Abstract
Recombination, along with sister chromatid cohesion, is used during meiosis to physically connect homologous chromosomes so that they can be segregated properly at the first meiotic division. Recombination is initiated by the introduction of programmed double strand breaks (DSBs) into the genome, a subset of which are processed into crossovers. In budding yeast, the regulation of meiotic DSB repair is controlled by a meiosis-specific kinase called Mek1. Mek1 kinase activity promotes recombination between homologs, rather than sister chromatids, as well as the processing of recombination intermediates along a pathway that results in synapsis of homologous chromosomes and the distribution of crossovers throughout the genome. In addition, Mek1 kinase activity provides a readout for the number of DSBs in the cell as part of the meiotic recombination checkpoint. This checkpoint delays entry into the first meiotic division until DSBs have been repaired by inhibiting the activity of the meiosis-specific transcription factor Ndt80, a site-specific DNA binding protein that activates transcription of over 300 target genes. Recent work has shown that Mek1 binds to Ndt80 and phosphorylates it on multiple sites, including the DNA binding domain, thereby preventing Ndt80 from activating transcription. As DSBs are repaired, Mek1 is removed from chromosomes and its activity decreases. Loss of the inhibitory Mek1 phosphates and phosphorylation of Ndt80 by the meiosis-specific kinase, Ime2, promote Ndt80 activity such that Ndt80 transcribes its own gene in a positive feedback loop, as well as genes required for the completion of recombination and entry into the meiotic divisions. Mek1 is therefore the key regulator of meiotic recombination in yeast.
Keywords: yeast, meiosis, recombination, Mek1, Ndt80, checkpoint, double strand break repair
Meiosis is a fundamental biological process necessary for sexual reproduction. This unique cell division reduces the chromosome number of a cell in a highly specific way to make gametes that contain a single copy of each homolog. Fusion of two haploid gametes then restores the diploid state, keeping the chromosome number constant from generation to generation. The reduction in chromosome number during meiosis results when duplicated chromosomes undergo two consecutive rounds of chromosome segregation without an intervening S phase. For many organisms such as budding yeast and humans, which have 16 and 23 pairs of homologous chromosomes, respectively, sorting chromosomes into gametes is a daunting task. To determine which chromosomes are homologs, recombination is used to generate crossovers between the non-sister chromatids of homologous chromosomes. These crossovers, in combination with sister chromatid cohesion, physically connect homologs, allowing them to align and segregate properly at the first meiotic division (PETRONCZKI et al. 2003).
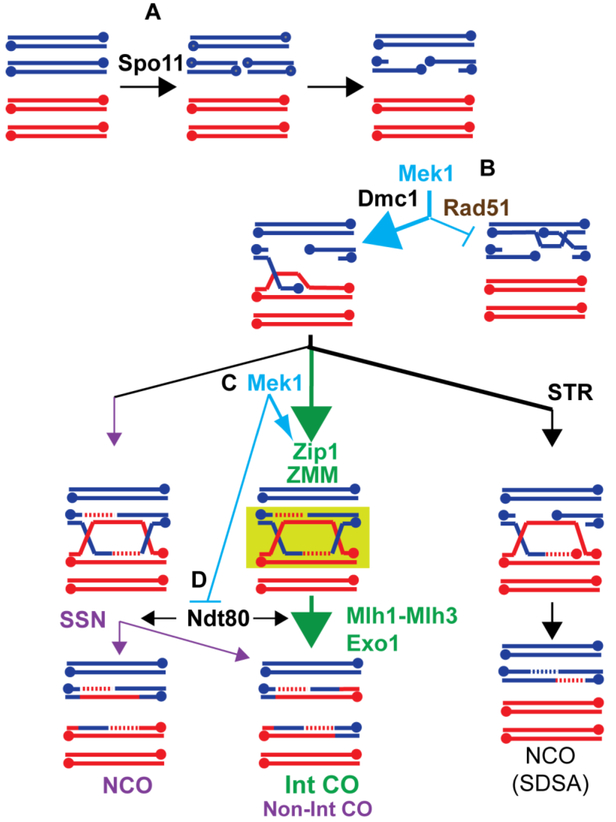
Recombination is initiated by double strand breaks (DSBs) that are deliberately created in preferred regions of the genome called “hotspots” by a highly conserved protein called Spo11 (Figure 1A) (KEENEY et al. 2014). The 5’ ends of the DSBs are resected and the 3’ single strand tails are bound by the mitotic recombinase, Rad51, and a meiosis-specific recombinase, Dmc1 (HUNTER 2007; BROWN et al. 2015). In combination with accessory factors, the resulting nucleoprotein filament mediates “strand invasion”, which involves searching for the homologous sequence on a non-sister chromatid, locally denaturing the duplex and annealing to the complementary strand to create a displacement (D)-loop. Further processing of this intermediate results in a double Holliday junction that can be resolved to form either a crossover or noncrossover (see below).
Figure 1. Mek1 regulates multiple steps during meiotic recombination to promote the formation of interhomolog crossovers.
(A) Homologous chromosomes (indicated by red and blue) replicate to make pairs of identical sister chromatids. Spo11 introduces a DSB on one of the four chromatids. The 5’ ends of the DSB are then resected to generated 3’ single stranded tails (3’ ends are indicated by dots). (B) The 3’ ends are bound by the Rad51 and Dmc1 recombinases which mediate strand invasion of an homologous duplex. Mek1 activity ensures that the bulk of strand invasion events occur via the homolog using Dmc1, in part by preventing Rad51 from interacting with its accessory factor, Rad54. (C) Mek1 promotes the ZMM pathway of crossover formation by enabling Cdc7-Dbf4 to phosphorylate a conserved region of the C-terminus of the transverse filament protein, Zip1. The green box indicates the protection of the double Holliday junction from disassembly or dissolution, resulting in biased resolution to form interfering crossovers (Int-CO). (D) Mek1 phosphorylates the Ndt80 transcription factor, keeping it inactive while double Holliday junction formation is occurring. Activation of Ndt80 results in the production of the polo-like kinase Cdc5, which triggers Holliday junction resolution.
Because it is critical for the survival of a species that every pair of homologs gets at least one crossover, many safeguards have evolved to ensure this outcome. First, more DSBs are introduced into the genome during meiosis than the number of crossovers required (for example, budding yeast undergoes ~160 DSBs but has only 16 pairs of homologs while mice have 250–300 DSBs for 20 pairs of homologs) (PAN et al. 2011; KEENEY et al. 2014). Second, multiple mechanisms act to promote recombination between homologs, as opposed to sister chromatids. Third, there is a specialized recombination pathway for generating crossovers that are distributed throughout the genome. Finally, DSBs that do not become crossovers are repaired either as non-crossovers or by sister chromatid recombination before chromosomes segregate at the first meiotic division to prevent broken chromosomes from making aneuploid gametes. In budding yeast, the regulation of recombination and its coordination with meiotic progression are under the control of the meiosis-specific kinase, Mek1 (also known as Mre4) (ROCKMILL AND ROEDER 1991; LEEM AND OGAWA 1992; WU et al. 2010; HOLLINGSWORTH 2016; CHEN et al. 2018).
