Abstract
Allogeneic hematopoietic cell transplantation (alloHCT) remains the sole curative therapy for patients with CLL leading to 40–45% long-term survival. The impact of donor KIR genotype on outcomes of unrelated donor (URD) alloHCT for CLL is unknown.
Methods:
We examined 573 adult (URD)-CLL recipient pairs. KIR genotype (presence/absence) was determined for each donor and comprehensive modeling of interactions with recipient HLA class I loci (KIR ligands) was used to evaluate their effect on relapse and survival.
Results:
Recipients had a median age of 56 years, and most were not in remission (65%). Both 8/8 HLA-matched (81%) or 7/8 HLA matched grafts (19%) were studied. Factors associated with improved OS were reduced intensity conditioning (HR of death 0.76) and good performance status (HR 0.46), while allo-HCT in non-remission (HR 1.96) and mismatched donors (HR 2.01) increased mortality. No models demonstrated a relationship between donor KIR genotype and transplant outcomes. Cox regression models comparing donors with A/A vs B/x KIR haplotypes and those with KIR gene content scores of 0 vs 1 vs ≥2 yielded similar rates of non-relapse mortality, relapse, acute graft-versus-host disease (GVHD), and chronic GVHD and the same progression-free survival (PFS) and overall survival (OS). Relapse risk was not different with grafts from donors with KIR3DL1 transplanted into HLA C1/1 vs C2 recipients.
Conclusion:
This large analysis failed to demonstrate an association between unrelated donor KIR genotype and transplant outcome for patients with CLL and thus KIR genotyping should not be used as a donor selection criterion in this setting.
Keywords: chronic lymphocytic leukemia, KIR, allogeneic transplantation, genotype, NK cells
INTRODUCTION
Chronic lymphocytic leukemia (CLL), the most common leukemia in adults, is a biologically heterogenous disease. High-risk CLL harbors adverse genetic aberrations including mutations in chromosomes 17 or 11 or the tumor suppressor gene TP53. Clinically, high-risk CLL exhibits a relapsing course with short and incomplete remissions after standard chemotherapy. High-risk CLL can be temporarily controlled with the small molecule inhibitors ibrutinib, idelalisib, or venetoclax; however, allogeneic donor transplantation (alloHCT) is the only known curative therapy1. AlloHCT can cure about 40–50% of patients with CLL in part by inducing a potent donor-derived graft-versus-leukemia (GVL) response. Donor T cells contribute to GVL by recognition of minor antigen peptides for which the donor and patient are disparate presented by self-human leukocyte antigens (HLA).2,3 Disease progression in CLL may be influenced by both HLA homozygosity and haplotype bias.4 However, donor-derived NK cells can also generate potent alloreactivity toward tumor cells and may play a significant role in eradicating residual tumor after alloHCT.5
NK cells kill malignant cells by direct or antibody-dependent cellular cytotoxicity.6 NK cell recognition and killing of target cells is controlled by the balance of signals from both inhibitory and activating NK cell receptors.6 These include inhibitory and activating killer cell immunoglobulin-like receptors (KIRs) that interact with “self” class I HLA-C1, HLA-C2, and Bw4 to regulate NK cell education and target killing. Inhibitory KIR include KIR2DL2/3 (specific for HLA-C1 epitopes), 2DL1 (specific for HLA-C2), and 3DL1 (specific for HLA-Bw4).5,6 The ligands for activating KIR (2DS1, 2DS2, 2DS3, 2DS5 and 3DS1) are less well characterized but include HLA-A11 (2DS2), HLA-C2 (2DS1) and HLA-F (3DS1). KIR genes are organized into A or B haplotypes combine to form genotypes A/A or B/x (A/B or B/B).6 Haplotype B contains variable numbers of activating KIR genes, while group A has a fixed number of inhibitory and fewer activating KIR genes. About 70% of the Caucasian population has at least one KIR B haplotype. NK cell recognition and killing of target cells is controlled by the balance of signals from both inhibitory and activating NK cell receptors.6 KIR genes and HLA genes segregate independently on different chromosomes (19 and 6, respectively), even fully HLA-matched allogeneic donors may have a high frequency of NK cells mismatched between KIR and their cognate HLA class I ligands.
