisabeth et al., 2014). These studies point to the existence of selective host-microbial interactions allowing for the establishment of some species in particular tissues.
Consistent with this expectation, some molluscan species have often been used as model systems for the study of the ecology and evolution of host-microbe symbioses. The symbiotic association between the bobtail squid Euprymna scolopes and Vibrio fischeri is an example of partner fidelity in which the symbiont is acquired each generation by horizontal transmission through a selective immune response from the host (McFall-Ngai, 2014). Although microbiome sequencing approaches have been proposed as a tool to discover other examples of host-symbiont coevolution, most phylosymbiotic approaches are not sensitive enough to demonstrate partner fidelity or co-adaptation (Douglas and Werren, 2016). An initial study of 16S rDNA sequence diversity of a few isolates of Teredinibacter turnerae, the symbionts of shipworms (wood-feeding bivalves), did not detect evidence of host-specificity, which could only be detected after analyzing six bacterial genes (Altamia et al., 2014). It is unclear how these diverse and dynamic microbial communities in bivalves affect the fitness of the host and vice versa. A diverse but functionally resilient microbial community may support hosts and allow them to acclimate to changes in their environment (McKenney et al., 2018); e.g., bacterial isolates able to metabolize domoic acid, a potent neurotoxin associated with harmful algal blooms of Pseudo-nitzschia spp. have been isolated from blue mussels, M. edulis, and soft-shell clams Mya arenaria (Stewart et al., 1998). These microbial symbionts may be one of the multiple mechanisms allowing bivalves to survive the toxic effects of harmful algal blooms and pollutants and minimize toxin accumulation in their tissues (Carrier and Reitzel, 2017; Milan et al., 2018) (see also Section 2.3).
The interactions between host immune responses and their associated microbiomes are being increasingly investigated within the context of disease (Lozupone, 2018). It is hypothesized that the microbiome may be one factor in the modulation of their host immune response and disease phenotype (Douglas, 2018). As an example, microbial community diversity in Sydney rock oysters was found to be affected when the host is infected with macro-parasites (Green and Barnes, 2010). The microbial diversity of the hepatopancreas of Pacific oysters was also reported to be affected by hypoxia, potentially reflecting the effect of environmental stress on the immune response of these oysters (Khan et al., 2018). Microbial dysbiosis in the marine environment and aquaculture is strongly associated with mortality events and disease (Egan and Gardiner, 2016). In response, high-throughput sequencing of microbiomes has been used as a tool for disease management of bivalve species of aquaculture interest (Bentzon-Tilia et al., 2016; Gómez-Chiarri et al., 2015a). Additional research is needed to decipher this crosstalk between the bivalve host’s immune receptors and the microbiome.
Bivalves also play key roles in disease transmission to other species. As filter feeders, bivalves can act as biofilters, and therefore alter disease transmission by either decreasing concentrations of pathogens in the water through filtration and digestion (sinks) or concentrating and amplifying pathogens able to survive and, in cases, proliferate within their tissues (sources) (Burge et al., 2016). As an example, species of the Vibrio genus could cause a decrease in the survival of C. virginica larvae at a concentration of as low as 103 CFU per ml (Paillard et al., 2004). Oysters and their vibrio populations have been proposed as a model to study disease dynamics in wild animals within ecological and evolutionary frameworks, a model that could improve understanding on how polymicrobial assemblages and microbial-microbial interactions may affect disease initiation and progression in a variable environment (Le Roux et al. 2016). Increased understanding of microbe-host interactions would also allow developing and/or optimizing strategies to manipulate bivalve microbiome and to prevent/treat the accumulation of human pathogens. Two studies on changes in the composition of the microbiome of mussels after depuration have revealed a decrease in bacterial diversity and evenness, but the studies differ on the outcome regarding changes in the relative abundance of Proteobacteria (Rubiolo et al., 2018; Vezzulli et al., 2018).
Better tools are needed to identify critical interactions among the host, microbes, and the environment. While the 16S rRNA sequencing has been extremely useful for estimating the population structure of the microbiome community, linking the 16S rRNA gene sequencing data with the functional potentials and activities of the microbiome is still largely unexplored and remains a challenging undertake. In a recent study, a computational procedure was applied to infer the abundance of denitrification genes based on the distribution of 16S rRNA genes and a database of taxonomically verified function genes (Arfken et al., 2017). This approach, however, assumes the conservation of function genes among taxonomically related species and hence may overlook the genome-level variations within the same taxonomic unit. Another mostly unexplored area in the microbiome study of marine invertebrate is the application of metagenome and metatranscriptome sequencing (Martínez-Porchas and Vargas-Albores, 2015). Unlike the 16S rRNA based amplicon sequencing, the metagenome and metatranscriptome approaches target the totality of all the coding genes and gene transcripts, respectively. For example, a metatranscriptomic analysis of the gills and its microbes in a deep-sea mussel, Bathymodiolus azoricus, shows microbial functional acclimation to pressure changes (Barros et al., 2014). The further application of metagenome and metatranscriptome approaches to studying diverse marine invertebrates will likely bring new insights into the functional interactions both within and among the microbiome, the oyster host, and the surrounding environment.
Another area of future studies is to look for the presence, composition, and function of core microbiomes among different tissue types and host genotypes (King et al., 2012). While a few studies have suggested the presence of core microbiomes within different tissue types, the identity and relative abundance of the core microbiome requires further exploration. The temporal and spatial variation of core microbiome could potentially be linked to the variations in the host genotype, host physiology and the environmental conditions. The genotype-phenotype associations of the microbiome and host could be further explored with genome-scale modeling (Steffensen et al., 2016). The models could help establish physiological understanding of the individual organisms in the microbiome and help identify auxotrophy among different microbes and the host. Genome-scale modeling has been proven useful for the prediction of changes in activity under diverse environmental conditions (Dufault-Thompson et al., 2017). It could effectively integrate the metagenomic and metatranscriptomic data into creating a holistic view of the microbiome-host-environment interactions.
Moreover, questions remain about how microbes are acquired by the host and the impact of the microbiome composition and function on the biology and health of the host. In bivalves, metatranscriptomic studies will help identify potential obligate symbionts, which could not have been readily identified in bivalves due to the lack of cell cultures that would help in isolation (Carrier and Reitzel, 2017). To characterize the nature of these associations (mutualistic, commensal, pathogenic), it would be necessary to collect metatranscriptomic data at different states of the host, from health to disease, and in a variety of environments. These data may solve mysteries regarding symbiont colonization and persistence, microbiome effects on host genome evolution, roles in digestion and nutrition, defense mechanisms, and immune responses.
2.2.3. Alternatives to Antibiotics
Marine life, with its incredible biodiversity, has become the focus of intensive research as a unique source of natural products which could potentially find cost-effective biotechnological applications as drugs, cosmetics, and nutraceuticals (Anjum et al., 2017). The most relevant biotechnological applications of engineered proteins from Mollusca, the largest phylum of marine metazoans, include the 2014 approval by the Food and Drug Administration of a cone snail-derived peptide (ziconotide, for sale under the name Prialt) as a commercial drug for chronic pain therapy. Other exciting products currently under development include a “medical glue” inspired by mussel adhesive byssal proteins (Barrett et al., 2013) and artificial cancer cell-binding lectins which may find an application as diagnostics or therapeutic tools (Terada et al., 2017). The interdisciplinary paradigm that emerged from these innovations, if applied more broadly, could provide important results in the discovery of novel bioactive peptides with some potential applications. Antimicrobial peptides (AMPs), fundamental effectors of the innate immune system, have attracted considerable attention from pharmaceutical companies as alternative antibiotics for the treatment of human and farmed animal diseases (Galdiero et al., 2015; Giuliani et al., 2007; Hancock and Sahl, 2006). The focus on AMP research arises from their broad spectrum of activity and effectiveness against multidrug-resistant bacterial pathogenic strains which cannot be targeted by conventional antibiotic treatments (Vizioli and Salzet, 2002), a growing issue that, according to some experts, may lead to a “post-antibiotic era” (Kosikowska and Lesner, 2016). These properties derive from the universal cationic nature of most AMPs that, in spite of considerable diversity regarding primary sequence and 3D structure, interact with negatively charged bacterial cell wall or membrane components, leading to the elimination of invading microbes by lytic mechanisms. Consequently, the number of changes that would be required to counteract the complex mode of action of AMPs while maintaining cell membrane function and structure are thought to make the development of AMP resistance quite unlikely (Zasloff, 2002).
As filter-feeding organisms, marine bivalves are exposed continuously to high loads of bacteria, viruses and other potentially pathogenic microorganisms (Salazar and Sunagawa, 2017; Suttle, 2007), in addition to those generally associated with their tissues as symbionts (Section 2.2.2). Over time, selective pressure is thought to have favored the development of efficient and effective mechanisms of defense towards infections, which involve the production of a plethora of bioactive defense molecules (Hughes and Fenical, 2010), far superior in abundance and diversity than other molluscan classes (Gerdol, 2017). Although bivalve AMPs have been traditionally considered as humoral factors associated with circulating immune cells (hemocytes) (Mitta et al., 2000), in recent years a growing number of studies have revealed their importance in mucosal surfaces (i.e., mantle and gills), which represent the primary interface of contact with the external environment (Allam and Pales Espinosa, 2016). At the same time, it is now clear that bivalve AMPs do not merely act as pathogen-killing effectors but are more intimately linked to the modulation of host-microbiome interactions (Destoumieux-Garzon et al., 2016) and that they may cover accessory cytokine-like functions (Balseiro et al., 2011).
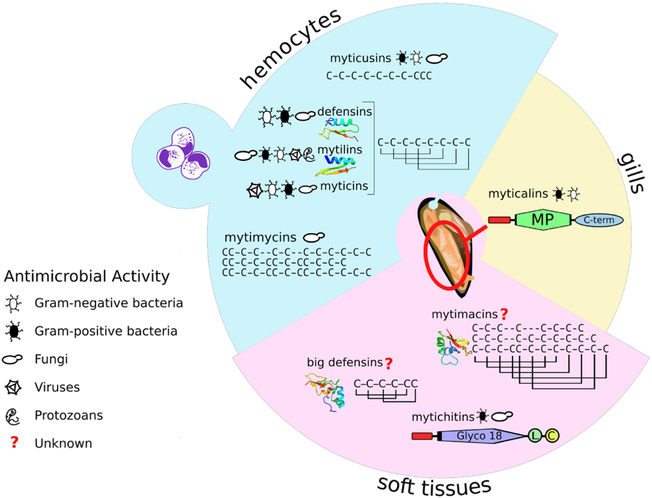
Since the isolation of the first bivalve AMPs in mid ‘90s (Hubert et al., 1996), several dozen peptides pertaining to different families have been characterized in various bivalve species, with those with commercial importance (i.e., mussels, oysters, clams, and scallops) as the main subject of research (Li et al., 2011b). Mussels (Mytilus spp.) in particular, represent a striking example of AMP diversity, as their arsenal of antibiotic peptides currently includes defensins, big defensins, mytilins, myticins, mytimacins, mytimycins, myticusins, myticalins, and mytichitins, but possibly others yet to be discovered (Gerdol and Venier, 2015) (Fig. 3). Interestingly, although multiple pathogens and disease conditions have been described in mussels (Boehs et al., 2010; Robledo et al., 1994), both natural beds and aquacultured mussels species appear to indicate a superior resilience to pathogens compared to other cultured bivalves (Gauthier-Clerc et al., 2013; Romero et al., 2014). Within this context, mussel AMPs may represent good candidates for the development of biotechnological products aimed at the improvement of aquaculture practices in bivalve species more susceptible to infection and disease. The vast majority of known bivalve AMPs are rich in cysteine residues engaged in multiple disulfide bonds, which enable a compact structure, often corresponding to a “CSαβ motif” (Dias Rde and Franco, 2015). On the other hand, AMP classes widespread in different domains of life, such as alpha-helical peptides, are seemingly absent in bivalves or, as in the case of linear peptides with compositional biases (i.e., enriched in positively charged amino acids), have only been reported on a few occasions (Gueguen et al., 2009; Leoni et al., 2017).
Fig. 3. Summary of the known antimicrobial peptides produced by mussels (Mytilus spp.).
Antimicrobial activity (defined as an experimentally determined Minimum Inhibitory Concentration <32 μM), tissue specificity and three-dimensional structure (whenever known) are indicated for each AMP family. The typical cysteine array and disulfide connectivity of cysteine-rich AMPs are also displayed. Different members of the mytimacin and mytimycin family are characterized by different cysteine arrays. The disulfide connectivity of mytimycin and myticusins has not been elucidated yet. MP: mature peptide; C-term: anionic C-terminal extension; Glyco 18: glycoside hydrolase, family 18, domain; L: linker region; C: chitinase-like domain of mytichitin.
Over the past two decades, bivalve AMP research has been mainly directed towards two interconnected areas of interests, the first dealing with the role of these effector molecules in the context of the bivalve immune system, the second focused on their possible application in the treatment of human and animal diseases. While the reader is directed to recent reviews for an overview of the biological role of bivalve AMPs in bivalve immunity (Bachére et al., 2015; Gerdol and Venier, 2015), we will here briefly outline the progress and current limitations of the use of bivalve AMPs as therapeutics for human health.
The number of functionally characterized bivalve AMPs displaying significant in vitro activity against biomedically relevant strains of Gram-positive and Gram-negative bacteria, fungi and, less frequently, protozoans and viruses, is steadily growing (Gueguen et al., 2009; Leoni et al., 2017; Yang et al., 2000). Interestingly, while bivalves are hosts of several metazoan parasites (e.g., nematodes, cestodes, trematodes and, in some cases, copepods), no defense molecule active against these targets has been described to date, in stark contrast with arthropods, where a few multifunctional AMPs active against parasitic nematodes (e.g., cecropins) have been described (Chalk et al., 1995). The promising results obtained with standard Minimal Inhibitory Concentration assays, evidencing biological activities at concentrations as low as 0.01 μM for oyster defensins against Gram-positive bacteria (Bachére et al., 2015), prompted the design of a few engineered AMP variants, and the evaluation of their potential for biotechnological applications.
The mussel AMP mytilin has been the target of multiple studies. In 2004, a synthetic variant was tested in vitro against the shrimp white spot syndrome virus, evidencing a significant reduction of mortality in shrimps challenged with viral particles pre-incubated with the AMP (Dupuy et al., 2004). Subsequent studies enabled the identification of fragment adopting a beta-hairpin structure as the minimal active peptide (Roch et al., 2008). Two synthetic peptides derived from the sequence of Mytilus coruscus were subsequently produced and tested with success against some Gram-positive, Gram-negative bacteria and fungi, also displaying high stability in human serum (Liu et al., 2010). Investigations carried out on the mussel defensin MGD1 led to the development a synthetic cyclized nonapeptide (Romestand et al., 2003), highly active against the protozoan parasites Trypanosoma brucei and Leishmania major, also capable of preventing HIV-1 viral infection in MAGIC-5B cells (Roch et al., 2004). Moreover, the rational design of a novel peptide, named Ap-S, based on an AMP isolated from the hemolymph of the Chilean scallop A. purpuratus enabled the creation of a synthetic antifungal peptide showing a broader spectrum of activity compared to the native peptide (Arenas et al., 2009). However, none of these studies has led to significant developments to date, and a more recent work, which revealed the high biotechnological potential of mussel myticin C as a potent antiviral against Herpes simplex viruses (−1 and −2) infection in vitro, still awaits confirmation from in vivo testing (Novoa et al., 2016).
The current lack of marketable bivalve AMP-derived molecules is in line with the inherent difficulties in the engineering and production of peptide-based therapeutics. Globally, only a handful AMPs isolated from various animal sources are currently undergoing phase II/III clinical trials (Mahlapuu et al., 2016), which highlights the multiple issues that hamper the development of these molecules as valid alternatives to antibiotics (Hancock and Sahl, 2006). First, relevant discrepancies are often observed between in vitro and in vivo test. This is in part due to the critical influence of environmental and physiological conditions on AMP activity (e.g., pH in the case of mussel myticins) (Domeneghetti et al., 2015); in part also due to the scarce knowledge on their mode of action, which, among all bivalve AMPs, has only been firmly established for oyster defensins (Schmitt et al., 2010). AMPs might also be susceptible to degradation by endogenous proteases and generally display short half-life in vivo, which might prevent their intravenous and oral administration, and even sensibly reduce their effectiveness as topical antibiotics (Vlieghe et al., 2010).
Although AMPs generally bear little risk due to their amino acidic nature and lack of dangerous metabolites, some potential safety concerns have been reported, in particular whenever high concentrations are required to reach a critical biological action. For example, mytilin A displayed significant antiparasitic activity and inhibition of Herpes simplex virus type I replication only at concentrations, potentially cytotoxic for human cells (Carriel-Gomes et al., 2007; Löfgren et al., 2008).
Besides the limitations mentioned above, high manufacturing costs currently represent the major obstacle towards the commercial scalability of AMP, limiting their competitiveness compared to other small non-peptidic drugs (Bray, 2003). Direct isolation of AMPs from bivalve biomass is usually impractical, and chemical synthesis, frequently applied for laboratory testing, results in costs incompatible with large-scale applications. The methodology that holds the best promises is probably the production of recombinant AMPs in heterologous microbial systems (Ingham and Moore, 2007). Although several bivalve AMPs have been produced in Escherichia coli (Bachére et al., 2015), Pichia pastoris is emerging as the preferred system as it offers many advantages over bacteria, such as the inclusion of post-translational modifications typical of eukaryotic cells (Cereghino and Cregg, 2000). Some attempts have been made at setting up large-scale production of recombinant bivalve AMPs in yeast, producing conflicting results. For example, while large quantities of mytichitin-A could be obtained with this method, the recombinant protein lost some of the biological properties of the native AMP, most notably its activity towards Gram-negative bacteria (Meng et al., 2016).
Altogether, these considerations highlight that, in spite of the enormous potential offered by marine biodiversity for AMP discovery, the pharmaceutical industry currently displays a preference for technology-intensive methods that usually lead to a fallback to traditional and more accessible resources. It is the prevailing opinion that new strategies are required to exploit the biotechnological potential of the marine environment, increasing the success of drug discovery (Desbois, 2014). The emergence of multidrug resistance in bacteria might provide the appropriate framework for a necessary investment in this field, which would see bivalves as primary targets of research.
AMPs are not the only secretory components of the bivalve immune system that hold biotechnological potential for applications aimed at the improvement of human health. Indeed, the production of antimicrobial effectors and the regulation of immune response at the cellular and systemic levels depend on the activity of cytokines, small glycoproteins that guarantee a fast and effective immune response even at a very low concentration in body fluids. Due to their role, human cytokines and their receptors have been the target of intensive biomedical research and several analogs, derivatives and competitive inhibitors have been developed for the treatment of multiple pathologies, including cancer, AIDS, asthma and autoimmune disorders (Fry and Mackall, 2002; Gordon et al., 1996; Leonardi et al., 2008; Noble and Goa, 1997; Vatrella et al., 2014).
Cytokines have long been thought to be a prerogative of organism endowed with an adaptive immune system until the first evidence supporting the existence of invertebrate cytokine-like factors started to surface in the early 2000’s (Beschin et al., 2001). Little progress has been made in bivalves since mostly due to the elusive nature of such molecules and the lack of sequence similarity with well-characterized vertebrate interleukins). Functional studies have so far targeted a few evolutionarily conserved cytokines (i.e. interleukin-17, the macrophage migration inhibitory factor MIF and the allograft inflammatory factor AIF-1), whereas research is still at its very early stages for others which have been discovered very recently (for a review on the subject, see (Gerdol et al., 2018)).
Large uncertainties remain about the ability of bivalve cytokines to interact with the highly divergent cytokine receptors expressed by human cells To the best of our knowledge, just two studies have been able to confirm in vitro the immunomodulatory effects of exogenous bivalve cytokines on vertebrate cell lines. In detail, it has been revealed that recombinant MIF protein from the scallop Azumapecten farreri could induce sheep fibroblast migration into scraped spaces, suggesting potential usefulness for wound healing (Li et al., 2011a). A second study was able to demonstrate the interaction of the oyster Tumor Necrosis Factor with TNF receptors in the human cell line HEK293T through the activation of a reporter gene (Qu et al., 2017). Our knowledge of bivalve cytokines is still minimal compared to AMPs, and this lack of knowledge most certainly currently hampers the development of biotechnological products at this stage. Yet, the multifaceted immunomodulatory properties of cytokines may lead to surprising innovations once these obstacles are overcome in the years to come.
2.3. Harmful Algal Bloom (HAB): Toxin Impacts, Metabolism, and Detoxification
A worldwide increase in the frequency, magnitude, and distribution of harmful algal blooms was first recognized in the late 20th-century (Anderson, 1989; Smayda, 1989). While this increase has been attributed to any one of many causes depending on location and the algal species involved, the trend is predicted to continue as climate change increasingly impacts these blooms (Hallegraeff, 2010). Many of these HAB species contain potent biotoxins which may be concentrated by filter-feeding molluscan shellfish and subsequently vectored to humans and/or bioaccumulated through food chains (Landsberg, 2002). Although the specific HAB toxin differs depending on the algal species involved, which in turn can vary with location, bloom timing and the shellfish species involved, these blooms constitute a significant current and future threat to both cultured and natural harvests of filter-feeding molluscan shellfish (Shumway, 1990).
Bivalves can serve as vectors for biotoxin transfer to humans via both natural harvest and aquaculture activities, especially oysters and mussels (Farabegoli et al., 2018). Although filter-feeding bivalves have a low sensitivity to phycotoxins, they can accumulate high concentrations of these toxins (Haberkorn et al., 2010a; Haberkorn et al., 2010b) and thus are often routinely monitored for these toxins (Anderson, 2009). Originally thought to be exempt from accumulating toxins (see (Shumway, 1990) for a historical review), oysters and mussels can accumulate a variety of algal toxins, including saxitoxin(s), brevetoxin(s), domoic acid, and okadaic acid. These toxins can be subsequently vectored to humans via consumption of contaminated oysters and result in syndromes which include paralytic shellfish poisoning (PSP), neurotoxic shellfish poisoning (NSP), amnesic shellfish poisoning (ASP), and diarrhetic shellfish poisoning (DSP). These algal toxins represent a variety of chemical structures ranging from tetrohydropurines to ladder polyethers, and the amino acid domoic acid, and hence exposure results in a variety of in vitro physiological effects in humans due to differences in the model of action of individual toxins (see (Baden and Trainer., 1993) for review). A commonality, however, is the ability of bivalves to bioaccumulate and tolerate these algal toxins while only experiencing sublethal impacts themselves. Bivalves also can metabolize and biotransform toxins in toxin analogs via complex chemical and enzymatic conversion mechanisms (Manfrin et al., 2012).