Mek1 regulates interhomolog bias as well as recombination pathway choice
When a 3’ end is searching for homology, there are three possible templates: the sister chromatid or the two chromatids of the homolog. Because only interhomolog crossovers connect chromosomes, strand invasion during meiosis is biased to occur between the homologs (LAO AND HUNTER 2010). The key player in regulating interhomolog bias is Mek1 (Figure 1B). In the absence of Mek1 kinase activity, DSBs are repaired using the sister chromatid (NIU et al. 2005; KIM et al. 2010). While this repair produces intact chromosomes, the homologs are not connected, and random chromosome segregation results in high levels of aneuploidy and inviable spores (ROCKMILL AND ROEDER 1991; LEEM AND OGAWA 1992). One way that Mek1 promotes interhomolog recombination is by inhibition of Rad51 (Figure 1B). In vegetative cells, Rad51 is used to repair DSBs and this repair is biased to occur between sister chromatids (KADYK AND HARTWELL 1992; BZYMEK et al. 2010). In meiotic cells, the Rad51 protein (but not its activity) is required for interhomolog bias, while Dmc1 is responsible for most of the DSB repair that occurs (SCHWACHA AND KLECKNER 1997; LAO et al. 2008; CLOUD et al. 2012). Rad54 is an accessory factor that binds to Rad51 and promotes its strand invasion activity (PETUKHOVA et al. 1999). Mek1 inhibits the Rad51-Rad54 interaction in two different ways: (1) it directly phosphorylates threonine (T) 132 on Rad54, thereby reducing the affinity of Rad54 for Rad51 (NIU et al. 2009) and (2) it phosphorylates and stabilizes Hed1, a meiosis-specific protein that binds to Rad51, thereby excluding Rad54 (TSUBOUCHI AND ROEDER 2006; BUSYGINA et al. 2008; CALLENDER et al. 2016). Removing these constraints on Rad51 activity only reduces interhomolog bias by two-fold, as Dmc1 itself can inhibit Rad51 (HONG et al. 2013; LAO et al. 2013; LIU et al. 2014). However, when the hed1Δ RAD54-T132A mutations are combined with a hypomorphic allele of DMC1 that is delayed in filament formation, but otherwise behaves like wild type, interhomolog bias is reduced 8-fold and spore viability is reduced (LIU et al. 2014). These results argue that Mek1 inhibition helps prevent Rad51 from competing with Dmc1 to repair DSBs. It should be noted that the presence of DMC1 is not sufficient to ensure interhomolog recombination, as inactivation of Mek1 results in intersister recombination even in DMC1 cells (GOLDFARB AND LICHTEN 2010; KIM et al. 2010). Therefore, in addition to the inhibition of Rad51 and the presence of Dmc1, there must be additional mechanisms controlled by Mek1 that promote interhomolog recombination. For example, it has been proposed that Mek1 antagonizes sister chromatid cohesion locally around DSBs to make it easier to invade a homolog, although how this occurs is unknown (KIM et al. 2010).
After interhomolog strand invasion, there are different ways in which the resulting D-loop can be processed (Figure 1). One pathway, called synthesis dependent strand annealing (SDSA), occurs when the extended invading strand is disassembled from the D-loop by the STR protein complex containing Sgs1 (an ortholog of Bloom helicase in humans), Topoisomerase III and Rmi1 (ALLERS AND LICHTEN 2001; MCMAHILL et al. 2007; OH et al. 2007; DE MUYT et al. 2012; KAUR et al. 2015; TANG et al. 2015). The extended strand then anneals to the 3’ end on the other side of the break, resulting in a noncrossover. The major pathway for crossover formation in budding yeast occurs via a group of functionally distinct proteins known collectively as the ZMM proteins (BÖRNER et al. 2004; LYNN et al. 2007). The ZMM pathway of recombination leads to the stable association of homologous chromosomes by formation of a meiosis-specific chromosome structure called the synaptonemal complex (see below) (BÖRNER et al. 2004). Crossovers generated by this pathway are distributed throughout the genome by a mysterious process known as “interference” (KOHL AND SEKELSKY 2013). D-loop intermediates processed using the ZMM pathway are protected from disassembly by STR (DE MUYT et al. 2012) (Figure 1). Instead D-loops are stabilized to form “single end invasion” (SEI) intermediates (HUNTER AND KLECKNER 2001). Extension of the invading strand by DNA synthesis displaces the complementary strand of the invaded duplex, which then anneals to the other side of the break to form a double Holliday junction (SCHWACHA AND KLECKNER 1995). Resolution of ZMM-mediated double Holliday junctions is biased to generate crossovers (ALLERS AND LICHTEN 2001; BÖRNER et al. 2004).
A key member of the ZMM group is Zip1, which is the transverse filament protein that connects condensed pairs of sister chromatids together to form the synaptonemal complex (SYM et al. 1993; DONG AND ROEDER 2000; BÖRNER et al. 2004). In addition to “zippering up the chromosomes”, Zip1 also plays an early role in the ZMM pathway that is regulated by Mek1 (Figure 1C)(CHEN et al. 2015). Mek1 is required for phosphorylation of a conserved region in the C-terminus of Zip1 by the Cdc7-Dbf4 (DDK) cell cycle kinase (CHEN et al. 2015). Phosphorylation of this region can be prevented using the zip1–4A mutant which substitutes alanines for the serines that are phosphorylated. Phenotypic characterization of zip1–4A has revealed that phosphorylation of the Zip1 C-terminus is necessary for ZMM-mediated crossovers and synapsis (CHEN et al. 2015). The double Holliday junctions undergo unbiased resolution by structure-selective endonucleases (SSNs) such as Mus81-Mms4 to give both crossovers and non-crossovers (DE MUYT et al. 2012; CHEN et al. 2015). Finally, Mek1 indirectly controls the timing of Holliday junction resolution by regulating the activity of the meiosis-specific transcription factor, Ndt80 (Figure 1D)(see below). NDT80 is required to transcribe the polo-like kinase CDC5, which then triggers Holliday junction resolution (SOURIRAJAN AND LICHTEN 2008; MATOS et al. 2011). Mek1 therefore regulates all the steps of recombination in yeast.
Mek1 controls the meiotic recombination checkpoint to provide time for interhomolog recombination
In vegetative cells, the unexpected appearance of a DSB triggers the “DNA damage checkpoint” (HARRISON AND HABER 2006). This checkpoint delays cell cycle progression to provide time for DSBs to be repaired and works using the Tel1 and Mec1 kinases (ATM and ATR in mammals, respectively). These kinases are recruited to DSBs where they phosphorylate an adaptor protein called Rad9. An FHA-domain containing effector kinase, Rad53, binds to phosphorylated Rad9 where it autoactivates and phosphorylates target proteins that prevent entry into mitosis.
