Abstract
Chromatin dynamics are central to the regulation of gene expression and genome stability, particularly in the presence of environmental signals or stresses that prompt rapid reprogramming of the genome to promote survival or differentiation. While numerous chromatin regulators have been implicated in modulating cellular responses to stress, gaps in our mechanistic understanding of chromatin-based changes during stress suggest additional proteins are likely critical to these responses and the molecular details underlying their activities are unclear in many cases. We recently identified a role for the relatively uncharacterized SET domain protein Set4 in promoting cell survival during oxidative stress in Saccharomyces cerevisiae. Set4 is a member of the Set3 subfamily of SET domain proteins which are defined by the presence of a PHD finger and divergent SET domain sequences. Here, we integrate our new observations on the function of Set4 with known roles for other related family members, including yeast Set3, fly UpSET and mammalian proteins MLL5 and SETD5. We discuss outstanding questions regarding the molecular mechanisms by which these proteins control gene expression and their potential contributions to cellular responses to environmental stress.
Keywords: chromatin, SET domain, PHD finger, stress responses, oxidative stress, gene expression, Set4, budding yeast
Introduction
In a changing environment, disruption of chromatin homeostasis has severe cellular consequences due to deregulated gene expression programs that are normally required for the proper functioning of stress response pathways (Chi, et al. 2010, D’Urso and Brickner 2017, Maze, et al. 2014, Soontorngun 2017, Tee and Reinberg 2014). Under stress conditions, cells are often dependent on a specific set of chromatin-modifying enzymes and chromatin remodelers to mount defenses and promote survival, although the precise mechanisms by which these proteins fine-tune gene expression in response to stress are not always clear. SET (Su(var)3–9, Enhancer-of-zeste, Trithorax) domain proteins are predominantly lysine methyltransferases that target both histone and non-histone proteins (Clarke 2013, Dillon, et al. 2005, Jaiswal, et al. 2017), and they are known to be critical to cell viability in the presence of diverse stresses (Green, et al. 2012, McDaniel, et al. 2017, Nadal-Ribelles, et al. 2015, Weiner, et al. 2012). There are 12 SET domain proteins in the budding yeast Saccharomyces cerevisiae, of which Set1 and Set2 are the most well-characterized as H3K4 and H3K36 methyltransferases, respectively (Jaiswal, et al. 2017, Porras-Yakushi, et al. 2006). Six of the SET domain proteins are primarily non-histone protein methyltransferases and appear to be enriched for activity on ribosomal proteins (Porras-Yakushi, et al. 2006). Set5 and Set6 are orthologous to the metazoan SMYD proteins (Calpena, et al. 2015); Set5 has been characterized as an H4 K5, K8, and K12 methyltransferase (Green, et al. 2012) implicated in gene expression control with Set1 (Jezek, et al. 2017, Martín, et al. 2014), whereas Set6 has no defined substrates or biological functions. The remaining SET domain proteins are subfamily members Set3 and Set4, of which Set3 is known to primarily function in gene repression through a mechanism thought to be independent of its SET domain (Kim and Buratowski 2009, Kim, et al. 2012), however functional and molecular roles for Set4 have not been well-described. Intriguingly, the expression of SET4 had been reported to be upregulated under stress conditions, including during stationary phase (Aragon, et al. 2008) and anaerobic growth (Lai, et al. 2006). These observations prompted us to test the hypothesis that Set4 regulates stress response pathways through a chromatin-based mechanism.
Stress defense and the chromatin regulator Set4
In our recent work, we showed that Set4 is a chromatin-associated protein that has a specific role in protecting cells during oxidative stress (Tran, et al. 2018). Genetic analyses demonstrated that cells lacking Set4 showed increased sensitivity to hydrogen peroxide (H2O2) treatment and overexpression of Set4 provided resistance to H2O2 treatment. These phenotypes also correlated with levels of intracellular reactive oxygen species (ROS): set4Δ cells showed increased ROS levels following H2O2 exposure, and endogenous ROS were decreased upon overexpression of Set4. Interestingly, we also observed that high levels of Set4 were toxic to cells grown under normal conditions, indicating the possibility that the abundance of Set4 may be carefully regulated in the cell to achieve survival under different environmental conditions.
