Abstract
Duplication of the genome poses one of the most significant threats to genetic integrity, cellular fitness and organismal health. Therefore, numerous mechanisms have evolved that maintain replication fork stability in the face of DNA damage and allow faithful genome duplication. The fission yeast BRCT-domain containing protein Brc1, and its budding yeast orthologue Rtt107, has emerged as a “hub” factor that integrates multiple replication fork protection mechanisms. Notably, the cofactors and pathways through which Brc1, Rtt107 and their human orthologue (PTIP) act appeared largely distinct. This either represents true evolutionary functional divergence, or perhaps an incomplete genetic and biochemical analysis of each protein. In this regard we recently showed that like Rtt107, Brc1 supports key functions of the Smc5-Smc6 complex; including its recruitment into DNA repair foci, chromatin association and SUMO ligase activity. Furthermore, fission yeast lacking the Nse5-Nse6 genome stability factor were found to exhibit defects in Smc5-Smc6 function, similar to but more severe than those in cells lacking Brc1. Here we place these findings in context with the known functions of Brc1, Rtt107 and Smc5-Smc6, present data suggesting a role for acetylation in Smc5-Smc6 chromatin loading, and discuss wider implications for genome stability.
Keywords: Brc1, Smc5, Smc6, SUMO, Rad18, Nse5, Nse6, PTIP, Rtt107, replication stress
Introduction
The essential Smc5-Smc6 complex is related to the cohesin and condensin structural maintenance of chromosomes (SMC) complexes, but is uniquely equipped with E3 ligase activities for the post-translational modifiers SUMO and ubiquitin (Aragon 2018; Diaz and Pecinka 2018; Kakui and Uhlmann 2018; Matityahu and Onn 2018; Uhlmann 2016). Briefly, Smc5-Smc6 consists of an essential hexameric core complex assembled by the association of the stable sub-complexes Smc5/Smc6/Nse2 with Nse1/Nse3/Nse4 (Figure 1, (Aragon 2018; Diaz and Pecinka 2018; Uhlmann 2016)). In addition, we identified a third sub-complex of Smc5-Smc6 containing Nse5 and Nse6 (Pebernard et al. 2006), which appear functionally conserved across species despite extreme divergence in primary structure (Figure 1, (Duan et al. 2009; Raschle et al. 2015; Yan et al. 2013)).
Figure 1. Smc5-Smc6 Subunits and Architecture.
Yeast Smc5-Smc6 subunits organized by known position in complex (Nse5-Nse6 position uncertain). Table shows yeast and human subunits grouped by the stable sub-complexes formed. Protein functions and families indicated: SMC (structural maintenance of chromosomes, SUMO/Ub E3 ligases tag proteins with SUMO or ubiquitin, respectively; MAGE (melanoma-associated antigen), and “kleisin” bridges Smc5/Smc6 ATPase head groups (reviewed in (Aragon 2018; Diaz and Pecinka 2018)).
Smc5-Smc6 remains the most mechanistically enigmatic of the SMC complexes, but it is of fundamental importance to cellular processes such as genome replication, mitotic and meiotic chromosome segregation, DNA repair through homologous recombination, and viral restriction (Aragon 2018; Diaz and Pecinka 2018; Hang and Zhao 2016; Rai and Laloraya 2017; Uhlmann 2016; Wani et al. 2018). Of note, the SUMO ligase activity of Smc5-Smc6 supports several of the foregoing processes by SUMOylating target proteins that include components of the Smc5-Smc6 complex (“auto-SUMOylation” (Andrews et al. 2005; Mahendrawada et al. 2017; Pebernard et al. 2008; Zhao and Blobel 2005)), the cohesin complex, the BLM/Sgs1 RecQ-like helicase, and telomere protection proteins e.g. TRF1/2 (Aragon 2018; Diaz and Pecinka 2018). The above functions of Smc5-Smc6 are amply described in excellent recent reviews (Aragon 2018; Diaz and Pecinka 2018; Hang and Zhao 2016; Uhlmann 2016), so will not be detailed herein. Instead, we discuss our data on the role of the Nse5-Nse6 sub-complex and its interacting partner Brc1 in the focal accumulation, chromatin association and SUMO ligase activity of Smc5-Smc6 at DNA lesions.
Materials and Methods
Yeast strains and growth conditions
Standard fission yeast methods were performed as described previously (Moreno et al. 1991). Strains used in this study are listed in Table 1.
Table 1.
List of yeast strains used in this study.
| Strain | Genotypei | Source or reference |
|---|---|---|
| NB5931 | h+ nse4-Flag-His:kanR | (Zabrady et al. 2016) |
| NB5953 | h+ pJK148-Padh-ura4:leu1 | this study |
| NB5954 | h+ pJK148-Padh-nts1-ura4:leu1 | this study |
| NB5955 | h+ nse6::kanMX6 pJK148-Padh-ura4:leu1 | this study |
| NB5956 | h+ nse6::kanMX6 pJK148-Padh-nts1-ura4:leu1 | this study |
| NB5959 | h+ smc6-74 | (Verkade et al. 1999) |
| NB6009 | h- nse4-Flag-His:kanR pJK148-Padh-ura4:leu1 | this study |
| NB6010 | h- nse4-Flag-His:kanR pJK148-Padh-nts1-ura4:leu1 | this study |
| NB6004 | h- nse4-Flag-His:kanR nse6::natMX6 pJK148-Padh-ura4:leu1 | this study |
| NB6005 | h- nse4-Flag-His:kanR nse6::natMX6 pJK148-Padh-nts1- ura4:leu1 | this study |
| NB5963 | h- nse4-Flag-His:kanR nse6::natMX6 | this study |
| NB5993 | h- smc6-74 pJK148-Padh-ura4:leu1 | this study |
| NB5994 | h- smc6-74 pJK148-Padh-nts1-ura4:leu1 | this study |
All strains are of ura4-D18 leu1-32 background genotype. Double colons represent knockouts; single colons represent tagging.