Although Spo11-generated DSBs are programmed to occur during meiosis, they are just as lethal as a break in a mitotic cell if left unrepaired. Meiotic DSB repair is therefore highly regulated using meiosis-specific modifications of the DNA damage checkpoint (LYDALL et al. 1996; SUBRAMANIAN AND HOCHWAGEN 2014). The meiotic recombination checkpoint delays meiotic prophase to provide time for interhomolog recombination to occur. Given its role in regulating partner and recombination pathway choice, it is not surprising that the effector kinase regulating the meiotic recombination checkpoint is Mek1 (XU et al. 1997; PEREZ-HIDALGO et al. 2003). Similar to Rad53, Mek1 also contains an FHA domain. MEK1’s role in this checkpoint was first established using the rad50S mutant which makes DSBs whose ends are not resected and therefore cannot be repaired (ALANI et al. 1990). The unrepaired DSBs trigger the checkpoint, causing a delay in meiotic progression. Deletion of MEK1 eliminates the rad50S progression delay, indicating it is required for the meiotic recombination checkpoint (XU et al. 1997).
Mek1 activation occurs in the context of a meiosis-specific structure formed along sister chromatids called the axial element
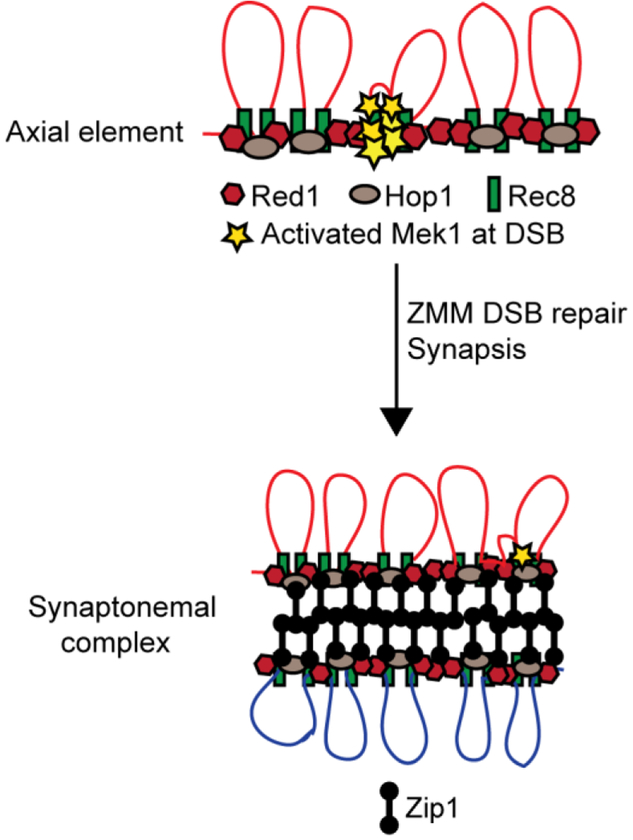
One of the key differences between the DNA damage and meiotic recombination checkpoints is that the latter occurs in the context of a meiosis-specific structure called the axial element (Figure 2). Axial elements are generated when condensation of sister chromatids forms chromatin loops that are tethered along a protein core (BLAT AND KLECKNER 1999). In yeast, axial elements contain the meiosis-specific Hop1 and Red1 proteins, as well as cohesin complexes containing the meiosis-specific kleisin subunit, Rec8 (HOLLINGSWORTH et al. 1990; SMITH AND ROEDER 1997; BLAT AND KLECKNER 1999; KLEIN et al. 1999; LORENZ et al. 2004; PANIZZA et al. 2011; BROWN et al. 2018). Hop1 contains an evolutionarily conserved sequence called the HORMA domain that mediates both Hop1 homoligomerization and heteroligomerization with Red1 (ARAVIND AND KOONIN 1998; DE LOS SANTOS AND HOLLINGSWORTH 1999; WOLTERING et al. 2000; WEST et al. 2018).
Figure 2. Cartoon of an axial element and synaptonemal complex from budding yeast.
After recruitment of Tel1 and Mec1 to Spo11 breaks, these checkpoint kinases phosphorylate Hop1 as the adaptor protein instead of Rad9 (CARBALLO et al. 2008). Mek1 binds to phosphorylated Hop1 via its FHA domain where it activates itself in trans (NIU et al. 2007; CARBALLO et al. 2008). The C-terminus of Hop1 is required for Mek1 activation by promoting Mek1 oligomerization (NIU et al. 2005). This function of Hop1 can be bypassed by ectopic dimerization of Mek1 using GST (NIU et al. 2005). Ectopic dimerization does not bypass the need for Mek1 to bind to phospho-Hop1 via its FHA domain, but it does enhance Mek1 activity (NIU et al. 2005; WU et al. 2010). Activation of Mek1 also requires RED1, as well as the Hop1-Red1 interaction (NIU et al. 2007). These requirements for Mek1 activation explain why hop1 and red1 mutants resemble mek1Δ in being defective in interhomolog bias, resulting in inviable spores (HOLLINGSWORTH AND BYERS 1989; ROCKMILL AND ROEDER 1990; SCHWACHA AND KLECKNER 1994; KIM et al. 2010).
How is it that DSBs, which occur in the chromatin loops, activate a kinase that is localized to the chromosome axis? Several discoveries led to the hypothesis that hotspot sequences within chromatin loops are tethered to the axial elements where DSBs are then generated (Figure 2). First, an axis protein called Mer2, which is required for making DSBs, is phosphorylated by DDK as the replication fork passes during premeiotic S-phase (COOL AND MALONE 1992; ROCKMILL et al. 1995; SASANUMA et al. 2008; WAN et al. 2008; MURAKAMI AND KEENEY 2014). Phosphorylated Mer2 recruits Mei4 and Rec114 which brings Spo11 to the axes (LI et al. 2006; SASANUMA et al. 2008). In addition, Mer2 binds to Spp1 which in turn interacts with trimethylated histones adjacent to DSB hotspots, thereby recruiting hotspot sequences to the axes to be cleaved by Spo11 (ACQUAVIVA et al. 2013; SOMMERMEYER et al. 2013; ADAM et al. 2018). DSBs are therefore generated on the axial elements. Chromatin-immunoprecipitation experiments using an antibody specific for phosphorylated Histone H3 threonine 11, a substrate of Mek1, has shown that Mek1 kinase activity is highest on chromosome axes and correlates with the presence of Hop1 and Red1. Interestingly, Mek1 phosphorylation of histone H3 can spread for several kilobases around a DSB (KNIEWEL et al. 2017).