Set4 contains a PHD finger, which is commonly a methyl-lysine effector domain (Shi, et al. 2007), and a SET domain, which usually provides catalytic lysine methyltransferase activity (Dillon, et al. 2005). These domains are frequently found in chromatin regulators and have been linked to the regulation of gene expression in numerous contexts. A large-scale study of potential gene expression regulators in yeast grown under standard laboratory conditions identified differentially expressed genes in set4Δ cells (Kemmeren, et al. 2014) that were enriched for functional roles in oxidation-reduction processes and iron ion homeostasis. Follow-up experiments using quantitative PCR showed that a subset of stress-response genes is attenuated following H2O2 treatment in cells lacking Set4, and overexpression of Set4 under normal conditions results in improper activation of stress-response genes. In addition, chromatin immunoprecipitation experiments revealed that Set4 localizes to chromatin in response to oxidative stress, and that it can be found in promoter regions of stress-response genes which show Set4-dependent regulation (Tran, et al. 2018). Altogether, these findings support a model in which Set4 associates with chromatin in response to stress to modulate gene expression and protects cells when challenged by oxidative insults.
Another recent work (Serratore, et al. 2018) also identified a role for Set4 in condition-dependent gene expression control. These authors reported wide-spread changes to the transcriptome of set4Δ cells compared to wildtype cells grown under conditions of hypoxia. Targeted investigation of ergosterol biosynthetic genes revealed that Set4 promotes repression of these genes in hypoxia, likely through a direct mechanism, as Set4 was shown to bind to their promoters in hypoxic conditions. Interestingly, expression of SET4 was substantially increased in hypoxia compared to aerobic growth, providing further support that the expression of SET4 is also subject to stress-dependent regulation (Lai, et al. 2006, Serratore, et al. 2018). In our studies of oxidative stress, cells were subjected to stress for relatively short timescales (usually 30 minutes), which are unlikely to induce drastic changes to protein levels. Unexpectedly, we observed decreased levels of SET4 mRNA following 30 minute-treatment with H2O2 (Tran, et al. 2018), although changes in protein level were not detected (unpublished data). This leaves open the possibility that Set4 abundance may also be modulated by ROS over longer timescales and underscores the importance of elucidating the signals that govern SET4 expression levels under different stress conditions. Nonetheless, combined with our observations of increased chromatin association during acute oxidative stress (Tran, et al. 2018), these data argue that multiple levels of regulation act on Set4 to calibrate its abundance and localization in response to different types of cellular stress.
The budding yeast paralogs Set3 and Set4
The budding yeast proteins Set3 and Set4 are paralogs, and represent the only two proteins in the yeast genome containing both a PHD finger and a SET domain (Figure 1A), a common signature of chromatin regulators in mammalian systems. Set3 is the defining member of the Set3 complex, or Set3C, which contains two histone deacetylases (HDACs), Hos2 and Hst1, and other regulatory subunits (Pijnappel, et al. 2001; Figure 1B). The PHD finger of Set3 has been reported to bind to both H3K4me2 and H3K4me3 in vitro (Gatchalian, et al. 2017, Kim and Buratowski 2009, Shi, et al. 2007), although Set3C primarily localizes to the 5’ transcribed regions of genes in cells, which are enriched for H3K4me2 (Kim and Buratowski 2009, Kim, et al. 2012). The interaction of the Set3 PHD finger with H3K4me2 recruits Set3C and its associated HDAC activity to chromatin, which modulates expression through multiple mechanisms, including repressing and activating genes that have overlapping noncoding transcripts and repressing transcription from cryptic initiation sites (Kim, et al. 2012). The activity of Set3 is therefore critical under conditions of changing environmental conditions when rapid changes in gene repression or activation are required.
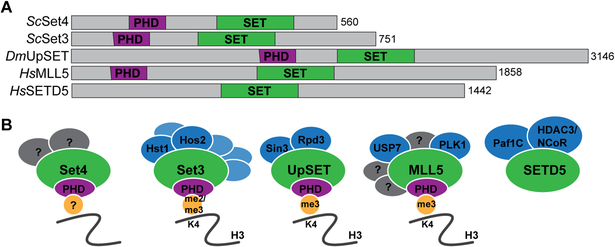
Figure 1.
