Abstract
The bacterial hexameric helicase known as Rho is an archetypal sequence-specific transcription terminator that typically halts the synthesis of a defined set of transcripts, particularly those bearing cytosine-rich 3’ untranslated regions. However, under conditions of translational stress, Rho can also terminate transcription at cytosine-poor sites when assisted by the transcription factor NusG. Recent structural, biochemical, and computational studies of the Rho•NusG interaction in Escherichia coli have helped establish how NusG reprograms Rho activity. NusG is found to be an allosteric activator of Rho that directly binds to the ATPase motor domain of the helicase and facilitates closure of the Rho ring around non-ideal (purine-rich) target RNAs. The manner in which NusG acts on Rho helps to explain how the transcription terminator is excluded from acting on RNA polymerase by exogenous factors, such as the antitermination protein NusE, the NusG paralog RfaH, and RNA polymerase-coupled ribosomes. Collectively, an understanding of the link between NusG and Rho provides new insights into how transcriptional and translational fidelity are maintained during gene expression in bacteria.
Introduction
The appropriate functioning of a cell depends upon the accurate and timely expression of genetic information through the biochemical process of transcription. Transcriptional control is exerted at multiple levels, including initiation, elongation, and termination. Transcription termination mechanisms can be broadly grouped into two categories, known as factor-independent and factor-dependent termination. The efficiency of factor-independent (also known as “intrinsic”) termination is dictated by the sequence of the nascent RNA itself. By contrast, factor-dependent termination requires the action of one or more secondary protein- or nucleic acid-based elements.
The Rho transcription termination factor is an ATP-dependent, RecA-type hexameric helicase that terminates the synthesis of a wide variety of genes in bacteria. In organisms such as E. coli, Rho classically recognizes specific classes of 3’ untranslated regions in transcripts known as Rho utilization (or ‘rut’) sequences. Rho-dependent termination begins with the binding of the helicase to cytosine-rich elements within a rut site (Morgan et al., 1985; Yu et al., 2000). The initial interaction with the transcript occurs between the RNA and a set OB folds located within the N-terminus of each Rho subunit (Bogden et al., 1999; Dolan et al., 1990); these RNA-binding domains are referred to as the ‘primary’ binding site of Rho. Next, using an open-ring, or ‘lock washer,’ helicase configuration that permits nucleic-acid entry (Skordalakes and Berger, 2003; Yu et al., 2000). Rho engages the transcript in its central pore through a set of ‘secondary’ RNA binding sites (Miwa et al., 1995). In the presence of ATP, RNA binding to the pore triggers the isomerization of Rho into a closed-ring and catalytically active translocase (Thomsen and Berger, 2009; Thomsen et al., 2016). Rho then moves 5’➔3’ along the transcript, remaining tethered to its rut site, until it terminates transcription upon encountering RNA polymerase (Brennan et al., 1987; Park and Roberts, 2006; Schwartz et al., 2007). Rho-dependent termination has been extensively reviewed elsewhere (Bidnenko and Bidnenko, 2018; Mitra et al., 2017; Peters et al., 2011; RaySoni et al., 2016).
Although Rho is an efficient terminator of transcripts bearing 3’ rut sites, Rho can also terminate transcripts lacking a rut element with the assistance of a transcription factor called NusG (Downing et al., 1990; Peters et al., 2012). NusG is notable for its evolutionary kinship to the eukaryotic Spt5 family of proteins (Tomar and Artsimovitch, 2013)), which collectively serve as adapters that bridge RNA polymerase to other, dissociable, regulatory factors. Although it has been established that NusG acts as a flexible bridge between RNA polymerase and Rho (interacting with these factors by its N-terminal and C-terminal domains, respectively (Mooney et al., 2009)), the significance of these interactions for controlling transcription termination have been unclear. NusG is required for Rho-dependent termination of cytosine-poor, weak rut sequences (Peters et al., 2012), suggesting that the transcription factor impacts one or more sub-steps of Rho’s catalytic cycle; however, there has been no discernable effect of NusG on Rho activity in vitro in the absence of RNA polymerase despite decades of study (Burns and Richardson, 1995; Chalissery et al., 2011; Nehrke et al., 1993; Pasman and von Hippel, 2000; Valabhoju et al., 2016).