Mek1 activity provides a readout for the presence of DSBs
For the meiotic recombination checkpoint to monitor DSB repair, it must be able to detect the presence and amount of DSBs in the cell. Mek1 activity correlates with both DSB levels and the rate of meiotic progression, suggesting it is the sensor for the checkpoint (PRUGAR et al. 2017). In DSB-defective mutants (as well as mek1Δ, hop1Δ and red1Δ), Mek1 is inactive and meiotic progression occurs faster than in wild type due to the absence of the checkpoint (MALONE et al. 2004; NIU et al. 2007). In dmc1Δ strains, DSBs are formed but unrepaired due to the inhibition of Rad51 and the cells arrest in meiotic prophase with high levels of DSBs (BISHOP et al. 1992; HUNTER AND KLECKNER 2001; NIU et al. 2005). Mek1 kinase activity (detected using an antibody specific for the Mek1-phosphorylated form of Hed1) persists at a high level in dmc1Δ strains as well (CALLENDER et al. 2016; PRUGAR et al. 2017). In contrast, deletion of NDT80, which is needed to exit from meiotic prophase, arrests cells at the pachytene stage when synapsis is complete (XU et al. 1995; WINTER 2012). By this stage most DSBs have been processed either into noncrossovers or double Holliday junctions awaiting resolution (ALLERS AND LICHTEN 2001). Furthermore, synapsis downregulates, but does not abolish, Spo11 activity, resulting in a low number of DSBs (1–10/cell) (THACKER et al. 2014; SUBRAMANIAN et al. 2016) (Figure 2). In an ndt80Δ diploid, Hed1 phosphorylation is reduced, but not eliminated, indicating a decrease in Mek1 activity that is consistent with the removal of Mek1 from chromosomes that occurs as homologs synapse (SUBRAMANIAN et al. 2016; PRUGAR et al. 2017). Another example of the correlation between DSB levels and the activity of the meiotic recombination checkpoint is the meiotic progression delay observed for the zip1Δ and zip1–4A mutants. Both of these mutants are defective in synapsis and therefore in turning down Spo11 activity, resulting in increased numbers of DSBs compared to wild type (SYM et al. 1993; THACKER et al. 2014; CHEN et al. 2015). However, for reasons that are unclear, the zip1–4A diploid exhibits significantly higher levels of DSBs compared to the zip1Δ (CHEN et al. 2015). This mutant also enters into the meiotic divisions more slowly than zip1Δ. Taken together, these results indicate that meiotic progression in yeast is regulated using Mek1 kinase activity as a readout for the number of DSBs. Furthermore, they argue against the presence of a synapsis checkpoint in yeast. The spo11Δ, zip1Δ and zip1–4A mutants are all defective in synapsis but exhibit differences in meiotic progression that mirror instead the number of DSBs.
The target of the meiotic recombination checkpoint is the meiosis-specific transcription factor, Ndt80
Ndt80 is a transcriptional activator that can be divided in three domains: an N-terminal DNA binding domain, a C-terminal activation domain, and the region in the middle (LAMOUREUX et al. 2002; MONTANO et al. 2002; SOPKO et al. 2002). Ndt80 binds to a specific DNA sequence called the middle sporulation element (MSE) in the promoters of >300 target genes to activate their transcription (HEPWORTH et al. 1995; OZSARAC et al. 1997; CHU AND HERSKOWITZ 1998). One of these genes is CDC5, which, together with DDK, is sufficient to trigger Holliday junction resolution and degradation of Red1, resulting in the destruction of the SC (SOURIRAJAN AND LICHTEN 2008; OKAZ et al. 2012; ARGUNHAN et al. 2017; PRUGAR et al. 2017; TSUBOUCHI et al. 2018). Another Ndt80 target is the CLB1 cyclin, which together with Cdc28 forms the cyclin-dependent kinase (CDK) needed for Meiosis I spindle formation and meiotic progression (SHUSTER AND BYERS 1989; BENJAMIN et al. 2003). When cells enter meiosis, a meiosis-specific E3 ligase degrades mitotic regulators like Cdc5 and Clb1, thereby making meiotic progression dependent upon their transcription by Ndt80 (OKAZ et al. 2012). Compelling evidence showing that Ndt80 is the target of the meiotic recombination checkpoint was the discovery of a 57 amino acid domain within the middle region of the protein that is required for the meiotic recombination checkpoint delay/arrest in mutants such as zip1Δ and dmc1Δ (WANG et al. 2011). Deletion of this “bypass checkpoint” (bc) domain creates a version of Ndt80 (NDT80-bc) which functions as a transcriptional activator but no longer responds to the meiotic recombination checkpoint (WANG et al. 2011). Mek1 signals the presence of DSBs and Ndt80 receives the signal—the question is how?
Mek1 coordinates exit from meiotic prophase with DSB repair by directly inhibiting the transcriptional activity of Ndt80
The answer to this question was recently revealed by the discovery that Mek1 binds and negatively regulates the Ndt80 transcription factor in response to the level of DSBs in the cell (CHEN et al. 2018). The transcriptional regulation of the NDT80 gene itself is key to understanding how it functions in the checkpoint (WINTER 2012). In vegetative cells, transcription of early meiotic genes such as SPO11, RED1, HOP1, MEK1 and DMC1 is repressed by the Ume6 complex (STRICH et al. 1994; GOLDMARK et al. 2000).
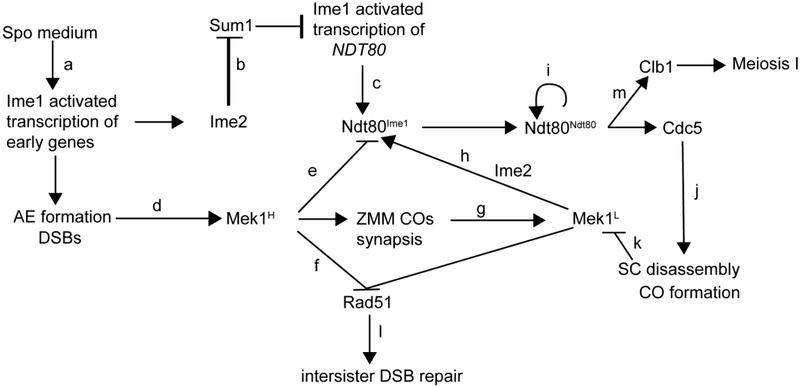
When diploid cells are transferred to sporulation (Spo) medium which lacks nitrogen and uses a non-fermentable carbon source such as acetate, the Ume6 repressor is replaced by the transcriptional activator Ime1 and early gene expression proceeds (Figure 3a)(PAK AND SEGALL 2002; MALLORY et al. 2007). The NDT80 promoter is also repressed by Ume6, but has an additional repressor bound at MSE sites within the promoter called Sum1 (XIE et al. 1999). In meiotic cells, Sum1 must be removed before Ime1 can activate transcription of the NDT80 gene (PAK AND SEGALL 2002). This removal requires phosphorylation by Ime2, which is encoded by an early gene (Figure 3b). The need to transcribe and translate IME2, as well as the fact that phosphorylation of Sum1 by DDK and CDK is also necessary to eliminate repression, means that transcription of NDT80 is delayed relative to the other early genes (Figure 3c) (AHMED et al. 2009; SHIN et al. 2010; LO et al. 2012). This delay provides time for making axial elements, generating DSBs and activating Mek1 (Figure 3d) (CHEN et al. 2018).
Figure 3. Model for how Mek1 coordinates DSB repair with meiotic progression in budding yeast.
See text for description of the indicated steps.