The vast majority of algal toxicological studies have focused on human cell lines and model vertebrate systems (Manfrin et al., 2012). The variety of genomic, genetic, behavioral, and pathological responses documented in bivalves, as well as the sublethal nature of the majority of these impacts in response to algal biotoxin exposure, suggests that bivalves are a potentially suitable model organism for toxin metabolism, including modification and detoxification. Additionally, the complete genome of the several bivalve species has been assembled (Gómez-Chiarri et al., 2015b; Powell et al., 2018; Wang et al., 2017a; Zhang et al., 2012). This makes bivalve species excellent choices to potentially serve as a model system/organism to both study the impacts of these toxins and to serve as potential tools for human health monitoring by developing sensitive, cost-effective bioindicators or cell markers (e.g., cell damage and detoxification responses) of toxin presence, especially at low algal cell concentrations. The following summary is a brief overview of the variety of sublethal impacts of these algal toxins on oyster species, with the goal of fostering and advancing the potential use of oysters as model systems for the study of the impacts and monitoring of these toxins.
Impacts of algal toxins on bivalves can be grouped into either behavioral, pathological, genetic, or genomic effects (Fig. 4). As bivalves have no adaptive immunity to algal toxin exposure, their responses are limited to their innate immune responses to stressors (e.g., pathogens, parasite and physical injury (Janeway, 1994)). Bivalves take up and accumulate algal cells as food, and often exhibit short-term behavioral responses to toxin exposure. If toxic cells are not immediately rejected as pseudofeces (Hégaret et al., 2007b; Mafra et al., 2010), toxins accumulate in the bivalve digestive gland (Bardouil et al., 1993; García et al., 2015; Guéguen et al., 2008; Haberkorn et al., 2011), and immediate behavioral effects include changes in valve activity (Basti et al., 2009; Borcier et al., 2017; Haberkorn et al., 2011; Mat et al., 2013; Nagai et al., 2006; Tran et al., 2010), respiration (Shumway and Terry, 1987), and reduced clearance, and filtration rates (Hégaret et al., 2007b; Lassus et al., 1999; Lesser and Shumway, 1993; Wildish et al., 1998). These negative impacts extend to embryonic and larval stages and include less active spermatozoa (Haberkorn et al., 2010b), decreased sperm viability (Rolton et al., 2015) and impairment of larval survival and reproductive development (Basti et al., 2016; Gainey Jr and Shumway, 1988), and clearance rates (Leverone et al., 2007). Extreme behavior effects such as paralysis of the adductor muscle in C. virginica have been reported on exposure to the dinoflagellate Alexandrium fundyense (Hégaret et al., 2007a). Some bivalve adaptive mechanisms are possible when feeding on toxic algae however. M. edulis was able to alter absorption kinetics of biochemical compounds and digestive gland enzymes on exposure to Alexandrium catenella, allowing exploitation of the toxic algae as a food source with minimal impacts (Fernández-Reiriz et al., 2008).
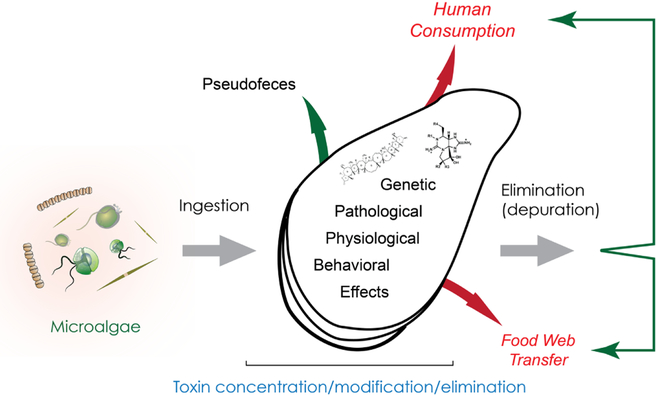
Fig. 4. Transfer routes and effects of toxins in bivalves.
HAB species containing biotoxins are ingested through the filter-feeding process. Some species are rejected in the pseudofeces. However, the ingestion of the HAB species may result in the concentration and modification of the toxins, resulting in genetic, pathological, physiological, and behavioral effects. Accumulated toxins are vectored to humans and/or bioaccumulated through the food chain. Interestingly, the bivalves also can eliminate (depurate) the toxins. Some drawings modified from IAN (http://ian.umces.edu/).
Although toxins accumulate primarily in the digestive gland, toxins do directly affect the bivalve immune systems, resulting in inflammation and impaired immune function (Hégaret and Wikfors, 2005a; Hégaret and Wikfors, 2005b). Immune response in bivalves is mediated by hemocytes and exposure to algal toxins directly affect bivalve hemocytes. Hégaret et al. (Hégaret and Wikfors, 2005a; Hégaret and Wikfors, 2005b) demonstrated changes in the number and proportion of hemocytes in C. virginica on exposure to the dinoflagellate Prorocentrum minimum. M. edulis, conversely, experiences diapedesis of hemocytes into the intestine when exposed to P. minimum, aggregating the P. minimum cells and potentially them to minimize M. edulis tissue damage (Galimany et al., 2008). Changes in oyster hemocyte counts and viability are commonly reported upon exposure to algal toxins (Basti, 2011; Haberkorn et al., 2010b; Hegaret et al., 2011; Medhioub et al., 2013; Mello et al., 2013), however there are some exceptions. Hégarett et al. (Hégaret et al., 2007a) reported that the toxic dinoflagellates A. fundyense and Alexandrium catenella had no impacts on C. virginica hemocyte morphology or function. However, toxin production is highly variable in some toxic algal genera (Burkholder and Glibert, 2006), which may explain reported difference in hemocyte impacts. Additional immune-related responses to toxin exposure include hemocyte phagocytosis (Ford et al., 2008; Mello et al., 2013), higher mucus production in gills (Haberkorn et al., 2010b), and histopathological lesions (Pearce et al., 2005) including myopathies in the adductor muscle (Haberkorn et al., 2010b). In vivo exposure of Mytilus galloprovincialis to Prorocentrum lima causes oxidative DNA damage resulting in early genotoxicity in hemocytes (Prego-Faraldo et al., 2016). In addition to the toxins, there is new evidence that bioactive extracellular compounds produced by the microalgae also have an effect on valve-activity behavior and hemocyte mobilization (Castrec et al., 2018).
Recent research on genetic and genomic responses of bivalves to algal toxins suggests that these responses may provide a promising approach to identify biomarkers of toxin contamination. Mat et al. (Mat et al., 2018) report that toxin load is a primary driver of transcriptomic variation; the whole transcriptome of the digestive gland of C. gigas exposed to Alexandrium minutum exhibited differences in transcript abundance between oysters with different toxin loads (1,098 transcripts) rather than between exposed and non-exposed oysters (16 transcripts); only 70 of 29,000 oyster genes described transcripts modeled the toxin load. A. minutum also disrupts circadian rhythmicity in the cryptochrome gene (CgCry) in exposed C. gigas, and gene transcription remains at a constant low level throughout a daily cycle with exposure (Tran et al., 2015a) and also provokes activation of oyster antioxidant system at the transcription level (Fabioux et al., 2015; García-Lagunas et al., 2013). Other oyster genes have also been shown to be upreuglated by toxin exposure, including pro-apoptotic genes (Bax and Bax-like) implicated in the mitochondrial pathway, two caspase executor genes (caspase-3 and caspase-7) and five (Bcl2, BI-1, IAP1, IAP7B and Hsp70) inhibitors of apoptosis-related genes (Medhioub et al., 2013) as well as genes involved in antioxidant defense (copper/zinc superoxide dismutase), cell detoxification (glutathione S-transferase, and cytochrome P450), intermediate immune response activation (lipopolysaccharide and beta-glucan binding protein), and stress responses (glutamine synthetase) (Garcíia-Lagunas et al., 2013). Overexpression of these genes indicates the activation of a protective mechanism, whose answer depends on both toxic cell concentration and exposure time (García-Lagunas et al., 2013). Exposition of M. galloprovincialis to P. lima downregulates the expression of genes involved in antioxidant stress (Prego-Faraldo et al., 2017). Similarly, transcriptomic profiles in response to the experimental exposition of mussels to okadaic acid results in an increase in proteasomal activity, molecular transport, cell cycle regulation, energy production, and immune activity in the digestive gland and gill tissues (Suárez-Ulloa et al., 2015).
All of these impacts will vary depending on biological and environmental variables at the time of exposure, including the algal species, the toxicity of the clones and the specific toxin(s) involved, the oyster species, its life cycle stage, nutritional status and size, as well as local environment conditions which may favor bloom development and exposure periods. Interactions between algal toxins and other stressors also have impacts on oysters. Lassudrie et al. (Lassudrie et al., 2015) demonstrated increased dermo susceptibility in C. virginica infested with the trematode Bucephalus sp. when exposed to A. fundyense. Conversely, exposure to the dinoflagellate Alexandrium catenatum reduced the prevalence of herpes infection in C. gigas, and the oysters accumulated fewer toxins (Lassudrie et al., 2015). Whether the host-pathogen interaction was modified by behavioral, physiological or direct interaction effects is unknown. While extended exposure to toxic algae may ultimately result in reduced bivalve survival and growth (Shumway 1990), the variety, extent and commonality of the sublethal impacts of toxin exposure, which include genomic, genetic, behavioral and pathological effects (Fig. 4), suggest that bivalves have the potential to serve as a model system for the study of direct and indirect impacts of algal toxin on biological functions on a variety of timescales.
2.4. Microplastics in the Ocean
The ocean and other aquatic environments are considered sinks for anthropogenic contamination. Microplastics have recently gained attention and notoriety in society and the mass media beyond the reason why they were engineered. The release of plastics into the environment during manufacture, transport, and as waste is increasing by approximately 25 million tons a year (Mato et al., 2001; Waite et al., 2018). Since plastics are engineered to have a slow rate of degradation, they build up in the environment making them a hazard for future generations (Galloway and Lewis, 2016; Law et al., 2010). These plastics do not just stay in their original form, the ocean breaks them down into millions of microplastics (MP) through mechanical degradation such as wave action or chemical processes like photodegradation, thermal degradation, and biodegradation (Law and Thompson, 2014; Smith et al., 2018b; Waite et al., 2018). Technically, MPs are defined as any plastic less than 5 mm, but plastics less than 1 μm have also been reported in the environment (Smith et al., 2018b). Their origin comes from clothing fibers and microbeads in cosmetic and hygiene products being deposited into the ocean by rivers, tides, runoff, cargo dumps and even lost fishing gear (Law and Thompson, 2014). The most prevalent plastic chemicals found in ocean debris are polypropylene, polyethylene, and polyvinylchloride (PVC) (Wright et al., 2013). In addition to the chemical toxicity, MCs are a dynamic mixture of chemical and microbial contaminants that bind to them changing toxicity and bioavailability (Galloway et al., 2017; Law and Thompson, 2014). Studies have found high concentrations of harmful chemicals (polychlorinated biphenyl, dichlorodiphenyldichloroethylene, and non-phenols) in polypropylene resin pellets in both ambient seawater and ocean sediments (Mato et al., 2001). These pellets are highly absorbent and could absorb other harmful chemicals beyond those tested (Mato et al., 2001). The MPs are causing issues for trophic systems, as they are small enough to be eaten by zooplankton (Law et al., 2010; Law and Thompson, 2014). Since many zooplankton species are not highly selective in their food choice, subsequently MPs can be passed from one organism to the next through the trophic web (Law et al., 2010; Law and Thompson, 2014; Wright et al., 2013).
The complexity of current systems in the ocean makes very challenging to determine the exact concentration of MPs (Law and Thompson, 2014); however, there are report of MPs in bivalves is increasing worldwide (Kanhai et al., 2018; Li et al., 2018a; Li et al., 2018b; Naji et al., 2018; Santana et al., 2016). Marine snow contributes to mobilize MPs away from the surface into deep environments (Porter et al., 2018). High-density plastics, such as PVC, float down to sediments (Wright et al., 2013) allowing them to be consumed by benthic organisms, like bivalves. Aggregates of microbeads form in upper layers of the ocean via currents and sink to the sediment regions. On the seafloor, bivalves are more likely to ingest these aggregates rather than loose plastics, which incorporates MPs into the food web (Wright et al., 2013). It is estimated that the sediment in coastal waters contains a range of 2-30 MPs per 250 mL (Law and Thompson, 2014). Plastics in the range of the 16.5 MP pieces per individual have been reported in oysters (C. virginica) (Waite et al., 2018) and 1.1 - 6.4 MPs per individual in blue mussels (M. edulis) (Li et al., 2018b). As bivalves accumulate MPs, higher trophic levels are at risk for exposure to the chemicals found in MPs. Globally, 6.7% of human protein consumed is seafood (Smith et al., 2018b). These MPs have the potential to be both toxic and cause physical issues; however, the full extent of this risk must be studied further.
Some studies have found that MPs can cause physical blockages, chemical changes, and biological change in European flat oysters (Green, 2016). While bivalves do not have the highest amount of selectivity for trapping food particles, they are capable of selecting their food particles using various criteria, including size (Pales Espinosa and Allam, 2018; Wright et al., 2013) (see also Section 2.2.1). This mucus generally allows bivalves to effectively capture particles as small as 3-4 μm and some smaller particles as well (Wright et al., 2013). Indeed, mussels (M. edulis and Perna viridis) are more likely to consume smaller MPs such as microbeads instead of larger microfibers (Qu et al., 2018). Besides, MPs can also adhere to muscle, mantle, and other bivalve organs (Kolandhasamy et al., 2018). Many MPs are within the 1-4 μm range and therefore, bivalves are incredibly susceptible to ingesting them. Bivalves can’t digest MPs easily, and they build up in tubules and the digestive organs, sometimes persisting for 48 hours (Wright et al., 2013). The build-up of these MPs can cause clogging of the bivalve digestive tract and decrease the ability to feed (Wright et al., 2013). As the microplastics weaken these animals, they may make them more susceptible to predation, causing bioaccumulation of the toxins in the food web. In addition to the incorporation into the food web and the risk for human consumption (Van Cauwenberghe and Janssen, 2014), the evidence is building that MPs also affect the physiology of the bivalves.
Experimental exposure of sediment-dwelling bivalves to environmental relevant MPs results in changes of the energy reserve tissues directly (Bour et al., 2018) or indirectly (Xu et al., 2017). Exposures of (Mytilus galloprovincialis) to polycyclic aromatic hydrocarbons commonly found in MPs led to bioaccumulation in digestive glands and gills (Pittura et al., 2018). Prolonged exposure of oysters to polystyrene microspheres resulted in significant decreases in oocyte number and diameter, and sperm velocity as well (Sussarellu et al., 2016). The larval development of bivalves can also be impacted by leachate from MPs (Gandara e Silva et al., 2016). Nano polystyrene particles reduce the filtering activity of mussels (Wegner et al., 2012). Interestingly, mussels exposed to PVC particles do not result in physiological effects (Santana et al., 2018). MPs can also affect the benthic communities associated with bivalves (Green, 2016). The literature on the effect of MPs on the immune system of marine macrofauna is very minimal. Corals under the acute MP exposure exhibit immune system and detoxification repression (Tang et al., 2018). When exposed to MPs, M. galloprovincialis had MPs in hemocyte cells that exhibited destabilized lysosomal membranes (Pittura et al., 2018). It has been shown that MPs (<1 mm) translocated into the circulatory system of mussels where they enter in direct contact with hemocytes (Browne et al., 2008). MPs exposure of M. galloprovincialis results in disruption of the homeostasis associated with the production of stress and immune-related proteins (Detree and Gallardo-Escarate, 2018).
Microplastics have become prevalent not only in our environment but in the food we eat and even ourselves. As they accumulate in the ocean, they are ingested by bivalves and moved up through the food web. The effects of these toxins are still mostly unknown due to the highly variable compounds and concentrations found in these plastics. What we do know is that bivalves are one of the primary organisms that can transfer them into humans. As a result, further study of the toxicity of MPs in bivalves is critical to determine the risks to the bivalve community, marine ecosystems and humans. Finally, using bivalves as bioindicators can play an essential role in understanding pollution levels, bioavailability and the ecological risks of contaminants (Su et al., 2018).
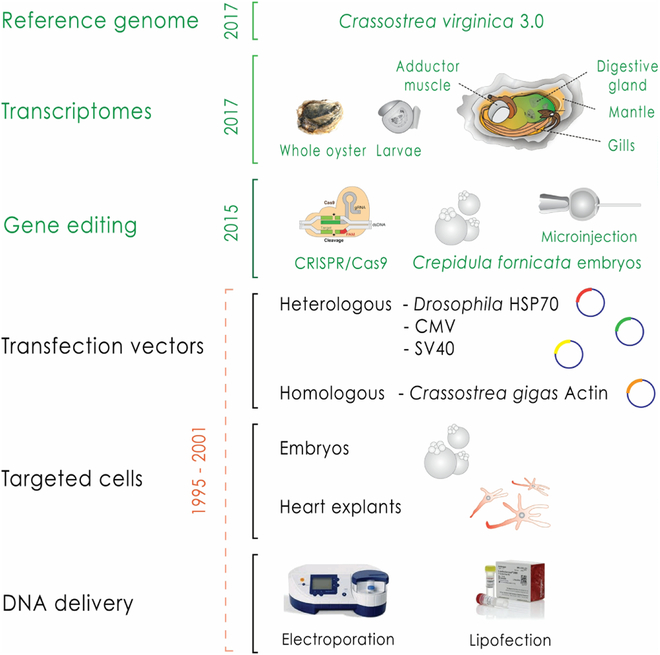
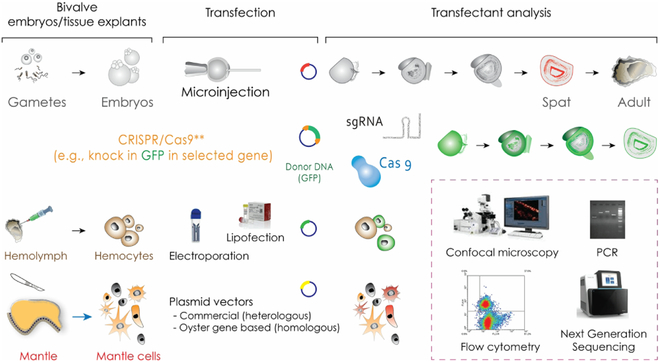
2.5. Regeneration and Stem Cells
Regeneration is the replacement of lost tissue with new following damage or wounding (Wells and Watt, 2018). Although sharing similarities with developmental growth or asexual reproduction, regeneration is, therefore, a distinct process at both the cellular and molecular level (Bely and Nyberg, 2010; Erickson and Echeverri, 2018; Simoes and Riley, 2018; Wells and Watt, 2018). Regeneration within the body can occur at different, e.g., cellular, tissue, organ, and limb levels. However in the current context of regenerative medicine in humans its focus is usually on critical organs such as the brain, central nervous system, or heart, which if critically damaged or wounded do not regenerate and at very best repair with scarring that highly limits functional recovery (Bely and Nyberg, 2010; Erickson and Echeverri, 2018; Wells and Watt, 2018). Mammals and birds are largely incapable of larger organs/ limb regeneration (Bely and Nyberg, 2010; Erickson and Echeverri, 2018). However, across the animal kingdom, there is high variability in a species ability to regenerate lost body parts (Bely and Nyberg, 2010; Erickson and Echeverri, 2018).
In evolutionary terms, the ability to regenerate all body parts is common to metazoan lineages and some of lophotrochozoan and deuterostomes (Bely and Nyberg, 2010). Platyhelminths and tunicates can regenerate from small body fragments (Bely and Nyberg, 2010; Gehrke and Srivastava, 2016). A number of lizards can regenerate their tail but not limbs (Alibardi and Toni, 2005; Bely and Nyberg, 2010), while amphibians like salamanders can regenerate their tails, limbs, and jaws (Alibardi and Toni, 2005; Bely and Nyberg, 2010; Erickson and Echeverri, 2018; Yin et al., 2015). The reasons why some animals are capable of regenerating while others not, combined with an explosion in research on stem cells; cells capable of differentiating into any tissue, has led to a renaissance in research in regenerative medicine.
Although they represent one of the most abundant phyla, little research has been done in bivalves concerning regenerative pathways (Bely and Nyberg, 2010; Rinkevich, 2011). Bivalves including oysters do appear to be very capable of repair and regenerate at least shell and shell and mantle (Acosta-Salmon and Southgate, 2006; Huning et al., 2016; Mamangkey and Southgate, 2009; Takahashi et al., 2012). Although there is no evidence that regeneration of the whole body can occur from simple small fragments (Bely and Nyberg, 2010). Working with vertebrate research models requires to follow the policy on institutional care and use of laboratory animals (e.g., Institutional Animal Care and Use Committee, IACUC in the USA; Directive 2010/63/EU of the European Parliament). Given the availability and ethical use of invertebrates over vertebrates such as bivalves, we would argue these represent not only a hugely underdeveloped phylum for this research but also a less restricted phylum for animal experimentation since with the exception of cephalopods in Europe, no IACUC is yet required. It is also worth noting that although not studied directly for regenerative capacity the unique qualities of some marine bivalve products, most notably from the mussel, have inspired polymers that are being trialed for potential uses in regenerative studies in mammals (Kaushik et al., 2015). Mussels secrete a sticky glue that allows them it to stick to rocks and solid surfaces (Kaushik et al., 2015). This protein glue, made up of proteins called mussel adhesive proteins rapidly hardens into a water-resistant adhesive (Kaushik et al., 2015). Products based upon mussel adhesive proteins have been tested for diverse applications such as tissue glue, a suture free wound closure glue (Mehdizadeh et al., 2012), which may reduce tissue scarring (Jeon et al., 2017), as well as structurally inspired microspheres, nanofiber and 3D printed matrices that can been developed as drug delivery systems for bone repair (Wei et al., 2018b), wound healing (Kim et al., 2017; Wang et al., 2016b), and cancer drug delivery (Ma et al., 2016). Similarly, nacre (mother of pearl) is being tested as a biodegradable material for bone grafting, bone scaffolding, and surgical screws (Gerhard et al., 2017; Libouban et al., 2016). Powdered extract nacre has also been shown to stimulate mammalian fibroblast migration (Lopez et al., 2000). Since bivalves represent approximately 23% of all the named marine organisms, there is undoubtedly a considerable potential for novel marine or marine-inspired products to form the basis of therapies that translate into human medicine.