Unrelated donors with KIR B/x genotype were demonstrated to protect against relapse and improve survival in a cohort of patients with acute myeloid leukemia (AML), especially when donors had centromeric KIR B genes.7,8,9 Relapse protection was enhanced in recipients who had one or two C1-bearing HLA-C allotypes compared to C2 homozygous recipients. Donor KIR B/x genotype and higher numbers of B genes have also been associated with anti-tumor responses in cohort of patients with non-Hodgkin lymphoma (NHL) and pediatric acute lymphoblastic leukemia (ALL).10,11 Here, we studied whether unrelated donor KIR gene content influences GVL reactions against CLL by evaluating the incidence of relapse and other outcomes after allo-HCT in a large cohort of B-cell CLL patients reported to the Center for International Blood and Marrow Transplant Research (CIBMTR).
PATIENTS AND METHODS
We studied 573 HCT recipients with CLL and their corresponding donors transplanted between 1995–2014 at 106 different centers. All HCTs used URD grafts identified through the National Marrow Donor Program (NMDP). Donor DNA samples were available in the NMDP/CIBMTR Biorepository. Stored donor samples were genotyped for the presence or absence of KIR genes. Outcome data were collected from the CIBMTR. Patient-related variables in the analysis included age, sex, coexistence of disease or organ impairment, Karnofsky and comorbidity scores, donor-recipient HLA match status and recipient HLA-based KIR ligands. Disease-related characteristics including presence/absence of chromosome 17p, immunoglobulin heavy-chain variable (IGHV) mutation status; Rai stage, lactate dehydrogenase levels, presence of bulky adenopathy at the time of HCT and fludarabine responsiveness were examined in the CIBMTR database; these variables were missing for many subjects and not included in the analysis. Disease status at transplantation was recorded by a center as complete response (CR), partial response (PR), partial response nodal (nPR), stable disease (SD) and progressive disease (PD). Transplant-related variables included the type of preparative regimen (myeloablative [MA], reduced-intensity conditioning [RIC], or non-myeloablative [NMA]), graft source (bone marrow versus peripheral blood), use of antithymocyte globulin and/or alemtuzumab, type of graft-versus-host disease (GVHD) prophylaxis, number of lines of therapy and transplant period. The study protocol was approved by the Institutional Review Board of the NMDP in accordance with the Declaration of Helsinki.
KIR genotyping
Donor KIR genotyping was performed via gene specific or group specific multiplex polymerase chain reaction and next generation sequencing (Illumina, CA) for 16 individual killer immunoglobulin receptor (KIR) genes: KIR2DL1–5, KIR2DS1–5, KIR3DL1–3, KIR3DS1, KIR2DP1 and KIR3DP1. Sequences were analyzed using an American Society of Histocompatibility and Immunogenetics approved multiplex KIR typing algorithm developed by Histogenetics (using IMGT KIR database v2.6.0), and each KIR gene is defined as present or absent. We identified A and B KIR haplotypes and assigned donor genotypes (A/A or B/x). For B/x donors, we further determined whether their B-defining genes were in the centromeric or telomeric part of the KIR locus, or in both (Cen AA vs B/x, Tel AA vs B/x).10 The KIR B–gene motifs content score (KIR B-score) for each donor was calculated as the sum of centromeric and telomeric gene-content motifs containing B haplotype–defining genes (values ranged from 0–4).8
Outcome Definitions
Overall survival (OS) was defined as time from HCT to death from any cause. Relapse and progression-free survival (PFS) were defined per CIBMTR criteria.12 Non-relapse mortality (NRM) was defined as death in absence of relapse. Acute (Grades II-IV, III-IV) GVHD were defined by the Glucksberg scale.13 Chronic GVHD (cGVHD) was defined according to the Seattle criteria.14. There was incomplete reporting in a small subset of the population (1%) that affected the cohort evaluable for aGVHD, relapse, PFS and NRM. Six cases from the B/x group had a missing relapse status and one case from the AA group had missing aGVHD status, therefore the total sample size for OS differs from aGVHD, relapse and NRM.
Statistical Analysis
In univariate analysis, we evaluated PFS and OS using Kaplan-Meier estimates. Relapse, NRM and aGVHD were evaluated using the cumulative incidence function. In multivariable analysis, Cox proportional hazard models were used for the following end-points: OS, PFS, relapse, NRM, aGVHD II-IV, aGVHD III-IV and cGVHD. All clinical variables including donor source, GVHD prophylaxis, conditioning regimen, HLA-match, time from diagnosis to transplant, disease status, in vivo T-cell depletion, recipient age and KPS were tested for the proportional hazard assumption and adjusted as needed through stratification. Stepwise forward-backward selection was performed to build Cox regression models with a threshold of 0.05 for model entry.