NusG overrides Rho’s dependence on primary site ligands to promote ring closure and termination of cytosine-poor transcripts
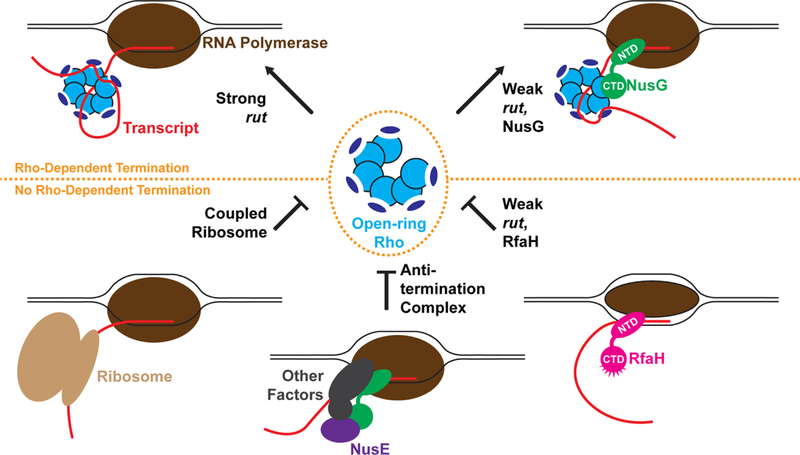
In considering how NusG might directly or indirectly impact Rho function, it is interesting to note that cytosine-rich primary site ligands (such as would be present in a strong rut site) are potent stimulators of Rho ATPase activity (Richardson, 1982). The mechanistic basis for this stimulation had remained unclear until the relatively recent development of a biochemical assay that allows for the tracking of Rho ring state (i.e., open vs. closed) in solution. Using this assay, it was discovered that RNAs capable of tightly binding to Rho’s primary site (i.e., pyrimidine-rich sequences) markedly potentiate Rho ring closure (Lawson et al., 2016). This observation indicates that Rho’s sequence specificity as a transcription termination factor arises in part from RNA sequence-dependent conformational control of the helicase (Figure 1, top left). How the primary-site occupancy is transmitted from the RNA-bound OB folds of Rho to their associated RecA ATPase motor domains to drive ring closure is poorly understood, but may involve the linker that connects these two functional elements (Hu and Artsimovitch, 2017).
Figure 1:
Regulation of Rho-dependent termination. Rho efficiently halts the synthesis of transcripts that bear a strong rut sequence (upper left), and also at weak rut sites when assisted by NusG (upper right). Rho-dependent termination is inhibited by coupled ribosomes (bottom left), antitermination complexes (bottom middle), and the NusG paralog RfaH (bottom right).
Given that cytosine-rich transcripts can directly promote Rho ring closure, it is reasonable to ask whether NusG might assist Rho’s action on cytosine- or pyrimidine-poor transcripts by helping to overcome the ring closure defect associated with binding to such ligands (Valabhoju et al., 2016). Using the aforementioned ring closure assay, it was found that NusG could indeed drive the nucleotide-dependent closure of Rho around non-ideal (purine-rich) RNAs in the absence of primary site ligands, and that the C-terminal domain (‘CTD’) of NusG was sufficient for this activity (Lawson et al., 2018). A crystal structure of Rho in complex with NusG CTD revealed that NusG engages three α-helices appended to the C-terminus of Rho’s RecA domain. Using long-timescale molecular dynamics simulations to explore internal rearrangements within Rho that occur between open- and closed-ring states, a conserved allosteric network was discovered within the helicase that resists ring closure and is remodeled by the binding of NusG. Mutagenesis of this allosteric network yielded hyperactive Rho constructs capable of terminating transcription at cytosine-poor rut sites in the absence of NusG. Collectively, these findings indicate that while Rho has evolved to exhibit a preference for cytosine-rich transcripts, its sequence specificity is sufficiently pliable that it can be overridden with assistance from NusG (Figure 1, top right).
At first glance, an indiscriminate terminator of transcription (as Rho would seem to transform into upon encountering NusG) could be considered undesirable in the context of regulating gene expression. This potential problem is likely alleviated (at least in part) by the fact that translation is physically coupled to transcription in bacteria (Miller et al., 1970), which conceivably could allow ribosomes to block Rho from accessing RNA to terminate transcription (Figure 1, bottom left). For example, the dual-purpose transcription/translation factor NusE (also known as S10), which has been recognized to bind to the same surface of NusG as Rho, could occlude Rho from accessing a transcribing polymerase that is accompanied by an actively translating ribosome (Burmann et al., 2010). Alternatively, Rho might be blocked by direct ribosomal sequestration of the nascent transcript from RNA polymerase, as seen in the so-called “expressome” arrangement (Kohler et al., 2017). It is intriguing to speculate that these various coupling interactions (and means of occluding Rho) might map to different stages of translation, such as initiation or elongation; real-time monitoring of coupling will be required to resolve this question. However, in cases of translational stress such as amino acid starvation or non-optimal codon usage (Mitra et al., 2017; Ruteshouser and Richardson, 1989), the rate of translation by a ribosome may not always keep pace with transcript synthesis by RNA polymerase, presenting Rho with an opportunity to access NusG and terminate transcription. NusG is also required for the Rho-dependent termination of widespread antisense transcription (Peters et al., 2012; Peters et al., 2009), which is consistent with the Rho•NusG complex acting as an attenuator of transcripts that lack translational potential.