Since regeneration in bivalves is highly likely to share similarities with other species, it is possible to learn about this potential, from looking at these common pathways shared in animal regeneration (Agata et al., 2007; Bely and Nyberg, 2010; Erickson and Echeverri, 2018; Wells and Watt, 2018). Moreover, with advances in genome sequencing and RNA-Seq technology, it is likely that species-specific and regeneration-specific gene expression pathways will only become easier to study. Unsurprisingly, when comparing events occurring in tissue regeneration, there is considerable tissue remodeling associated with an early immune response (Kimura et al., 2003; Li et al., 2016a; Ramirez-Gómez et al., 2008); growth factor up-regulation and signaling e.g. fibroblast growth factor (FGF) and Wnt (Lengfeld et al., 2009; Lin and Slack, 2008), and the production of matrix metalloproteinases (Quinones et al., 2002; Yokoyama, 2008), along with re-epithelialization (Rousselle et al., 2018; Takeo et al., 2015). The Wnt signaling pathways, in particular, appear critical for early blastema (the regenerative seed) development (Lengfeld et al., 2009; Lin and Slack, 2008). Searching the recently published genomic data from C. gigas, for example, indicates that homologs for all these pathways are very likely to be present in this and other related bivalve species. Interestingly, the cloned C. gigas epithelial growth factor receptor transfected into mouse myoblast cells was able to stimulate proliferation showing a remarkable level of conservation between molluscan and mammalian growth factor signaling systems (Sun and Irvine, 2014). Additionally, innervation of the regenerating tissue also appears a relatively common early step driving the process (Kumar et al., 2007; Monaghan et al., 2009). One caveat that must be noted from these studies is that many, if not all, of these events, are also shared by wound healing processes in mammals that do not exhibit regeneration (Fathke et al., 2006; Houschyar et al., 2015). An important question, therefore, is what distinguishes wound healing (or even scar-free wound healing) from regeneration? One answer may lie in the presence, numbers, and location of different types of stem cells. Regeneration is initiated by the formation of a blastema (Satoh et al., 2015; Stocum, 2017). This blastema can be formed of adult stem cells, de-differentiated cells of mature tissue (vertebrate limb regeneration; repair in jellyfish), or by a combination of both (Satoh et al., 2015; Stocum, 2017). One factor that appears particularly important in this trans-differentiation process and achieving stem-like biology is the P-Element induced wimpy testis protein (Piwi) and its homolog Cniwi (van Wolfswinkel, 2014).
Ever since the cloning of the first mammal “Dolly the sheep” (Gurdon, 2017) there has been a huge interest and development in stem cell research for regenerative medicine. The “stem cells” are self- replicating, undifferentiated master cells that are capable of differentiating and producing daughter cells that make up every different cellular and tissue type in the body (Rognoni and Watt, 2018). One focus has been on embryonic stem cells, but these do not account for regenerative capacity once an animal is developed and born. Hence there has been interest in the stem cell populations that may exist in juveniles and adults (adult stem cells), that allow for regeneration of tissue after wounding. A number of adult stem cell populations, although relatively rare, have been found in different tissue in mammals, with most demonstrating a more limited ability to produce cells along certain lineages such as the hematopoietic stem cells (Kiel and Morrison, 2008; Rognoni and Watt, 2018). These mammalian adult ‘stem cells’ do not exhibit true stem cell totipotency; i.e., the ability to differentiate into all cell types of the ectoderm, mesoderm, and endoderm embryonic germ layers. These adult stem/ progenitor cell populations appear to have more of a role in tissue homeostasis and minor repairs rather than large-scale regenerative capacity (Abnave and Ghigo, 2018; Rognoni and Watt, 2018). These hurdles have not stopped researchers trying to isolate and expand these cells types so that they can be used in larger quantities to test for regenerative ability (Abnave and Ghigo, 2018; Rognoni and Watt, 2018).
Although the presence and activity of adult stem cells in bivalves has not been investigated in depth, many marine invertebrates, such as sponges are known to contain relatively large pools of adult stem cells), that are involved in their ability to regenerate through their lifetime (Bode et al., 2006; Rinkevich, 2011; Sun et al., 2007). Since they also represent some of the longest-lived animals, we believes represent a vast area of untapped stem cell research.
One of the limitations for studying stem cells outside of mammalian biology is the ability to identifying stem cells by either transcription factor or cell surface markers. Mammalian researchers have access to a substantial increasing resource of well-characterized specific antibodies that allow identification, isolation, and characterization (both in terms of stem cells and their lineages) of stem cells. At this moment in time, more resources are required to identify both the conserved proteins and antibodies that cross-react with these in bivalves to allow identification and isolation of these stem cell populations. Recent data does, however, allow a degree of hope for this, as there appears a degree of conservation between mammalian and at least oyster adult stem cells. C. gigas germline cells exhibit intense alkaline phosphatase activity, the transcription factor Sox2 and a cytoplasmic marker for germline tissue Vasa (Cavelier et al., 2017). C. gigas adult hemocytes were recently shown to be derived from Sox2 positive stem cells found in an irregularly folded structure of the gill epithelium (Jemaá et al., 2014). Moreover, recent evidence also indicates roles for the conserved hematopoietic transcription factors Tal- 1/Scl (Stem Cell Leukemia), GATA3, C-Myb, and c-kit (Song et al., 2016) in this hemocyte generation. Tal-1/Scl, GATA3, C-Myb, and c-kit are all transcription factors associated with leukocyte development in mammals (Labastie et al., 1998).
Another experimental approach to generate and investigate stem cells, and now routinely used in mammalian systems, but not yet bivalves, is the reprogramming of adult cells into induced pluripotent stem cells iPSc (Di Baldassarre et al., 2018; Rinkevich, 2011). Here adult cells are forced to either permanently or transiently express a combination of transcription factors that dedifferentiate the cells back to ones that closely resemble embryonic stem cells. Interestingly, one of the factors commonly used to induce iPSc is Sox2 (along with others such as Oct4, KLF4, cMyc and Nanog) (Di Baldassarre et al., 2018). To our knowledge, this has yet to be reported on in marine invertebrates, so it is not known whether these exact combinations will induce iPSc, but the approach using these transcription factors or similar homologs, offers a considerable potential to understand the biology and regenerative capacity of marine bivalves. With current technology it should be feasible as molluscan cells can be transfected with plasmid DNA and with the recent advances in genomic sequencing, species-specific homologs can be identified and cloned if necessary. One limitation to note does remain, in the lack of ability to grow longterm cell line cultures from marine bivalves. Although cells can be isolated and can be maintained alive for extended periods, they stop dividing and become quiescent relatively quickly (Rinkevich, 2011). As yet only one molluscan cell line Bge exists from the land snail (Yoshino et al., 2013), so advances also need to be made here to allow the long-term culture of any derived cells. It is possible that understanding multipotency of cells and the ability to culture them may go hand in hand to potentiate both fields. Understanding, cell growth and differentiation in bivalves, will not only help understand processes of regeneration that may be applied to species such as humans that have poor regenerative capacity but ultimately inform the physiology and biology of bivalves themselves greatly.
2.6. Bivalve Transmissible Neoplasia (BTN)
Disseminated neoplasia, or hemic neoplasia, is a leukemia-like proliferation of cells (presumably of hemocyte origin) in the hemolymph of bivalves that eventually disseminates through the tissues of the organism, leading to death, and it has been reported in at least 15 different bivalve species (Barber, 2004; Carballal et al., 2015). The disease can occur at stable enzootic levels, but outbreaks have been reported in multiple species, including soft-shell clams (Mya arenaria), in which massive mortality has been observed (Muttray et al., 2012). In at least four affected species (M. arenaria, Mytilus trossulus, Cerastoderma edule, and Polititapes aureus), the disease has been shown to be a transmissible clonal cancer—a lineage of cancer in which the cells themselves jump from one animal to the next as a natural allograft (Metzger et al., 2015; Metzger et al., 2016). Transmissible cancer had only been observed in the wild previously in Tasmanian devils (Pearse and Swift, 2006) and dogs (Murgia et al., 2006; Rebbeck et al., 2009). Transmissible cancer models can be used to investigate some oncogenic mechanisms, including cancer evolution, metastasis, and the role of transposable elements in oncogenesis, as well as mechanisms of resistance of the host itself to cancer engraftment.
Transmissible cancer cells have been used to investigate classical oncogenic proteins such as p53 and myc. The presence of a LINE1 insertion immediately upstream of c-myc in all known cases of the canine transmissible venereal tumor (CTVT) provides evidence that transposons can generate oncogenic mutations that are selected for in cancer (Katzir et al., 1987; Katzir et al., 1985). The role of many other well-known cancer proteins (including p53 and telomerase) have been studied in CTVT (Murchison, 2008) and devil facial tumor disease (DFTD) (Deakin and Belov, 2012). In bivalves, the clam neoplasia model has been used to investigate the conserved role of p53 in invertebrate cancers, showing that mortalin sequesters p53 in the cytoplasm of neoplastic cells (Power et al., 2006; Walker et al., 2011). These models may also allow us to identify oncogenes or tumor suppressors that are either conserved in human cancers or are novel players in independent pathways.
Bivalve transmissible neoplasia (BTN) offers a novel model to examine the process of cancer evolution. These cancers represent extreme cases of metastasis (Lazebnik and Parris, 2015) in which cancers have acquired the ability to outlive their hosts and continue to evolve as they repeatedly engraft into new hosts. Their evolution, therefore, represents exaggerated versions of adaptations that occur in common primary tumors as well as adaptations to unique challenges (Greaves and Maley, 2012; Nowell, 1976; Ujvari et al., 2016). While investigation of a single cancer lineage may give us some insight into possible effects of retrotransposons (as in the case of LINE-c-myc in CTVT), the finding of multiple independent lineages of BTN increases our ability to understand the conserved processes involved in the multiple steps from healthy cell to neoplasia, to metastasis, to transmission to another organism, and finally to a widespread successful transmissible cancer lineage.
The transmissible cancer lineage in soft-shell clams is marked by a massive amplification of a retrotransposon called Steamer, from about 2-10 copies per haploid genome in healthy animals to >150 in neoplastic cells (Arriagada et al., 2014). By definition, retrotransposon activity induces chromosomal mutations, but the extent of their role in cancer mutations and the mechanisms and pathways that they affect are not fully known. Development of genomic resources will allow us to identify the locations of these neoplasia-associated transposon insertion sites in the soft-shell clam neoplasia and will allow for testing whether different transposable elements are amplified in the other BTN lineages. Additionally, in soft-shell clams, BrdU injection has been reported to induce new transmissible cancers experimentally (Oprandy and Chang, 1983; Taraska and Anne Bottger, 2013), and this may provide many new oncogenic events for study. The soft-shell clam is, therefore, a model organism that would allow for direct testing of the role of transposable elements in oncogenesis and transmission.
In addition to the standard hallmarks of cancer (Hanahan and Weinberg, 2000), transmissible cancer cells also evade the host immune system, which could otherwise identify them as coming from a foreign individual. Normal cancers have mutations that can form neoantigens, so they too must escape immune recognition and destruction, and the extreme phenotype of transmissible cancers allows for investigation of the mechanisms by which hosts allow or reject cancer cells. Bivalves do not possess an MHC-based histocompatibility system, but BTN does not appear to cross species barriers readily. Multiple experiments have shown that disseminated neoplasia can be transplanted from one individual to another within the same species (McLaughlin et al., 1992; Taraska and Anne Bottger, 2013; Weinberg et al., 1997), but cross-species transplantation has been unsuccessful. In three of the four bivalve species analyzed, each is affected by one or more cancer independent lineages derived from within the same species (Metzger et al., 2015; Metzger et al., 2016). In the one exception (Polititapes aureus) cancer appears to be a transmissible lineage derived from a closely related species from the same family (Venerupis corrugate), which, interestingly, may itself have evolved resistance to cancer. These observations suggest that there is some mechanism of resistance to cancer lineages in bivalves, which may be a type of self/non-self-recognition system that clears cancer or some host factor required for cancer cell engraftment which can be mutated in the host, or some other unknown pathway. These contagious cancers are essentially acting as massive natural experiments in which outbred organisms are being challenged with a clonal cancer. Understanding the mechanisms of resistance that have been selected for may have profound impacts on human health.
Also, primary cell cultures from marine bivalves have been used in basic research for decades; still, the attempts to establish a cell line have been unsuccessful (Rinkevich, 2011; Yoshino et al., 2013). Most efforts to generate cell lines from bivalves were based on explants transferred into culture media for spontaneous immortalization derived from stochastic genetic events. Most invertebrate cells stop dividing 24–72 h after isolation (Rinkevich, 2011). BTN may provide a useful immortalized cell line itself; understanding the mechanisms behind BTN’s survival and continued growth may provide the clues for immortalizing cells from marine bivalves. The development of bivalve cell lines would provide a simple model for studying numerous aspects of complex biological systems.
While the study on the transmissible nature of bivalve disseminated neoplasia is new, and many questions remain to be answered, the research on this cancer model has the potential to yield new insights into human cancer development and evolution as well as mechanisms by which humans can block cancer engraftment and growth.
3. Resources and Tools
Over the last decade, there has been an enormous sequencing effort in species of interest in the aquatic environment; early advances focused on finfish with zebrafish (Danio rerio) becoming a model organism for numerous human diseases and conditions (Sarasamma et al., 2017). Bivalve species are also benefiting from the latest technologies for genome sequencing (Abdelrahman et al., 2017) (Table 1). P. fucata and C. gigas genomes encode for 23,257 and 28,027 predicted genes respectively (Takeuchi et al., 2012; Zhang et al., 2012). Initial insights into the Mediterranean mussel (M. galloprovincialis) genome indicate a gene enrichment of gene ontology categories related to multixenobiotic resistance, glutamate biosynthetic process, and the maintenance of ciliary structures (Murgarella et al., 2016a). The number of predicted genes (39,493 protein-coding genes, 4,230 non-coding) for eastern oyster genome (684 Mb; C_virginica-3.0; GCF_002022765.2) is much larger than for P. fucata and C. gigas genomes (Gómez-Chiarri et al., 2015b). In addition to the genome, the eastern oyster research consortium is now focusing in comparing C. virginica genomes from multiples sites in the USA (Gómez-Chiarri, unpublished work). Next-generation sequencing offers an unique opportunity not only to understand the general process of innate defenses in invertebrates but also as a essential tool for comparative immunology of vertebrates and invertebrates (Schultz and Adema, 2017).
Table 1.
Published molluscan genome assemblies. The access to the latest sequences technologies together with the continuous decrease in the price is resulting in an increasing number of genomes publicly available. Modified from (Takeuchi, 2017).
| Species | Common name | Genome size (Mb) |
Total scaffold length (Mb) |
Scaffolds | Scaffold N50 (Kb) |
Reference |
|---|---|---|---|---|---|---|
| Crassostrea virginica | Eastern oyster | 578-675 | 669 | 10+1* | 1,900 | (Gómez-Chiarri, unpublished work) |
| Saccostrea glomerata | Sydney Rock oyster | 788.1 | 746.5 | 10,101 | 804,2 | (Powell et al., 2018) |
| Bathymodiolus platifrons | Deep sea mussel | 1,630 | 1,660 | 65,664 | 343.4 | (Sun et al., 2017) |
| Modiolus philippinarum | Mussel | 2,210 | 2,630 | 74,575 | 100,2 | (Sun et al., 2017) |
| Patinopecten yessoensis | Scallop | 1,440 | 987.6 | 82,731 | 803.6 | (Wang et al., 2017a) |
| Argopecten irradians | Bay scallop | 990 | 700.3 | 217,310 | 6.8 | (Du et al., 2017) |
| Ruditapes philippinarum | Manila clam | 1,370 | 2,571 | 223,851 | 48.4 | (Mun et al., 2017) |
| Mytilus galloprovincialis | Mediterranean mussel | 1,600 | 1,599 | 1,746,447 | 2.6 | (Murgarella et al., 2016b) |
| Pinctada fucata martensii | Pearl oyster | - | 990.6 | 8,621 | 324.3 | (Du et al., 2017) |
| Pinctada fucata | Pearl oyster | 1,150 | 815.3 | 29,306 | 167.3 | (Takeuchi et al., 2012;Takeuchi et al., 2016) |
| Crassostrea gigas | Pacific oyster | 637 | 558.6 | 11,969 | 401.3 | (Zhang et al., 2012) |
10 chromosomes and mitochondrial DNA
Significant economic losses derived from infectious diseases in the aquaculture industry, as well as interest in the development of selected strains with faster growth, drove pioneering efforts in the 1990s for developing systems for delivering genetic material into bivalves of commercial interest (Boulo et al., 1996; Buchanan, 1999; Buchanan et al., 2001; Cadoret et al., 1999; Powers et al., 1994-1997). These efforts, however, were hindered by the low efficiency of the transfection methods (1% for embryos, and not reported for primary cultures), as well as the lack of information on the molecular basis for the traits of interest. Also, the lack of transcriptomes and a draft genome precluded the placement of the genes in context to define promoter regions to build homologous transfection vectors. Therefore, in following decades, the oyster research community focused efforts on the development of genomic tools needed to identify traits of commercial, biological, and ecological interest (Abdelrahman et al., 2017). Taking full advantage of the oyster genome and transcriptomes and other approaches (Powers et al., 1995; Tsai et al., 1997) requires to build and validate the genetic toolbox for making the oyster a genetically tractable system for both basic and applied research (Fig. 5). Along the same lines, immortalized cell lines provide the simplest model for studying numerous aspects of complex biological systems. Without cell lines, the analysis of cell biology is limited by the availability and lifetime of cells and primary cultures, the access to tissue donors or animals, and the genetic background variability from sample to sample. Clones can be grown indefinitely in culture, providing unlimited and standardized material that allows for reproducibility on genetically identical cells and in tightly controlled experimental conditions and datasets that are comparable across laboratories. Growing cells in vitro is also a cost-effective way to produce proteins of interest similar to those found in multicellular organisms. Despite decades of extensive research efforts by multiple laboratories around the world and the urgent need to create new cell lines from recalcitrant species in phyla underrepresented or not represented, there is not yet a single cell line available from marine invertebrates (reviewed in (Rinkevich, 2005; Yoshino et al., 2013)).
Fig. 5. Overview of available resources towards a genetically tractable system in bivalves.
Most attempts to introducing genetic material in oysters date from two decades ago and focused on the Pacific oyster. With the genomes of commercial and non-commercial bivalves being sequenced, the minimum tools and resources for building a genetically tractable system are already available. Indeed, CRISPR/Cas9 has been recently achieved in the gastropod Crepidula fornicata (common slipper shell).
Primary cell cultures from marine bivalves have been used in basic research for decades; still, the attempts to establish a cell line have been unsuccessful (Rinkevich, 2011; Yoshino et al., 2013). Most attempts to generate cell lines from bivalves were based on explants transferred into culture media for spontaneous immortalization derived from stochastic genetic events. However, invertebrate cells stop dividing 24–72 h after isolation (Rinkevich, 2011) although primary cell cultures can remain in culture for longer periods of time with no proliferation (2.5–22 mo (Chen and Wang, 1999; Daugavet and Blinova, 2015; Odintsova et al., 2010)). Targeted cells for deriving cell lines are variable (Table 1) with the heart being one of the first choices because it is easily accessible after cutting the pericardium which isolates it from the pallial liquid content—hence, reducing the risk of microbial contamination when transferred to culture medium. Using the mantle, a single individual can generate a large number of explants (Daugavet and Blinova, 2015), and there is some evidence of cell propagation (Fang et al., 2008). Cell lines from hemocytes are especially appealing since they are the first line of defense against pathogens.
Hemocytes are also involved in transport and wound repair, and several protozoan parasites specifically residing in this cell type (e.g., Perkinsus spp., Bonamia spp. (Fernandez Robledo et al., 2014)), and they are also used for testing for environmental contaminants (reviewed in (Barrick et al., 2018)). Also crucial, the knowledge of the bivalve innate immune system is still lagging behind vertebrates and arthropods even when dealing with similar variety of pathogens (Cunningham and Robledo, 2015). Over decades, research in numerous labs has resulted in an in-depth knowledge of the biology of Perkinsus spp. (Fernandez Robledo et al., 2014). This genetically tractable protozoan parasite is becoming a model system for protozoan parasites in bivalves (Cold et al., 2016; Fernandez-Robledo et al., 2008) and it has potential for medical applications (Cold et al., 2017; Wijayalath et al., 2014). Mechanistic studies at the cellular level would also benefit from cell lines and a genetically tractable system.
Primary cell cultures, derived from explants or circulating cells transferred into a culture medium, lead to cell lines when they lose their limited proliferative capability and start replicating. Interfering with mechanisms that trigger cellular senescence is a strategy for cell immortalization. One involves the critical shortening of the telomeres by interfering with the end-replication during chromosome replication (Baker et al., 2017; Campisi, 2005; Loaiza and Demaria, 2016). The second involves cell cycle arrest, which is controlled by two tumor suppressor pathways (Ahuja et al., 2005; Lundberg et al., 2000; Ozer, 2000). The over-expression of hTERT into the cells has been used to immortalize cells from multiple vertebrate species (He et al., 2016; Veitonmaki et al., 2003; Wang et al., 2017b), but to our knowledge, it has never been attempted in bivalves. Interestingly, the telomeric sequence (TTAGGG)n target by the enzyme is present in numerous bivalves including Crassostrea spp. (Cross et al., 2005; Guo and Allen Jr, 1997; Huang et al., 2007; Koroleva et al., 2015; Perez-García et al., 2010; Plohl et al., 2002). Simian Virus 40 T antigen (SV40 T) is a simple and reliable agent for the immortalization of many different cell types by interfering with the inhibition of the p53 and Rb-family of tumor suppressor. The small T antigen’s action on the pp2A phosphatase p53 is a transcription factor and tumor suppressor involved in cell cycle arrest, apoptosis, or senescence in damaged cells; it is often mutated in human cancer (Chen and Wang, 1999), and it has been associated with cancers in bivalves (Barber, 2004; Farcy et al., 2008; Holbrook et al., 2009; Walker et al., 2011).