The main objective was to test for association of each donor KIR genotype with relapse and PFS. Each KIR variable was forced into each outcome model and tested separately with the adjustment for the selected clinical risk factors. Interactions between each KIR variable and the adjusted clinical variables were tested, and none were statistically significant at the 0.05 nominal level. To adjust for multiple testing, a significance level of 0.01 was used for declaring a significant association of donor KIR genotypes. To test for donor-recipient KIR-KIR ligand genetic interactions, we conducted a comprehensive analysis using previously established models: A) Donor KIR A/A vs B/x genotype;15 B) Donor KIR Cen AA vs AB vs BB;8 C) KIR B-gene score 0 vs 1–4;8 D) Missing ligand in recipients (HLA-C1/x vs C2);16,17 E) HLA C1/C1 vs C2/2 vs C2/C1 in the presence of activating KIR2DS1.18 F) Missing Bw4 ligand for inhibitory KIR3DL1;16,17 All p values were 2-sided. SAS version 9.4 (SAS Institute, Cary, NC) was used for all analyses.
RESULTS
Patients, disease and transplant characteristics
Median age at allo-HCT of the 573 CLL patients was 56 years (range 21–74), and 31% were older than 60 years of age; 78% were male, and 64% had a good performance status (KPS>90% [Table 1]). Median time from diagnosis to HCT was 4.9 years (range 1 month–26 years). Most patients were not in remission (65%). Data on 17p chromosome abnormalities were available for 27% of patients; 52 patients (9%) harbored chromosome 17p deletion. Most recipients received RIC/NMA (77%) and about a third had in vivo T cell depletion. About one third were CMV seropositive. Tacrolimus-based GVHD prophylaxis was the most frequently used (60%). The frequency of KIR ligands in recipients was as follows: HLA-C1 82%, HLA-C2 57% and HLA-Bw4 58%. Donor KIR genotype reflected the general pattern in the Caucasian population: 67% (n=386) were B/x genotype (only 4% had B/B), and 23% were A/A genotype (n=183). Almost all donors carried KIR3DL1 (96%), and 36% carried the activating KIR2DS1. The majority of donor-recipient pairs were 8/8 high-resolution, allele-level matched at HLA-A, -B, -C, -DRB1 and -DQB1 (n=379, 66%) and the remainder were mismatched at one HLA allele. Donor KIR genotypes were similar in the HLA matched and single allele mismatched HCTs.
Table 1.
Patient, Donor, Disease and Transplant Characteristics
| Characteristics of patients | N (%) |
|---|---|
| Number of patients | 573 |
| Age, median (range), years | 56 (21–74) |
| ≥ 60 | 184 (32) |
| Sex Male | 428 (75) |
| Race / ethnicity | |
| Caucasian | 530 (92) |
| African-American | 25 ( 4) |
| Asian | 1 (<1) |
| Multiple | 2 (<1) |
| Unknown | 15 ( 3) |
| Karnofsky performance score | |
| <80% | 44 ( 8) |
| 80–100% | 492 (86) |
| Missing | 37 ( 6) |
| HCT-CI | |
| 0–2 | 273 (47) |
| 3+ | 63 (11) |
| Not Available | 237 (41) |
| Donor age, median (range), years | 33 (19–60) |
| Lines of prior chemotherapy | |
| ≤3 | 223 (39) |
| >3 | 35 ( 6) |
| Unknown / missing | 315 (55) |
| Disease status | |