(A) Schematic indicating the protein domain organization of S. cerevisiae (Sc) Set3 and Set4, D. melanogaster (Dm) UpSET, and H. sapiens (Hs) MLL5 and SETD5. PHD fingers are shown in purple and SET domains are shown in green. The total number of amino acids is indicated for each protein. (B) Members of the Set3-Set4 SET domain subfamily are shown with known interacting partners and methyl-lysine binding activity of their PHD fingers. Known binding partners are shown in blue, with the primary catalytic or regulatory interactors labelled. Set4 is predicted to interact with other factors (shown in gray) that remain to be identified. MLL5 has known interactors, a subset of which are shown in blue, however a comprehensive characterization of its binding partners at chromatin has not been demonstrated, therefore it is predicted to bind other yet-to-be-determined factors, which are indicated in gray. SETD5 binding partners include the multi-subunit PAF1 and HDAC3/NCoR complexes, which are simplified in the diagram. References for depicted interactions are provided in the main text.
To date, the mechanistic role for Set3 within Set3C appears primarily linked to the methyl-lysine binding activity of its PHD finger (Kim and Buratowski 2009). A defining feature of both yeast Set3 and Set4, as well as its metazoan orthologs, is the noncanonical sequence within their SET domains. These proteins contain amino acids substitutions within the SET domain that are predicted to interfere with binding to the cofactor S-adenosylmethionine (SAM) and the target lysine (Dillon, et al. 2005, Mas-Y-Mas, et al. 2016). Set3 is not known to have histone methylation activity (Pijnappel, et al. 2001), and our investigation of Set4 also suggests it is not likely to be a histone methyltransferase (Tran, et al. 2018). While discussed further below in the context of other related proteins, the precise roles for the SET domains within Set3 and Set4 are currently unknown.
The architectural similarities between Set3 and Set4 suggest the possibility of functional redundancy between the two proteins. Intriguingly, we observed that loss of Set3 also leads to increased sensitivity to H2O2, and double set4Δ set3Δ cells show an even greater defect in survival following H2O2 treatment (Tran, et al. 2018). These results suggest that both proteins promote survival during oxidative stress, as has been shown for other paralogs linked to oxidative stress survival in yeast (Matsuo, et al. 2017). However, based on the additive phenotype of the double mutant, Set3 and Set4 likely act through distinct pathways. Cells without Set3 are also sensitive to the transcription inhibitor mycophenolic acid, however set4Δ cells did not show similar sensitivity, nor did the double mutant show a dissimilar phenotype from that of set3Δ cells. While these data suggest partially overlapping biological roles for Set3 and Set4 in oxidative stress defense, their molecular distinctions highlight independent roles and different mechanisms to promote stress defense. Primarily, Set3 is more abundant than Set4 (Ghaemmaghami, et al. 2003), and its well-documented to interact with H3K4 methyl marks, whereas Set4 does not appear to show similar activity in vitro (Gatchalian, et al. 2017, Shi, et al. 2007). However, based on its known function in regulating the dynamics of transcriptional responses under changing nutrient conditions (Kim, et al. 2012), the role for Set3 in gene expression regulation during oxidative stress warrants further investigation, and new mechanistic insights into Set4 activity at chromatin would provide opportunities for defining potential stress-responsive roles for each of these proteins.
The Set3-Set4 subfamily of SET domain proteins in metazoans
The Drosophila protein UpSET and mammalian proteins MLL5 and SETD5 are homologous to Set3 and Set4 over their SET domains, and, with the exception of SETD5, also contain a PHD finger (Figure 1A). The PHD finger of MLL5 binds the H3K4me3 mark (Ali, et al. 2013, Lemak, et al. 2013), and fly UpSET also recognizes H3K4me3 (Ali, et al. 2013). Phenotypes associated with the loss of UpSET in flies vary due to differences in mutant fly lines used, but the loss of UpSET is most likely lethal (McElroy, et al. 2017), whereas alleles likely to be hypomorphic cause female sterility and have enhanced homeotic transformation phenotypes, predicted to be due to decreased silencing of homeotic genes (Rincon-Arano, et al. 2012). UpSET loss-of-function led to disrupted position effect variegation and an altered heterochromatin landscape, with decreased silencing (McElroy, et al. 2017). Similar to Set3, UpSET is known to interact with an HDAC complex, Rpd3 and Sin3 (Rincon-Arano, et al. 2012), and stabilize this complex at transcriptionally-active genes, thereby controlling histone acetylation levels and chromatin accessibility, as well as limiting acetylation to promoters and regions of active transcription (Rincon-Arano, et al. 2012; Figure 1B). The SET domain of UpSET is also reported to lack catalytic activity, and while UpSET is largely thought to mediate protein-protein interactions at chromatin (McElroy, et al. 2017, Rincon-Arano, et al. 2012), the molecular details underlying its function are still unclear.