Spot assays
Fission yeast were grown at 32 °C to logarithmic phase (optical density at 600 nm [OD600] of 0.6 to 0.8), spotted on YES agar plates supplemented with the relevant drug in 5-fold dilutions from a starting OD600 of 0.5, and then incubated at 30 °C or 32 °C for 3 to 5 days.
Chromatin Immunoprecipitation
Method was adapted from (Nelson et al. 2006). Briefly, exponentially growing cell cultures (untreated, treated with 15 mM HU or 0.005% MMS 6 hrs) were fixed with 1% formaldehyde for 25 min, quenched with glycine. Cells were broken with beads in Lysis buffer (50 mM HEPES, 1 mM EDTA, 150 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate; pH 7.6), supplemented with protease inhibitor cocktail (Roche) and 2 mM PMSF. Chromatin extract was sheared with the Bioruptor Pico (Diagenode). Immunoprecipitation was done with 10 μg of FLAG antibody (F3165, Sigma) for 2 hrs followed by incubation with protein G dynabeads (Invitrogen) for 1 hr at 4 °C. Dynabeads were washed 6 times with IP buffer (50 mM Tris-HCl, 1% Triton X-100, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, pH 7.5). Input and IP samples were incubated with 10% Chelex 100 (BioRad) and treated with Proteinase K. Quantitative PCR was performed with the resulting DNA samples using SensiFAST SYBR No-ROX Kit (Bioline). Primer sequences for telo2R and STE1 were as previously published (Hayashi et al. 2007; Pebernard et al. 2008; Tapia-Alveal and O’Connell 2011). DNA recovery was calculated using 2−ΔCt method and data, representative of 4 independent repeats, are presented as a percentage of input DNA.
Function of the Nse5-Nse6 Sub-Complex
We identified Nse5-Nse6 as an interactor and key cofactor of Smc5-Smc6 in both mitotic and meiotic nuclear divisions (Figure 1, (Pebernard et al. 2006; Wehrkamp-Richter et al. 2012)). The primary structures of Nse5 and Nse6 are not significantly conserved beyond closely related fission yeast species. However, based on their predicted structural composition, we proposed that they were functionally related to budding yeast YML023C (Nse5) and KRE29 (Nse6) that are also sequence orphans and components of the Smc5-Smc6 complex (Pebernard et al. 2006). Supporting functional conservation, we identified the SUMO-targeted Ubiquitin Ligase (STUbL) complex through its physical interaction with fission yeast Nse5 (Perry et al. 2008; Prudden et al. 2007), an interaction that is conserved in the highly diverged budding yeast (Horigome et al. 2016).
Recently, a human protein called FAM178A was found to interact with SMC5-SMC6 and exhibit weak similarity to yeast Nse6 (Raschle et al. 2015). Interestingly, FAM178A (renamed SLF2) interacts with the RAD18 cofactor BRCTx (renamed SLF1), forming a complex that is apparently orthologous to Nse5-Nse6 (Figure 1, (Raschle et al. 2015)).
Importantly, despite critically supporting Smc5-Smc6 functions, Nse5-Nse6 is non-essential in fission yeast but essential in other organisms where tested (Aragon 2018; Diaz and Pecinka 2018). Thus, phenotypic analysis of fission yeast lacking Nse5-Nse6 provides a unique clean separation-of-function within the Smc5-Smc6 complex. We hypothesized that Nse5-Nse6 supports the recruitment and/or loading of Smc5-Smc6 on chromatin, as described for the cohesin loader Scc2-Scc4 (Litwin and Wysocki 2018; Pebernard et al. 2006; Uhlmann 2016). We recently tested this hypothesis using ChIP-qPCR to determine Smc5-Smc6 chromatin association at some of its previously identified binding hotspots (Pebernard et al. 2008).
Strikingly, cells lacking Nse5-Nse6 showed an ~5–10 fold reduction in chromatin occupancy of Smc5-Smc6 at sites including centromeres, telomeres and tDNAs, in the presence or absence of replication stress (see Figure 2 and (Oravcova et al. 2018)). This provided the first unequivocal evidence for Nse5-Nse6 acting as an Smc5-Smc6 recruitment factor during both the unchallenged cell cycle, and in response to replication stress. Moreover, because Smc5-Smc6 is essential, and residual loading is detected in the absence of Nse5-Nse6, an alternative recruitment pathway likely exists.