| CR / PR / nPR | 193 (34) |
| Stable/Progressive / untreated | 372 (65) |
| Missing | 8 ( 1) |
| Time from diagnosis to transplant, median (range) months URD HLA match | 59 (1–323) |
| 8/8 | 466 (81) |
| 7/8 | 107 (19) |
| Graft source | |
| Bone marrow | 118 (21) |
| Peripheral blood | 455 (79) |
| Donor-recipient CMV status | |
| Recipient positive | 166(29) |
| Recipient negative / donor positive | 214 (37) |
| Recipient negative / donor negative | 161 (28) |
| Conditioning regimen intensity | |
| Myeloablative | 131 (23) |
| RIC / NMA | 438 (77) |
| Unknown | 4 (<1) |
| TBI in conditioning | 220 (38) |
| In vivo T cell depletion | |
| None | 356 (62) |
| Anti-thymocyte globulin | 150 (26) |
| Alemtuzumab | 67 (12) |
| GVHD prophylaxis | |
| Tac+ MMF ± other(s) | 141 (25) |
| Tac MTX ± other(s) and Tac ± other(s) | 251 (43) |
| CSA + MTX or MMF ± other(s) | 111 (19) |
| CSA ± other(s) | 64 (12) |
| Other(s) | 6 ( 1) |
| Year of transplant | |
| 1995–1999 | 29 ( 5) |
| 2000–2004 | 146 (25) |
| 2005–2009 | 299 (52) |
| 2010–2014 | 99 (17) |
| Recipient expressing HLA C1 | 467 (82) |
| Recipient expressing HLA C2 | 325 (57) |
| Recipient expressing HLABw4 | 331 (58) |
| Donor KIR Genotype | |
| KIR AA | 187 (33) |
| KIR Bx | 386 (67) |
| Donor with KIR2DS1 | 205 (36) |
| Donor with KIR3DL1 | 549 (96) |
Abbreviations: HCT-CI hematopoietic cell transplantation – comorbidity index, URD unrelated donor, ATG, antithymocyte globulin; CSA, cyclosporin A; CR, complete response; nPR, nodal partial response; PR, partial response; RIC reduced intensity conditioning, NMA non-myeloablative conditioning, TBI total body irradiation, MMF, mycophenolate mofetil; MTX, methotrexate; Tac, tacrolimus
Effect of KIR genotype on transplant outcomes
In this CLL cohort, transplants from donors with KIR B/x and A/A genotypes yielded similar rates of relapse, PFS (Figure 1) and OS after allo-HCT. After adjusting for important clinical variables, multivariate analysis confirmed that donor KIR genotype (AA vs B/x), KIR B-score and individual Cen KIR B-gene localization also conferred similar rates of NRM, aGVHD II-IV, aGVHD III-IV, cGVHD, relapse, PFS and OS (Table 2; Figure 1). Comprehensive modeling evaluating missing ligand, missing C1 ligand (HLA-C1 vs -C2), missing Bw4 ligand for inhibitory KIR3DL1 vs ligand present and HLA C1/C1 vs C2/2 vs C2/C1 in the presence of activating KIR2DS1 did not reveal significant effects on the main transplant outcomes (Table 2). In addition, no differences on outcome based on KIR A vs KIR B/x donors when the HLA-matched (n=367) or HLA-mismatched transplants (n=169) were analyzed separately (data not shown). In aggregate, none of the models tested showed any statistically significant association between donor KIR genotype and transplant outcomes.
Figure 1.
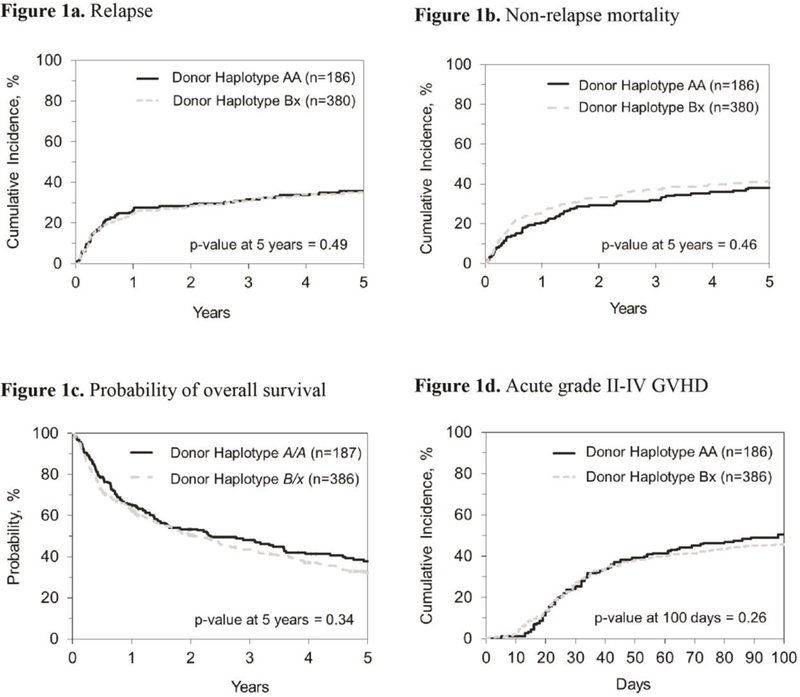
Unadjusted cumulative incidence of relapse (1a), non-relapse mortality (1b), probability of overall survival (1c) and acute grades II-IV graft-versus-host disease (1d) by KIR genotype.