In vegetative yeast cells, a single DSB is sufficient to arrest cells, but in meiosis, a threshold number of DSBs is required to trigger the meiotic recombination checkpoint (CALLENDER AND HOLLINGSWORTH 2010; GRAY et al. 2013). A threshold amount of DSBs is also necessary to activate sufficient Mek1 to impose interhomolog bias, as evidenced by the fact that early in meiotic prophase DSBs are repaired preferentially using sister chromatids (JOSHI et al. 2015). By the time Ime1-activated NDT80 transcription occurs, sufficient Mek1 has been activated (indicated as Mek1H in Figure 3) to inhibit the low level of Ndt80 protein that results from Ime1 (Ndt80Ime1) (Figure 3e). In addition, Mek1H inhibits Rad51 activity and promotes interhomolog bias (Figure 3f).
Recent work has shown that the mechanism by which Mek1 inhibits Ndt80 is direct (CHEN et al. 2018). Mek1 binds to a conserved five amino acid sequence (RPSKR) within the Ndt80 bc domain and phosphorylates the transcription factor at multiple sites within the DNA binding domain and the middle region (CHEN et al. 2018). The consensus phosphorylation site of Mek1 is RXXT/S and Ndt80 contains 10 such sites (MOK et al. 2010; SUHANDYNATA et al. 2016). Preventing phosphorylation using alanine substitutions at these sites, as well as a few non-consensus sites (ndt80–10AMS), results in partial bypass of the dmc1Δ arrest (CHEN et al. 2018). Nearly complete checkpoint bypass occurs when the RPSKR sequence is deleted from the bc domain in NDT80, suggesting that binding of Mek1 to Ndt80 allows phosphorylation of non-consensus sites in the ndt80–10AMS mutant.
Four of the Mek1 consensus sites in the DNA binding domain are located immediately adjacent to the DNA sugar-phosphate backbone when bound to an MS (LAMOUREUX et al. 2002; MONTANO et al. 2002; CHEN et al. 2018). A compelling hypothesis therefore is that negative charges on the phosphates prevent DNA binding by repelling the negatively charged DNA sugar-phosphate backbone. Consistent with this idea, substitution of the DNA binding domain consensus sites with negatively charged aspartic acid creates a constitutively inactive form of Ndt80 in both dmc1Δ and DMC1 diploids (CHEN et al. 2018). The ndt80–6D mutant enters meiosis but arrests in meiotic prophase and is unable to activate transcription of itself, CDC5 or CLB1 (CHEN et al. 2018). The amino acid asparagine has a nearly identical side chain as aspartic acid but is not negatively charged. The NDT80–6N mutant sporulates efficiently, indicating that it is the negative charge which is inhibiting the activity of ndt80–6D. Negative charges disrupt both specific and non-specific DNA binding by recombinant Ndt80 DNA binding domain in vitro (CHEN et al. 2018). Finally, Mek1 directly phosphorylates at least one of the DNA binding domain consensus sites in vitro.
As DSBs are repaired by the ZMM pathway resulting in synapsis, Mek1 is removed from chromosomes and Mek1 kinase activity goes down (indicated as Mek1L in Figure 3)(Figure 3g) (SUBRAMANIAN et al. 2016; PRUGAR et al. 2017). The combination of Ime2 phosphorylation and reduced level of Mek1 phosphorylation activates Ndt80 (Figure 3h), which binds to MSEs in its own promoter to activate transcription of the NDT80 gene in a positive feedback loop (indicated Ndt80Ndt80 in Figure 3i). Ndt80 also promotes transcription of CDC5, which, in addition to triggering Holliday junction resolution, works in conjunction with DDK to promote degradation of Red1 (Figure 3j), resulting in the complete inactivation of Mek1 (Figure 3k). Rad51 is then able to repair any remaining DSBs (Figure 3l) while Ndt80-activated transcription of CLB1 allows entry into Meiosis I (Figure 3m) (ARGUNHAN et al. 2017; PRUGAR et al. 2017)..
Ndt80 activity is controlled by an antagonistic interaction between Mek1 and Ime2
It was known for over 20 years that MEK1 is required for the meiotic recombination checkpoint and that the target is Ndt80 (XU et al. 1997; CHU AND HERSKOWITZ 1998). Yet the simple model that Mek1 negatively regulates Ndt80 by phosphorylating the transcription factor was ignored until recently. A major reason is because the inhibitory phosphorylation by Mek1 is obscured by phosphorylation from a second kinase, Ime2. During wild-type meiosis, Ndt80 exhibits a robust mobility shift that is dependent upon phosphorylation by Ime2 (SOPKO et al. 2002; BENJAMIN et al. 2003; SHUBASSI et al. 2003). In checkpoint arrested cells, hyperphosphorylation of Ndt80 is lost, and only a low level of Ime1-dependent Ndt80 protein is observed (TUNG et al. 2000; SHUBASSI et al. 2003; CHEN et al. 2018). Ndt80 hyperphosphorylation is restored in the absence of Mek1 kinase activity (TUNG et al. 2000; CHEN et al. 2018). These observations were interpreted to mean that (1) Ndt80 is not a substrate of Mek1 and (2) Ime2 phosphorylation activates Ndt80 and the checkpoint works by preventing this activating phosphorylation (TUNG et al. 2000). One problem with this simple interpretation, however, is that there is no checkpoint in the absence of MEK1, and so Ndt80 in active and phosphorylated by Ime2. Therefore, whether Ndt80 is phosphorylated by Mek1 cannot be determined simply by looking at Ndt80 mobility in the presence or absence of Mek1 activity.
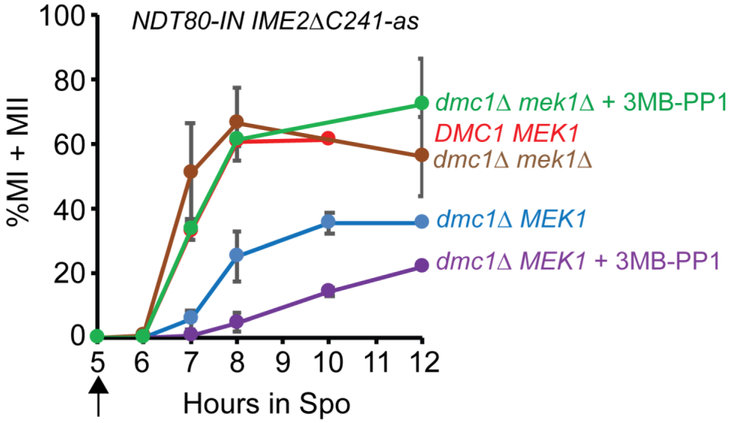
Recently, (CHEN et al. 2018) showed that inactive Ndt80 is phosphorylated in dmc1Δ-arrested cells independently of IME2, raising the possibility that this phosphorylation is due to Mek1. The dogma is that Ime2 phosphorylation of Ndt80 activates the transcription factor, although there are studies which show that Ime2 phosphorylation is not required for Ndt80 to activate transcription (SOPKO et al. 2002; BENJAMIN et al. 2003). We propose an alternative hypothesis that Ime2 phosphorylation indirectly activates Ndt80 by promoting the removal of the inhibitory Mek1 phosphates. This idea was tested using a dmc1Δ IME2ΔC241-as NDT80-IN diploid. The NDT80-IN genotype consists of NDT80 under the control of the GAL1 promoter in combination with a GAL4-estrogen receptor (GAL4-ER) fusion that allows induction of NDT80 transcription by addition of estradiol to the Spo medium (BENJAMIN et al. 2003; CARLILE AND AMON 2008). Using a heterologous promoter allows the production of similar levels of NDT80 whether or not the checkpoint is active (CHEN et al. 2018). IME2ΔC241-as contains a truncation of a C-terminal inhibitory domain, resulting in a constitutively active kinase, as well as an analog-sensitive mutation that allows inactivation of the kinase using 3-MB-PP1 (BENJAMIN et al. 2003; SARI et al. 2008; JIN et al. 2015). After NDT80 induction with estradiol, approximately 40% of the dmc1Δ IME2ΔC241-as NDT80-IN cells active Ime2ΔC241 entered the meiotic divisions, in contrast to a dmc1Δ NDT80-IN diploid which exhibited a nearly complete prophase arrest, as expected if the meiotic recombination checkpoint is working (Figure 4)(CHEN et al. 2018). One explanation for the partial checkpoint bypass is that the truncated, constitutively active Ime2ΔC241-as protein has enhanced stability compared to wild type and thus may be able to counteract the inhibitory Mek1 phosphates in a way that normal levels of Ime2 cannot. Inhibiting Ime2ΔC241-as with 3MB-PP1 decreases the rate of meiotic progression, confirming that the partial bypass requires Ime2 kinase activity (Figure 4)(CHEN et al. 2018).