The pioneering efforts in the 1990’s to develop systems to deliver genetic material into oyster primary cell cultures and embryos included lipofection, electroporation, and microinjection (Table 3, Fig. 5). Lipofection of C. gigas heart primary cell cultures with plasmids coding for the luciferase gene under transcriptional control of several heterologous promoters (Drosophila hsp70 promoter, cytomegalovirus, CMV, and SV40 early promoters) resulted in enzymatic activity in the transfectants (Boulo et al., 1996). The Drosophila hsp70 was the stronger promoter, and the expression was inducible in a similar way to that observed in Drosophila (Boulo et al., 1996). A promoter including a 1,670 bp fragment upstream from the actin ORF from C. gigas was also validated (Cadoret et al., 1999). Transfection by electroporation of 3-hour-postfertilization C. virginica embryos was achieved using the commercial plasmid pS65T-C1, which carries the genes for neor (a bacterial gene that confers Neomycin resistance) and rsGFP (red-shifted Green Fluorescence Protein) under the SV40 and the immediate-early promoter from human CMV (Buchanan et al., 2001). PCR and expression of green fluorescent protein were observed in transfected embryos (20% viability after transfection) (Buchanan et al., 2001). Interestingly, hemocytes expressed the same plasmid when adult oysters were injected with the plasmid mixed with dendrimers (Buchanan et al., 2001). Recent developments in genome editing are providing new opportunities in basic and applied biological research. Recently, the genome of the gastropod Crepidula fornicata (the Atlantic slipper snail) was edited by microinjecting CRISPR/Cas9 mRNA, donor DNA, and sgRNA in embryos; the targeted gene was endogenous β-catenin, and the knock-in was the mCherry coding sequence (Perry and Henry, 2015). From a total of 280 embryos injected, a total of 30 (11%) had visible mCherry fluorescence.
Table 3.
Vectors and transfection methods used with bivalves.
| Organism | Target | Vector | Transfection | Reference |
|---|---|---|---|---|
| Crepidula fornicate (Gastropoda) | Embryos | Cas9 RNA, sgRNA, donor DNA | Microinjection | (Perry and Henry, 2015) |
| Crassostrea virginica | Adults | pS65T-C1 | Dendrimer | (Buchanan et al., 2001) |
| C. virginica | Embryos | pS65T-C1 | Electroporation | (Buchanan et al., 2001) |
| Crassostrea gigas | Heart explants | pGL3-basic* | Lipofection | (Cadoret et al., 1999) |
| C. gigas | Heart explants | pDr-luc, pC-luc, pCMV-L, pSV-luc | Lipofection | (Boulo et al., 1996) |
Based on Crassostrea gigas actin gene.
Ultimately, the generated cell lines, as well as the optimization of genetic tools and methodologies for manipulating the genome would make oysters a genetically tractable and appealing experimental system (Fig. 6). Achieving this goal would provide a critical resource for addressing multiple aspects of the oyster physiology, genetics, toxicology, virus infection dynamics, and host-parasite interactions. New cell lines could contribute to the prediction that “an ever-expanding breadth of model systems may be a hallmark of future cell biology” (Goldstein and King, 2016).
Fig. 6. Genetic material delivery approaches in bivalves.
In the absence of cell lines of marine invertebrates, embryos are the only stage with proliferating cells necessary for testing knocking in genes using CRISPR/Cas9 approaches. Available all year round, hemocytes and mantle explants are useful to test plasmid constructs carrying fluorescence tags. For delivering the genetic material, microinjection, electroporation, and lipofection are techniques that have been successfully used in the past. Confocal microscopy, flow cytometry, enzymatic activity, and PCR/sequencing can be used to characterize the transfectants.
4. Conclusions
The phylum Mollusca comprises around 200,000 extant species inhabiting many habitats. The bivalve diversity in size, anatomical structure, behavior, and habitat have fascinated scientists for centuries. Bivalves also are an important food source for humans. Here, we highlight several aspects of the bivalve biology that has the potential to impact human health. The last decade has seen the generation of draft genomes for most of the species with heavy aquaculture implantation and the field is now navigating the post-genomic era. Bivalve genomes are exceptionally rich in predicted genes, arguably derived from having evolved in an aquatic environment subjected to multiple environmental changes. In the review, we focus on aspects that have already received the attention of the scientific community; still, we believe that the biology and adaptation of these organisms hide clues to undercover mechanisms and strategies that have a direct application to human health. Bivalves as a model system is already a reality; however, to be able to attract students and junior scientists and engage funding agencies and foundations, we think that like in other well-established model organisms, developing a genetically tractable model system and cell lines are two much-needed tools.
Table 2.
Representative primary cell cultures derived from bivalves. DG: Digestive gland.
| Species | Tissue origin | Base medium | Year | Reference |
|---|---|---|---|---|
| Mytilus edulis | Hemocytes | L-15 | 2018 | (Barrick et al., 2018) |
| M. edulis | Mantle edge | L-15 | 2015 | (Daugavet and Blinova, 2015) |
| Mytilus trossulus | Embryos | L-15 | 2012 | (Odintsova et al., 2010) |
| Dreissena polymorpha | Hemocytes, heart, DG | L-15 | 2011 | (Parolini et al., 2011) |
| Mya arenaria | Disseminated Neoplasia | Eagle’s MEM | 2009 | (Walker et al., 2009) |
| Crassostrea virginica | Hemocytes | ASW/DME:HAMS F12 | 2009 | (Alavi et al., 2009) |
| Dreissena polymorpha | DG, gill, gonad | RPMI 1640, L-15 | 2009 | (Quinn et al., 2009) |
| Crassostrea gigas | Heart, mantle, DG | L-15 | 1999 | (Chen and Wang, 1999) |
| Pinctada fucata | Mantle edge | Balanced salt solution | 1998 | (Awaji and Suzuki, 1998) |
| C. gigas | Heart | L-15 | 1996 | (Boulo et al., 1996) |
| Ostrea edulis | Heart | L-15 | 1995 | (Renault et al., 1995) |
| C. gigas | Heart | L-15 | 1993 | (Wen et al., 1993) |
Bivalves as a model system for human health.
Shells in bone grafts and biomaterial design.
Mucosal immunity in invertebrates.
HABs and bivalves to learn about elimination of toxins.
Stem cells in invertebrates.
Oncogenic mechanisms and bivalve transmissible neoplasia.
Acknowledgements
This study was supported by institutional funds from Bigelow Laboratory for Ocean Sciences, by the Saltonstall-Kennedy Foundation Grant/NOAA # NA15NMF4270303 (JAFR), National Science Foundation #1701480 (JAFR, RY, ANT), National Institutes of Health K22 CA226047 (MJM), National Science Foundation Graduate Research Fellowship under Grant # 1244657 (RJS), the National Science Foundation Established Program to Stimulate Competitive Research under Cooperative Agreement Number OIA-1655221 (YZ, MGC, RJS) and the RI Research Alliance Collaborative Research Grant #AWD05730 (YZ, MGC).
Footnotes
Declarations of interest: none.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelrahman H, ElHady M, Alcivar-Warren A, Allen S, Al-Tobasei R, Bao L, Beck B, Blackburn H, Bosworth B, Buchanan J, Chappell J, Daniels W, Dong S, Dunham R, Durland E, Elaswad A, Gomez-Chiarri M, Gosh K, Guo X, Hackett P, Hanson T, Hedgecock D, Howard T, Holland L, Jackson M, Jin Y, Kahlil K, Kocher T, Leeds T, Li N, Lindsey L, Liu S, Liu Z, Martin K, Novriadi R, Odin R, Palti Y, Peatman E, Proestou D, Qin G, Reading B, Rexroad C, Roberts S, Salem M, Severin A, Shi H, Shoemaker C, Stiles S, Tan S, Tang KF, Thongda W, Tiersch T, Tomasso J, Prabowo WT, Vallejo R, van der Steen H, Vo K, Waldbieser G, Wang H, Wang X, Xiang J, Yang Y, Yant R, Yuan Z, Zeng Q, Zhou T, 2017. Aquaculture genomics, genetics and breeding in the United States: current status, challenges, and priorities for future research. BMC Genomics. 18, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abele D, Brey T, Philipp E, 2009. Bivalve models of aging and the determination of molluscan lifespans. Exp Gerontol 44, 307–315. [DOI] [PubMed] [Google Scholar]
- Abnave P, Ghigo E, 2018. Role of the immune system in regeneration and its dynamic interplay with adult stem cells. Semin Cell Dev Biol. [DOI] [PubMed] [Google Scholar]
- Acosta-Salmon H, Southgate PC, 2006. Wound healing after excision of mantle tissue from the Akoya pearl oyster, Pinctada fucata. Comp Biochem Physiol A Mol Integr Physiol 143, 264–268. [DOI] [PubMed] [Google Scholar]
- Agata K, Saito Y, Nakajima E, 2007. Unifying principles of regeneration I: Epimorphosis versus morphallaxis. Dev Growth Differ 49, 73–78. [DOI] [PubMed] [Google Scholar]
- Aguilera F, McDougall C, Degnan BM, 2017. Co-option and de novo gene evolution underlie molluscan shell diversity. Mol. Biol. Evol. 34, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja D, Saenz-Robles MT, Pipas JM, 2005. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 24, 7729–7745. [DOI] [PubMed] [Google Scholar]
- Alavi MR, Fernández Robledo JA, Vasta GR, 2009. Development of an in vitro assay to examine intracellular survival of Perkinsus marinus trophozoites upon phagocytosis by oyster (Crassostrea virginica and Crassostrea ariakensis) hemocytes. J Parasitol. 95, 900–907. [DOI] [PubMed] [Google Scholar]
- Alibardi L, Toni M, 2005. Wound keratins in the regenerating epidermis of lizard suggest that the wound reaction is similar in the tail and limb. J Exp Zool A Comp Exp Biol 303, 845–860. [DOI] [PubMed] [Google Scholar]
- Allam B, Carden WE, Ward JE, Ralph G, Winnicki S, Pales Espinosa E, 2013. Early host-pathogen interactions in marine bivalves: Evidence that the alveolate parasite Perkinsus marinus infects through the oyster mantle during rejection of pseudofeces. J Invertebr Pathol 113, 26–34. [DOI] [PubMed] [Google Scholar]
- Allam B, Paillard C, 1998. Defense factors in clam extrapallial fluids. DIS Aquat Organ. 33, 123–128. [Google Scholar]
- Allam B, Pales Espinosa E, 2015. Mucosal immunity in mollusks, in: Beck B, Peatman E (Eds.), Mucosal Health in Aquaculture. Academic Press, pp. 325–370. [Google Scholar]
- Allam B, Pales Espinosa E, 2016. Bivalve immunity and response to infections: Are we looking at the right place? Fish & shellfish immunology 53, 4–12. [DOI] [PubMed] [Google Scholar]
- Allam B, Pales Espinosa E, Tanguy A, Jeffroy F, Le Bris C, Paillard C, 2014. Transcriptional changes in Manila clam (Ruditapes philippinarum) in response to Brown Ring Disease. Fish & shellfish immunology 41,2–11. [DOI] [PubMed] [Google Scholar]
- Allam B, Raftos D, 2015. Immune responses to infectious diseases in bivalves. J. Invertebr. Pathol. 131, 121–136. [DOI] [PubMed] [Google Scholar]
- Almeida MJ, Pereira L, Milet C, Haigle J, Barbosa M, Lopez E, 2001. Comparative effects of nacre water-soluble matrix and dexamethasone on the alkaline phosphatase activity of MRC-5 fibroblasts. J. Biomed. Mater. Res. 57, 306–312. [DOI] [PubMed] [Google Scholar]
- Altamia MA, Wood N, Fung JM, Dedrick S, Linton EW, Concepcion GP, Haygood MG, Distel DL, 2014. Genetic differentiation among isolates of Teredinibacter turnerae, a widely occurring intracellular endosymbiont of shipworms. Mol Ecol 23, 1418–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, 1989. Toxic algal blooms and red tides: a global perspective, in: Okaichi T, D.M.A.T.N. (Ed.), Red Tides: Biology, Environmental Science and Toxicology. Elsevier Science Publishing Co., N.Y., pp. 1–16. [Google Scholar]
- Anderson DM, 2009. Approaches to monitoring, control and management of harmful algal blooms (HABs). Ocean Coast Manag 52, 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjum K, Abbas SQ, Akhter N, Shagufta BI, Shah SAA, Hassan SSU, 2017. Emerging biopharmaceuticals from bioactive peptides derived from marine organisms. Chem Biol Drug Des 90, 12–30. [DOI] [PubMed] [Google Scholar]
- Arenas G, Guzman F, Cardenas C, Mercado L, Marshall SH, 2009. A novel antifungal peptide designed from the primary structure of a natural antimicrobial peptide purified from Argopecten purpuratus hemocytes. Peptides 30, 1405–1411. [DOI] [PubMed] [Google Scholar]
- Arfken A, Song B, Bowman JS, Piehler M, 2017. Denitrification potential of the eastern oyster microbiome using a 16S rRNA gene based metabolic inference approach. PLoS One 12, e0185071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arivalagan J, Yarra T, Marie B, Sleight VA, Duvernois-Berthet E, Clark MS, Marie A, Berland S, 2017. Insights from the shell proteome: Biomineralization to adaptation. Mol. Biol. Evol. 34, 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriagada G, Metzger MJ, Muttray AF, Sherry J, Reinisch C, Street C, Lipkin WI, Goff SP, 2014. Activation of transcription and retrotransposition of a novel retroelement, Steamer, in neoplastic hemocytes of the mollusk Mya arenaria. Proc. Natl. Acad. Sci. USA. 111, 14175–14180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmani K, Petton B, Le Grand J, Mounier J, Robert R, Nicolas J-L, 2016. Establishment of microbiota in larval culture of Pacific oyster, Crassostrea gigas. Aquaculture 464, 434–444. [Google Scholar]
- Awaji M, Suzuki T, 1998. Monolayer formation and DNA synthesis of the outer epithelial cells from pearl oyster mantle in coculture with amebocytes. In Vitro Cell Dev. Biol. Anim. 34, 486–491. [DOI] [PubMed] [Google Scholar]
- Bachére E, Rosa RD, Schmitt P, Poirier AC, Merou N, Charriere GM, Destoumieux-Garzon D, 2015. The new insights into the oyster antimicrobial defense: Cellular, molecular and genetic view. Fish Shellfish Immunol. 46, 50–64. [DOI] [PubMed] [Google Scholar]
- Baden DG, Trainer VL, 1993. Mode of action of toxins of seafood poisoning, in: Falconer IR (Ed.), Algal Toxins in Seafood and Drinking Water. Academic Press, N.Y., pp. 49–74. [Google Scholar]
- Baker DJ, Alimirah F, van Deursen JM, Campisi J, Hildesheim J, 2017. Oncogenic senescence: a multi-functional perspective. Oncotarget 8, 27661–27672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balseiro P, Falco A, Romero A, Dios S, Martinez-Lopez A, Figueras A, Estepa A, Novoa B, 2011. Mytilus galloprovincialis myticin C: a chemotactic molecule with antiviral activity and immunoregulatory properties. PLoS One 6, e23140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin E, Israely T, Fine M, Loya Y, Rosenberg E, 2001. Role of endosymbiotic zooxanthellae and coral mucus in the adhesion of the coral-bleaching pathogen Vibrio shiloi to its host. FEMS Microbiol Lett 199, 33–37. [DOI] [PubMed] [Google Scholar]
- Barber BJ, 2004. Neoplastic diseases of commercially important marine bivalves. Aquat. Living Resour. 17, 449–466. [Google Scholar]
- Bardouil M, Bohec M, Cormerais M, Bougrier S, Patrick L, 1993. Experimental study of the effect of a toxic microalgal diet on feeding of the oyster Crassostrea gigas Thunberg. J shellfish Res. 12, 417–422. [Google Scholar]
- Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, Stotland A, Wolkowicz R, Cutting AS, Doran KS, Salamon P, Youle M, Rohwer F, 2013. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci U S A. 110, 10771–10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett DG, Bushnell GG, Messersmith PB, 2013. Mechanically robust, negative-swelling, mussel-inspired tissue adhesives. Adv Healthc Mater 2, 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick A, Guillet C, Mouneyrac C, Chatel A, 2018. Investigating the establishment of primary cultures of hemocytes from Mytilus edulis. Cytotechnology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros I, Divya B, Martins I, Vandeperre F, Santos R, Bettencourt R, 2014. Post-capture immune gene expression studies in the deep-sea hydrothermal vent mussel Bathymodiolus azoricus acclimatized to atmospheric pressure. Fish Shellfish Immun. 42, 159–170. [DOI] [PubMed] [Google Scholar]
- Basti L, 2011. Effects of the toxic dinoflagellate Heterocapsa circularisquama on larvae of the pearl oyster Pinctada fucata martensii (Dunker, 1873). J Shellfish Res. 30, 177–186. [Google Scholar]
- Basti L, Nagai K, Go J, Okano S, Oda T, Tanaka Y, Nagai S, 2016. Lethal effects of ichthyotoxic raphidophytes, Chattonella marina, C. antiqua, and Heterosigma akashiwo, on post-embryonic stages of the Japanese pearl oyster, Pinctada fucata martensii. Harmful Algae 59, 112–122. [DOI] [PubMed] [Google Scholar]
- Basti L, Nagai K, Shimasaki Y, Oshima Y, Honjo T, Segawa S, 2009. Effects of the toxic dinoflagellate Heterocapsa circularisquama on the valve movement behaviour of the Manila clam Ruditapes philippinarum. Aquaculture 291,41–47. [Google Scholar]
- Bely AE, Nyberg KG, 2010. Evolution of animal regeneration: re-emergence of a field. Trends Ecol Evol 25, 161–170. [DOI] [PubMed] [Google Scholar]
- Beninger PG, Lynn JW, Dietz TH, Silverman H, 1997. Mucociliary transport in living tissue: The two-layer model confirmed in the mussel Mytilus edulis L. Biol Bull 193, 4–7. [DOI] [PubMed] [Google Scholar]
- Bentzon-Tilia M, Sonnenschein EC, Gram L, 2016. Monitoring and managing microbes in aquaculture - Towards a sustainable industry. Microb Biotechnol 9, 576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beschin A, Bilej M, Torreele E, De Baetselier P, 2001. On the existence of cytokines in invertebrates. Cell Mol Life Sci 58, 801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blay C, Planes S, Ky CL, 2017. Donor and recipient contribution to phenotypic traits and the expression of biomineralisation genes in the pearl oyster model Pinctada margaritifera. Sci. Rep. 7, 2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode A, Salvenmoser W, Nimeth K, Mahlknecht M, Adamski Z, Rieger RM, Peter R, Ladurner P, 2006. Immunogold-labeled S-phase neoblasts, total neoblast number, their distribution, and evidence for arrested neoblasts in Macrostomum lignano (Platyhelminthes, Rhabditophora). Cell Tissue Res 325, 577–587. [DOI] [PubMed] [Google Scholar]
- Boehs G, Villalba A, Ceuta LO, Luz JR, 2010. Parasites of three commercially exploited bivalve mollusc species of the estuarine region of the Cachoeira river (Ilheus, Bahia, Brazil). J INVertebr Pathol. 103, 43–47. [DOI] [PubMed] [Google Scholar]
- Borcier E, Morvezen R, Boudry P, Miner P, Charrier G, Laroche J, Hégaret H, 2017. Effects of bioactive extracellular compounds and paralytic shellfish toxins produced by Alexandrium minutum on growth and behaviour of juvenile great scallops Pecten maximus. Aquat Toxicol 184, 142–154. [DOI] [PubMed] [Google Scholar]
- Boulo V, Cadoret JP, Le Marrec F, Dorange G, Miahle E, 1996. Transient expression of luciferase reporter gene after lipofection in oyster (Crassostrea gigas) primary cell cultures. Mol Mar Biol Biotechnol 5, 167–174. [PubMed] [Google Scholar]
- Bour A, Haarr A, Keiter S, Hylland K, 2018. Environmentally relevant microplastic exposure affects sediment-dwelling bivalves. Environ Pollut 236, 652–660. [DOI] [PubMed] [Google Scholar]
- Bray BL, 2003. Large-scale manufacture of peptide therapeutics by chemical synthesis. Nat Rev Drug Discov 2, 587–593. [DOI] [PubMed] [Google Scholar]
- Brion A, Zhang G, Dossot M, Moby V, Dumas D, Hupont S, Piet MH, Bianchi A, Mainard D, Galois L, Gillet P, Rousseau M, 2015. Nacre extract restores the mineralization capacity of subchondral osteoarthritis osteoblasts. J. Struct. Biol. 192, 500–509. [DOI] [PubMed] [Google Scholar]
- Browne MA, Dissanayake A, Galloway TS, Lowe DM, Thompson RC, 2008. Ingested microscopic plastic translocates to the circulatory system of the mussel, Mytilus edulis (L). Environ Sci Technol 42, 5026–5031. [DOI] [PubMed] [Google Scholar]
- Brun NT, Ross NW, Boghen AD, 2000. Changes in the electrophoretic profiles of gill mucus proteases of the eastern oyster Crassostrea virginica in response to infection by the turbellarian Urastoma cyprinae. J Invertebr Pathol 75, 163–170. [DOI] [PubMed] [Google Scholar]
- Buchanan JT, 1999. Production of transgenic eastern oysters, The Department of Oceanography and Coastal Sciences. Louisiana State University and Agricultural and Mechanical College, USA, pp. 1–255. [Google Scholar]
- Buchanan JT, Nickens AD, Cooper RK, Tiersch TR, 2001. Transfection of eastern oyster (Crassotrea virginica) embryos. Mar Biotechnol. (NY) 3, 322–335. [DOI] [PubMed] [Google Scholar]