Table 2.
Overall p-values for impact of KIR genotype on main transplant outcomes using models shown. All p≥0.02. (Hazard ratios are shown in supplemental Table 2).
| KIR Variants | OS | PFS | Relapse | NRM | *aGVHD II-IV | cGVHD |
|---|---|---|---|---|---|---|
| Model A: Haplotype AA vs Bx | 0.54 | 0.68 | 0.91 | 0.44 | 0.71 | 0.24 |
| Model B: Centromeric AA vs AB vs BB | 0.84 | 0.69 | 0.82 | 0.57 | 0.73 | 0.23 |
| Model C: B-gene score (0 vs1 vs 2vs 3–4) | 0.70 | 0.87 | 0.72 | 0.61 | 0.92 | 0.39 |
| Model D: Missing recipient HLA C2 | 0.59 | 0.4.3 | 0.54 | 0.21 | 0.19 | 0.24 |
| Missing recipient HLA C1 | 0.92 | 0.81 | 0.64 | 0.63 | 0.33 | 0.95 |
| Missing recipient HLA Bw4 | 0.50 | 0.53 | 0.02 | 0.22 | 0.82 | 0.86 |
| Model E: Donor KIR2DS1 present in C1/C1 vs C2/x recipient | 0.15 | 0.58 | 0.86 | 0.55 | 0.82 | 0.83 |
| Model F: Donor KIR3DL1 present in Bw4 recipient vs missing Bw4 recipient | 0.95 | 0.91 | 0.76 | 0.61 | 0.82 | 0.90 |
aGVHD III-IV results non-significant and similar to aGVHD II-IV; data not shown.
Abbreviations: KIR killer immunoglobulin-like receptors, OS overall survival, PFS progression-free survival, NRM non-relapse mortality, GVHD graft-versus-host disease,
Clinical factors affecting transplant outcomes
At median follow-up of 84 months (range 6–218 months), the probability of OS and PFS at 5-years for the entire cohort were 34% (95% confidence interval (CI): 30–38%) and 22% (95% CI: 19–36%), respectively. Cumulative incidence of relapse and NRM were 40% (95% CI: 36–45%) and 37% (95% CI: 33–42%), respectively. The main cause of death was disease recurrence (n=190; 49% [Supplemental Table 1]). In multivariate analysis, factors associated with improved OS included RIC regimen (hazard ratio [HR] of death 0.76; p=0.05), Karnofsky score >/=80% (HR 0.46; p=0.0006), 8/8 HLA-matched URD (HR 0.5; p=0.004) and GVHD prophylaxis using tacrolimus with methotrexate as compared to tacrolimus with mycophenolate (HR 0.64; p=0.0016). Shorter OS was associated with disease not in remission at the time of transplant as compared to CR/PR (HR 1.49; p=0.003), >3 lines of prior therapy (HR 2.54; p=<0.001) and use of alemtuzumab (HR 1.46; 0.049; Table 3).
Table 3.
Multivariate analysis of clinical factors affecting overall survival and relapse
| Overall Survival | Relapse | |||
|---|---|---|---|---|
| Variables | HR (range) | p-value | HR (range) | p-value |
| In vivo T cell depletion | ||||
| Anti-thymocyte globulin* | 1.00 | 0.023 | 1.00 | 0.001 |
| Alemtuzumab | 1.46 (1.0–2.13) | 0.049 | 1.10 (0.71–1.70) | 0.66 |
| No in vivo depletion | 0.89 (0.7–1.14) | 0.362 | 0.57 (0.4–0.81) | 0.0014 |
| Conditioning regimen | ||||
| Myeloablative* | 1.00 | 0.049 | NS | NS |
| RIC | 0.76 (0.57–1.0) | 0.05 | ||
| NMA | 0.70 (0.52–0.94) | 0.02 | ||
| Disease status | ||||
| CR/PR/nPR* | 1.0 | 1.0 | ||
| no CR/PR/nPR | 1.49.(1.14–7.56) | 0.0035 | 1.66 (1.13–2.45) | 0.010 |
| Donor type | ||||
| 8/8 HLA matched URD* | 1.0 | NS | NS | |
| 7/8 HLA matched URD | 1.56 (1.22–2.01) | 0.0004 | ||
| GVHD prophylaxis | ||||
| Tac+−others* | 1.00 | 0.0130 | NS | NS |
| Tac+ MTX or MMF+− other(s) | 0.64 (0.49–0.85) | 0.0016 | ||
| CSA +/−others | 0.80 (0.61–1.05) | 0.1117 | ||
| Karnofsky score | ||||
| <80%* | 1.0 | 1.0 | ||
| 80–100% | 0.46 (0.32–0.66) | 0.0001 | 0.42 (0.26–0.69) | 0.0005 |
| Number of chemotherapy line | ||||
| 3* | 1.0 | 1.0 | ||
| >3 | 2.54 (1.64–3.94) | 0.0001 | 1.82 (0.99–3.34) | 0.053 |
reference category, Abbreviations: OS overall survival, GVHD graft-versus-host disease, ATG, antithymocyte globulin; CSA, cyclosporin A; CR, complete response; NS, not significant, not included in multivariate model for outcome; nPR, nodal partial response; PR, partial response; MMF, mycophenolate mofetil; MTX, methotrexate; Tac, tacrolimus, RIC reduced intensity regimen, NMA, non-myeloablative.