Antitermination complexes hinder Rho-dependent termination by preventing Rho from accessing NusG
If a Rho•NusG complex might be capable of terminating the transcription of any RNAs that lack a closely-associated ribosome, it follows that cells would probably need a secondary means of preventing Rho activity on certain subsets of RNAs, such as those that are never translated (e.g., ribosomal RNAs). To this end, bacteria deploy antitermination complexes that block Rho from acting on specific RNAs. The mechanistic basis of this Rho inhibition has remained poorly understood, but by comparing how NusG engages Rho with the structure of NusG in a λN antitermination complex (Said et al., 2017), it was realized that these antitermination factors (and more specifically, NusE) sterically preclude Rho from accessing NusG (Figure 1, bottom center) (Lawson et al., 2018). Thus, anti-termination factors appear to counteract Rho function from terminating transcription at least in part by blocking a key partner, NusG, from associating with the helicase. It is worth noting that the field’s understanding of antitermination factors is rapidly evolving, as the Nus factors were recently demonstrated to regulate genes other than rRNAs (Baniulyte et al., 2017), and novel antitermination factors such as LoaP are still being discovered (Goodson et al., 2017).
The NusG paralog RfaH lacks Rho binding residues, explaining functional specialization of these factors
In E. coli, NusG has several known paralogs that appear to participate in the regulation of specific transcriptional events (Artsimovitch and Landick, 2002; Goodson et al., 2017). One such paralog, RfaH, does not associate with Rho, despite a high degree of shared structural similarity between its C-terminal domain and that of NusG (Figure 1, bottom right) (Burmann et al., 2012). Guided by the Rho•NusG structure, five residues were identified on RfaH that, when replaced with the corresponding residues from NusG, rendered RfaH capable of binding to Rho. Surprisingly, this chimeric RfaH construct with NusG residues appeared to bind Rho with higher affinity than wild-type NusG, suggesting that the affinity of NusG for Rho has been tuned by evolution to facilitate competition with other factors such as antitermination complexes. RfaH constructs with high affinity for Rho may be useful for stabilizing other transcription termination complexes for high-resolution structural studies.
Conclusions and Future Directions
Far from being a simple functional transition, the formation of a closed-ring and translocation-competent Rho assembly from an open-ring loading state can now be appreciated as a highly-regulated event controlled by proteinaceous factors such as NusG or cytosine-rich nucleic acids. It remains to be seen whether the allosteric network utilized by NusG to promote Rho ring closure overlaps with, or is distinct from, the network that communicates RNA binding to Rho’s primary sites with the helicase’s RecA domains. Despite the focus here on ring closure around a client RNA, there are numerous other sub-steps of Rho-dependent termination that merit further exploration as possible regulatory points, including the recruitment of Rho to rut sites, the translocation rate and processivity of Rho, and the interaction of Rho with RNA polymerase. It is possible that NusG may also exert an effect on Rho beyond the promotion of ring closure, which could be addressed by the development of assays that track discrete sub-steps of Rho-dependent termination.
Aside from NusG, there are several other intriguing factors that also affect Rho action. One such regulator is YaeO, which may inhibit Rho by directly competing with primary site RNAs (Gutierrez et al., 2007). Another is Hfq, a hexameric RNA chaperone that is reported to inhibit Rho through an unknown mechanism (Rabhi et al., 2011). The bacteriophage P4 protein Psu is third antagonist of Rho that merits further study (Pani et al., 2006) – it would not be surprising if additional phage-derived Rho inhibitors are discovered given the central role that Rho plays in the silencing of xenogenes (Cardinale et al., 2008). Rho-dependent termination is also regulated by RNA sequence and secondary structure, as demonstrated by the recent discovery of an anti-Rho RNA element termed RARE (Sevostyanova and Groisman, 2015) and the realization that sRNAs can control rut accessibility (Sedlyarova et al., 2016). It will be important to determine whether Rho binds to a specific site on RNA polymerase (Epshtein et al., 2007), or instead simply translocates until it applies a level of force to the RNA exit channel sufficient to induce hypertranslocation and/or shearing of RNA/DNA interactions (Park and Roberts, 2006; Ray-Soni et al., 2016); the latter mechanism would explain how Rho can terminate the transcription of phage polymerases that are structurally distinct from bacterial RNA polymerases (Pasman and von Hippel, 2000). Additional studies will be needed to address such questions in the future.