- Bulgheresi S, Schabussova I, Chen T, Mullin NP, Maizels RM, Ott JA, 2006. A new C-type lectin similar to the human immunoreceptor DC-SIGN mediates symbiont acquisition by a marine nematode. Appl Environ Microbiol 72, 2950–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge CA, Closek CJ, Friedman CS, Groner ML, Jenkins CM, Shore-Maggio A, Welsh JE, 2016. The use of filter-feeders to manage disease in a changing world. Integr Comp Biol 56, 573–587. [DOI] [PubMed] [Google Scholar]
- Burkholder JM, Glibert PM, 2006. Intraspecific variability: an important consideration in forming generalisations about toxigenic algal species. African J Mar Sci 28, 177–180. [Google Scholar]
- Burreson EM, Ford SE, 2004. A review of recent information on the Haplosporidia, with special reference to Haplosporidium nelsoni (MSX disease). Aquat Living Resour 17, 499–517. [Google Scholar]
- Cadoret JP, Debon R, Cornudella L, Lardans V, Morvan A, Roch P, Boulo V, 1999. Transient expression assays with the proximal promoter of a newly characterized actin gene from the oyster Crassostrea gigas. FEBS letters 460, 81–85. [DOI] [PubMed] [Google Scholar]
- Campisi J, 2005. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120, 513–522. [DOI] [PubMed] [Google Scholar]
- Carballal MJ, Barber BJ, Iglesias D, Villalba A, 2015. Neoplastic diseases of marine bivalves. J Invertebr Pathol 131,83–106. [DOI] [PubMed] [Google Scholar]
- Carriel-Gomes MC, Kratz JM, Barracco MA, Bachere E, Barardi CR, Simoes CM, 2007. In vitro antiviral activity of antimicrobial peptides against herpes simplex virus 1, adenovirus, and rotavirus. Mem Inst Oswaldo Cruz 102, 469–472. [DOI] [PubMed] [Google Scholar]
- Carrier TJ, Reitzel AM, 2017. The hologenome across environments and the implications of a host-associated microbial repertoire. Front Microbiol 8, 802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriker MR, 2009. The Shell and Ligament, in: Kennedy VSN, R. I. E.; Eble AF (Ed.), The Eastern Oyster Crassostrea virginica. Maryland Sea Grant College, College Park, Maryland, USA, pp. 75–168. [Google Scholar]
- Castrec J, Soudant P, Payton L, Tran D, Miner P, Lambert C, Le Goic N, Huvet A, Quillien V, Boullot F, Amzil Z, Hegaret H, Fabioux C, 2018. Bioactive extracellular compounds produced by the dinoflagellate Alexandrium minutum are highly detrimental for oysters. Aquat Toxicol 199, 188–198. [DOI] [PubMed] [Google Scholar]
- Catapane EJ, Stefano G, Aiello E, 1978. Pharmacological study of the reciprocal dual innervation of the lateral ciliated gill epithelium by the CNS of Mytilus edulis (Bivalvia). J Exp Biol 74, 101–113. [Google Scholar]
- Cavelier P, Cau J, Morin N, Delsert C, 2017. Early gametogenesis in the Pacific oyster: new insights using stem cell and mitotic markers. J Exp Biol 220, 3988–3996. [DOI] [PubMed] [Google Scholar]
- Cereghino JL, Cregg JM, 2000. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev 24, 45–66. [DOI] [PubMed] [Google Scholar]
- Chagot D, Boulo V, Hervio D, Mialhe E, Bachere E, Mourton C, Grizel H, 1992. Interactions between Bonamia ostreae (Protozoa: Ascetospora) and hemocytes of Ostrea edulis and Crassostrea gigas (Mollusca: Bivalvia): Entry mechanisms. J Invertebr Pathol 59, 241–249. [Google Scholar]
- Chalk R, Townson H, Ham PJ, 1995. Brugia pahangi: the effects of cecropins on microfilariae in vitro and in Aedes aegypti. Exp Parasitol 80, 401–406. [DOI] [PubMed] [Google Scholar]
- Chauhan A, Wafula D, Lewis DE, Pathak A, 2014. Metagenomic assessment of the eastern oyster-associated microbiota. Genome announcements 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SN, Wang CS, 1999. Establishment of cell lines derived from oyster, Crassostrea gigas Thunberg and hard clam, Meretrix lusoria Roding. Methods Cell Sci 21, 183–192. [DOI] [PubMed] [Google Scholar]
- Cima F, Ballarin L, Gasparini F, Burighel P, 2006. External amebocytes guard the pharynx entry in a tunicate (Ascidiacea). Developmental and comparative immunology 30, 463–472. [DOI] [PubMed] [Google Scholar]
- Cold ER, Freyria NJ, Martínez Martínez J, Fernández Robledo JA, 2016. An agar-based method for plating marine protozoan parasites of the genus Perkinsus. PLoS One 11, e0155015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cold ER, Vasta GR, Fernández Robledo JA, 2017. Transient expression of Plasmodium berghei MSP8 and HAP2 in the marine protozoan parasite Perkinsus marinus. J Parasitol. 103, 118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross I, Díaz E, Sánchez I, Rebordinos L, 2005. Molecular and cytogenetic characterization of Crassostrea angulata chromosomes. Aquaculture 247, 135–144. [Google Scholar]
- Cunningham C, Robledo JA, 2015. Molluscan immunology. Fish Shellfish Immunol 46, 1. [DOI] [PubMed] [Google Scholar]
- Dahl SF, Thiel J, Allam B, 2010. Field performance and QPX disease progress in cultured and wild-type strains of Mercenaria mercenaria in New York waters. J Shellfish Res 29, 83–90. [Google Scholar]
- Daugavet MA, Blinova MI, 2015. Culture of mussel Mytiuls edulis I. mantle cells. Tsitologiia 57, 153–161. [PubMed] [Google Scholar]
- De Wit P, Durland E, Ventura A, Langdon CJ, 2018. Gene expression correlated with delay in shell formation in larval Pacific oysters (Crassostrea gigas) exposed to experimental ocean acidification provides insights into shell formation mechanisms. BMC Genomics. 19, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JE, Belov K, 2012. A comparative genomics approach to understanding transmissible cancer in Tasmanian devils. Annu Rev Genomics Hum Genet 13, 207–222. [DOI] [PubMed] [Google Scholar]
- Desbois AP, 2014. How might we increase success in marine-based drug discovery? Expert Opin Drug Discov 9, 985–990. [DOI] [PubMed] [Google Scholar]
- Destoumieux-Garzon D, Rosa RD, Schmitt P, Barreto C, Vidal-Dupiol J, Mitta G, Gueguen Y, Bachere E, 2016. Antimicrobial peptides in marine invertebrate health and disease. Philos Trans R Soc Lond B Biol Sci 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detree C, Gallardo-Escarate C, 2018. Single and repetitive microplastics exposures induce immune system modulation and homeostasis alteration in the edible mussel Mytilus galloprovincialis. Fish Shellfish Immunol 83, 52–60. [DOI] [PubMed] [Google Scholar]
- Di Baldassarre A, Cimetta E, Bollini S, Gaggi G, Ghinassi B, 2018. Human-induced pluripotent stem cell technology and cardiomyocyte generation: Progress and clinical applications. Cells 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias Rde O, Franco OL, 2015. Cysteine-stabilized alphabeta defensins: From a common fold to antibacterial activity. Peptides 72, 64–72. [DOI] [PubMed] [Google Scholar]
- Domeneghetti S, Franzoi M, Damiano N, Norante R, El Halfawy NM, Mammi S, Marin O, Bellanda M, Venier P, 2015. Structural and antimicrobial features of peptides related to Myticin C, a special defense molecule from the Mediterranean mussel Mytilus galloprovincialis. J Agric Food Chem 63, 9251–9259. [DOI] [PubMed] [Google Scholar]
- Douglas AE, 2018. Fundamentals of Microbiome Science: How Microbes Shape Animal Biology. University Press, Princeton. [Google Scholar]
- Douglas AE, Werren JH, 2016. Holes in the Hologenome: Why Host-Microbe Symbioses Are Not Holobionts. mBio 7, e02099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Song K, Wang J, Cong R, Li L, Zhang G, 2017. Draft genome and SNPs associated with carotenoid accumulation in adductor muscles of bay scallop (Argopecten irradians). J Genomics 5, 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubilier N, Bergin C, Lott C, 2008. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol 6, 725–740. [DOI] [PubMed] [Google Scholar]
- Ducklow Hugh W, Mitchell R, 1979. Bacterial populations and adaptations in the mucus layers on living corals. Limnol Oceanogr 24, 715–725. [Google Scholar]
- Dufault-Thompson K, Jian H, Cheng R, Li J, Wang F, Zhang Y, 2017. A Genome-Scale Model of Shewanella piezotolerans Simulates Mechanisms of Metabolic Diversity and Energy Conservation. mSystems 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour SC, 2005. Gill anatomy and the evolution of symbiosis in the bivalve family Thyasiridae. Biol Bull 208, 200–212. [DOI] [PubMed] [Google Scholar]
- Duncan HE, Edberg SC, 1995. Host-microbe interaction in the gastrointestinal tract. Crit Rev Microbiol 21,85–100. [DOI] [PubMed] [Google Scholar]
- Dupuy JW, Bonami JR, Roch P, 2004. A synthetic antibacterial peptide from Mytilus galloprovincialis reduces mortality due to white spot syndrome virus in palaemonid shrimp. J Fish Dis 27, 57–64. [DOI] [PubMed] [Google Scholar]
- Ebran N, Julien S, Orange N, Saglio P, Lemaitre C, Molle G, 1999. Pore-forming properties and antibacterial activity of proteins extracted from epidermal mucus of fish. Comp Biochem Physiol A Mol Integr Physiol 122, 181–189. [DOI] [PubMed] [Google Scholar]
- Egan S, Gardiner M, 2016. Microbial Dysbiosis: Rethinking Disease in Marine Ecosystems. Front Microbiol 7, 991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elisabeth NH, Caro A, Cesaire T, Mansot JL, Escalas A, Sylvestre MN, Jean-Louis P, Gros O, 2014. Comparative modifications in bacterial gill-endosymbiotic populations of the two bivalves Codakia orbiculata and Lucina pensylvanica during bacterial loss and reacquisition. FEMS Microbiol Ecol 89, 646–658. [DOI] [PubMed] [Google Scholar]
- Erickson JR, Echeverri K, 2018. Learning from regeneration research organisms: The circuitous road to scar free wound healing. Dev Biol 433, 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabioux C, Sulistiyani Y, Haberkorn H, Hegaret H, Amzil Z, Soudant P, 2015. Exposure to toxic Alexandrium minutum activates the detoxifying and antioxidant systems in gills of the oyster Crassostrea gigas. Harmful Algae 48, 55–62. [DOI] [PubMed] [Google Scholar]
- Fang Z, Feng Q, Chi Y, Xie L, Zhang R, 2008. Investigation of cell proliferation and differentiation in the mantle of Pinctada fucata (Bivalve, Mollusca). Marine Biology 153, 745–754. [Google Scholar]
- FAO, 2018. http://www.fao.org/3/a-i7989t.pdf.
- Farabegoli F, Blanco L, Rodriguez LP, Vieites JM, Cabado AG, 2018. Phycotoxins in marine shellfish: Origin, occurrence and effects on humans. Mar Drugs 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcy E, Fleury C, Lelong C, Dubos MP, Voiseux C, Fievet B, Lebel JM, 2008. Molecular cloning of a new member of the p53 family from the Pacific oyster Crassostrea gigas and seasonal pattern of its transcriptional expression level. Mar Environ Res 66, 300–308. [DOI] [PubMed] [Google Scholar]
- Fathke C, Wilson L, Shah K, Kim B, Hocking A, Moon R, Isik F, 2006. Wnt signaling induces epithelial differentiation during cutaneous wound healing. BMC Cell Biol 7, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feis ME, John U, Lokmer A, Luttikhuizen PC, Wegner KM, 2018. Dual transcriptomics reveals co-evolutionary mechanisms of intestinal parasite infections in blue mussels Mytilus edulis. Mol Ecol 27, 1505–1519. [DOI] [PubMed] [Google Scholar]
- Feng C, Ghosh A, Amin MN, Bachvaroff TR, Tasumi S, Pasek M, Banerjee A, Shridhar S, Wang LX, Bianchet MA, Vasta GR, 2015. Galectin CvGal2 from the eastern oyster (Crassostrea virginica) displays unique specificity for ABH blood group oligosaccharides and differentially recognizes sympatric Perkinsus species. Biochemistry 54, 4711–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández Robledo JA, Vasta GR, Record NR, 2014. Protozoan parasites of bivalve molluscs: Literature follows culture. PLoS One 9, e100872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Reiriz MJ, Navarro JM, Contreras AM, Labarta U, 2008. Trophic interactions between the toxic dinoflagellate Alexandrium catenella and Mytilus chilensis: feeding and digestive behaviour to long-term exposure. Aquat Toxicol 87, 245–251. [DOI] [PubMed] [Google Scholar]
- Fernández-Robledo JA, Lin Z, Vasta GR, 2008. Transfection of the protozoan parasite Perkinsus marinus. Mol Bioch Parasitol 157, 44–53. [DOI] [PubMed] [Google Scholar]
- Ferreira JG, Hawkins AJS, Bricker SB, 2007. Management of productivity, environmental effects and profitability of shellfish aquaculture — the Farm Aquaculture Resource Management (FARM) model. Aquaculture 264, 160–174. [Google Scholar]
- Ford S,E, Bricelj VM, Lambert C, Paillard C, 2008. Deleterious effects of a nonPST bioactive compound(s) from Alexandrium tamarense on bivalve hemocytes. Mar Biol 154, 241–253. [Google Scholar]
- Ford SE, Chintala MM, Bushek D, 2002. Comparison of in vitro-cultured and wild-type Perkinsus marinus. I. Pathogen virulence. Dis Aquat Organ 51, 187–201. [DOI] [PubMed] [Google Scholar]
- Fry TJ, Mackall CL, 2002. Interleukin-7 and immunorestoration in HIV: beyond the thymus. J Hematother Stem Cell Res 11,803–807. [DOI] [PubMed] [Google Scholar]
- Funabara D, Ohmori F, Kinoshita S, Koyama H, Mizutani S, Ota A, Osakabe Y, Nagai K, Maeyama K, Okamoto K, Kanoh S, Asakawa S, Watabe S, 2014. Novel genes participating in the formation of prismatic and nacreous layers in the pearl oyster as revealed by their tissue distribution and RNA interference knockdown. PLoS One 9, e84706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi T, Schwarzinger C, Miksik I, Smrz M, Beran A, 2009. Molluscan shell evolution with review of shell calcification hypothesis. Comp Biochem Physiol B Biochem Mol Biol 154, 351–371. [DOI] [PubMed] [Google Scholar]
- Gainey LF Jr, Shumway SE, 1988. Physiological effects of Protogonyaulax tamarensis on cardiac activity in bivalve molluscs. Comp Bioch Physiol Part C, Comp 91, 159–164. [Google Scholar]
- Galdiero S, Falanga A, Berisio R, Grieco P, Morelli G, Galdiero M, 2015. Antimicrobial peptides as an opportunity against bacterial diseases. Curr Med Chem 22, 1665–1677. [DOI] [PubMed] [Google Scholar]
- Galimany E, Sunila I, Hégaret H, Ramon M, Wikfors G,H,, 2008. Pathology and immune response of the blue mussel (Mytilus edulis L.) after an exposure to the harmful dinoflagellate Prorocentrum minimum. Harmful Algae 7, 630–638. [Google Scholar]
- Galloway TS, Cole M, Lewis C, 2017. Interactions of microplastic debris throughout the marine ecosystem. Nat Ecol Evol 1, 116. [DOI] [PubMed] [Google Scholar]
- Galloway TS, Lewis CN, 2016. Marine microplastics spell big problems for future generations. Proc Natl Acad Sci U S A. 113, 2331–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandara e Silva PP, Nobre CR, Resaffe P, Pereira CDS, Gusmao F, 2016. Leachate from microplastics impairs larval development in brown mussels. Water Research 106, 364–370. [DOI] [PubMed] [Google Scholar]
- Garcia C, Perez F, Contreras C, Figueroa D, Barriga A, López-Rivera A, Araneda OF, Contreras HR, 2015. Saxitoxins and okadaic acid group: accumulation and distribution in invertebrate marine vectors from Southern Chile. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 32, 984–1002. [DOI] [PubMed] [Google Scholar]
- Garcia-Lagunas N, Romero-Geraldo R, Hernandez-Saavedra NY, 2013. Genomics study of the exposure effect of Gymnodinium catenatum, a paralyzing toxin producer, on Crassostrea gigas’ defense system and detoxification genes. PLoS One 8, e72323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier-Clerc S, Boily I, Fournier M, Lemarchand K, 2013. In vivo exposure of Mytilus edulis to living enteric bacteria: a threat for immune competency? Environ Sci Pollut Res Int 20, 612–620. [DOI] [PubMed] [Google Scholar]
- Gehrke AR, Srivastava M, 2016. Neoblasts and the evolution of whole-body regeneration. Curr Opin Genet Dev 40, 131–137. [DOI] [PubMed] [Google Scholar]
- Gerdol M, 2017. Immune-related genes in gastropods and bivalves: a comparative overview. Invertebr. Surviv. J. 14, 95–111. [Google Scholar]
- Gerdol M, Gómez-Chiarri M, Castillo MG, Figueras A, Fiorito G, Moreira R, Novoa B, Pallavicini A, Ponte G, Roumbedakis K, Venier P, Vasta GR, 2018. Immunity in Molluscs: Recognition and Effector Mechanisms, with a Focus on Bivalvia, in: Cooper ELE (Ed.), Advances in Comparative Immunology. Springer International Publishing, Cham, pp. 225–341. [Google Scholar]
- Gerdol M, Venier P, 2015. An updated molecular basis for mussel immunity. Fish Shellfish Immunol. 46, 17–38. [DOI] [PubMed] [Google Scholar]
- Gerhard EM, Wang W, Li C, Guo J, Ozbolat IT, Rahn KM, Armstrong AD, Xia J, Qian G, Yang J, 2017. Design strategies and applications of nacre-based biomaterials. Acta Biomater 54, 21–34. [DOI] [PubMed] [Google Scholar]
- Giuliani A, Pirri G, Nicoletto S, 2007. Antimicrobial peptides: an overview of a promising class of therapeutics, Open Life Sciences, p. 1. [Google Scholar]
- Goldstein B, King N, 2016. The duture of xell viology: Emerging model organisms. Trends Cell Biol 26, 818–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Chiarri M, Guo X, Tanguy A, He Y, Proestou D, 2015a. The use of -omic tools in the study of disease processes in marine bivalve mollusks. J. Invertebr. Pathol. 131, 137–154. [DOI] [PubMed] [Google Scholar]
- Gómez-Chiarri M, Warren WC, Guo X, Proestou D, 2015b. Developing tools for the study of molluscan immunity: The sequencing of the genome of the eastern oyster, Crassostrea virginica. Fish Shellfish Immunol 46, 2–4. [DOI] [PubMed] [Google Scholar]
- Gordon MS, McCaskill-Stevens WJ, Battiato LA, Loewy J, Loesch D, Breeden E, Hoffman R, Beach KJ, Kuca B, Kaye J, Sledge GW Jr., 1996. A phase I trial of recombinant human interleukin-11 (neumega rhIL-11 growth factor) in women with breast cancer receiving chemotherapy. Blood 87, 3615–3624. [PubMed] [Google Scholar]
- Greaves M, Maley CC, 2012. Clonal evolution in cancer. Nature 481,306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DS, 2016. Effects of microplastics on European flat oysters, Ostrea edulis and their associated benthic communities. Environ Pollut 216, 95–103. [DOI] [PubMed] [Google Scholar]
- Green TJ, Barnes AC, 2010. Bacterial diversity of the digestive gland of Sydney rock oysters, Saccostrea glomerata infected with the paramyxean parasite, Marteilia sydneyi. J Appl Microbiol 109, 613–622. [DOI] [PubMed] [Google Scholar]
- Guéguen M, Michele B, Baron R, Patrick L, Philippe T, Julie M, Amzil Z, 2008. Detoxification of Pacific oyster Crassostrea gigas fed on diets of Skeletonema costatum with and without silt, following PSP contamination by Alexandrium minutum. Aquatic Liv Res 21. [Google Scholar]
- Gueguen Y, Bernard R, Julie F, Paulina S, Delphine DG, Franck V, Philippe B, Evelyne B, 2009. Oyster hemocytes express a proline-rich peptide displaying synergistic antimicrobial activity with a defensin. Mol Immunol 46, 516–522. [DOI] [PubMed] [Google Scholar]
- Guo X, Allen SK Jr, 1997. Fluorescence in situ hybridization of vertebrate telomere sequence to chromosome ends of the Pacific oyster Crassostrea gigas Thunberg. J Shellfish Res 6, 87–89. [Google Scholar]
- Gurdon JB, 2017. Nuclear transplantation, the conservation of the genome, and prospects for cell replacement. FEBS J. 284, 211–217. [DOI] [PubMed] [Google Scholar]
- Haberkorn H, Lambert C, Go’i’c N, Guéguen Minerbe M, Moal J, Palacios E, Patrick L, Soudant P, 2010a. Effects of Alexandrium minutum exposure upon physiological and hematological variables of diploid and triploid oysters, Crassostrea gigas. Harmful Algae 97, 96–108. [DOI] [PubMed] [Google Scholar]
- Haberkorn H, Lambert C, Go’i’c N, Moal J, Suquet M, Guéguen Minerbe M, Sunila I, Soudant P, 2010b. Effects of Alexandrium minutum exposure on nutrition-related processes and reproductive output in oysters Crassostrea gigas. Harmful Algae 9, 427–439. [Google Scholar]
- Haberkorn H, Tran D, Massabuau JC, Ciret P, Savar V, Soudant P, 2011. Relationship between valve activity, microalgae concentration in the water and toxin accumulation in the digestive gland of the Pacific oyster Crassostrea gigas exposed to Alexandrium minutum. Mar Pollut Bull 62, 1191–1197. [DOI] [PubMed] [Google Scholar]
- Hallegraeff G, 2010. Ocean climate change, phytoplankton community responses, and harmful algal blooms: A formidable predictive challenge. Journal of phycology 46, 220–235. [Google Scholar]