Figure 4. Ime2 is not required to activate Ndt80 in the absence of MEK1.
Diploids containing NDT80-IN IME2ΔC241-as and either DMC1 MEK1 (yLJ92), dmc1Δ MEK1 (NH2451) or dmc1Δ mek1Δ (NH2478) were incubated in Spo medium for five hours at which time a final concentration of 1 μM estradiol was added to induce transcription of NDT80 (indicated by the arrow). For the dmc1Δ and dmc1Δ mek1Δ diploids, the cultures were split in half and the Ime2ΔC241-as inhibitor, 3MB-PP1, was add to a concentration of 50 μM to one half. Cells were fixed at the indicated timepoints and stained with 4’6-diamidino-2-phenylindole (DAPI) and examined by fluorescent microscopy to count the number of mono-, bi- (MII) and tetra-nucleate (MII) cells. Two hundred cells were counted for each timepoint. Error bars indicate the standard deviation for n ≥ 3, or the range when n = 2. For yLJ92, n = 1, NH2451, n = 4, NH2478, n = 2.
To ask whether the role of Ime2 phosphorylation in Ndt80 activation is direct or indirect, the same experiment was performed in an isogenic diploid homozygous for mek1Δ. In this diploid, no inhibitory Mek1 phosphates are present on Ndt80 and, when Ime2ΔC241-as is active (i.e. no inhibitor), meiotic progression occurred with similar kinetics to a DMC1 IME2ΔC241-as NDT80-IN diploid (Figure 4). If Ime2 phosphorylation directly promotes the transcriptional activity of Ndt80, then inhibition of Ime2ΔC241-as in the dmc1Δ IME2ΔC241-as mek1Δ NDT80-IN diploid should slow down meiotic progression. If, instead, Ime2 helps remove the inhibitory Mek1 phosphates, there should be no difference with or without inhibitor, since no inhibitory phosphates are present due to the mek1Δ. The latter result was observed, suggesting that an important role for Ime2 is to counteract Mek1 phosphorylation on Ndt80. One possibility is that Ime2 phosphorylation recruits phosphatases to the transcription factor that remove the Mek1 phosphates. When Mek1 levels are above a threshold level, the necessary inhibitory phosphates are maintained. However, lowering the amount of active Mek1, or increasing the amount of active Ime2, tips the balance so that Ndt80 is able to bind to the MSEs in its own promoter to start a positive feedback loop.
In summary, by using the same kinase, Mek1, to regulate both DSB repair and the checkpoint that monitors that repair, the cell is able to ensure that meiotic progression does not occur until all of the potentially lethal DSBs have been fixed.
Strains
The experiment shown in Figure 3 used the following SK1 isogenic diploids:

Acknowledgements
The authors thank Lauren Bednor, Xiangyu Chen, Aaron Neiman and Andrew Zeisel for helpful comments on the manuscript and the members of the Hollingsworth lab for helpful discussions. This work was supported by National Institutes of Health R01 grant to N. M. H., GM050717.
References
- Acquaviva L, Szekvolgyi L, Dichtl B, Dichtl BS, de C La Roche Saint Andre et al. , 2013. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339: 215–218. [DOI] [PubMed] [Google Scholar]
- Adam C, Guerois R, Citarella A, Verardi L, Adolphe F et al. , 2018. The PHD finger protein Spp1 has distinct functions in the Set1 and the meiotic DSB formation complexes. PLoS Genet 14: e1007223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed NT, Bungard D, Shin ME, Moore M and Winter E, 2009. The Ime2 protein kinase enhances the disassociation of the Sum1 repressor from middle meiotic promoters. Mol. Cell. Biol 29: 4352–4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E, Padmore R and Kleckner N, 1990. Analysis of wildtype and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61: 419–436. [DOI] [PubMed] [Google Scholar]
- Allers T, and Lichten M, 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Aravind L, and Koonin EV, 1998. The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem Sci 23: 284–286. [DOI] [PubMed] [Google Scholar]
- Argunhan B, Leung WK, Afshar N, Terentyev Y, Subramanian VV et al. , 2017. Fundamental cell cycle kinases collaborate to ensure timely destruction of the synaptonemal complex during meiosis. EMBO J 36: 2488–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin KR, Zhang C, Shokat KM and Herskowitz I, 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev 17: 1524–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L and Kleckner N, 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation and cell cycle progression. Cell 69: 439–456. [DOI] [PubMed] [Google Scholar]
- Blat Y, and Kleckner N, 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98: 249–259. [DOI] [PubMed] [Google Scholar]
- Börner GV, Kleckner N and Hunter N, 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- Brown MS, Grubb J, Zhang A, Rust MJ and Bishop DK, 2015. Small Rad51 and Dmc1 complexes often co-occupy both ends of a meiotic DNA double strand break. PLoS Genet 11: e1005653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Jarosinska OD and Lorenz A, 2018. Genetic interactions between the chromosome axis-associated protein Hop1 and homologous recombination determinants in Schizosaccharomyces pombe. Curr Genet 64: 1089–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busygina V, Sehorn MG, Shi IY, Tsubouchi H, Roeder GS et al. , 2008. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev 22: 786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M, Thayer NH, Oh SD, Kleckner N and Hunter N, 2010. Double Holliday junctions are intermediates of DNA break repair. Nature 464: 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callender TL, and Hollingsworth NM, 2010. Mek1 suppression of meiotic double-strand break repair is specific to sister chromatids, chromosome autonomous and independent of Rec8 cohesin complexes. Genetics 185: 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callender TL, Laureau R, Wan L, Chen X, Sandhu R et al. , 2016. Mek1 down regulates Rad51 activity during yeast meiosis by phosphorylation of Hed1. PLoS Genet 12: e1006226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo JA, Johnson AL, Sedgwick SG and Cha RS, 2008. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 132: 758–770. [DOI] [PubMed] [Google Scholar]
- Carlile TM, and Amon A, 2008. Meiosis I is established through division-specific translational control of a cyclin. Cell 133: 280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Gaglione R, Leong T, Bednor L, de los Santos T et al. , 2018. Mek1 coordinates meiotic progression with DNA break repair by directly phosphorylating and inhibiting the yeast pachytene regulator Ndt80. PloS Genetics 14(11): e1007832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Suhandynata RT, Sandhu R, Rockmill B, Mohibullah N et al. , 2015. Phosphorylation of the synaptonemal complex protein Zip1 regulates the crossover/noncrossover decision during yeast meiosis. PLoS Biology 13: e1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, and Herskowitz I, 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1: 685–696. [DOI] [PubMed] [Google Scholar]
- Cloud V, Chan Y-L, Grubb J, Budke B and Bishop DK, 2012. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337: 1222–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cool M, and Malone RE, 1992. Molecular and genetic analysis of the yeast early meiotic recombination genes REC102 and REC107/MER2. Mol Cell Biol 12: 1248–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T, and Hollingsworth NM, 1999. Red1p, a MEK1-dependent phosphoprotein that physically interacts with Hop1p during meiosis in yeast. J Biol Chem 274: 1783–1790. [DOI] [PubMed] [Google Scholar]