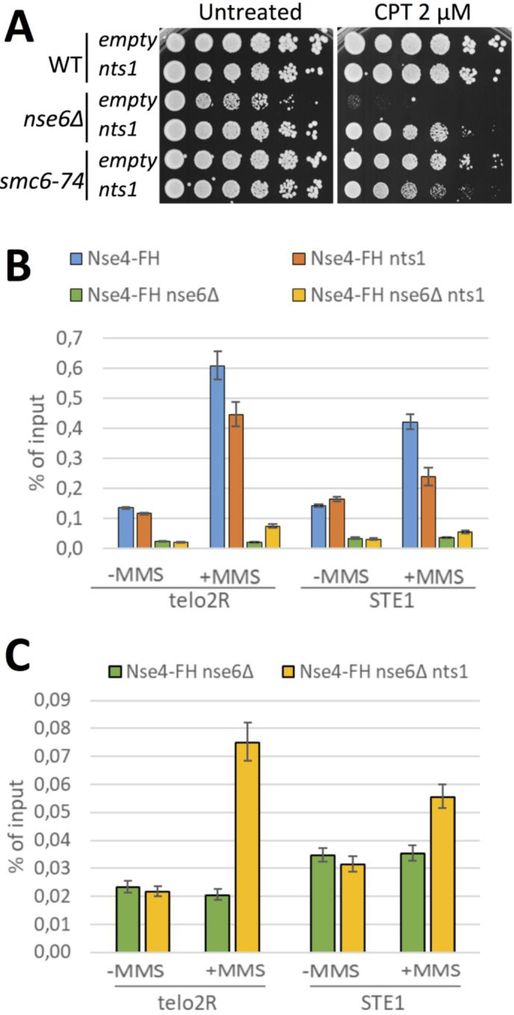
Figure 2. Nts1, a Potent and Specific Suppressor of nse6Δ.
A. pAdh1-nts1 or empty vector (pAdh1) stably integrated at the leu1 locus in the indicated strains. Cells were grown at 30°C for 3 days in the presence or absence of camptothecin (CPT). B. ChIP-qPCR against Nse4-FlagHis in the indicated strains at telomeric loci (as described in (Oravcova et al. 2018)). C. As in panel “B” but showing only nse6Δ background. Note 10-fold lower scale on Y-axis. Data represent ≥ 3 independent experiments.
In an attempt to identify such a pathway, we recently screened for specific dosage suppressors of the phenotypes caused by Nse5 or Nse6 deletion (Zilio et al. 2014). This approach returned Nts1 as a highly specific suppressor of nse5Δ or nse6Δ cells (Figure 2A and (Zilio et al. 2014)). Strikingly however, Nts1 expression aggravated the phenotypes of hypomorphic alleles of the essential Smc5-Smc6 core complex (e.g. smc6-74, Figure 2A and (Zilio et al. 2014)). This result underscores the separation of function between Nse5-Nse6 and the core Smc5-Smc6 complex.
Interestingly, Nts1 overexpression in wild-type cells significantly reduced Smc5-Smc6 chromatin association during replication stress, whereas in nse6Δ cells it had the opposite effect (Figure 2B,C). Whilst the mechanism is currently unknown, these data fit with the Nts1-induced sensitization of smc6–74 cells but partial rescue of nse6Δ cells undergoing replication stress (Figure 2A and (Zilio et al. 2014)). Nts1 is part of a novel Clr6 (HDAC1)-based histone deacetylase complex (Zilio et al. 2014), suggesting that modulation of the acetylation state of chromatin or Smc5-Smc6 subunits could positively influence the loading of Smc5-Smc6 in the absence of Nse5-Nse6. Indeed, the association of condensin with chromosomes is impacted by local chromatin structure and, the acetylation state of cohesin’s SMC coiled-coils affects its loading (Kulemzina et al. 2016; Robellet et al. 2017), both of which could be regulated by an Nts1-like HDAC.
Overall, Nse5-Nse6 is emerging as a functionally conserved cofactor for Smc5-Smc6 recruitment. Consistent with this, budding yeast hypomorphic for Nse5 (nse5ts1) show reduced localization of Smc5-Smc6 to stalled replication forks, but nse5ts1 also appeared to impact the integrity of the Smc5-Smc6 complex (Bustard et al. 2016). The authors assessed complex stability by co-immunoprecipitation of Nse6 with Smc5 in nse5ts1 cells (Bustard et al. 2012). Given that Nse5 and Nse6 form a sub-complex this is not surprising, and unlikely affects core complex stability, as previously determined using in vitro Smc5-Smc6 complex reconstitution (Pebernard et al. 2006).
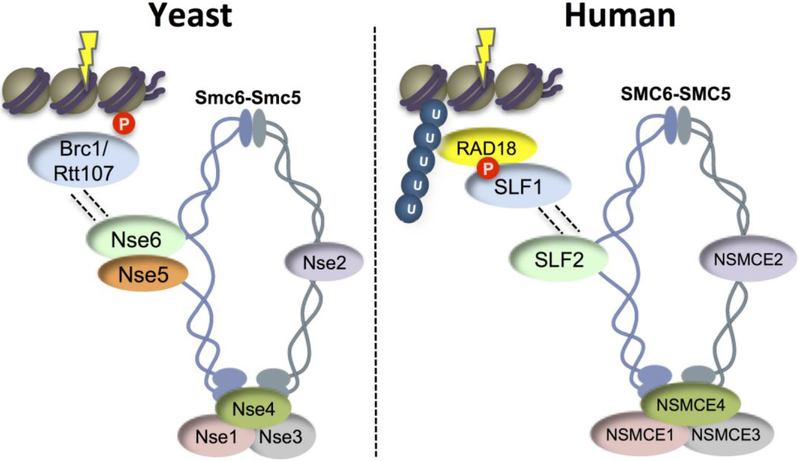
In human cells, the SLF1-SLF2 complex recruits SMC5-SMC6 to stalled replication forks (Raschle et al. 2015). Whereas SLF2 contains a highly diverged Nse6-like domain, SLF1 contains ankyrin repeats and a BRCT domain, which are not found in Nse5 from either fission or budding yeasts. The SLF1 BRCT domain binds phosphorylated RAD18, which in turn uses its UBZ domain to recognize ubiquitin deposited at DNA lesions to drive SMC5-SMC6 recruitment (Figure 3, (Raschle et al. 2015)). As described below, this mechanism appears to be both an interesting “amalgam” of, and departure from, the mechanisms used in fission and budding yeasts.