Cumulative incidence of aGVHD II-IV at 100 days was 47% (95% CI: 43–51%), aGVHD III-IV at 100 days was 19% (95% CI: 16–23%) and cGVHD occurred at 1-year in 50% (95% CI: 45–54%) patients. GVHD contributed to death in 45 patients (12%).
DISCUSSION
The importance of donor KIR genotype in affecting outcome after URD allo-HCT has been clearly demonstrated in AML and NHL, where URD KIR B/x genotype confers an independent reduction in relapse and improved PFS.6,7 In the NHL cohort, relapse protection was strongest in HLA-matched transplants. Here we studied the potential contribution of donor NK cells to graft-versus-leukemia responses in a large cohort of CLL patients. We tested all published models, none of which demonstrated any significant association between donor KIR genotype or KIR/KIR-ligand genetic polymorphisms on alloHCT outcomes for CLL, in contrast to reported associations in AML, NHL and pediatric ALL. We evaluated HLA-matched and HLA-mismatched subsets in our CLL cohort, but found no significant effect of KIR genotype associated with HLA matching. A similar lack of significant association was observed in adult ALL, but not in pediatric ALL. 8,11 These results support the conclusion that donor NK cell-host interactions are specific to disease biology and the lineage of the underlying leukemia. We speculate that in CLL, T cell mediated GVL may be more potent than NK cell mediated allo-reactivity and thus play more of a role on clinical outcome.
The role of NK cell interactions with HLA KIR ligands in CLL is supported by studies in the non-transplant setting. Karabon et al. examined autologous KIR/HLA gene combinations in almost 200 CLL patients and reported that PFS differed by HLA C allele.19 The 10-year PFS was 75% for C1/C1 patients, 58% for C2/C2 patients and 43% for C1/C2 patients. Among HLA-C2/C2 patients, PFS was significantly higher in the patients carrying the activating KIR2DS1 (77% vs 34%). However in our study, although 205 (36%) allogeneic donors had KIR2DS1 we found no significant differences in relapse or PFS based on recipient HLA class C1 vs C2. Another non-transplant study in CLL showed a striking reduction in the frequency and viability of autologous NK cells expressing inhibitory KIR2DL1 (for which the ligand is HLA-C2) and/or KIR3DL1 (for which the ligand is HLA-Bw4); an effect which progressed over time.20 The NK cells in untreated CLL patients exhibited enhanced susceptibility to activation-induced cell death and poor degranulation when exposed to targets and rituximab. Unrelated donors carrying KIR2DL1 and/or KIR3DL1, contained in KIR A haplotypes, did not confer improved protection from relapse or better survival. In our analysis, genotype A/A vs B/x donors conferred the same survival regardless of C2 or B4w ligand expression in recipients. These findings suggest that the contribution of NK cell KIR polymorphisms to graft-versus-leukemia alloreactivity is weak and likely surpassed by other immunologic variables or stronger clinical factors such as HLA matching, disease status and in vivo T cell depletion. For example, CLL cells have been shown to over-express HLA-E, the natural ligand for the inhibitory receptor NKG2A expressed on NK-cells.21 NKG2A/HLA-E interaction has been shown to be a mechanism of tumor evasion in CLL patients and may be a mechanism relevant for immune escape from GVL.22 Recent studies showed that blocking NKG2A on NK cells from patients with CLL was sufficient to restore their direct cytotoxicity against HLA-E-expressing targets. This mechanism needs to be studied further in CLL where it may outweigh any KIR-mediated effects.