Acknowledgements
This work was supported by G. Harold and Leila Y. Mathers Foundation and the National Institute of General Medical Sciences (R37–071747), to J.M.B. M.R.L. gratefully acknowledges support from the A.P. Giannini Foundation.
References
- Artsimovitch I, and Landick R (2002). The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109, 193–203. [DOI] [PubMed] [Google Scholar]
- Baniulyte G, Singh N, Benoit C, Johnson R, Ferguson R, Paramo M, Stringer AM, Scott A, Lapierre P, and Wade JT (2017). Identification of regulatory targets for the bacterial Nus factor complex. Nat Commun 8, 2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidnenko E, and Bidnenko V (2018). Transcription termination factor Rho and microbial phenotypic heterogeneity. Curr Genet 64, 541–546. [DOI] [PubMed] [Google Scholar]
- Bogden CE, Fass D, Bergman N, Nichols MD, and Berger JM (1999). The structural basis for terminator recognition by the Rho transcription termination factor. Mol Cell 3, 487–493. [DOI] [PubMed] [Google Scholar]
- Brennan CA, Dombroski AJ, and Platt T (1987). Transcription termination factor rho is an RNA-DNA helicase. Cell 48, 945–952. [DOI] [PubMed] [Google Scholar]
- Burmann BM, Knauer SH, Sevostyanova A, Schweimer K, Mooney RA, Landick R, Artsimovitch I, and Rosch P (2012). An alpha helix to beta barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell 150, 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, and Rosch P (2010). A NusE:NusG complex links transcription and translation. Science 328, 501–504. [DOI] [PubMed] [Google Scholar]
- Burns CM, and Richardson JP (1995). NusG is required to overcome a kinetic limitation to Rho function at an intragenic terminator. Proc Natl Acad Sci U S A 92, 4738–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, and Nudler E (2008). Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science 320, 935–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalissery J, Muteeb G, Kalarickal NC, Mohan S, Jisha V, and Sen R (2011). Interaction surface of the transcription terminator Rho required to form a complex with the C-terminal domain of the antiterminator NusG. J Mol Biol 405, 49–64. [DOI] [PubMed] [Google Scholar]
- Dolan JW, Marshall NF, and Richardson JP (1990). Transcription termination factor rho has three distinct structural domains. J Biol Chem 265, 5747–5754. [PubMed] [Google Scholar]
- Downing WL, Sullivan SL, Gottesman ME, and Dennis PP (1990). Sequence and transcriptional pattern of the essential Escherichia coli secE-nusG operon. J Bacteriol 172, 1621–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, and Nudler E (2007). An allosteric path to transcription termination. Mol Cell 28, 991–1001. [DOI] [PubMed] [Google Scholar]
- Goodson JR, Klupt S, Zhang C, Straight P, and Winkler WC (2017). LoaP is a broadly conserved antiterminator protein that regulates antibiotic gene clusters in Bacillus amyloliquefaciens. Nat Microbiol 2, 17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez P, Kozlov G, Gabrielli L, Elias D, Osborne MJ, Gallouzi IE, and Gehring K (2007). Solution structure of YaeO, a Rho-specific inhibitor of transcription termination. J Biol Chem 282, 23348–23353. [DOI] [PubMed] [Google Scholar]
- Hu K, and Artsimovitch I (2017). A Screen for rfaH Suppressors Reveals a Key Role for a Connector Region of Termination Factor Rho. MBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler R, Mooney RA, Mills DJ, Landick R, and Cramer P (2017). Architecture of a transcribing-translating expressome. Science 356, 194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson MR, Dyer K, and Berger JM (2016). Ligand-induced and small-molecule control of substrate loading in a hexameric helicase. Proc Natl Acad Sci U S A 113, 13714–13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson MR, Ma W, Bellecourt MJ, Artsimovitch I, Martin A, Landick R, Schulten K, and Berger JM (2018). Mechanism for the Regulated Control of Bacterial Transcription Termination by a Universal Adaptor Protein. Mol Cell 71, 911–922.e914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OL Jr., Hamkalo BA, and Thomas CA Jr. (1970). Visualization of bacterial genes in action. Science 169, 392–395. [DOI] [PubMed] [Google Scholar]
- Mitra P, Ghosh G, Hafeezunnisa M, and Sen R (2017). Rho Protein: Roles and Mechanisms. Annu Rev Microbiol 71, 687–709. [DOI] [PubMed] [Google Scholar]
- Miwa Y, Horiguchi T, and Shigesada K (1995). Structural and functional dissections of transcription termination factor rho by random mutagenesis. J Mol Biol 254, 815–837. [DOI] [PubMed] [Google Scholar]