Figure 3. Smc5-Smc6 Recruitment Mechanisms.
In yeast, Brc1 and Rtt107 recruits Smc5-Smc6, likely through Nse5-Nse6, to DNA lesions decorated with gamma-H2A (Leung et al. 2011; Oravcova et al. 2018). Brc1 and Rtt107 use their BRCTs 1–4 to interact with Nse5-Nse6 and BRCTs 5–6 to bind gamma-H2A (Leung et al. 2011; Oravcova et al. 2018). In human cells, SLF2 (Nse6-like) binds SLF1, which binds phosphorylated RAD18 that in turn binds ubiquitin via its UBZ domain (Raschle et al. 2015).
Brc1 Supports Smc5-Smc6 Foci Formation at DNA Lesions
Brc1, a six BRCT domain-containing protein, critically supports resistance to replication stress in fission yeast (Reubens et al. 2017; Sheedy et al. 2005; Williams et al. 2010). It shares this role and its content of six BRCT domains with Rtt107 of budding yeast and human PTIP (Hang and Zhao 2016; Ray Chaudhuri et al. 2016). Furthermore, the C-terminal BRCTs of Brc1, PTIP and Rtt107 bind gamma-H2A enriched chromatin around DNA lesions, enabling them to recruit DNA repair factors through their amino terminal BRCTs (Li et al. 2012; Manke et al. 2003; Williams et al. 2010). Despite these similarities, Brc1, Rtt107 and PTIP appeared to “scaffold” distinct partner proteins to execute their replication fork protection roles (Hang and Zhao 2016).
Brc1 was initially isolated as a dosage suppressor of smc6–74 (Verkade et al. 1999). Elegant genetic analyses showed that smc6–74 suppression by Brc1 requires multiple nucleases and a non-catalytic “platform” function of the postreplication repair factor Rad18 (Lee et al. 2007; Sheedy et al. 2005). The combination of a BRCT domain factor (Brc1) and Rad18 in supporting Smc5-Smc6 function presents an intriguing parallel to the human SMC5-SMC6 recruitment factors i.e. SLF1 and RAD18 (Figure 3, (Raschle et al. 2015)). However, the fission yeast genetic analyses of Brc1 indicated that it acted as a bypass suppressor of Smc5-Smc6 dysfunction.
Nonetheless, through biochemical and cell biology approaches we recently revealed that there is a more intimate connection between Brc1 and Smc5-Smc6 than thought. Smc5-Smc6 forms foci in response to replication stress such as hydroxyurea (HU) and methyl methanesulphonate (MMS, (Pebernard et al. 2008)). Brc1 also forms foci in response to replication stress (Lee et al. 2013; Williams et al. 2010) and of note; Brc1 is recruited to the same regions of the fission yeast heterochromatic centromeres as Smc5-Smc6 in the presence of HU (Lee et al. 2013; Pebernard et al. 2008). Moreover, brc1Δ and nse6Δ cells are hypersensitive to the microtubule poison thiabendazole, which together with their similar chromosome mis-seggregation phenotypes points to potentially overlapping roles for Brc1 and Smc5-Smc6 in centromere function (Lee et al. 2013; Pebernard et al. 2008).
Intriguingly, we found that Smc5-Smc6 foci depend on both Brc1 and Nse5-Nse6, and identified a physical interaction between these factors (Oravcova et al. 2018). Moreover, using ChIP-qPCR, Brc1 was shown to promote Smc5-Smc6 chromatin association (like Nse5-Nse6) in response to replication stress. Together with gamma-H2A binding by Brc1, this suggests a model for Smc5-Smc6 recruitment to DNA lesions (Figure 3). The role of Brc1 in Smc5-Smc6 chromatin association echoes that of Rtt107 in budding yeast (Leung et al. 2011), which was unforeseen given the largely distinct synthetic genetic interaction profiles of brc1Δ and rtt107Δ (Sanchez et al. 2015). Such conservation of Brc1 and Rtt107 function between these highly divergent species also raises the possibility that PTIP may have an SMC5-SMC6 related role that remains to be uncovered.
Surprisingly, in contrast to human SMC5-SMC6 recruitment we found that fission yeast Rad18 is not required for Smc5-Smc6 foci formation or chromatin association, despite recently being shown to form a complex with Brc1 (Reubens et al. 2017). Rad18 and Smc5-Smc6 both interact with Brc1’s amino-terminal BRCT domains, so these interactions may be mutually exclusive (Oravcova et al. 2018; Reubens et al. 2017). If so, Brc1 acts through distinct pathways to promote Smc5-Smc6 activity during replicative stress. In this regard, Rtt107 scaffolds at least three distinct complexes through its amino-terminal BRCTs (Hang and Zhao 2016).
Activation of the Smc5-Smc6 SUMO Ligase During Replication Stress
The Smc5-Smc6 SUMO ligase is activated during replication stress, especially during MMS treatment, which can be detected as “auto-SUMOylation” of its subunits e.g. Nse4, and modification of certain replication proteins e.g. Mcm6 (Andrews et al. 2005; Hang et al. 2015; Pebernard et al. 2008). The mechanisms behind MMS-induced Smc5-Smc6 activation were not totally clear.