Our study confirmed previously established disease-specific prognostic factors for CLL, such as lack of remission and >3 lines of therapy pre-transplant. Notable host-related factors associated with increased mortality include low KPS, but not patient age. We also showed that mortality was increased by MA conditioning, donor-recipient HLA mismatch, in vivo T-cell depletion with alemtuzumab and GVHD prophylaxis using tacrolimus/MMF compared to tacrolimus/MTX.
In conclusion, our large retrospective analysis testing all published models of donor KIR and KIR/KIR-ligand interactions did not identify any effects on outcome to warrant their use in donor selection from the available HLA matched URD for patients with CLL.
Supplementary Material
Highlights.
Allogeneic hematopoietic transplantation (alloHCT) is the sole curative therapy for chronic lymphocytic leukemia (CLL).
Allogeneic donor Killer Immunoglobulin-like Receptor (KIR) genotype does not impact CLL patient outcomes.
KIR genotype should not be used as donor selection criteria in alloHCT for CLL.
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Center for Advancing Translational Sciences and the National Institutes of Health Award Number UL1TR000114 (VB) and NCI P01 111412 (DJW, TW, SGEM, ET, MH, SRS, LAG, ML, PP, JSM, SC). This work was supported in part by NIH P30 CA77598 utilizing the Masonic Cancer Center, University of Minnesota Oncology Medical Informatics and Services shared resource. The CIBMTR (TW, MH and SRS) is supported primarily by Public Health Service Grant/Cooperative Agreement 5U24CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); a Grant/Cooperative Agreement 1U24HL138660 from NHLBI and NCI; a contract HHSH250201700006C with Health Resources and Services Administration (HRSA/DHHS); three Grants N00014–17-1–2388, N00014–17-1–2850 and N00014–18-1–2045 from the Office of Naval Research; and grants from Adaptive Biotechnologies; *Amgen, Inc.; Anonymous donation to the Medical College of Wisconsin; Astellas Pharma US; Atara Biotherapeutics, Inc.; Be the Match Foundation; *bluebird bio, Inc.; *Bristol Myers Squibb Oncology; *Celgene Corporation; *Chimerix, Inc.; *CytoSen Therapeutics, Inc.; Fred Hutchinson Cancer Research Center; Gamida Cell Ltd.; Gilead Sciences, Inc.; HistoGenetics, Inc.; Immucor; *Incyte Corporation; Janssen Scientific Affairs, LLC; *Jazz Pharmaceuticals, Inc.; Karius, Inc.; Karyopharm Therapeutics, Inc.; *Kite Pharma, Inc.; Medac, GmbH; *Mediware; The Medical College of Wisconsin; *Merck & Co, Inc.; *Mesoblast; MesoScale Diagnostics, Inc.; Millennium, the Takeda Oncology Co.; *Miltenyi Biotec, Inc.; Mundipharma EDO; National Marrow Donor Program; Novartis Pharmaceuticals Corporation; PCORI; *Pfizer, Inc; *Pharmacyclics, LLC; PIRCHE AG; *Sanofi Genzyme; *Seattle Genetics; Shire; Spectrum Pharmaceuticals, Inc.; St. Baldrick’s Foundation; Swedish Orphan Biovitrum, Inc.; *Takeda Oncology; and University of Minnesota. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, Health Resources and Services Administration (HRSA) or any other agency of the U.S. Government.
*Corporate Members
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTERESTS
The authors report no relevant conflicts of interest.