MLL5 has been linked to a number of physiological pathways in mammalian systems, including cell cycle regulation (Cheng, et al. 2008, Deng, et al. 2004, Liu, et al. 2012, Sebastian, et al. 2009), hematopoietic stem cell proliferation and function (Heuser, et al. 2009, Madan, et al. 2009, Tasdogan, et al. 2016, Zhang, et al. 2009), and spermatogenesis (Yap, et al. 2011). MLL5 function is also predicted to be independent of catalytic activity, as methyltransferase activity has not been detected in vitro (Madan, et al. 2009, Mas-Y-Mas, et al. 2016, Sebastian, et al. 2009), the SET domain is not able to bind the methyl-donor SAM, and the structure revealed that the channel for SAM and target lysine binding is occluded (Mas-Y-Mas, et al. 2016). The loss of MLL5 has been linked to changes in H3K4 methylation levels, although the molecular mechanism through which this occurs is unresolved (Sebastian, et al. 2009). Although MLL5 has been reported to have a diversity of interacting partners (Ding, et al. 2015, Liu, et al. 2012, Zhou, et al. 2013), the extent to which MLL5 stably interacts with partners at chromatin, or potentially has context-dependent binding partners to mediate responses at chromatin is not yet clear and warrants further investigation (Figure 1B). Notably, loss of MLL5 is associated with hematopoietic stem/progenitor cell (HSPCs) malfunction due to elevated intracellular ROS levels and increased DNA damage (Tasdogan, et al. 2016), indicating that it also plays a critical role in oxidative stress responses.
Loss of mouse or human SETD5 is associated with developmental defects, intellectual disability, and autism spectrum disorders (Deliu, et al. 2018, Grozeva, et al. 2014, Kuechler, et al. 2015, Osipovich, et al. 2016). SETD5 has recently been reported to play a critical role in the regulation of neurodevelopmental gene expression and also modulates gene expression dynamics in context-fear-conditioned mice relative to controls (Deliu, et al. 2018). SETD5 has been found to interact with the HDAC3/NCoR and PAF1 complexes, and appears to be specifically associated with altering histone acetylation patterns near transcription start sites (Deliu, et al. 2018, Osipovich, et al. 2016), leading to altered RNA polymerase II occupancy patterns and aberrant expression of neurodevelopmental genes (Deliu, et al. 2018). As observed for related SET domain family members, SETD5 does not catalyze methylation on histones in vitro (Deliu, et al. 2018). Altogether, these findings on SETD5 suggests that it relies on similar molecular mechanisms as Set3, UpSET, and MLL5 to maintain critical gene expression programs in response to developmental cues.
The intriguing observation that yeast Set3 and mammalian MLL5 and SETD5, in addition to Set4, have been linked to responses to changing environmental conditions or stress signaling suggest that this subfamily of the SET domain proteins may play a specific role in governing proper responses to exogenous signals and stressors. The mechanistic details that have been uncovered to date for some of these proteins underscore their primary role in stabilizing HDAC activity at chromatin, although at distinct subsets of genes and regions of the genome. Similarly, in other chromatin-modifying complexes, such as NuA4, non-catalytic subunits have been identified as critical regulators of histone modification activity and chromatin localization (Searle and Pillus 2018, Searle, et al. 2017). We anticipate that further investigation of this SET domain subfamily will reveal the extent to which these proteins are environmentally-controlled and elucidate the range of mechanisms that promote their association with specific chromatin regions and their ability to modulate transcription and histone modification both dependent and independent of environmental signals. The extensive genetic and molecular tools available in the S. cerevisiae system will provide an excellent framework for further defining mechanistic details and functional roles for these proteins.