We had shown that Nse4 SUMOylation in MMS treated cells is dependent on Nse2 (Pebernard et al. 2008). Using Nse4 as a reporter, we recently showed that both Brc1 and Nse5-Nse6 are also required for full activation of the Smc5-Smc6 SUMO ligase (Oravcova et al. 2018). We propose that MMS-induced foci formation facilitates the trans-SUMOylation of Nse4 and of other subunits due to the increased local concentration of the complex. Indeed, a recent in vitro analysis indicated that budding yeast Nse4 is SUMOylated by Nse2 in trans, as opposed to standard E3 ligase “auto-sumoylation” (Pichler et al. 2017; Varejao et al. 2018). Notably, Rtt107 also promotes the SUMOylation of Smc5-Smc6 targets in MMS treated budding yeast, revealing a conserved mechanism (Hang et al. 2015).
Interestingly, whereas deletion of either Brc1 or Nse5-Nse6 abrogates Smc5-Smc6 foci formation in MMS, Nse4 SUMOylation is reduced in brc1Δ cells but abolished in nse6Δ cells (Oravcova et al. 2018). This difference correlates well with the Smc5-Smc6 DNA binding defect, which is much more severe in nse6Δ versus brc1Δ cells (Oravcova et al. 2018). Of relevance, DNA binding by Smc5 was recently shown to activate the Nse2 SUMO ligase (Varejao et al. 2018). In addition, Smc5 ATPase activity is required for both the DNA association and activation of Smc5-Smc6’s SUMO ligase (Bermudez-Lopez et al. 2015; Kanno et al. 2015).
In aggregate, the above data point to a sequential recruitment, loading, and activation model: (i) Brc1/Rtt107-mediated “concentration” around DNA lesions, (ii) Smc5-Smc6 ATPase mediated DNA loading assisted by Nse5-Nse6 and (iii) DNA and ATPase induced conformational changes in Smc5 that are transmitted to bound Nse2, promoting SUMOylation of its targets. This model provides a rationale for the selective SUMOylation of chromatin-associated targets by Smc5-Smc6 including cohesin subunits, telomere factors TRF1/2, the RecQ family helicase Sgs1, replication proteins and transcription machinery (Albuquerque et al. 2013; Aragon 2018; Diaz and Pecinka 2018; Hang et al. 2015).
The activation of Smc5-Smc6 SUMO ligase by Brc1/Rtt107 and Nse5-Nse6 will likely contribute to the recently described role of Smc5-Smc6 in regulating the Sgs1-Top3-Rmi1 (STR) complex (Bermudez-Lopez et al. 2016; Bonner et al. 2016). STR acts on DNA recombination structures to mitigate potential genome instability arising from a failure to remove covalent linkages between chromosomes and excessive crossover recombination outcomes, amongst other roles (Bermudez-Lopez and Aragon 2017; Mimitou and Symington 2011). SUMO-modified Smc5-Smc6 recruits STR into DNA repair foci via non-covalent SUMO-interacting motifs (SIMs) in STR, followed by Smc5-Smc5 dependent SUMOylation and “activation” of STR (Bermudez-Lopez et al. 2016; Bonner et al. 2016). Thus, given the defective foci formation and SUMOylation activity of Smc5-Smc6 in Brc1/Rtt107 and Nse5-Nse6 deleted cells, STR mis-regulation will likely contribute to their DNA repair defects.
In addition, by SUMOylating the cohesin subunit Scc1, Smc5-Smc6 promotes the establishment of cohesin-mediated sister chromatid cohesion and facilitates recombination-based repair (Almedawar et al. 2012; McAleenan et al. 2012; Wu et al. 2012). In a seemingly contrasting role, Smc5-Smc6 also promotes the prophase removal of cohesin from chromosome arms, but a role for SUMOylation in this function is untested (Tapia-Alveal et al. 2014; Tapia-Alveal et al. 2010). Clearly, the foregoing roles of Smc5-Smc6 in the cohesin cycle appear contradictory i.e. establishment versus removal. This may be due to context specific interactions or additional posttranslational modifications, and requires further study. The role of Brc1/Rtt107 in supporting the chromatin recruitment and activation of the Smc5-Smc6 SUMO ligase further complicates the picture, but also provides additional avenues through which to dissect Smc5-Smc6 functions in chromosome dynamics.
Concluding Remarks
The SUMO ligase activity of Smc5-Smc6 is important during the cellular response to replication stress, and is promoted by Brc1/Rtt107 in the highly diverged fission and budding yeasts (Hang et al. 2015; Oravcova et al. 2018). Perhaps then, PTIP will play a similar role in human cells, but it is possible that alternative pathways have evolved that render this role redundant (Raschle et al. 2015). In this respect, although Nse5-Nse6 appears functionally conserved across species, the primary structures of its subunits have rapidly diverged beyond recognition by standard homology searches (Bustard et al. 2012; Leung et al. 2011; Oravcova et al. 2018; Pebernard et al. 2006; Raschle et al. 2015).
The formation of SUMO-dependent foci of Smc5-Smc6, which recruit and concentrate the DNA repair machinery via SUMO interactions e.g. the STR complex, is an intriguing parallel to PML nuclear bodies (PML NBs) of human cells (Bermudez-Lopez and Aragon 2017; Bonner et al. 2016; Hang et al. 2015; Oravcova et al. 2018; Pebernard et al. 2008). PML NBs are dynamic phase-separated sub-nuclear sites that also concentrate and integrate the SUMO and DNA repair pathways, including the human STR homologue, BTR (Chung et al. 2012; Dellaire and Bazett-Jones 2007; Nagai et al. 2011)). PML NB dynamics are regulated by the SUMO-targeted ubiquitin ligase (STUbL) RNF4, and in yeast, STUbL interacts with the Smc5-Smc6 complex through Nse5 (Hakli et al. 2005; Hazbun et al. 2003; Lallemand-Breitenbach et al. 2008; Prudden et al. 2007; Tatham et al. 2008). STUbL regulates the chromatin residence and/or stability of multiple poly-SUMOylated target proteins, which interestingly include the STR/BTR complex (Bohm et al. 2015; Nie and Boddy 2016).