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J et al. , 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Molecular cell 46: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, and Roeder GS, 2000. Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. The Journal of cell biology 148: 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T, and Lichten M, 2010. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biology 8: e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark JP, Fazzio TG, Estep PW, Church GM and Tsukiyama T, 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103: 423–433. [DOI] [PubMed] [Google Scholar]
- Gray S, Allison RM, Garcia V, Goldman AS and Neale MJ, 2013. Positive regulation of meiotic DNA double-strand break formation by activation of the DNA damage checkpoint kinase Mec1(ATR). Open Biol 3: 130019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JC, and Haber JE, 2006. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet 40: 209–235. [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Ebisuzaki LK and Segall J, 1995. A 15-base-pair element activates the SPS4 gene midway through sporulation in Saccharomyces cerevisiae. Mol. Cell Biol 15: 3934–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, 2016. Mek1/Mre4 is a master regulator of meiotic recombination in budding yeast. Microbial Cell 3: 129–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, and Byers B, 1989. HOP1: a yeast meiotic pairing gene. Genetics 121: 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, Goetsch L and Byers B, 1990. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell 61: 73–84. [DOI] [PubMed] [Google Scholar]
- Hong S, Sung Y, Yu M, Lee M, Kleckner N et al. , 2013. The logic and mechanism of homologous recombination partner choice. Mol Cell 51: 440–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N, 2007. Meiotic Recombination. Springer-Verlag, Heidelberg. [Google Scholar]
- Hunter N, and Kleckner N, 2001. The single-end invasion: an asymmetric intermediate at the double- strand break to double-holliday junction transition of meiotic recombination. Cell 106: 59–70. [DOI] [PubMed] [Google Scholar]
- Jin L, Zhang K, Xu Y, Sternglanz R and Neiman AM, 2015. Sequestration of mRNAs modulates the timing of translation during meiosis in budding yeast. Mol Cell Biol 35: 3448–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N, Brown MS, Bishop DK and Borner GV, 2015. Gradual implementation of the meiotic recombination program via checkpoint pathways controlled by global DSB levels. Mol Cell 57: 797–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, and Hartwell LH, 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, De Muyt A and Lichten M, 2015. Top3-Rmi1 DNA single-strand decatenase is integral to the formation and resolution of meiotic recombination intermediates. Mol Cell 57: 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Lange J and Mohibullah N, 2014. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu Rev Genet 48: 187–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KP, Weiner BM, Zhang L, Jordan A, Dekker J et al. , 2010. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143: 924–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SBC, Michaelis C et al. , 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements and recombination during meiosis. Cell 98: 91–103. [DOI] [PubMed] [Google Scholar]
- Kniewel R, Murakami H, Liu Y, Hollingsworth NM and Keeney S, 2017. Histone H3 threonine 11 phosphorylation is catalyzed directly by the meiosis-specific kinase Mek1 and provides a molecular readout for Mek1 activity in vivo. bioRXiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl KP, and Sekelsky J, 2013. Meiotic and mitotic recombination in meiosis. Genetics 194: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoureux JS, Stuart D, Tsang R, Wu C and Glover JN, 2002. Structure of the sporulation-specific transcription factor Ndt80 bound to DNA. EMBO J. 21: 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Cloud V, Huang CC, Grubb J, Thacker D et al. , 2013. Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet 9: e1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, and Hunter N, 2010. Trying to avoid your sister. PLoS Biol 8: e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Oh SD, Shinohara M, Shinohara A and Hunter N, 2008. Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol. Cell 29: 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem S-H, and Ogawa H, 1992. The MRE4 gene encodes a novel protein kinase homologue required for meiotic recombination in Saccharomyces cerevisiae. Nucl. Acids Res 20: 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hooker GW and Roeder GS, 2006. Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics 173: 1969–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gaines WA, Callender T, Busygina V, Oke A et al. , 2014. Down-regulation of Rad51 activity during meiosis in yeast prevents competition with Dmc1 for repair of double-strand breaks. PLoS Genet 10: e1004005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H-C, Kunz RC, Marullo A, Gygi SP and Hollingsworth NM, 2012. Cdc7-Dbf4 is a gene-specific regulator of meiotic transcription in yeast. Mol. Cell. Bio 32: 541–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Wells JL, Pryce DW, Novatchkova M, Eisenhaber F et al. , 2004. S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. J Cell Sci 117: 3343–3351. [DOI] [PubMed] [Google Scholar]
- Lydall D, Nikolsky Y, Bishop DK and Weinert T, 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383: 840–843. [DOI] [PubMed] [Google Scholar]
- Lynn A, Soucek R and Borner GV, 2007. ZMM proteins during meiosis: crossover artists at work. Chromosome Res 15: 591–605. [DOI] [PubMed] [Google Scholar]
- Mallory MJ, Cooper KF and Strich R, 2007. Meoisis-specific destruction of the Ume6p repressor by the Cdc20-directed APC/C. Mol. Cell 27: 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone RE, Haring SJ, Foreman KE, Pansegrau ML, Smith SM et al. , 2004. The signal from the initiation of meiotic recombination to the first division of meiosis. Eukaryot Cell 3: 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Blanco MG, Maslen S, Skehel JM and West SC, 2011. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147: 158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahill MS, Sham CW and Bishop DK, 2007. Synthesis-dependent strand annealing in meiosis. PLoS Biol 5: e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok J, Kim PM, Lam HY, Piccirillo S, Zhou X et al. , 2010. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci Signal 3: ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano SP, Cote ML, Fingerman I, Pierce M, Vershon AK et al. , 2002. Crystal structure of the DNA-binding domain from Ndt80, a transcriptional activator required for meiosis in yeast. Proc Natl Acad Sci U S A 99: 14041–14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, and Keeney S, 2014. DDK links replication and recombination in meiosis. Cell Cycle 13: 3621–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Li X, Job E, Park C, Moazed D et al. , 2007. Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol Cell Biol 27: 5456–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J et al. , 2005. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell 16: 5804–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Wan L, Busygina V, Kwon Y, Allen JA et al. , 2009. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol Cell 36: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY, Taylor AF, Smith GR et al. , 2007. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okaz E, Arguello-Miranda O, Bogdanova A, Vinod PK, Lipp JJ et al. , 2012. Meiotic prophase requires proteolysis of M phase regulators mediated by the meiosis-specific APC/CAma1. Cell 151: 603–618. [DOI] [PubMed] [Google Scholar]