- Han SY, Lee JR, Kwon YK, Jo MJ, Park SJ, Kim SC, Lee HS, Ku SK, 2007. Ostreae Testa prevent ovariectomy-induced bone loss in mice by osteoblast activations. J Ethnopharmacol 114, 400–405. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA, 2000. The hallmarks of cancer. Cell 100, 57–70. [DOI] [PubMed] [Google Scholar]
- Hancock RE, Sahl HG, 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol 24, 1551–1557. [DOI] [PubMed] [Google Scholar]
- He S, Li Y, Chen Y, Zhu Y, Zhang X, Xia X, Sun H, 2016. Immortalization of pig fibroblast cells by transposon-mediated ectopic expression of porcine telomerase reverse transcriptase. Cytotechnology 68, 1435–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegaret H, da Silva PM, Wikfors GH, Haberkorn H, Shumway SE, Soudant P, 2011. In vitro interactions between several species of harmful algae and haemocytes of bivalve molluscs. Cell Biol Toxicol 27, 249–266. [DOI] [PubMed] [Google Scholar]
- Hégaret H, Wikfors G, 2005a. Time-dependent changes in hemocytes of eastern oysters, Crassostrea virginica, and northern bay scallops, Argopecten irradians irradians, exposed to a cultured strain of Prorocentrum minimum. Harmful Algae 4, 187–199. [Google Scholar]
- Hégaret H, Wikfors G, Soudant P, Lambert C, Shumway SE, Berard J-B, Patrick L, 2007a. Toxic dinoflagellates (Alexandrium fundyense and A. catenella) have minimal apparent effect on oyster hemocytes. Mar Bioll 152, 441–447. [Google Scholar]
- Hégaret H, Wikfors GH, 2005b. Effects of natural and field-simulated blooms of the dinoflagellate Prorocentrum minimum upon hemocytes of eastern oysters, Crassostrea virginica, from two different populations. Harmful Algae 4, 201–209. [Google Scholar]
- Hégaret H, Wikfors GH, Shumway SE, 2007b. Diverse feeding responses of five species of bivalve mollusc when exposed to three species of harmful algae. J Shellfish Res 26, 549–559. [Google Scholar]
- Holbrook LA, Butler RA, Cashon RE, Van Beneden RJ, 2009. Soft-shell clam (Mya arenaria) p53: a structural and functional comparison to human p53. Gene 433, 81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houschyar KS, Momeni A, Pyles MN, Maan ZN, Whittam AJ, Siemers F, 2015. Wnt signaling induces epithelial differentiation during cutaneous wound healing. Organogenesis 11,95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Li S, Liu Y, Liu C, Xie L, Zhang R, 2018. Hemocytes in the extrapallial space of Pinctada fucata are involved in immunity and biomineralization. Sci. Rep. 8, 4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Hu X, Hu J, Zhang L, Wang S, Lu W, Bao Z, 2007. Mapping of ribosomal DNA and (TTAGGG)n telomeric sequence by FISH in the bivalve Patinopecten yessoensis (Jay, 1857). J Molluscan Stud 73, 393–398. [Google Scholar]
- Hubert F, Noel T, Roch P, 1996. A member of the arthropod defensin family from edible Mediterranean mussels (Mytilus galloprovincialis). Eur J Biochem 240, 302–306. [DOI] [PubMed] [Google Scholar]
- Hughes CC, Fenical W, 2010. Antibacterials from the sea. Chemistry 16, 12512–12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huning AK, Lange SM, Ramesh K, Jacob DE, Jackson DJ, Panknin U, Gutowska MA, Philipp EE, Rosenstiel P, Lucassen M, Melzner F, 2016. A shell regeneration assay to identify biomineralization candidate genes in mytilid mussels. Mar Genomics 27, 57–67. [DOI] [PubMed] [Google Scholar]
- Ingham AB, Moore RJ, 2007. Recombinant production of antimicrobial peptides in heterologous microbial systems. Biotechnol Appl Biochem 47, 1–9. [DOI] [PubMed] [Google Scholar]
- Ivanina AV, Dickinson GH, Matoo OB, Bagwe R, Dickinson A, Beniash E, Sokolova IM, 2013. Interactive effects of elevated temperature and CO2 levels on energy metabolism and biomineralization of marine bivalves Crassostrea virginica and Mercenaria mercenaria. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 166, 101–111. [DOI] [PubMed] [Google Scholar]
- Ivanina AV, Falfushynska HI, Beniash E, Piontkivska H, Sokolova IM, 2017. Biomineralization-related specialization of hemocytes and mantle tissues of the Pacific oyster Crassostrea gigas. J. Exp. Biol. 220, 3209–3221. [DOI] [PubMed] [Google Scholar]
- Janeway CA, 1994. The role of microbial pattern recognition in self: nonself discrimination in innate and adaptive immunity, in: Hoffmann JA, Janeway CA, Natori S (Ed.), Phylogenetic perspectives in immunity. The Insect Host Defence. RG Landes Company, Austin, TX, pp. 115–122. [Google Scholar]
- Jemaà M, Morin N, Cavelier P, Cau J, Strub JM, Delsert C, 2014. Adult somatic progenitor cells and hematopoiesis in oysters. J Exp Biol 217, 3067–3077. [DOI] [PubMed] [Google Scholar]
- Jeon EY, Choi BH, Jung D, Hwang BH, Cha HJ, 2017. Natural healing-inspired collagen-targeting surgical protein glue for accelerated scarless skin regeneration. Biomaterials 134, 154–165. [DOI] [PubMed] [Google Scholar]
- Kanhai DK, Gardfeldt K, Lyashevska O, Hassellov M, Thompson RC, O’Connor I, 2018. Microplastics in sub-surface waters of the Arctic Central Basin. Mar Pollut Bull 130, 8–18. [DOI] [PubMed] [Google Scholar]
- Katzir N, Arman E, Cohen D, Givol D, Rechavi G, 1987. Common origin of transmissible venereal tumors (TVT) in dogs. Oncogene 1,445–448. [PubMed] [Google Scholar]
- Katzir N, Rechavi G, Cohen JB, Unger T, Simoni F, Segal S, Cohen D, Givol D, 1985. “Retroposon” insertion into the cellular oncogene c-myc in canine transmissible venereal tumor. Proc Natl Acad Sci U S A. 82, 1054–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik NK, Kaushik N, Pardeshi S, Sharma JG, Lee SH, Choi EH, 2015. Biomedical and clinical importance of mussel-inspired polymers and materials. Mar Drugs 13, 6792–6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan B, Clinton SM, Hamp TJ, Oliver JD, Ringwood AH, 2018. Potential impacts of hypoxia and a warming ocean on oyster microbiomes. Mar Environ Res 139, 27–34. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Morrison SJ, 2008. Uncertainty in the niches that maintain haematopoietic stem cells. Nat Rev Immunol 8, 290–301. [DOI] [PubMed] [Google Scholar]
- Kim BJ, Cheong H, Choi ES, Yun SH, Choi BH, Park KS, Kim IS, Park DH, Cha HJ, 2017. Accelerated skin wound healing using electrospun nanofibrous mats blended with mussel adhesive protein and polycaprolactone. J Biomed Mater Res A 105, 218–225. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Madhavan M, Call MK, Santiago W, Tsonis PA, Lambris JD, Del Rio-Tsonis K, 2003. Expression of complement 3 and complement 5 in newt limb and lens regeneration. J Immunol 170, 2331–2339. [DOI] [PubMed] [Google Scholar]
- King GM, Judd C, Kuske CR, Smith C, 2012. Analysis of stomach and gut microbiomes of the eastern oyster (Crassostrea virginica) from coastal Louisiana, USA. PLoS One 7, e51475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleeman SN, Adlard RD, Lester RJ, 2002. Detection of the initial infective stages of the protozoan parasite Marteilia sydneyi in Saccostrea glomerata and their development through to sporogenesis. Int J Parasitol. 32, 767–784. [DOI] [PubMed] [Google Scholar]
- Kocot KM, Aguilera F, McDougall C, Jackson DJ, Degnan BM, 2016. Sea shell diversity and rapidly evolving secretomes: insights into the evolution of biomineralization. Front. Zool. 13, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolandhasamy P, Su L, Li J, Qu X, Jabeen K, Shi H, 2018. Adherence of microplastics to soft tissue of mussels: A novel way to uptake microplastics beyond ingestion. Sci Total Environ 610–611, 635–640. [DOI] [PubMed] [Google Scholar]
- Koren O, Rosenberg E, 2008. Bacteria associated with the bleached and cave coral Oculina patagonica. Microb Ecol 55, 523–529. [DOI] [PubMed] [Google Scholar]
- Koroleva AG, Evtushenko EV, Maximova NV, Vershinin AV, Sitnikova TY, Kirilchik SV, 2015. Length and structure of telomeric DNA in three species of Baikal gastropods (Caenogastropoda: Hydrobioidea: Benedictiidae). Russ J Genet 51,300–307. [PubMed] [Google Scholar]
- Kosikowska P, Lesner A, 2016. Antimicrobial peptides (AMPs) as drug candidates: a patent review (2003–2015). Expert Opin Ther Pat 26, 689–702. [DOI] [PubMed] [Google Scholar]
- Kumar A, Godwin JW, Gates PB, Garza-Garcia AA, Brockes JP, 2007. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318, 772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labastie MC, Cortes F, Romeo PH, Dulac C, Peault B, 1998. Molecular identity of hematopoietic precursor cells emerging in the human embryo. Blood 92, 3624–3635. [PubMed] [Google Scholar]
- Lamghari M, Berland S, Laurent A, Huet H, Lopez E, 2001. Bone reactions to nacre injected percutaneously into the vertebrae of sheep. Biomaterials 22, 555–562. [DOI] [PubMed] [Google Scholar]
- Landsberg JH, 2002. The effects of harmful algal blooms on aquatic organisms. Rev Fish Sci 10, 113–390. [Google Scholar]
- Lassudrie M, Wikfors GH, Sunila I, Alix JH, Dixon MS, Combot D, Soudant P, Fabioux C, Hegaret H, 2015. Physiological and pathological changes in the eastern oyster Crassostrea virginica infested with the trematode Bucephalus sp. and exposed to the toxic dinoflagellate Alexandrium fundyense. J Invertebr Pathol 126, 51–63. [DOI] [PubMed] [Google Scholar]
- Lassus P, Bardouil M, Beliaeff B, Masselin P, Naviner M, Truquet P, 1999. Effect of a continuous supply of the toxic dinoflagellate Alexandrium minutum Halim on the feeding behavior of the Pacific oyster (Crassostrea gigas Thunberg). J Shellfish Res. 18, 211–216. [Google Scholar]
- Lau YT, Gambino L, Santos B, Espinosa EP, Allam B, 2018. Transepithelial migration of mucosal hemocytes in Crassostrea virginica and potential role in Perkinsus marinus pathogenesis. J Invertebr Pathol. [DOI] [PubMed] [Google Scholar]
- Lau YT, Sussman L, Pales Espinosa E, Katalay S, Allam B, 2017. Characterization of hemocytes from different body fluids of the eastern oyster Crassostrea virginica. Fish Shellfish Immunol. 71, 372–379. [DOI] [PubMed] [Google Scholar]
- Law KL, Moret-Ferguson S, Maximenko NA, Proskurowski G, Peacock EE, Hafner J, Reddy CM, 2010. Plastic accumulation in the North Atlantic subtropical gyre. Science 329, 1185–1188. [DOI] [PubMed] [Google Scholar]
- Law KL, Thompson RC, 2014. Oceans. Microplastics in the seas. Science 345, 144–145. [DOI] [PubMed] [Google Scholar]
- Lazebnik Y, Parris GE, 2015. Comment on: ‘guidelines for the use of cell lines in biomedical research’: human-to-human cancer transmission as a laboratory safety concern. Br J Cancer 112, 1976–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Jung SK, Chang YH, Kwak HS, 2017. Highly bioavailable nanocalcium from oyster shell for preventing osteoporosis in rats. Int. J. Food Sci. Nutr. 68, 931–940. [DOI] [PubMed] [Google Scholar]
- Lengfeld T, Watanabe H, Simakov O, Lindgens D, Gee L, Law L, Schmidt HA, Ozbek S, Bode H, Holstein TW, 2009. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev Biol 330, 186–199. [DOI] [PubMed] [Google Scholar]
- Leonardi CL, Kimball AB, Papp KA, Yeilding N, Guzzo C, Wang Y, Li S, Dooley LT, Gordon KB, investigators P.s., 2008. Efficacy and safety of ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with psoriasis: 76-week results from a randomised, double-blind, placebo-controlled trial (PHOENIX 1). Lancet 371, 1665–1674. [DOI] [PubMed] [Google Scholar]
- Leoni G, De Poli A, Mardirossian M, Gambato S, Florian F, Venier P, Wilson DN, Tossi A, Pallavicini A, Gerdol M, 2017. Myticalins: A novel multigenic family of linear, cationic antimicrobial peptides from marine mussels (Mytilus spp.). Mar Drugs 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser M, Shumway SE, 1993. Effects of toxic dinoflagellates on clearance rates and survival in juvenile bivalve molluscs. J Shellfish Res. 12, 377–382. [Google Scholar]
- Leverone JR, Shumway SE, Blake NJ, 2007. Comparative effects of the toxic dinoflagellate Karenia brevis on clearance rates in juveniles of four bivalve molluscs from Florida, USA. Toxicon 49, 634–645. [DOI] [PubMed] [Google Scholar]
- Li F, Huang S, Wang L, Yang J, Zhang H, Qiu L, Li L, Song L, 2011a. A macrophage migration inhibitory factor like gene from scallop Chlamys farreri: Involvement in immune response and wound healing. Developmental and comparative immunology 35, 62–71. [DOI] [PubMed] [Google Scholar]
- Li H, Parisi MG, Parrinello N, Cammarata M, Roch P, 2011b. Molluscan antimicrobial peptides, a review from activity-based evidences to computer-assisted sequences. Invert Surv J. 8. [Google Scholar]
- Li H, Zhang B, Fan S, Liu B, Su J, Yu D, 2017a. Identification and differential expression of biomineralization genes in the mantle of pearl oyster Pinctada fucata. Mar. Biotechnol. (NY) 19, 266–276. [DOI] [PubMed] [Google Scholar]
- Li HL, Gee P, Ishida K, Hotta A, 2016a. Efficient genomic correction methods in human iPS cells using CRISPR-Cas9 system. Methods (San Diego, Calif 101,27–35. [DOI] [PubMed] [Google Scholar]
- Li HX, Ma LS, Lin L, Ni ZX, Xu XR, Shi HH, Yan Y, Zheng GM, Rittschof D, 2018a. Microplastics in oysters Saccostrea cucullata along the Pearl River Estuary, China. Environ Pollut 236, 619–625. [DOI] [PubMed] [Google Scholar]
- Li J, Green C, Reynolds A, Shi H, Rotchell JM, 2018b. Microplastics in mussels sampled from coastal waters and supermarkets in the United Kingdom. Environ Pollut 241,35–44. [DOI] [PubMed] [Google Scholar]
- Li S, Huang J, Liu C, Liu Y, Zheng G, Xie L, Zhang R, 2016b. Interactive effects of seawater acidification and elevated temperature on the transcriptome and biomineralization in the pearl oyster Pinctada fucata. Environ. Sci. Technol. 50, 1157–1165. [DOI] [PubMed] [Google Scholar]
- Li S, Liu C, Huang J, Liu Y, Zhang S, Zheng G, Xie L, Zhang R, 2016c. Transcriptome and biomineralization responses of the pearl oyster Pinctada fucata to elevated CO2 and temperature. Sci. Rep. 6, 18943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu Y, Huang J, Zhan A, Xie L, Zhang R, 2017b. The receptor genes PfBMPR1B and PfBAMBI are involved in regulating shell biomineralization in the pearl oyster Pinctada fucata. Sci. Rep. 7, 9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu Y, Liu C, Huang J, Zheng G, Xie L, Zhang R, 2016d. Hemocytes participate in calcium carbonate crystal formation, transportation and shell regeneration in the pearl oyster Pinctada fucata. Fish Shellfish Immunol 51,263–270. [DOI] [PubMed] [Google Scholar]
- Liao H, Mutvei H, Hammarstrom L, Wurtz T, Li J, 2002. Tissue responses to nacreous implants in rat femur: an in situ hybridization and histochemical study. Biomaterials 23, 2693–2701. [DOI] [PubMed] [Google Scholar]
- Libouban H, Pascaretti-Grizon F, Camprasse G, Camprasse S, Chappard D, 2016. In vivo erosion of orthopedic screws prepared from nacre (mother of pearl). Orthop Traumatol Surg Res 102, 913–918. [DOI] [PubMed] [Google Scholar]
- Lin G, Slack JM, 2008. Requirement for Wnt and FGF signaling in Xenopus tadpole tail regeneration. Dev Biol 316, 323–335. [DOI] [PubMed] [Google Scholar]
- Liu G, Huan P, Liu B, 2017. A SoxC gene related to larval shell development and co-expression analysis of different shell formation genes in early larvae of oyster. Dev. Genes Evol. 227, 181–188. [DOI] [PubMed] [Google Scholar]
- Liu J, Yang D, Liu S, Li S, Xu G, Zheng G, Xie L, Zhang R, 2015. Microarray: a global analysis of biomineralization-related gene expression profiles during larval development in the pearl oyster, Pinctada fucata. BMC Genomics. 16, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Wu M, Zhou S, Gao P, Lu T, Wang R, Shi G, Liao Z, 2010. Designation, solid-phase synthesis and antimicrobial activity of Mytilin derived peptides based on Mytilin-1 from Mytilus coruscus. Sheng Wu Gong Cheng Xue Bao 26, 550–556. [PubMed] [Google Scholar]
- Liu Z, Wang L, Yan Y, Zheng Y, Ge W, Li M, Wang W, Song X, Song L, 2018. D1 dopamine receptor is involved in shell formation in larvae of Pacific oyster Crassostrea gigas. Dev. Comp. Immunol. 84, 337–342. [DOI] [PubMed] [Google Scholar]
- Loaiza N, Demaria M, 2016. Cellular senescence and tumor promotion: Is aging the key? Biochim Biophys Acta. 1865, 155–167. [DOI] [PubMed] [Google Scholar]
- Löfgren SE, Miletti LC, Steindel M, Bachere E, Barracco MA, 2008. Trypanocidal and leishmanicidal activities of different antimicrobial peptides (AMPs) isolated from aquatic animals. Exp Parasitol 118, 197–202. [DOI] [PubMed] [Google Scholar]
- Lokmer A, Goedknegt MA, Thieltges DW, Fiorentino D, Kuenzel S, Baines JF, Wegner KM, 2016. Spatial and temporal dynamics of Pacific oyster hemolymph microbiota across multiple scales. Front Microbiol 7, 1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez E, Le Faou A, Borzeix S, Berland S, 2000. Stimulation of rat cutaneous fibroblasts and their synthetic activity by implants of powdered nacre (mother of pearl). Tissue Cell 32, 95–101. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, 2018. Unraveling interactions between the microbiome and the host immune system to decipher mechanisms of disease. mSystems 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg AS, Hahn WC, Gupta P, Weinberg RA, 2000. Genes involved in senescence and immortalization. Curr Opin Cell Biol 12, 705–709. [DOI] [PubMed] [Google Scholar]
- Ma H, Luo J, Sun Z, Xia L, Shi M, Liu M, Chang J, Wu C, 2016. 3D printing of biomaterials with mussel-inspired nanostructures for tumor therapy and tissue regeneration. Biomaterials 111, 138–148. [DOI] [PubMed] [Google Scholar]
- Mack DR, Michail S, Wei S, McDougall L, Hollingsworth MA, 1999. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol 276, G941–950. [DOI] [PubMed] [Google Scholar]
- Mafra LL Jr., Bricelj VM, Fennel K, 2010. Domoic acid uptake and elimination kinetics in oysters and mussels in relation to body size and anatomical distribution of toxin. Aquat Toxicol 100, 17–29. [DOI] [PubMed] [Google Scholar]
- Mahlapuu M, Hakansson J, Ringstad L, Bjorn C, 2016. Antimicrobial peptides: An emerging category of therapeutic agents. Front Cell Infect Microbiol 6, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamangkey NG, Southgate PC, 2009. Regeneration of excised mantle tissue by the silver-lip pearl oyster, Pinctada maxima (Jameson). Fish Shellfish Immunol 27, 164–174. [DOI] [PubMed] [Google Scholar]
- Manfrin C, De Moro G, Torboli V, Venier P, Pallavicini A, Gerdol M, 2012. Physiological and molecular responses of bivalves to toxic dinoflagellates. Invert Surv J 9, 184–199. [Google Scholar]
- Mardones-Toledo DA, Montory JA, Joyce A, Thompson RJ, Diederich CM, Pechenik JA, Mardones ML, Chaparro OR, 2015. Brooding in the Chilean oyster Ostrea chilensis: unexpected complexity in the movements of brooded offspring within the mantle cavity. PLoS One 10, e0122859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, Arivalagan J, Matheron L, Bolbach G, Berland S, Marie A, Marin F, 2017. Deep conservation of bivalve nacre proteins highlighted by shell matrix proteomics of the Unionoida Elliptio complanata and Villosa lienosa. J. R. Soc. Interface 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin F, Le Roy N, Marie B, 2012. The formation and mineralization of mollusk shell. Front. Biosci. 4, 1099–1125. [DOI] [PubMed] [Google Scholar]
- Mark SD, Peter B, 1999. Role of mucus trails and trail-following in the behaviour and nutrition of the periwinkle Littorina littorea. Mar Ecol Prog Ser 179, 247–257. [Google Scholar]
- Martínez-Porchas M, Vargas-Albores F, 2015. Microbial metagenomics in aquaculture: a potential tool for a deeper insight into the activity. Rev Aquacult 9, 42–56. [Google Scholar]
- Mat AM, Haberkorn H, Bourdineaud JP, Massabuau JC, Tran D, 2013. Genetic and genotoxic impacts in the oyster Crassostrea gigas exposed to the harmful alga Alexandrium minutum. Aquat Toxicol 140–141,458–465. [DOI] [PubMed] [Google Scholar]
- Mat AM, Klopp C, Payton L, Jeziorski C, Chalopin M, Amzil Z, Tran D, Wikfors GH, Hegaret H, Soudant P, Huvet A, Fabioux C, 2018. Oyster transcriptome response to Alexandrium exposure is related to saxitoxin load and characterized by disrupted digestion, energy balance, and calcium and sodium signaling. Aquat Toxicol. 199, 127–137. [DOI] [PubMed] [Google Scholar]
- Mato Y, Isobe T, Takada H, Kanehiro H, Ohtake C, Kaminuma T, 2001. Plastic resin pellets as a transport medium for toxic chemicals in the marine environment. Environ Sci Technol 35, 318–324. [DOI] [PubMed] [Google Scholar]