Conclusions and outlook
Our recent investigation of the role for Set4 in protecting cells during oxidative stress (Tran, et al. 2018) identifies a biological function for this largely-uncharacterized chromatin regulator, and highlights the potential for the Set3-Set4 SET domain subfamily to contribute to defense pathways in the presence of oxidative insults, particularly in light of our observations regarding SET3 mutants (Tran, et al. 2018), and previous reports that MLL5-deficient cells show increased ROS (Tasdogan, et al. 2016). There remain numerous mechanistic questions regarding how Set4 regulates gene expression to promote cell survival during stress, including determining the contribution of the PHD finger and identifying the molecular signals that promote Set4 chromatin localization under stress. Additionally, while our evidence to date indicates that Set4 lacks intrinsic catalytic activity, it may interact with other chromatin-modifying enzymes to control gene expression, similar to Set3 and the metazoan orthologs. The underlying function of the SET domain within Set4 and orthologous proteins still remains to be determined. The amino acid substitutions within these noncanonical domains are very well-conserved (Madan, et al. 2009, Mas-Y-Mas, et al. 2016, Tran, et al. 2018), indicating the domain may play a still undiscovered catalytic role, or that it provides important structural elements to the protein. Further investigation of the biochemical roles for Set4 and related proteins, as well as additional structure-function analysis of the SET domains within these proteins, will shed light on the functional contribution of these domains to this conserved subfamily of proteins and the mechanisms by which they regulate gene expression and calibrate cellular responses to environmental stress.
Acknowledgements:
The authors acknowledge support from National Institutes of Health grant R01GM124342 to E.M.G.
References
- Ali M, Rincón-Arano H, Zhao W, Rothbart SB, Tong Q, Parkhurst SM, Strahl BD, Deng LW, Groudine M, Kutateladze TG (2013) Molecular basis for chromatin binding and regulation of MLL5. Proc Natl Acad Sci U S A 110: 11296–11301 doi: 10.1073/pnas.1310156110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon AD, Rodriguez AL, Meirelles O, Roy S, Davidson GS, Tapia PH, Allen C, Joe R, Benn D, Werner-Washburne M (2008) Characterization of differentiated quiescent and nonquiescent cells in yeast stationary-phase cultures. Mol Biol Cell 19: 1271–1280 doi: 10.1091/mbc.E07-07-0666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calpena E, Palau F, Espinós C, Galindo MI (2015) Evolutionary History of the Smyd Gene Family in Metazoans: A Framework to Identify the Orthologs of Human Smyd Genes in Drosophila and Other Animal Species. PLoS One 10: e0134106 doi: 10.1371/journal.pone.0134106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Liu J, Zhou SH, Wang XN, Chew JF, Deng LW (2008) RNA interference against mixed lineage leukemia 5 resulted in cell cycle arrest. Int J Biochem Cell Biol 40: 2472–2481 doi: 10.1016/j.biocel.2008.04.012 [DOI] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG (2010) Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10: 457–469 doi: 10.1038/nrc2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SG (2013) Protein methylation at the surface and buried deep: thinking outside the histone box. Trends Biochem Sci 38: 243–252 doi: 10.1016/j.tibs.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Urso A, Brickner JH (2017) Epigenetic transcriptional memory. Curr Genet 63: 435–439 doi: 10.1007/s00294-016-0661-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deliu E, Arecco N, Morandell J, Dotter CP, Contreras X, Girardot C, Käsper EL, Kozlova A, Kishi K, Chiaradia I, Noh KM, Novarino G (2018) Haploinsufficiency of the intellectual disability gene SETD5 disturbs developmental gene expression and cognition. Nat Neurosci 10.1038/s41593-018-0266-2 [DOI] [PubMed] [Google Scholar]