REFERENCES
- 1.Dreger P, Schetelig J, Andersen N, et al. Managing high-risk CLL during transition to a new treatment era: stem cell transplantation or novel agents? European Research Initiative on CLL (ERIC) and the European Society for Blood and Marrow Transplantation (EBMT). Blood 2014. December 18;124(26):3841–3849. doi: 10.1182/blood-2014-07-586826. Epub 2014 Oct 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown JR, Kim HT, Armand P, et al. Long-term follow-up of reduced-intensity allogeneic stem cell transplantation for chronic lymphocytic leukemia: prognostic model to predict outcome. Leukemia 2013. February;27(2):362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sobecks RM, Leis JF, Gale RP, et al. Outcomes of human leukocyte antigen-matched sibling donor hematopoietic cell transplantation in chronic lymphocytic leukemia: myeloablative versus reduced-intensity conditioning regimens. Biol Blood Marrow Transplant 2014. September;20(9):1390–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah N, Decker WK, Lapushin R, et al. HLA homozygosity and haplotype bias among patients with chronic lymphocytic leukemia: implications for disease control by physiological immune surveillance. Leukemia 2011;25(6):1036–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruggeri L, Capanni M, Urbani E, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science 2002;295(5562):2097–2100. [DOI] [PubMed] [Google Scholar]
- 6.Lanier LL. Natural killer cells: roundup. Immunol Rev 2006. December;214:5–8. Review. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, Heller G, Chewning J, Kim S, Yokoyama WM, Hsu KC. Hierarchy of the human natural killer cell response is determined by class and quantity of inhibitory receptors for self-HLA-B and HLA-C ligands. J Immunol 2007;179:5977–89. [DOI] [PubMed] [Google Scholar]
- 8.Cooley S, Weisdorf DJ, Guethlein LA, et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood 2010;116(14):2411–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooley S, Weisdorf DJ, Guethlein LA, et al. Donor Killer Cell Ig-like Receptor B Haplotypes, Recipient HLA-C1, and HLA-C Mismatch Enhance the Clinical Benefit of Unrelated Transplantation for Acute Myelogenous Leukemia. J Immunol 2014;192(10):4592–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachanova V, Weisdorf DJ, Wang T, et al. Donor KIR B Genotype Improves Progression-Free Survival of Non-Hodgkin Lymphoma Patients Receiving Unrelated Donor Transplantation. Biol Blood Marrow Transplant 2016;22(9):1602–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oevermann L, Michaelis SU, Mezger M, et al. KIR B haplotype donors confer a reduced risk of relapse after haploidentical transplantation in children with acute lymphoblastic leukemia. Blood 2014. [DOI] [PMC free article] [PubMed]
- 12.Flomenberg N, Baxter-Lowe LA, Confer D, et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood 2004;104:1923–1930. [DOI] [PubMed] [Google Scholar]
- 13.Glucksberg H, Storb R, Fefer A, et al. Clinical manifestations of graft-versus-host disease in human recipients of marrow from HL-A-matched sibling donors. Transplantation 1974;18:295–304. [DOI] [PubMed] [Google Scholar]
- 14.Shulman HM, Sullivan KM, Weiden PL, et al. Chronic graft-versus-host syndrome in man. A long-term clinicopathologic study of 20 Seattle patients. Am J Med 1980;69:204–217. [DOI] [PubMed] [Google Scholar]
- 15.Cooley S, Trachtenberg E, Bergemann TL, et al. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood 2009;113(3):726–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu KC, Gooley T, Malkki M, et al. KIR ligands and prediction of relapse after unrelated donor hematopoietic cell transplantation for hematologic malignancy. Biol Blood Marrow Transplant 2006;12(8):828–836. [DOI] [PubMed] [Google Scholar]
- 17.Miller JS, Cooley S, Parham P, et al. Missing KIR ligands are associated with less relapse and increased graft-versus-host disease (GVHD) following unrelated donor allogeneic HCT. Blood 2007;109(11):5058–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venstrom JM, Pittari G, Gooley TA, et al. HLA-C-dependent prevention of leukemia relapse by donor activating KIR2DS1. N Engl J Med 2012;367(9):805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karabon L, Jedynak A, Giebel S, Wolowiec D, Kielbinski M, Woszczyk D, et al. KIR/HLA gene combinations influence susceptibility to B-cell chronic lymphocytic leukemia and the clinical course of disease. Tissue Antigens 2011. August;78(2):129–138. [DOI] [PubMed] [Google Scholar]
- 20.MacFarlane AW 4th, Jillab M, Smith MR, et al. NK cell dysfunction in chronic lymphocytic leukemia is associated with loss of the mature cellsexpressing inhibitory killer cell Ig-like receptors. Oncoimmunology 2017. May 19;6(7):e1330235. doi: 10.1080/2162402X.2017.1330235. eCollection 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner B, da Silva Nardi F, Schramm S, et al. HLA-E allelic genotype correlates with HLA-E plasma levels and predicts early progression in chronic lymphocytic leukemia. Cancer 2017. March 1;123(5):814–823. doi: 10.1002/cncr.30427. Epub 2016 Nov 2. [DOI] [PubMed] [Google Scholar]
- 22.McWilliams EM, Mele JM, Cheney C, et al. Therapeutic CD94/NKG2A blockade improves natural killer cell dysfunction in chronic lymphocytic leukemia. Oncoimmunology 2016. September 9;5(10):e1226720 eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