- McDougall C, Degnan BM, 2018. The evolution of mollusc shells. Wiley Interdiscip Rev Dev Biol. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, 2014. Divining the essence of symbiosis: insights from the squid-vibrio model. PLoS Biol 12, e1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney EA, Koelle K, Dunn RR, Yoder AD, 2018. The ecosystem services of animal microbiomes. Mol Ecol 27, 2164–2172. [DOI] [PubMed] [Google Scholar]
- McLaughlin SM, Farley CA, Hetrick FM, 1992. Transmission studies of sarcoma in the soft-shell clam, Mya arenaria. In Vivo 6, 367–370. [PubMed] [Google Scholar]
- Medhioub W, Ramondenc S, Vanhove AS, Vergnes A, Masseret E, Savar V, Amzil Z, Laabir M, Rolland JL, 2013. Exposure to the neurotoxic dinoflagellate, Alexandrium catenella, induces apoptosis of the hemocytes of the oyster, Crassostrea gigas. Mar Drugs 11,4799–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdizadeh M, Weng H, Gyawali D, Tang LP, Yang J, 2012. Injectable citrate-based mussel-inspired tissue bioadhesives with high wet strength for sutureless wound closure. Biomaterials 33, 7972–7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello DF, Silva P.M.d., Barracco MA, Soudant P, Hégaret H, 2013. Effects of the dinoflagellate Alexandrium minutum and its toxin (saxitoxin) on the functional activity and gene expression of Crassostrea gigas hemocytes. Harmful Algae 26, 45–51. [Google Scholar]
- Meng DM, Dai HX, Gao XF, Zhao JF, Guo YJ, Ling X, Dong B, Zhang ZQ, Fan ZC, 2016. Expression, purification and initial characterization of a novel recombinant antimicrobial peptide Mytichitin-A in Pichia pastoris. Protein Expr Purif 127, 35–43. [DOI] [PubMed] [Google Scholar]
- Metzger MJ, Reinisch C, Sherry J, Goff SP, 2015. Horizontal transmission of clonal cancer cells causes leukemia in soft-shell clams. Cell 161,255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger MJ, Villalba A, Carballal MJ, Iglesias D, Sherry J, Reinisch C, Muttray AF, Baldwin SA, Goff SP, 2016. Widespread transmission of independent cancer lineages within multiple bivalve species. Nature 534, 705–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan M, Carraro L, Fariselli P, Martino ME, Cavalieri D, Vitali F, Boffo L, Patarnello T, Bargelloni L, Cardazzo B, 2018. Microbiota and environmental stress: how pollution affects microbial communities in Manila clams. Aquat Toxicol 194, 195–207. [DOI] [PubMed] [Google Scholar]
- Mitta G, Vandenbulcke F, Roch P, 2000. Original involvement of antimicrobial peptides in mussel innate immunity. FEBS letters 486, 185–190. [DOI] [PubMed] [Google Scholar]
- Monaghan JR, Epp LG, Putta S, Page RB, Walker JA, Beachy CK, Zhu W, Pao GM, Verma IM, Hunter T, Bryant SV, Gardiner DM, Harkins TT, Voss SR, 2009. Microarray and cDNA sequence analysis of transcription during nerve-dependent limb regeneration. BMC Biol 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount AS, Wheeler A, Paradkar RP, Snider D, 2004. Hemocyte-mediated shell mineralization in the eastern oyster. Science 304, 297–300. [DOI] [PubMed] [Google Scholar]
- Mun S, Kim YJ, Markkandan K, Shin W, Oh S, Woo J, Yoo J, An H, Han K, 2017. The whole-genome and transcriptome of the Manila clam (Ruditapes philippinarum). Genome Biol Evol 9, 1487–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, 2008. Clonally transmissible cancers in dogs and Tasmanian devils. Oncogene 27 Suppl 2, S19–30. [DOI] [PubMed] [Google Scholar]
- Murgarella M, Puiu D, Novoa B, Figueras A, Posada D, Canchaya C, 2016a. Correction: a first insight into the genome of the filter-feeder mussel Mytilus galloprovincialis. PloS one 11, e0160081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgarella M, Puiu D, Novoa B, Figueras A, Posada D, Canchaya C, 2016b. A first insight into the genome of the filter-feeder mussel Mytilus galloprovincialis. PLoS One 11, e0151561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia C, Pritchard JK, Kim SY, Fassati A, Weiss RA, 2006. Clonal origin and evolution of a transmissible cancer. Cell 126, 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muttray A, Reinisch C, Miller J, Ernst W, Gillis P, Losier M, Sherry J, 2012. Haemocytic leukemia in Prince Edward Island (PEI) soft shell clam (Mya arenaria): spatial distribution in agriculturally impacted estuaries. Sci Total Environ 424, 130–142. [DOI] [PubMed] [Google Scholar]
- Mydlarz LD, Jones LE, Harvell CD, 2006. Innate immunity, environmental drivers, and disease ecology of marine and freshwater invertebrates. Annu. Rev. Ecol. Evol. Syst. 37, 251–288. [Google Scholar]
- Nagai K, Honjo T, Go J, Yamashita H, Seok Jin O, 2006. Detecting the shellfish killer Heterocapsa circularisquama (Dinophyceae) by measuring bivalve valve activity with a Hall element sensor. Aquaculture 255, 395–401. [Google Scholar]
- Nagashima Y, Kikuchi N, Shimakura K, Shiomi K, 2003. Purification and characterization of an antibacterial protein in the skin secretion of rockfish Sebastes schlegeli. Comp Biochem Physiol C Toxicol Pharmacol 136, 63–71. [DOI] [PubMed] [Google Scholar]
- Naji A, Nuri M, Vethaak AD, 2018. Microplastics contamination in molluscs from the northern part of the Persian Gulf. Environ Pollut 235, 113–120. [DOI] [PubMed] [Google Scholar]
- Newell RI, 2004. Ecosystem influences of natural and cultivated populations of suspension-feeding bivalve molluscs: a review. J Shellfish Res 23, 51–62. [Google Scholar]
- Noble S, Goa KL, 1997. Aldesleukin (recombinant interleukin-2). BioDrugs 7, 394–422. [DOI] [PubMed] [Google Scholar]
- Novoa B, Romero A, Alvarez AL, Moreira R, Pereiro P, Costa MM, Dios S, Estepa A, Parra F, Figueras A, 2016. Antiviral activity of Myticin C peptide from mussel: an ancient defense against Herpesviruses. J Virol 90, 7692–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC, 1976. The clonal evolution of tumor cell populations. Science 194, 23–28. [DOI] [PubMed] [Google Scholar]
- Nunes-Alves C, 2014. Parasite biology: Divide and conquer. Nat Rev Microbiol 12, 192–192. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ, 2003. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl Environ Microbiol 69, 3932–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ, 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci U S A. 97, 10231–10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odintsova NA, Dyachuk VA, Nezlin LP, 2010. Muscle and neuronal differentiation in primary cell culture of larval Mytilus trossulus (Mollusca: Bivalvia). Cell Tissue Res 339, 625–637. [DOI] [PubMed] [Google Scholar]
- Ohmori F, Kinoshita S, Funabara D, Koyama H, Nagai K, Maeyama K, Okamoto K, Asakawa S, Watabe S, 2018. Novel isoforms of N16 and N19 families implicated for the Nnacreous layer formation in the pearl oyster Pinctada fucata. Mar. Biotechnol. (NY) 20, 155–167. [DOI] [PubMed] [Google Scholar]
- Oliveira DV, Silva TS, Cordeiro OD, Cavaco SI, Simes DC, 2012. Identification of proteins with potential osteogenic activity present in the water-soluble matrix proteins from Crassostrea gigas nacre using a proteomic approach. Sci. World J. 2012, 765909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprandy JJ, Chang PW, 1983. 5-bromodeoxyuridine induction of hematopoietic neoplasia and retrovirus activation in the soft-shell clam, Mya arenaria. J Invertebr Pathol 42, 196–206. [DOI] [PubMed] [Google Scholar]
- Ozer HL, 2000. SV40-mediated immortalization. Prog Mol Subcell Biol 24, 121–153. [DOI] [PubMed] [Google Scholar]
- Paillard C, Le Roux F, Borrego JJ, 2004. Bacterial disease in marine bivalves, a review of recent studies: Trends and evolution. Aquat. Living Resour. doi: 10.1051/alr:2004054. [DOI] [Google Scholar]
- Pales Espinosa E, Allam B, 2018. Reverse genetics demonstrate the role of mucosal C-type lectins in food particle selection in the oyster Crassostrea virginica. J Exp Biol 221. [DOI] [PubMed] [Google Scholar]
- Pales Espinosa E, Corre E, Allam B, 2014. Pallial mucus of the oyster Crassostrea virginica regulates the expression of putative virulence genes of its pathogen Perkinsus marinus. Int J Parasitol. 44, 305–317. [DOI] [PubMed] [Google Scholar]
- Pales Espinosa E, Koller A, Allam B, 2016. Proteomic characterization of mucosal secretions in the eastern oyster, Crassostrea virginica. J Proteomics 132, 63–76. [DOI] [PubMed] [Google Scholar]
- Pales Espinosa E, Perrigault M, Allam B, 2010. Identification and molecular characterization of a mucosal lectin (MeML) from the blue mussel Mytilus edulis and its potential role in particle capture. Comp Biochem Physiol A Mol Integr Physiol 156, 495–501. [DOI] [PubMed] [Google Scholar]
- Pales Espinosa E, Winnicki S, Allam B, 2013. Early host-pathogen interactions in a marine bivalve: Crassostrea virginica pallial mucus modulates Perkinsus marinus growth and virulence. Dis Aquat Organ 104, 237–247. [DOI] [PubMed] [Google Scholar]
- Parfrey LW, Moreau CS, Russell JA, 2018. Introduction: The host-associated microbiome: Pattern, process and function. Mol Ecol 27, 1749–1765. [DOI] [PubMed] [Google Scholar]
- Parolini M, Quinn B, Binelli A, Provini A, 2011. Cytotoxicity assessment of four pharmaceutical compounds on the zebra mussel (Dreissena polymorpha) haemocytes, gill and digestive gland primary cell cultures. Chemosphere 84, 91–100. [DOI] [PubMed] [Google Scholar]
- Pearce I, Handlinger JH, Hallegraeff GM, 2005. Histopathology in Pacific oyster (Crassostrea gigas) spat caused by the dinoflagellate Prorocentrum rhathymum. Harmful Algae 4, 61–74. [Google Scholar]
- Pearse AM, Swift K, 2006. Allograft theory: transmission of devil facial-tumour disease. Nature 439, 549. [DOI] [PubMed] [Google Scholar]
- Pechenik J, 2000. Biology of the Invertebrates. McGraw-Hill, Boston, MA. [Google Scholar]
- Perez-Garcia C, Guerra-Varela J, Moran P, Pasantes JJ, 2010. Chromosomal mapping of rRNA genes, core histone genes and telomeric sequences in Brachidontes puniceus and Brachidontes rodriguezi (Bivalvia, Mytilidae). BMC Genet 11, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-López A, Behnsen J, Nuccio SP, Raffatellu M, 2016. Mucosal immunity to pathogenic intestinal bacteria. Nat Rev Immunol 16, 135–148. [DOI] [PubMed] [Google Scholar]
- Perrigault M, Tran D, 2017. Identification of the molecular clockwork of the oyster Crassostrea gigas. PloS one 12, e0169790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry KJ, Henry JQ, 2015. CRISPR/Cas9-mediated genome modification in the mollusc, Crepidula fornicata. Genesis 53, 237–244. [DOI] [PubMed] [Google Scholar]
- Petersen JM, Osvatic J, 2018. Microbiomes In Natura: Importance of Invertebrates in Understanding the Natural Variety of Animal-Microbe Interactions. mSystems 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce ML, 2016. The Microbiome of the Eastern Oyster, Crassostrea virginica (Gmelin, 1791): Temporal and Spatial Variation, Environmental Influences, and its Impact on Host Physiology. University of Connecticut, p. Doctoral Dissertations. [Google Scholar]
- Pierce ML, Ward JE, 2018. Microbial ecology of the Bivalvia, with an emphasis on the family Ostreidae. J Shellfish Res 37, 793–806. [Google Scholar]
- Pittura L, Avio CG, Giuliani ME, d’Errico G, Keiter SH, Cormier B, Gorbi S, Regoli F, 2018. Microplastics as vehicles of environmental PAHs to marine organisms: Combined chemical and physical hazards to the Mediterranean mussels, Mytilus galloprovincialis. Front Mar Sci 5, 103. [Google Scholar]
- Plohl M, Prats E, Martinez-Lage A, Gonzalez-Tizon A, Mendez J, Cornudella L, 2002. Telomeric localization of the vertebrate-type hexamer repeat, (TTAGGG)n, in the wedgeshell clam Donax trunculus and other marine invertebrate genomes. J Biol Chem. 277, 19839–19846. [DOI] [PubMed] [Google Scholar]
- Porter A, Lyons BP, Galloway TS, Lewis C, 2018. Role of marine snows in microplastic fate and bioavailability. Environ Sci Technol 52, 7111–7119. [DOI] [PubMed] [Google Scholar]
- Powell D, Subramanian S, Suwansa-Ard S, Zhao M, O’Connor W, Raftos D, Elizur A, 2018. The genome of the oyster Saccostrea offers insight into the environmental resilience of bivalves. DNA Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell SM, Chapman CC, Bermudes M, Tamplin ML, 2013. Dynamics of seawater bacterial communities in a shellfish hatchery. Microb Ecol 66, 245–256. [DOI] [PubMed] [Google Scholar]
- Power A, McCrickard B, Mitchell M, Covington E, Sweeney-Reeves M, Payne K, Walker R, 2006. Perkinsus marinus in coastal Georgia, USA, following a prolonged drought. Dis Aquat Organ 73, 151–158. [DOI] [PubMed] [Google Scholar]
- Powers DA, Kirby VL, Cole T, Hereford L, 1995. Electroporation as an effective means of introducing DNA into abalone (Haliotis rufescens) embryos. Mol Mar Biol Biotechnol 4, 369–375. [PubMed] [Google Scholar]
- Powers DA, Madsen Hereford L, Gomez-Chiarri M, 1994–1997. Isolation and characterization of an actin gene from abalone, in: The Board of Governors for Higher Education, S.O.R.I.a.P.P. (Ed.), USA. [Google Scholar]
- Prego-Faraldo MV, Valdiglesias V, Laffon B, Mendez J, Eirin-Lopez JM, 2016. Early genotoxic and cytotoxic effects of the toxic dinoflagellate Prorocentrum lima in the mussel Mytilus galloprovincialis. Toxins (Basel) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prego-Faraldo MV, Vieira LR, Eirin-Lopez JM, Mendez J, Guilhermino L, 2017. Transcriptional and biochemical analysis of antioxidant enzymes in the mussel Mytilus galloprovincialis during experimental exposures to the toxic dinoflagellate Prorocentrum lima. Mar Environ Res 129, 304–315. [DOI] [PubMed] [Google Scholar]
- Qu F, Xiang Z, Zhang Y, Li J, Xiao S, Zhang Y, Qin Y, Zhou Y, Yu Z, 2017. Molecular identification and functional characterization of a tumor necrosis factor (TNF) gene in Crassostrea hongkongensis. Immunobiology 222, 751–758. [DOI] [PubMed] [Google Scholar]
- Qu X, Su L, Li H, Liang M, Shi H, 2018. Assessing the relationship between the abundance and properties of microplastics in water and in mussels. Sci Total Environ 621,679–686. [DOI] [PubMed] [Google Scholar]
- Quinn B, Costello MJ, Dorange G, Wilson JG, Mothersill C, 2009. Development of an in vitro culture method for cells and tissues from the zebra mussel (Dreissena polymorpha). Cytotechnology 59, 121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones JL, Rosa R, Ruiz DL, Garcia-Arraras JE, 2002. Extracellular matrix remodeling and metalloproteinase involvement during intestine regeneration in the sea cucumber Holothuria glaberrima. Dev Biol 250, 181–197. [DOI] [PubMed] [Google Scholar]
- Ramírez-Gómez F, Ortiz-Pineda PA, Rojas-Cartagena C, SuEarez-Castillo EC, García-Arraras JE, 2008. Immune-related genes associated with intestinal tissue in the sea cucumber Holothuria glaberrima. Immunogenetics 60, 57–71. [DOI] [PubMed] [Google Scholar]
- Rayner C, Wilson R, 1997. Airway mucus: Basic mechanisms and clinical perspectives, Birkhauser verlag Basel. [Google Scholar]
- Rebbeck CA, Thomas R, Breen M, Leroi AM, Burt A, 2009. Origins and evolution of a transmissible cancer. Evolution 63, 2340–2349. [DOI] [PubMed] [Google Scholar]
- Renault T, Flaujac G, Le Deuff R-M, 1995. Isolation and culture of heart cells from the European flat oyster, Ostrea edulis. Methods in Cell Sci. 17, 199–205. [Google Scholar]
- Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P, 2001. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol 2, 361–367. [DOI] [PubMed] [Google Scholar]
- Rinkevich B, 2005. Marine invertebrate cell cultures: new millennium trends. Mar Biotechnol. (NY) 7, 429–439. [DOI] [PubMed] [Google Scholar]
- Rinkevich B, 2011. Cell cultures from marine invertebrates: new insights for capturing endless stemness. Mar Biotechnol. (NY) 13, 345–354. [DOI] [PubMed] [Google Scholar]
- Ritchie KB, 2006. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser 322, 1–14. [Google Scholar]
- Robledo JAF, Santarem MM, Figueras A, 1994. Parasite loads of rafted blue mussels (Mytilus galloprovincialis) in Spain with special reference to the copepod, Mytilicola intestinalis. Aquaculture 127, 287–302. [Google Scholar]
- Roch P, Beschin A, Bernard E, 2004. Antiprotozoan and antiviral activities of non-cytotoxic truncated and variant analogues of mussel defensin. Evid Based Complement Alternat Med 1, 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roch P, Yang Y, Toubiana M, Aumelas A, 2008. NMR structure of mussel mytilin, and antiviral-antibacterial activities of derived synthetic peptides. Dev Comp Immunol. 32, 227–238. [DOI] [PubMed] [Google Scholar]
- Rodrigues JR, Alves NM, Mano JF, 2017. Nacre-inspired nanocomposites produced using layer-bylayer assembly: Design strategies and biomedical applications. Mater. Sci. Eng. C. Mater. Biol. Appl. 76, 1263–1273. [DOI] [PubMed] [Google Scholar]
- Rognoni E, Watt FM, 2018. Skin cell heterogeneity in development, wound healing, and cancer. Trends Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolton A, Soudant P, Vignier J, Pierce R, Henry M, Shumway SE, Bricelj VM, Volety AK, 2015. Susceptibility of gametes and embryos of the eastern oyster, Crassostrea virginica, to Karenia brevis and its toxins. Toxicon 99, 6–15. [DOI] [PubMed] [Google Scholar]
- Romero A, Costa M, Forn-Cuni G, Balseiro P, Chamorro R, Dios S, Figueras A, Novoa B, 2014. Occurrence, seasonality and infectivity of Vibrio strains in natural populations of mussels Mytilus galloprovincialis. Dis Aquat Organ 108, 149–163. [DOI] [PubMed] [Google Scholar]
- Ross MH, 1974. The organization of the seminiferous epithelium in the mouse testis following ligation of the efferent ductules. A light microscopic study. Anat Rec 180, 565–579. [DOI] [PubMed] [Google Scholar]
- Roterman YR, Benayahu Y, Reshef L, Gophna U, 2015. The gill microbiota of invasive and indigenous Spondylus oysters from the Mediterranean Sea and northern Red Sea. Environ Microbiol Rep 7, 860–867. [DOI] [PubMed] [Google Scholar]
- Rousseau M, Pereira-Mouries L, Almeida MJ, Milet C, Lopez E, 2003. The water-soluble matrix fraction from the nacre of Pinctada maxima produces earlier mineralization of MC3T3-E1 mouse preosteoblasts. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 135, 1–7. [PubMed] [Google Scholar]
- Rousselle P, Montmasson M, Garnier C, 2018. Extracellular matrix contribution to skin wound re-epithelialization. Matrix Biol. [DOI] [PubMed] [Google Scholar]
- Rubiolo JA, Lozano-Leon A, Rodriguez-Souto R, Fol Rodriguez N, Vieytes MR, Botana LM, 2018. The impact of depuration on mussel hepatopancreas bacteriome composition and predicted metagenome. Antonie Van Leeuwenhoek. [DOI] [PubMed] [Google Scholar]
- Runnegar B, 1996. Early evolution of the Mollusca: the fossil record. Orig. Evol. Radiat. Mollusca, Oxford. [Google Scholar]
- Russell JA, Dubilier N, Rudgers JA, 2014. Nature’s microbiome: introduction. Mol Ecol 23, 1225–1237. [DOI] [PubMed] [Google Scholar]
- Russell JJ, Theriot JA, Sood P, Marshall WF, Landweber LF, Fritz-Laylin L, Polka JK, Oliferenko S, Gerbich T, Gladfelter A, Umen J, Bezanilla M, Lancaster MA, He S, Gibson MC, Goldstein B, Tanaka EM, Hu C-K, Brunet A, 2017. Non-model model organisms. BMC Biol 15, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell MW, et al. , 2015. Overview: The mucosal immune system. Mucosal Immunology. [Google Scholar]
- Salazar G, Sunagawa S, 2017. Marine microbial diversity. Curr Biol. 27, R489–R494. [DOI] [PubMed] [Google Scholar]
- Santana MF, Ascer LG, Custodio MR, Moreira FT, Turra A, 2016. Microplastic contamination in natural mussel beds from a Brazilian urbanized coastal region: Rapid evaluation through bioassessment. Mar Pollut Bull 106, 183–189. [DOI] [PubMed] [Google Scholar]
- Santana MFM, Moreira FT, Pereira CDS, Abessa DMS, Turra A, 2018. Continuous exposure to microplastics does not cause physiological effects in the cultivated mussel Perna perna. Arch Environ Contam Toxicol 74, 594–604. [DOI] [PubMed] [Google Scholar]
- Sarasamma S, Varikkodan MM, Liang ST, Lin YC, Wang WP, Hsiao CD, 2017. Zebrafish: A premier vertebrate model for biomedical research in Indian scenario. Zebrafish 14, 589–605. [DOI] [PubMed] [Google Scholar]