- Ozsarac N, Straffon MJ, Dalton HE and Dawes IW, 1997. Regulation of gene expression during meiosis in Saccharomyces cerevisiae: SPR3 is controlled by both ABF1 and a new sporulation control element. Mol Cell Biol 17: 1152–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, and Segall J, 2002. Regulation of the premiddle and middle phases of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol Cell Biol 22: 6417–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG et al. , 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A et al. , 2011. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146: 372–383. [DOI] [PubMed] [Google Scholar]
- Perez-Hidalgo L, Moreno S and San-Segundo PA, 2003. Regulation of meiotic progression by the meiosis-specific checkpoint kinase Mek1 in fission yeast. J Cell Sci 116: 259–271. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Siomos MF and Nasmyth K, 2003. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112: 423–440. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Van Komen S, Vergano S, Klein H and Sung P, 1999. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem 274: 29453–29462. [DOI] [PubMed] [Google Scholar]
- Prugar E, Burnett C, Chen X and Hollingsworth NM, 2017. Coordination of double strand break repair and meiotic progression in yeast by a Mek1-Ndt80 negative feedback loop. Genetics 206: 497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Engebrecht J, Scherthan H, Loidl J and Roeder GS, 1995. The yeast MER2 gene is required for chromosome synapsis and the initiation of meiotic recombination. Genetics 141: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, and Roeder GS, 1990. Meiosis in asynaptic yeast. Genetics 126: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, and Roeder GS, 1991. A meiosis-specific protein kinase homologue required for chromosome synapsis and recombination. Genes Dev. 5: 2392–2404. [DOI] [PubMed] [Google Scholar]
- Sari F, Heinrich M, Meyer W, Braus GH and Irniger S, 2008. The C-terminal region of the meiosis-specific protein kinase Ime2 mediates protein instability and is required for normal spore formation in budding yeast. J Mol Biol 378: 31–43. [DOI] [PubMed] [Google Scholar]
- Sasanuma H, Hirota K, Fukuda T, Kakusho N, Kugou K et al. , 2008. Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes Dev 22: 398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, and Kleckner N, 1994. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids. Cell 76: 51–63. [DOI] [PubMed] [Google Scholar]
- Schwacha A, and Kleckner N, 1995. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83: 783–791. [DOI] [PubMed] [Google Scholar]
- Schwacha A, and Kleckner N, 1997. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90: 1123–1135. [DOI] [PubMed] [Google Scholar]
- Shin ME, Skokotas A and Winter E, 2010. The Cdk1 and Ime2 protein kinases trigger exit from meiotic prophase in Saccharomyces cerevisiae by inhibiting the Sum1 transcriptional repressor. Mol. Cell. Biol 30: 2996–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubassi G, Luca N, Pak J and Segall J, 2003. Activity of phosphoforms and truncated versions of Ndt80, a checkpoint-regulated sporulation-specific transcription factor of Saccharomyces cerevisiae. Mol. Genet. Genomics 270: 324–336. [DOI] [PubMed] [Google Scholar]
- Shuster EO, and Byers B, 1989. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics 123: 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AV, and Roeder GS, 1997. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol 136: 957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermeyer V, Beneut C, Chaplais E, Serrentino ME and Borde V, 2013. Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol Cell 49: 43–54. [DOI] [PubMed] [Google Scholar]
- Sopko R, Raithatha S and Stuart D, 2002. Phosphorylation and maximal activity of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is dependent on Ime2. Mol Cell Biol 22: 7024–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourirajan A, and Lichten M, 2008. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev 22: 2627–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich R, Surosky RT, Steber C, Dubois E, Messenguy F et al. , 1994. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev 8: 796–810. [DOI] [PubMed] [Google Scholar]
- Subramanian VV, and Hochwagen A, 2014. The meiotic checkpoint network: step-by-step through meiotic prophase. Cold Spring Harb Perspect Biol 6: a016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian VV, MacQueen AJ, Vader G, Shinohara M, Sanchez A et al. , 2016. Chromosome synapsis alleviates Mek1-dependent suppression of meiotic DNA repair. PLoS Biol 14: e1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhandynata RT, Wan L, Zhou H and Hollingsworth NM, 2016. Identification of putative Mek1 substrates during meiosis in Saccharomyces cerevisiae using quantitative phosphoproteomics. PLoS One 11: e0155931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M, Engebrecht J and Roeder GS, 1993. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72: 365–378. [DOI] [PubMed] [Google Scholar]
- Tang S, Wu MK, Zhang R and Hunter N, 2015. Pervasive and essential roles of the Top3-Rmi1 decatenase orchestrate recombination and facilitate chromosome segregation in meiosis. Mol Cell 57: 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker D, Mohibullah N, Zhu X and Keeney S, 2014. Homologue engagement controls meiotic DNA break number and distribution. Nature 510: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Argunhan B and Tsubouchi T, 2018. Exiting prophase I: no clear boundary. Curr Genet 64: 423–427. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H, and Roeder GS, 2006. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev 20: 1766–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung KS, Hong EJ and Roeder GS, 2000. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc Natl Acad Sci U S A 97: 12187–12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Niu H, Futcher B, Zhang C, Shokat KM et al. , 2008. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev 22: 386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chang CY, Wu JF and Tung KS, 2011. Nuclear localization of the meiosis-specific transcription factor Ndt80 is regulated by the pachytene checkpoint. Mol. Biol. Cell 22: 1878–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AMV, Komives EA and Corbett KD, 2018. Conformational dynamics of the Hop1 HORMA domain reveal a common mechanism with the spindle checkpoint protein Mad2. Nucleic Acids Res 46: 279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter E, 2012. The Sum1/Ndt80 transcriptional switch and commitment to meiosis in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 76: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltering D, Baumgartner B, Bagchi S, Larkin B, Loidl J et al. , 2000. Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol. Cell. Biol 20: 6646–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HY, Ho HC and Burgess SM, 2010. Mek1 kinase governs outcomes of meiotic recombination and the checkpoint response. Curr. Bio 20: 1707–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Pierce M, Gailus-Durner V, Wagner M, Winter E et al. , 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18: 6448–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Ajimura M, Padmore R, Klein C and Kleckner N, 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol 15: 6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Weiner BM and Kleckner N, 1997. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 11: 106–118. [DOI] [PubMed] [Google Scholar]