Therefore, yeast, which do not have PML NBs, may form distinct phase-separated “repair foci” through Brc1/Rtt107 and SUMOylated Smc5-Smc6. Future analyses of Nse5-Nse6, Brc1, and Nts1 in relation to Smc5-Smc6 function will address this possibility, and also provide key insights into the regulation and disease suppressing activity of Smc5-Smc6 (Aragon 2018; Diaz and Pecinka 2018; van der Crabben et al. 2016).
Acknowledgements
This study was funded by NIH grants GM068608 and GM081840 awarded to M.N.B.
References
- Albuquerque CP, Wang G, Lee NS, Kolodner RD, Putnam CD, Zhou H (2013) Distinct SUMO ligases cooperate with Esc2 and Slx5 to suppress duplication-mediated genome rearrangements PLoS Genet 9:e1003670 doi: 10.1371/journal.pgen.1003670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almedawar S, Colomina N, Bermudez-Lopez M, Pocino-Merino I, Torres-Rosell J (2012) A SUMO-dependent step during establishment of sister chromatid cohesion Curr Biol 22:1576–1581 doi: 10.1016/j.cub.2012.06.046 [DOI] [PubMed] [Google Scholar]
- Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, Watts FZ (2005) Nse2, a component of the Smc5–6 complex, is a SUMO ligase required for the response to DNA damage Mol Cell Biol 25:185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon L (2018) The Smc5/6 Complex: New and Old Functions of the Enigmatic Long-Distance Relative Annu Rev Genet 52:89–107 doi: 10.1146/annurevgenet-120417-031353 [DOI] [PubMed] [Google Scholar]
- Bermudez-Lopez M, Aragon L (2017) Smc5/6 complex regulates Sgs1 recombination functions Curr Genet 63:381–388 doi: 10.1007/s00294-016-0648-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez-Lopez M et al. (2015) ATPase-dependent control of the Mms21 SUMO ligase during DNA repair PLoS Biol 13:e1002089 doi: 10.1371/journal.pbio.1002089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez-Lopez M, Villoria MT, Esteras M, Jarmuz A, Torres-Rosell J, Clemente-Blanco A, Aragon L (2016) Sgs1’s roles in DNA end resection, HJ dissolution, and crossover suppression require a two-step SUMO regulation dependent on Smc5/6 Genes Dev 30:1339–1356 doi: 10.1101/gad.278275.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm S, Mihalevic MJ, Casal MA, Bernstein KA (2015) Disruption of SUMO-targeted ubiquitin ligases Slx5-Slx8/RNF4 alters RecQ-like helicase Sgs1/BLM localization in yeast and human cells DNA Repair (Amst) 26:1–14 doi: 10.1016/j.dnarep.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner JN et al. (2016) Smc5/6 Mediated Sumoylation of the Sgs1-Top3-Rmi1 Complex Promotes Removal of Recombination Intermediates Cell Rep 16:368–378 doi: 10.1016/j.celrep.2016.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustard DE, Ball LG, Cobb JA (2016) Non-Smc element 5 (Nse5) of the Smc5/6 complex interacts with SUMO pathway components Biol Open 5:777–785 doi: 10.1242/bio.018440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustard DE et al. (2012) During replication stress, non-SMC element 5 (NSE5) is required for Smc5/6 protein complex functionality at stalled forks J Biol Chem 287:11374–11383 doi: 10.1074/jbc.M111.336263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung I, Osterwald S, Deeg KI, Rippe K (2012) PML body meets telomere: the beginning of an ALTernate ending? Nucleus 3:263–275 doi: 10.4161/nucl.20326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaire G, Bazett-Jones DP (2007) Beyond repair foci: subnuclear domains and the cellular response to DNA damage Cell Cycle 6:1864–1872 doi: 10.4161/cc.6.15.4560 [DOI] [PubMed] [Google Scholar]
- Diaz M, Pecinka A (2018) Scaffolding for Repair: Understanding Molecular Functions of the SMC5/6 Complex Genes (Basel) 9 doi: 10.3390/genes9010036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Yang Y, Chen YH, Arenz J, Rangi GK, Zhao X, Ye H (2009) Architecture of the Smc5/6 Complex of Saccharomyces cerevisiae Reveals a Unique Interaction between the Nse5–6 Subcomplex and the Hinge Regions of Smc5 and Smc6 J Biol Chem 284:8507–8515 doi:M809139200 [pii] 10.1074/jbc.M809139200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakli M, Karvonen U, Janne OA, Palvimo JJ (2005) SUMO-1 promotes association of SNURF (RNF4) with PML nuclear bodies Exp Cell Res 304:224–233 [DOI] [PubMed] [Google Scholar]
- Hang L, Zhao X (2016) The Rtt107 BRCT scaffold and its partner modification enzymes collaborate to promote replication Nucleus 7:346–351 doi: 10.1080/19491034.2016.1201624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang LE et al. (2015) Rtt107 Is a Multi-functional Scaffold Supporting Replication Progression with Partner SUMO and Ubiquitin Ligases Mol Cell 60:268–279 doi: 10.1016/j.molcel.2015.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M et al. (2007) Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast EMBO J 26:1327–1339 doi:7601585 [pii] 10.1038/sj.emboj.7601585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazbun TR et al. (2003) Assigning function to yeast proteins by integration of technologies Mol Cell 12:1353–1365 [DOI] [PubMed] [Google Scholar]