- Satoh A, Mitogawa K, Makanae A, 2015. Regeneration inducers in limb regeneration. Dev Growth Differ 57, 421–429. [DOI] [PubMed] [Google Scholar]
- Schill WB, Iwanowicz D, Adams C, 2017. Endozoicomonas dominates the gill and intestinal content microbiomes of Mytilus edulis from Barnegat Bay, New Jersey. J Shellfish Res. 36, 391–401. [Google Scholar]
- Schmitt P, Wilmes M, Pugniere M, Aumelas A, Bachere E, Sahl HG, Schneider T, Destoumieux-Garzon D, 2010. Insight into invertebrate defensin mechanism of action: oyster defensins inhibit peptidoglycan biosynthesis by binding to lipid II. J Biol Chem. 285, 29208–29216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz JH, Adema CM, 2017. Comparative immunogenomics of molluscs. Developmental and comparative immunology 75, 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yang S, Liu J, Xu H, Shi Z, Lin Z, Ying X, Guo P, Lin T, Yan S, Huang Q, Peng L, 2014. Engineering scaffolds integrated with calcium sulfate and oyster shell for enhanced bone tissue regeneration. ACS Appl. Mater. Interfaces 6, 12177–12188. [DOI] [PubMed] [Google Scholar]
- Shumway SE, 1990. A review of the effects of algal blooms on shellfish and aquaculture. J World Aquacult Soc 21, 104. [Google Scholar]
- Shumway SE, Terry LC, 1987. The effect of the toxic dinoflagellate Protogonyaulax tamarensis on the feeding and behaviour of bivalve molluscs. Aquat Toxicol 10, 9–27. [Google Scholar]
- Simoes FC, Riley PR, 2018. The ontogeny, activation and function of the epicardium during heart development and regeneration. Development 145. [DOI] [PubMed] [Google Scholar]
- Smayda TJ, 1989. Primary production and the global epidemic of phytoplankton blooms in the sea: a linkage?, in: Carpenter EMCVMBEJ (Ed.), Novel Phytoplankton Blooms. Springer, Berlin, Heidelberg, pp. 449–483. [Google Scholar]
- Smith CS, Ito M, Namba M, Nakaoka M, 2018a. Oyster aquaculture impacts Zostera marina epibiont community composition in Akkeshi-ko estuary, Japan. PLoS ONE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Love DC, Rochman CM, Neff RA, 2018b. Microplastics in Seafood and the Implications for Human Health. Curr Environ Health Rep 5, 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Wang L, Qiu L, Zhang H, 2010. Bivalve immunity, Invertebrate immunity. Springer, pp. 44–65. [DOI] [PubMed] [Google Scholar]
- Song X, Wang H, Chen H, Sun M, Liang Z, Wang L, Song L, 2016. Conserved hemopoietic transcription factor Cg-SCL delineates hematopoiesis of Pacific oyster Crassostrea gigas Fish Shellfish Immunol. 51, 180–188. [DOI] [PubMed] [Google Scholar]
- Southward EC, 1986. Gill symbionts in Thyasirids and other bivalve molluscs. J. Mar. Biol. Assoc. U. K. 66, 889–914. [Google Scholar]
- Steffensen JL, Dufault-Thompson K, Zhang Y, 2016. PSAMM: A portable system for the analysis of metabolic models. PLoS Comput Biol 12, e1004732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JE, Marks LJ, Gilgan MW, Pfeiffer E, Zwicker BM, 1998. Microbial utilization of the neurotoxin domoic acid: blue mussels (Mytilus edulis) and soft shell clams (Mya arenaria) as sources of the microorganisms. Can J Microbiol 44, 456–464. [PubMed] [Google Scholar]
- Stocum DL, 2017. Mechanisms of urodele limb regeneration. Regeneration (Oxf) 4, 159–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Cai H, Kolandhasamy P, Wu C, Rochman CM, Shi H, 2018. Using the Asian clam as an indicator of microplastic pollution in freshwater ecosystems. Environ Pollut 234, 347–355. [DOI] [PubMed] [Google Scholar]
- Suárez-Ulloa V, Fernández-Tajes J, Aguiar-Pulido V, Prego-Faraldo MV, Florez-Barros F, Sexto-Iglesias A, Mendez J, Eirin-Lopez JM, 2015. Unbiased high-throughput characterization of mussel transcriptomic responses to sublethal concentrations of the biotoxin okadaic acid. PeerJ 3, e1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Irvine KD, 2014. Control of growth during regeneration. Curr Top Dev Biol 108, 95–120. [DOI] [PubMed] [Google Scholar]
- Sun J, Zhang Y, Xu T, Zhang Y, Mu H, Zhang Y, Lan Y, Fields CJ, Hui JHL, Zhang W, Li R, Nong W, Cheung FKM, Qiu J-W, Qian P-Y, 2017. Adaptation to deep-sea chemosynthetic environments as revealed by mussel genomes. Nature Ecology &Amp; Evolution 1,0121. [DOI] [PubMed] [Google Scholar]
- Sun L, Song Y, Qu Y, Yu X, Zhang W, 2007. Purification and in vitro cultivation of archaeocytes (stem cells) of the marine sponge Hymeniacidon perleve (Demospongiae). Cell Tissue Res 328, 223–237. [DOI] [PubMed] [Google Scholar]
- Sussarellu R, Suquet M, Thomas Y, Lambert C, Fabioux C, Pernet ME, Le Goic N, Quillien V, Mingant C, Epelboin Y, Corporeau C, Guyomarch J, Robbens J, Paul-Pont I, Soudant P, Huvet A, 2016. Oyster reproduction is affected by exposure to polystyrene microplastics. Proc Natl Acad Sci U S A. 113, 2430–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA, 2007. Marine viruses--major players in the global ecosystem. Nat Rev Microbiol 5, 801–812. [DOI] [PubMed] [Google Scholar]
- Takahashi J, Takagi M, Okihana Y, Takeo K, Ueda T, Touhata K, Maegawa S, Toyohara H, 2012. A novel silk-like shell matrix gene is expressed in the mantle edge of the Pacific oyster prior to shell regeneration. Gene 499, 130–134. [DOI] [PubMed] [Google Scholar]
- Takeo M, Lee W, Ito M, 2015. Wound healing and skin regeneration. Cold Spring Harb Perspect Med 5, a023267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, 2017. Molluscan genomics: Implications for biology and aquaculture. Curr Mol Biol Rep 3, 297–305. [Google Scholar]
- Takeuchi T, Kawashima T, Koyanagi R, Gyoja F, Tanaka M, Ikuta T, Shoguchi E, Fujiwara M, Shinzato C, Hisata K, Fujie M, Usami T, Nagai K, Maeyama K, Okamoto K, Aoki H, Ishikawa T, Masaoka T, Fujiwara A, Endo K, Endo H, Nagasawa H, Kinoshita S, Asakawa S, Watabe S, Satoh N, 2012. Draft genome of the pearl oyster Pinctada fucata: a platform for understanding bivalve biology. DNA Res 19, 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Koyanagi R, Gyoja F, Kanda M, Hisata K, Fujie M, Goto H, Yamasaki S, Nagai K, Morino Y, Miyamoto H, Endo K, Endo H, Nagasawa H, Kinoshita S, Asakawa S, Watabe S, Satoh N, Kawashima T, 2016. Bivalve-specific gene expansion in the pearl oyster genome: implications of adaptation to a sessile lifestyle. Zoological Lett. 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Ni X, Zhou Z, Wang L, Lin S, 2018. Acute microplastic exposure raises stress response and suppresses detoxification and immune capacities in the scleractinian coral Pocillopora damicornis. Environ Pollut 243, 66–74. [DOI] [PubMed] [Google Scholar]
- Taraska NG, Anne Bottger S, 2013. Selective initiation and transmission of disseminated neoplasia in the soft shell clam Mya arenaria dependent on natural disease prevalence and animal size. J Invertebr Pathol 112, 94–101. [DOI] [PubMed] [Google Scholar]
- Tasumi S, Vasta GR, 2007. A galectin of unique domain organization from hemocytes of the eastern oyster (Crassostrea virginica) Is a receptor for the protistan parasite Perkinsus marinus. J Immunol 179, 3086–3098. [DOI] [PubMed] [Google Scholar]
- Terada D, Voet ARD, Noguchi H, Kamata K, Ohki M, Addy C, Fujii Y, Yamamoto D, Ozeki Y, Tame JRH, Zhang KYJ, 2017. Computational design of a symmetrical beta-trefoil lectin with cancer cell binding activity. Sci Rep. 7, 5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins-Schiffman E, Coffey WD, Hua W, Nunn BL, Dickinson GH, Roberts SB, 2014. Shotgun proteomics reveals physiological response to ocean acidification in Crassostrea gigas. BMC Genomics. 15, 951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabal Fernández N, Mazon-Suastegui JM, Vazquez-Juarez R, Ascencio-Valle F, Romero J, 2014. Changes in the composition and diversity of the bacterial microbiota associated with oysters (Crassostrea corteziensis, Crassostrea gigas and Crassostrea sikamea) during commercial production. FEMS Microbiol Ecol 88, 69–83. [DOI] [PubMed] [Google Scholar]
- Tran D, Ciutat A, Mat A, Massabuau JC, Hegaret H, Lambert C, Le Goic N, Soudant P, 2015a. The toxic dinoflagellate Alexandrium minutum disrupts daily rhythmic activities at gene transcription, physiological and behavioral levels in the oyster Crassostrea gigas. Aquat Toxicol 158, 41–49. [DOI] [PubMed] [Google Scholar]
- Tran D, Haberkorn H, Soudant P, Ciret P, Massabuau JC, 2010. Behavioral responses of Crassostrea gigas exposed to the harmful algae Alexandrium minutum. Aquaculture 298, 338–345. [Google Scholar]
- Tran NK, Kwon JE, Kang SC, Shim SM, Park TS, 2015b. Crassaostrea gigas oyster shell extract inhibits lipogenesis via suppression of serine palmitoyltransferase. Nat. Prod. Commun. 10, 349–352. [PubMed] [Google Scholar]
- Tsai HJ, Lai CH, Yang HS, 1997. Sperm as a carrier to introduce an exogenous DNA fragment into the oocyte of Japanese abalone (Haliotis divorsicolor suportexta). Transgenic Res 6, 85–95. [DOI] [PubMed] [Google Scholar]
- Tuomola EM, Ouwehand AC, Salminen SJ, 1999. The effect of probiotic bacteria on the adhesion of pathogens to human intestinal mucus. FEMS Immunol Med Microbiol 26, 137–142. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI, 2007. The human microbiome project. Nature 449, 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara A, Sugawara S, Watanabe K, Echigo S, Sato M, Yamaguchi T, Takada H, 2003. Constitutive expression of a bacterial pattern recognition receptor, CD14, in human salivary glands and secretion as a soluble form in saliva. Clin Diagn Lab Immunol 10, 286–292. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ujvari B, Papenfuss AT, Belov K, 2016. Transmissible cancers in an evolutionary context. Bioessays 38 Suppl 1, S14–23. [DOI] [PubMed] [Google Scholar]
- Van Cauwenberghe L, Janssen CR, 2014. Microplastics in bivalves cultured for human consumption. Environ Pollut 193, 65–70. [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel JC, 2014. Piwi and potency: PIWI proteins in animal stem cells and regeneration. Integr Comp Biol 54, 700–713. [DOI] [PubMed] [Google Scholar]
- Vasta GR, Feng C, Bianchet MA, Bachvaroff TR, Tasumi S, 2015. Structural, functional, and evolutionary aspects of galectins in aquatic mollusks: From a sweet tooth to the Trojan horse. Fish Shellfish Immunol. 46, 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatrella A, Fabozzi I, Calabrese C, Maselli R, Pelaia G, 2014. Dupilumab: a novel treatment for asthma. J Asthma Allergy 7, 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitonmaki N, Fuxe J, Hultdin M, Roos G, Pettersson RF, Cao Y, 2003. Immortalization of bovine capillary endothelial cells by hTERT alone involves inactivation of endogenous p16INK4A/pRb. FASEB J 17, 764–766. [DOI] [PubMed] [Google Scholar]
- Vethaak AD, Davies IM, Thain JE, Gubbins MJ, Martínez-Gómez C, Robinson CD, Moffat CF, Burgeot T, Maes T, Wosniok W, Giltrap M, Lang TF, Hylland K, 2017. Integrated indicator framework and methodology for monitoring and assessment of hazardous substances and their effects in the marine environment. Mar. Environ. Res. 124, 11–20. [DOI] [PubMed] [Google Scholar]
- Vezzulli L, Stagnaro L, Grande C, Tassistro G, Canesi L, Pruzzo C, 2018. Comparative 16SrDNA Gene-Based Microbiota Profiles of the Pacific Oyster (Crassostrea gigas) and the Mediterranean Mussel (Mytilus galloprovincialis) from a Shellfish Farm (Ligurian Sea, Italy). Microb Ecol 75, 495–504. [DOI] [PubMed] [Google Scholar]
- Villalba A, Reece KS, Ordas A, Casas SM, Figueras A, 2004. Perkinsosis in molluscs: A review. Aquat. Living Res. 17, 411–432. [Google Scholar]
- Vine NG, Leukes WD, Kaiser H, Daya S, Baxter J, Hecht T, 2004. Competition for attachment of aquaculture candidate probiotic and pathogenic bacteria on fish intestinal mucus. J Fish Dis 27, 319–326. [DOI] [PubMed] [Google Scholar]
- Vizioli J, Salzet M, 2002. Antimicrobial peptides versus parasitic infections? Trends Parasitol 18, 475–476. [DOI] [PubMed] [Google Scholar]
- Vlieghe P, Lisowski V, Martinez J, Khrestchatisky M, 2010. Synthetic therapeutic peptides: science and market. Drug Discov Today 15, 40–56. [DOI] [PubMed] [Google Scholar]
- Waddell SJ, de Andres MC, Tsimbouri PM, Alakpa EV, Cusack M, Dalby MJ, Oreffo RO, 2018. Biomimetic oyster shell-replicated topography alters the behaviour of human skeletal stem cells. J Tissue Eng 9, 2041731418794007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite HR, Donnelly MJ, Walters LJ, 2018. Quantity and types of microplastics in the organic tissues of the eastern oyster Crassostrea virginica and Atlantic mud crab Panopeus herbstii from a Florida estuary. Mar Pollut Bull 129, 179–185. [DOI] [PubMed] [Google Scholar]
- Walker C, Bottger SA, Mulkern J, Jerszyk E, Litvaitis M, Lesser M, 2009. Mass culture and characterization of tumor cells from a naturally occurring invertebrate cancer model: applications for human and animal disease and environmental health. Biol. Bull. 216, 23–39. [DOI] [PubMed] [Google Scholar]
- Walker CW, Van Beneden RJ, Muttray AF, Bottger SA, Kelley ML, Tucker AE, Thomas WK, 2011. p53 Superfamily proteins in marine bivalve cancer and stress biology. Adv Mar Biol. 59, 1–36. [DOI] [PubMed] [Google Scholar]
- Wang L, Song X, Song L, 2018a. The oyster immunity. Dev Comp Immunol. 80, 99–118. [DOI] [PubMed] [Google Scholar]
- Wang R, He J, Wang J, 2016a. Heterotrophic bacterial abundance and diversity in the farming environment and guts of the oyster Crassostrea hongkongensis. J Shellfish Res 35, 343–350. [Google Scholar]
- Wang S, Zhang J, Jiao W, Li J, Xun X, Sun Y, Guo X, Huan P, Dong B, Zhang L, Hu X, Sun X, Wang J, Zhao C, Wang Y, Wang D, Huang X, Wang R, Lv J, Li Y, Zhang Z, Liu B, Lu W, Hui Y, Liang J, Zhou Z, Hou R, Li X, Liu Y, Li H, Ning X, Lin Y, Zhao L, Xing Q, Dou J, Li Y, Mao J, Guo H, Dou H, Li T, Mu C, Jiang W, Fu Q, Fu X, Miao Y, Liu J, Yu Q, Li R, Liao H, Li X, Kong Y, Jiang Z, Chourrout D, Li R, Bao Z, 2017a. Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat Ecol Evol 1, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhang T, Wu C, Wang S, Wang Y, Li H, Wang N, 2017b. Immortalization of chicken preadipocytes by retroviral transduction of chicken TERT and TR. PLoS One 12, e0177348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen Z, Luo G, He W, Xu K, Xu R, Lei Q, Tan J, Wu J, Xing M, 2016b. In-situ-generated vasoactive intestinal peptide loaded microspheres in mussel-inspired polycaprolactone panosheets creating spatiotemporal releasing microenvironment to promote wound healing and angiogenesis. ACS Appl Mater Interfaces 8, 7411–7421. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yoshinaga T, Itoh N, 2018b. New insights into the entrance of Perkinsus olseni in the Manila clam, Ruditapes philippinarum. J Invertebr Pathol 153, 117–121. [DOI] [PubMed] [Google Scholar]
- Ward JE, Beninger PG, MacDonald BA, Thompson RJ, 1991. Direct observations of feeding structures and mechanisms in bivalve molluscs using endoscopic examination and video image analysis. Mar Biol 111,287–291. [Google Scholar]
- Wegner A, Besseling E, Foekema EM, Kamermans P, Koelmans AA, 2012. Effects of nanopolystyrene on the feeding behavior of the blue mussel (Mytilus edulis L.). Environ Toxicol Chem 31,2490–2497. [DOI] [PubMed] [Google Scholar]
- Wei L, Xu F, Wang Y, Cai Z, Yu W, He C, Jiang Q, Xu X, Guo W, Wang X, 2018a. The molecular differentiation of anatomically paired left and right mantles of the Pacific oyster Crassostrea gigas. Mar. Biotechnol. [DOI] [PubMed] [Google Scholar]
- Wei P, Yuan Z, Cai Q, Mao J, Yang X, 2018b. Bioresorbable microspheres with surface-loaded nanosilver and apatite as dual-functional injectable cell carriers for bone regeneration. Macromol Rapid Commun, e1800062. [DOI] [PubMed] [Google Scholar]
- Weinberg JR, Leavitt DF, Lancaster BA, Capuzzo JM, 1997. Experimental field studies with Mya arenaria (Bivalvia) on the induction and effect of hematopoietic neoplasia. J Invertebr Pathol 69, 183–194. [DOI] [PubMed] [Google Scholar]
- Wells JM, Watt FM, 2018. Diverse mechanisms for endogenous regeneration and repair in mammalian organs. Nature 557, 322–328. [DOI] [PubMed] [Google Scholar]
- Wen CM, Kou GH, Chen SN, 1993. Establishment of cell lines from the Pacific oyster. In vitro cellular & developmental biology. Animal 29A, 901–903. [DOI] [PubMed] [Google Scholar]
- Westbroek P, Marin F, 1998. A marriage of bone and nacre. Nature 392, 861–862. [DOI] [PubMed] [Google Scholar]
- Wijayalath W, Majji S, Kleschenko Y, Pow-Sang L, Brumeanu TD, Villasante EF, Vasta GR, Fernández-Robledo J-A, Casares S, 2014. Humanized HLA-DR4 mice fed with the protozoan pathogen of oysters Perkinsus marinus (Dermo) do not develop noticeable pathology but elicit systemic immunity. PLoS One 9, e87435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildish D, Lassus P, Martin J, Saulnier A, Bardouil M, 1998. Effect of the PSP-causing dinoflagellate, Alexandrium sp. on the initial feeding response of Crassostrea gigas. Aquat Living Resour 11,35–43. [Google Scholar]
- Wright SL, Thompson RC, Galloway TS, 2013. The physical impacts of microplastics on marine organisms: a review. Environ Pollut 178, 483–492. [DOI] [PubMed] [Google Scholar]
- Xing J, Pales Espinosa E, Perrigault M, Allam B, 2011. Identification, molecular characterization and expression analysis of a mucosal C-type lectin in the eastern oyster, Crassostrea virginica. Fish Shellfish Immunol 30, 851–858. [DOI] [PubMed] [Google Scholar]
- Xu XY, Lee WT, Chan AKY, Lo HS, Shin PKS, Cheung SG, 2017. Microplastic ingestion reduces energy intake in the clam Atactodea striata. Mar Pollut Bull 124, 798–802. [DOI] [PubMed] [Google Scholar]
- Yang YS, Mitta G, Chavanieu A, Calas B, Sanchez JF, Roch P, Aumelas A, 2000. Solution structure and activity of the synthetic four-disulfide bond Mediterranean mussel defensin (MGD-1). Biochemistry 39, 14436–14447. [DOI] [PubMed] [Google Scholar]
- Yin X, Li J, Salmon B, Huang L, Lim WH, Liu B, Hunter DJ, Ransom RC, Singh G, Gillette M, Zou S, Helms JA, 2015. Wnt signaling and its contribution to craniofacial tissue homeostasis. J Dent Res 94, 1487–1494. [DOI] [PubMed] [Google Scholar]
- Yokoyama H, 2008. Initiation of limb regeneration: the critical steps for regenerative capacity. Dev Growth Differ 50, 13–22. [DOI] [PubMed] [Google Scholar]
- Yoshino TP, Bickham U, Bayne CJ, 2013. Molluscan cells in culture: primary cell cultures and cell lines. Can J Zool 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M, 2002. Antimicrobial peptides of multicellular organisms. Nature 415, 389–395. [DOI] [PubMed] [Google Scholar]
- Zhang G, Brion A, Willemin AS, Piet MH, Moby V, Bianchi A, Mainard D, Galois L, Gillet P, Rousseau M, 2017. Nacre, a natural, multi-use, and timely biomaterial for bone graft substitution. J. Biomed. Mater. Res. A 105, 662–671. [DOI] [PubMed] [Google Scholar]
- Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H, Xiong Z, Que H, Xie Y, Holland PW, Paps J, Zhu Y, Wu F, Chen Y, Wang J, Peng C, Meng J, Yang L, Liu J, Wen B, Zhang N, Huang Z, Zhu Q, Feng Y, Mount A, Hedgecock D, Xu Z, Liu Y, Domazet-Loso T, Du Y, Sun X, Zhang S, Liu B, Cheng P, Jiang X, Li J, Fan D, Wang W, Fu W, Wang T, Wang B, Zhang J, Peng Z, Li Y, Li N, Wang J, Chen M, He Y, Tan F, Song X, Zheng Q, Huang R, Yang H, Du X, Chen L, Yang M, Gaffney PM, Wang S, Luo L, She Z, Ming Y, Huang W, Zhang S, Huang B, Zhang Y, Qu T, Ni P, Miao G, Wang J, Wang Q, Steinberg CE, Wang H, Li N, Qian L, Zhang G, Li Y, Yang H, Liu X, Wang J, Yin Y, Wang J, 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. [DOI] [PubMed] [Google Scholar]
- Zhao M, Shi Y, He M, Huang X, Wang Q, 2016. PfSMAD4 plays a role in biomineralization and can transduce bone morphogenetic protein-2 signals in the pearl oyster Pinctada fucata. BMC Dev. Biol. 16, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Takeuchi T, Luo YJ, Ishikawa A, Kobayashi T, Koyanagi R, Villar-Briones A, Yamada L, Sawada H, Iwanaga S, Nagai K, Satoh N, Endo K, 2018. Dual gene repertoires for larval and adult shells reveal molecules essential for molluscan shell formation. Mol Biol Evol, doi: 10.1093/molbev/msy1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuykov M, Pelletier E, Harper DAT, 2013. Bivalve mollusks in metal pollution studies: From bioaccumulation to biomonitoring. Chemosphere 93, 201–208. [DOI] [PubMed] [Google Scholar]