- Deng LW, Chiu I, Strominger JL (2004) MLL 5 protein forms intranuclear foci, and overexpression inhibits cell cycle progression. Proc Natl Acad Sci U S A 101: 757–762 doi: 10.1073/pnas.2036345100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SC, Zhang X, Trievel RC, Cheng X (2005) The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol 6: 227 doi: 10.1186/gb-2005-6-8-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Jiang W, Zhou P, Liu L, Wan X, Yuan X, Wang X, Chen M, Chen J, Yang J, Kong C, Li B, Peng C, Wong CC, Hou F, Zhang Y (2015) Mixed Lineage Leukemia 5 (MLL5) Protein Stability Is Cooperatively Regulated by O-GlcNac Transferase (OGT) and Ubiquitin Specific Protease 7 (USP7). PLoS One 10: e0145023 doi: 10.1371/journal.pone.0145023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchalian J, Ali M, Andrews FH, Zhang Y, Barrett AS, Kutateladze TG (2017) Structural Insight into Recognition of Methylated Histone H3K4 by Set3. J Mol Biol 429: 2066–2074 doi: 10.1016/j.jmb.2016.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 doi: 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Green EM, Mas G, Young NL, Garcia BA, Gozani O (2012) Methylation of H4 lysines 5, 8 and 12 by yeast Set5 calibrates chromatin stress responses. Nat Struct Mol Biol 19: 361–363 doi: 10.1038/nsmb.2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozeva D, Carss K, Spasic-Boskovic O, Parker MJ, Archer H, Firth HV, Park SM, Canham N, Holder SE, Wilson M, Hackett A, Field M, Floyd JA, Hurles M, Raymond FL, Consortium UK (2014) De novo loss-of-function mutations in SETD5, encoding a methyltransferase in a 3p25 microdeletion syndrome critical region, cause intellectual disability. Am J Hum Genet 94: 618–624 doi: 10.1016/j.ajhg.2014.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser M, Yap DB, Leung M, de Algara TR, Tafech A, McKinney S, Dixon J, Thresher R, Colledge B, Carlton M, Humphries RK, Aparicio SA (2009) Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood 113: 1432–1443 doi: 10.1182/blood-2008-06-162263 [DOI] [PubMed] [Google Scholar]
- Jaiswal D, Turniansky R, Green EM (2017) Choose Your Own Adventure: The Role of Histone Modifications in Yeast Cell Fate. J Mol Biol 429: 1946–1957 doi: 10.1016/j.jmb.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezek M, Gast A, Choi G, Kulkarni R, Quijote J, Graham-Yooll A, Park D, Green EM (2017) The histone methyltransferases Set5 and Set1 have overlapping functions in gene silencing and telomere maintenance. Epigenetics 12: 93–104 doi: 10.1080/15592294.2016.1265712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmeren P, Sameith K, van de Pasch LA, Benschop JJ, Lenstra TL, Margaritis T, O’Duibhir E, Apweiler E, van Wageningen S, Ko CW, van Heesch S, Kashani MM, Ampatziadis-Michailidis G, Brok MO, Brabers NA, Miles AJ, Bouwmeester D, van Hooff SR, van Bakel H, Sluiters E, Bakker LV, Snel B, Lijnzaad P, van Leenen D, Groot Koerkamp MJ, Holstege FC (2014) Large-scale genetic perturbations reveal regulatory networks and an abundance of gene-specific repressors. Cell 157: 740–752 doi: 10.1016/j.cell.2014.02.054 [DOI] [PubMed] [Google Scholar]
- Kim T, Buratowski S (2009) Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5’ transcribed regions. Cell 137: 259–272 doi: 10.1016/j.cell.2009.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Xu Z, Clauder-Münster S, Steinmetz LM, Buratowski S (2012) Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell 150: 1158–1169 doi: 10.1016/j.cell.2012.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuechler A, Zink AM, Wieland T, Lüdecke HJ, Cremer K, Salviati L, Magini P, Najafi K, Zweier C, Czeschik JC, Aretz S, Endele S, Tamburrino F, Pinato C, Clementi M, Gundlach J, Maylahn C, Mazzanti L, Wohlleber E, Schwarzmayr T, Kariminejad R, Schlessinger A, Wieczorek D, Strom TM, Novarino G, Engels H (2015) Loss-of-function variants of SETD5 cause intellectual disability and the core phenotype of microdeletion 3p25.3 syndrome. Eur J Hum Genet 23: 753–760 doi: 10.1038/ejhg.2014.