- Horigome C, Bustard DE, Marcomini I, Delgoshaie N, Tsai-Pflugfelder M, Cobb JA, Gasser SM (2016) PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL Genes Dev 30:931–945 doi: 10.1101/gad.277665.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakui Y, Uhlmann F (2018) SMC complexes orchestrate the mitotic chromatin interaction landscape Curr Genet 64:335–339 doi: 10.1007/s00294-017-0755-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Berta DG, Sjogren C (2015) The Smc5/6 Complex Is an ATP-Dependent Intermolecular DNA Linker Cell Rep 12:1471–1482 doi: 10.1016/j.celrep.2015.07.048 [DOI] [PubMed] [Google Scholar]
- Kulemzina I et al. (2016) A Reversible Association between Smc Coiled Coils Is Regulated by Lysine Acetylation and Is Required for Cohesin Association with the DNA Mol Cell 63:1044–1054 doi: 10.1016/j.molcel.2016.08.008 [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V et al. (2008) Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway Nat Cell Biol 10:547–555 doi:ncb1717 [pii] 10.1038/ncb1717 [DOI] [PubMed] [Google Scholar]
- Lee KM, Nizza S, Hayes T, Bass KL, Irmisch A, Murray JM, O’Connell MJ (2007) Brc1-mediated rescue of Smc5/6 deficiency: requirement for multiple nucleases and a novel Rad18 function Genetics 175:1585–1595 doi:genetics.106.067801 [pii] 10.1534/genetics.106.067801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Rozenzhak S, Russell P (2013) gammaH2A-binding protein Brc1 affects centromere function in fission yeast Mol Cell Biol 33:1410–1416 doi: 10.1128/MCB.01654-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung GP, Lee L, Schmidt TI, Shirahige K, Kobor MS (2011) Rtt107 Is Required for Recruitment of the SMC5/6 Complex to DNA Double Strand Breaks J Biol Chem 286:26250–26257 doi:M111.235200 [pii] 10.1074/jbc.M111.235200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu K, Li F, Wang J, Huang H, Wu J, Shi Y (2012) Structure of C-terminal tandem BRCT repeats of Rtt107 protein reveals critical role in interaction with phosphorylated histone H2A during DNA damage repair J Biol Chem 287:9137–9146 doi: 10.1074/jbc.M111.311860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin I, Wysocki R (2018) New insights into cohesin loading Curr Genet 64:53–61 doi: 10.1007/s00294-017-0723-6 [DOI] [PubMed] [Google Scholar]
- Mahendrawada L, Rai R, Kothiwal D, Laloraya S (2017) Interplay between Top1 and Mms21/Nse2 mediated sumoylation in stable maintenance of long chromosomes Curr Genet 63:627–645 doi: 10.1007/s00294-016-0665-4 [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB (2003) BRCT repeats as phosphopeptide-binding modules involved in protein targeting Science 302:636–639 doi: 10.1126/science.1088877 [DOI] [PubMed] [Google Scholar]
- Matityahu A, Onn I (2018) A new twist in the coil: functions of the coiled-coil domain of structural maintenance of chromosome (SMC) proteins Curr Genet 64:109–116 doi: 10.1007/s00294-017-0735-2 [DOI] [PubMed] [Google Scholar]
- McAleenan A et al. (2012) SUMOylation of the alpha-kleisin subunit of cohesin is required for DNA damage-induced cohesion Curr Biol 22:1564–1575 doi: 10.1016/j.cub.2012.06.045 [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2011) DNA end resection--unraveling the tail DNA Repair (Amst) 10:344–348 doi:S1568–7864(10)00413–1 [pii] 10.1016/j.dnarep.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe Meth Enzymol 194:795–823 [DOI] [PubMed] [Google Scholar]
- Nagai S, Davoodi N, Gasser SM (2011) Nuclear organization in genome stability: SUMO connections Cell Res 21:474–485 doi: 10.1038/cr.2011.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K (2006) Protocol for the fast chromatin immunoprecipitation (ChIP) method Nat Protoc 1:179–185 doi: 10.1038/nprot.2006.27 [DOI] [PubMed] [Google Scholar]
- Nie M, Boddy MN (2016) Cooperativity of the SUMO and Ubiquitin Pathways in Genome Stability Biomolecules 6:14 doi: 10.3390/biom6010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravcova M, Gadaleta MC, Nie M, Reubens MC, Limbo O, Russell P, Boddy MN (2018) Brc1 Promotes the Focal Accumulation and SUMO Ligase Activity of Smc5-Smc6 During Replication Stress Mol Cell Biol doi: 10.1128/MCB.00271-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard S, Schaffer L, Campbell D, Head SR, Boddy MN (2008) Localization of Smc5/6 to centromeres and telomeres requires heterochromatin and SUMO, respectively EMBO J 27:3011–3023 doi:emboj2008220 [pii] 10.1038/emboj.2008.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard S, Wohlschlegel J, McDonald WH, Yates JR 3rd, Boddy MN (2006) The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex Mol Cell Biol 26:1617–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JJ, Tainer JA, Boddy MN (2008) A SIM-ultaneous role for SUMO and ubiquitin Trends Biochem Sci 33:201–208 doi:S0968–0004(08)00061–3 [pii] 10.1016/j.tibs.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Pichler A, Fatouros C, Lee H, Eisenhardt N (2017) SUMO conjugation - a mechanistic view Biomol Concepts 8:13–36 doi: 10.1515/bmc-2016-0030 [DOI] [PubMed] [Google Scholar]