- Mooney RA, Schweimer K, Rosch P, Gottesman M, and Landick R (2009). Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol 391, 341–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan WD, Bear DG, Litchman BL, and von Hippel PH (1985). RNA sequence and secondary structure requirements for rho-dependent transcription termination. Nucleic Acids Res 13, 3739–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrke KW, Zalatan F, and Platt T (1993). NusG alters rho-dependent termination of transcription in vitro independent of kinetic coupling. Gene Expr 3, 119–133. [PMC free article] [PubMed] [Google Scholar]
- Pani B, Banerjee S, Chalissery J, Muralimohan A, Loganathan RM, Suganthan RB, and Sen R (2006). Mechanism of inhibition of Rho-dependent transcription termination by bacteriophage P4 protein Psu. J Biol Chem 281, 26491–26500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, and Roberts JW (2006). Role of DNA bubble rewinding in enzymatic transcription termination. Proc Natl Acad Sci U S A 103, 4870–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasman Z, and von Hippel PH (2000). Regulation of rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry 39, 5573–5585. [DOI] [PubMed] [Google Scholar]
- Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, and Landick R (2012). Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev 26, 2621–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Mooney RA, Kuan PF, Rowland JL, Keles S, and Landick R (2009). Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci U S A 106, 15406–15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Vangeloff AD, and Landick R (2011). Bacterial transcription terminators: the RNA 3’-end chronicles. J Mol Biol 412, 793–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabhi M, Espeli O, Schwartz A, Cayrol B, Rahmouni AR, Arluison V, and Boudvillain M (2011). The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rhodependent terminators. Embo j 30, 2805–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Soni A, Bellecourt MJ, and Landick R (2016). Mechanisms of Bacterial Transcription Termination: All Good Things Must End. Annu Rev Biochem 85, 319–347. [DOI] [PubMed] [Google Scholar]
- Richardson JP (1982). Activation of rho protein ATPase requires simultaneous interaction at two kinds of nucleic acid-binding sites. J Biol Chem 257, 5760–5766. [PubMed] [Google Scholar]
- Ruteshouser EC, and Richardson JP (1989). Identification and characterization of transcription termination sites in the Escherichia coli lacZ gene. J Mol Biol 208, 23–43. [DOI] [PubMed] [Google Scholar]
- Said N, Krupp F, Anedchenko E, Santos KF, Dybkov O, Huang YH, Lee CT, Loll B, Behrmann E, Burger J, et al. (2017). Structural basis for lambdaN-dependent processive transcription antitermination. Nat Microbiol 2, 17062. [DOI] [PubMed] [Google Scholar]
- Schwartz A, Margeat E, Rahmouni AR, and Boudvillain M (2007). Transcription termination factor rho can displace streptavidin from biotinylated RNA. J Biol Chem 282, 31469–31476. [DOI] [PubMed] [Google Scholar]
- Sedlyarova N, Shamovsky I, Bharati BK, Epshtein V, Chen J, Gottesman S, Schroeder R, and Nudler E (2016). sRNA-Mediated Control of Transcription Termination in E. coli. Cell 167, 111–121.e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevostyanova A, and Groisman EA (2015). An RNA motif advances transcription by preventing Rho-dependent termination. Proc Natl Acad Sci U S A 112, E6835–6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skordalakes E, and Berger JM (2003). Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell 114, 135–146. [DOI] [PubMed] [Google Scholar]
- Thomsen ND, and Berger JM (2009). Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell 139, 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ND, Lawson MR, Witkowsky LB, Qu S, and Berger JM (2016). Molecular mechanisms of substrate-controlled ring dynamics and substepping in a nucleic acid-dependent hexameric motor. Proc Natl Acad Sci U S A 113, E7691–e7700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar SK, and Artsimovitch I (2013). NusG-Spt5 proteins-Universal tools for transcription modification and communication. Chem Rev 113, 8604–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valabhoju V, Agrawal S, and Sen R (2016). Molecular Basis of NusG-mediated Regulation of Rho-dependent Transcription Termination in Bacteria. J Biol Chem 291, 22386–22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Horiguchi T, Shigesada K, and Egelman EH (2000). Three-dimensional reconstruction of transcription termination factor rho: orientation of the N-terminal domain and visualization of an RNA-binding site. J Mol Biol 299, 1279–1287. [DOI] [PubMed] [Google Scholar]