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai LC, Kosorukoff AL, Burke PV, Kwast KE (2006) Metabolic-state-dependent remodeling of the transcriptome in response to anoxia and subsequent reoxygenation in Saccharomyces cerevisiae. Eukaryot Cell 5: 1468–1489 doi: 10.1128/EC.00107-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemak A, Yee A, Wu H, Yap D, Zeng H, Dombrovski L, Houliston S, Aparicio S, Arrowsmith CH (2013) Solution NMR structure and histone binding of the PHD domain of human MLL5. PLoS One 8: e77020 doi: 10.1371/journal.pone.0077020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cheng F, Deng LW (2012) MLL5 maintains genomic integrity by regulating the stability of the chromosomal passenger complex through a functional interaction with Borealin. J Cell Sci 125: 4676–4685 doi: 10.1242/jcs.110411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V, Madan B, Brykczynska U, Zilbermann F, Hogeveen K, Döhner K, Döhner H, Weber O, Blum C, Rodewald HR, Sassone-Corsi P, Peters AH, Fehling HJ (2009) Impaired function of primitive hematopoietic cells in mice lacking the Mixed-Lineage-Leukemia homolog MLL5. Blood 113: 1444–1454 doi: 10.1182/blood-2008-02-142638 [DOI] [PubMed] [Google Scholar]
- Martín GM, King DA, Green EM, Garcia-Nieto PE, Alexander R, Collins SR, Krogan NJ, Gozani OP, Morrison AJ (2014) Set5 and Set1 cooperate to repress gene expression at telomeres and retrotransposons. Epigenetics 9: 513–522 doi: 10.4161/epi.27645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas-Y-Mas S, Barbon M, Teyssier C, Déméné H, Carvalho JE, Bird LE, Lebedev A, Fattori J, Schubert M, Dumas C, Bourguet W, le Maire A (2016) The Human Mixed Lineage Leukemia 5 (MLL5), a Sequentially and Structurally Divergent SET Domain-Containing Protein with No Intrinsic Catalytic Activity. PLoS One 11: e0165139 doi: 10.1371/journal.pone.0165139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo R, Mizobuchi S, Nakashima M, Miki K, Ayusawa D, Fujii M (2017) Central roles of iron in the regulation of oxidative stress in the yeast Saccharomyces cerevisiae. Curr Genet 63: 895–907 doi: 10.1007/s00294-017-0689-4 [DOI] [PubMed] [Google Scholar]
- Maze I, Noh KM, Soshnev AA, Allis CD (2014) Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat Rev Genet 15: 259–271 doi: 10.1038/nrg3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel SL, Hepperla AJ, Huang J, Dronamraju R, Adams AT, Kulkarni VG, Davis IJ, Strahl BD (2017) H3K36 Methylation Regulates Nutrient Stress Response in Saccharomyces cerevisiae by Enforcing Transcriptional Fidelity. Cell Rep 19: 2371–2382 doi: 10.1016/j.celrep.2017.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy KA, Jung YL, Zee BM, Wang CI, Park PJ, Kuroda MI (2017) upSET, the Drosophila homologue of SET3, Is Required for Viability and the Proper Balance of Active and Repressive Chromatin Marks. G3 (Bethesda) 7: 625–635 doi: 10.1534/g3.116.037788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal-Ribelles M, Mas G, Millán-Zambrano G, Solé C, Ammerer G, Chávez S, Posas F, de Nadal E (2015) H3K4 monomethylation dictates nucleosome dynamics and chromatin remodeling at stress-responsive genes. Nucleic Acids Res 43: 4937–4949 doi: 10.1093/nar/gkv220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipovich AB, Gangula R, Vianna PG, Magnuson MA (2016) Setd5 is essential for mammalian development and the co-transcriptional regulation of histone acetylation. Development 143: 4595–4607 doi: 10.1242/dev.141465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, Rigaut G, Séraphin B, Aasland R, Stewart AF (2001) The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev 15: 2991–3004 doi: 10.1101/gad.207401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porras-Yakushi TR, Whitelegge JP, Clarke S (2006) A novel SET domain methyltransferase in yeast: Rkm2-dependent trimethylation of ribosomal protein L12ab at lysine 10. J Biol Chem 281: 35835–35845 doi: 10.1074/jbc.M606578200 [DOI] [PubMed] [Google Scholar]