- Prudden J et al. (2007) SUMO-targeted ubiquitin ligases in genome stability EMBO J 26:4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Laloraya S (2017) Genetic evidence for functional interaction of Smc5/6 complex and Top1 with spatial frequency of replication origins required for maintenance of chromosome stability Curr Genet 63:765–776 doi: 10.1007/s00294-017-0680-0 [DOI] [PubMed] [Google Scholar]
- Raschle M et al. (2015) DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links Science 348:1253671 doi: 10.1126/science.1253671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A et al. (2016) Replication fork stability confers chemoresistance in BRCA-deficient cells Nature 535:382–387 doi: 10.1038/nature18325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubens MC, Rozenzhak S, Russell P (2017) Multi-BRCT Domain Protein Brc1 Links Rhp18/Rad18 and gammaH2A To Maintain Genome Stability during S Phase Mol Cell Biol 37 doi: 10.1128/MCB.00260-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robellet X, Vanoosthuyse V, Bernard P (2017) The loading of condensin in the context of chromatin Curr Genet 63:577–589 doi: 10.1007/s00294-016-0669-0 [DOI] [PubMed] [Google Scholar]
- Sanchez A, Roguev A, Krogan NJ, Russell P (2015) Genetic Interaction Landscape Reveals Critical Requirements for Schizosaccharomyces pombe Brc1 in DNA Damage Response Mutants G3 (Bethesda) 5:953–962 doi: 10.1534/g3.115.017251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy DM et al. (2005) Brc1-mediated DNA repair and damage tolerance Genetics 171:457–468 doi: 10.1534/genetics.105.044966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-Alveal C, Lin SJ, O’Connell MJ (2014) Functional interplay between cohesin and Smc5/6 complexes Chromosoma 123:437–445 doi: 10.1007/s00412-014-0474-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-Alveal C, O’Connell MJ (2011) Nse1-dependent recruitment of Smc5/6 to lesion-containing loci contributes to the repair defects of mutant complexes Mol Biol Cell 22:4669–4682 doi: 10.1091/mbc.E11-03-0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-Alveal C, Outwin EA, Trempolec N, Dziadkowiec D, Murray JM, O’Connell MJ (2010) SMC complexes and topoisomerase II work together so that sister chromatids can work apart Cell Cycle 9:2065–2070 [DOI] [PubMed] [Google Scholar]
- Tatham MH et al. (2008) RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation Nat Cell Biol 10:538–546 doi:ncb1716 [pii] 10.1038/ncb1716 [DOI] [PubMed] [Google Scholar]
- Uhlmann F (2016) SMC complexes: from DNA to chromosomes Nat Rev Mol Cell Biol doi: 10.1038/nrm.2016.30 [DOI] [PubMed] [Google Scholar]
- van der Crabben SN et al. (2016) Destabilized SMC5/6 complex leads to chromosome breakage syndrome with severe lung disease J Clin Invest 126:2881–2892 doi: 10.1172/JCI82890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varejao N, Ibars E, Lascorz J, Colomina N, Torres-Rosell J, Reverter D (2018) DNA activates the Nse2/Mms21 SUMO E3 ligase in the Smc5/6 complex EMBO J doi: 10.15252/embj.201798306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkade HM, Bugg SJ, Lindsay HD, Carr AM, O’Connell MJ (1999) Rad18 is required for DNA repair and checkpoint responses in fission yeast Mol Biol Cell 10:2905–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani S, Maharshi N, Kothiwal D, Mahendrawada L, Kalaivani R, Laloraya S (2018) Interaction of the Saccharomyces cerevisiae RING-domain protein Nse1 with Nse3 and the Smc5/6 complex is required for chromosome replication and stability Curr Genet 64:599–617 doi: 10.1007/s00294-017-0776-6 [DOI] [PubMed] [Google Scholar]
- Wehrkamp-Richter S, Hyppa RW, Prudden J, Smith GR, Boddy MN (2012) Meiotic DNA joint molecule resolution depends on Nse5-Nse6 of the Smc5-Smc6 holocomplex Nucleic Acids Res 40:9633–9646 doi: 10.1093/nar/gks713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Williams RS, Dovey CL, Guenther G, Tainer JA, Russell P (2010) gammaH2A binds Brc1 to maintain genome integrity during S-phase EMBO J 29:1136–1148 doi: 10.1038/emboj.2009.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Kong X, Ji Z, Zeng W, Potts PR, Yokomori K, Yu H (2012) Scc1 sumoylation by Mms21 promotes sister chromatid recombination through counteracting Wapl Genes Dev 26:1473–1485 doi: 10.1101/gad.193615.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S et al. (2013) Salicylic acid activates DNA damage responses to potentiate plant immunity Mol Cell 52:602–610 doi: 10.1016/j.molcel.2013.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabrady K et al. (2016) Chromatin association of the SMC5/6 complex is dependent on binding of its NSE3 subunit to DNA Nucleic Acids Res 44:1064–1079 doi: 10.1093/nar/gkv1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Blobel G (2005) A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization Proc Natl Acad Sci U S A 102:4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilio N et al. (2014) A novel histone deacetylase complex in the control of transcription and genome stability Mol Cell Biol 34:3500–3514 doi: 10.1128/MCB.00519-14 [DOI] [PMC free article] [PubMed] [Google Scholar]