- Rincon-Arano H, Halow J, Delrow JJ, Parkhurst SM, Groudine M (2012) UpSET recruits HDAC complexes and restricts chromatin accessibility and acetylation at promoter regions. Cell 151: 1214–1228 doi: 10.1016/j.cell.2012.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle NE, Pillus L (2018) Critical genomic regulation mediated by Enhancer of Polycomb. Curr Genet 64: 147–154 doi: 10.1007/s00294-017-0742-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle NE, Torres-Machorro AL, Pillus L (2017) Chromatin Regulation by the NuA4 Acetyltransferase Complex Is Mediated by Essential Interactions Between Enhancer of Polycomb (Epl1) and Esa1. Genetics 205: 1125–1137 doi: 10.1534/genetics.116.197830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S, Sreenivas P, Sambasivan R, Cheedipudi S, Kandalla P, Pavlath GK, Dhawan J (2009) MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc Natl Acad Sci U S A 106: 4719–4724 doi: 10.1073/pnas.0807136106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serratore ND, Baker KM, Macadlo LA, Gress AR, Powers BL, Atallah N, Westerhouse KM, Hall MC, Weake VM, Briggs SD (2018) A Novel Sterol-Signaling Pathway Governs Azole Antifungal Drug Resistance and Hypoxic Gene Repression in. Genetics 208: 1037–1055 doi: 10.1534/genetics.117.300554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, Howe L, Utz PJ, Kutateladze TG, Lugovskoy AA, Bedford MT, Gozani O (2007) Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem 282: 2450–2455 doi: 10.1074/jbc.C600286200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soontorngun N (2017) Reprogramming of nonfermentative metabolism by stress-responsive transcription factors in the yeast Saccharomyces cerevisiae. Curr Genet 63: 1–7 doi: 10.1007/s00294-016-0609-z [DOI] [PubMed] [Google Scholar]
- Tasdogan A, Kumar S, Allies G, Bausinger J, Beckel F, Hofemeister H, Mulaw M, Madan V, Scharfetter-Kochanek K, Feuring-Buske M, Doehner K, Speit G, Stewart AF, Fehling HJ (2016) DNA Damage-Induced HSPC Malfunction Depends on ROS Accumulation Downstream of IFN-1 Signaling and Bid Mobilization. Cell Stem Cell 19: 752–767 doi: 10.1016/j.stem.2016.08.007 [DOI] [PubMed] [Google Scholar]
- Tee WW, Reinberg D (2014) Chromatin features and the epigenetic regulation of pluripotency states in ESCs. Development 141: 2376–2390 doi: 10.1242/dev.096982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran K, Jethmalani Y, Jaiswal D, Green EM (2018) Set4 is a chromatin-associated protein, promotes survival during oxidative stress, and regulates stress response genes in yeast. J Biol Chem 293: 14429–14443 doi: 10.1074/jbc.RA118.003078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Chen HV, Liu CL, Rahat A, Klien A, Soares L, Gudipati M, Pfeffner J, Regev A, Buratowski S, Pleiss JA, Friedman N, Rando OJ (2012) Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol 10: e1001369 doi: 10.1371/journal.pbio.1001369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap DB, Walker DC, Prentice LM, McKinney S, Turashvili G, Mooslehner-Allen K, de Algara TR, Fee J, de Tassigny X, Colledge WH, Aparicio S (2011) Mll5 is required for normal spermatogenesis. PLoS One 6: e27127 doi: 10.1371/journal.pone.0027127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wong J, Klinger M, Tran MT, Shannon KM, Killeen N (2009) MLL5 contributes to hematopoietic stem cell fitness and homeostasis. Blood 113: 1455–1463 doi: 10.1182/blood-2008-05-159905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Wang Z, Yuan X, Zhou C, Liu L, Wan X, Zhang F, Ding X, Wang C, Xiong S, Yuan J, Li Q, Zhang Y (2013) Mixed lineage leukemia 5 (MLL5) protein regulates cell cycle progression and E2F1-responsive gene expression via association with host cell factor-1 (HCF-1). J Biol Chem 288: 17532–17543 doi: 10.1074/jbc.M112.439729 [DOI] [PMC free article] [PubMed] [Google Scholar]