Abstract
In this study, the volatile molecule profile of Streptococcus pneumoniae serotypes was evaluated using solid phase microextraction (SPME) and two dimensional gas chromatography time-of-flight mass spectrometry (GC × GC-TOFMS). Here, seven serotypes (6B, 14, 15, 18C, 19F, 9V, and 23F) were analyzed in an isogenic background. We identified 13 core molecules associated with all seven serotypes, and seven molecules thatwere differentially produced between serotypes. Serotype 14 was found to have the most distinct volatile profile, and could be discriminated from the other six serotypes in aggregate with an area under the curve (AUC) of 89%. This study suggests that molecules from S. pneumoniae culture headspace show potential for rapid serotype identification.
1. Introduction
Pneumococcal disease, a leading cause globally of childhood mortality, is caused by the organism Streptococcus pneumoniae. Infection with S. pneumoniae ranges from asymptomatic colonization to invasive disease, the severity of which is partially dependent on the serotype of the infecting strain [1]. There are over 90 serotypes of S. pneumoniae, defined by the capsular polysaccharide which surrounds the organism. Capsule type is an important virulence factor which protects the organism from host phagocytic engulfment [2,3], neutrophil extracellular traps (NETs) [4], defensins [5], and mucus clearance [6]. To date, vaccines are only available for 23 out of the 90 serotypes, 13 of which are effective in children [7]. Due to the process of “serotype replacement”, serotypes not currently covered by available vaccines are at risk for increased incidence, necessitating surveillance [8]. Current serotype identification methods, such as the Quellung reaction, are technically difficult, time consuming, and lack the ability to provide serotype specific identification within a clinically relevant time frame [9]. Rapid serotype identification could aid in the identification of invasive serotypes, help identify the best targets for future vaccines, and be used to evaluate vaccine efficacy.
Volatile biomarkers from in vitro cultures or ex vivo specimens (such as sputum or breath) have been widely studied for the purpose of identifying infectious agents. Gas-phase molecules are an appealing source of diagnostic biomarkers, given their potential for rapid detection with little or no sample preparation required. With regard to S. pneumoniae, a number of studies have evaluated the utility of volatile molecules for detecting this pathogen, using analytical platforms such as electronic nose [10–15], gas chromatography-mass spectrometry (GC-MS) [16–21], comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry (GC × GC-TOFMS) [22], secondary electrospray ionization-mass spectrometry (SESI-MS) [23], ion mobility spectrometry (IMS) [24], and selected ion flow tube-mass spectrometry (SIFT-MS) [25–28]. Although these studies have demonstrated proof-of-concept that S. pneumoniae may be identified based on its volatile profile, no previous study has assessed volatile molecules associated with different capsular serotypes of this organism.
In the present study, SPME and GC × GC-TOFMS [29] was used for the identification of volatile molecules produced by different S. pneumoniae serotypes in an isogenic background. Here, we identified 13 core molecules associated with all seven serotypes evaluated (6B, 9V, 14, 15, 18C, 19F, 23F), and seven molecules that were differentially produced between serotypes. Serotype 14 was found to have the most distinct volatile profile, and could be discriminated from the other six serotypes in aggregate with an area under the curve (AUC) of 89%. This study serves as a proof-of-concept that volatile molecules may be used for the rapid detection of S. pneumoniae serotypes.
2. Materials and methods
2.1. Bacterial strains
Eleven capsule switch mutants of Streptococcus pneumoniae were kindly provided by the Institute for Infectious Diseases (University of Bern, Bern, Switzerland). Details on the construction of the capsule switch mutants is described elsewhere [30]. In short, for the wild type clinical isolate S. pneumoniae strain 106.66, the capsule operon was removed and replaced with the capsule operons from eleven other strains. A total of eleven capsule switch mutants belonging to seven serotypes were analyzed (Supplementary Table S1).
2.2. Bacterial culturing
Single colonies of each S. pneumoniae strain were pre-cultured without shaking at 37 °C in a 5% CO2 atmosphere to an OD600 of 0.5 in 10 mL Brain Heart Infusion broth +5% Fetal Calf Serum (Difco, Detroit, MI, BHI + FCS). Starter cultures were spun down, the culture supernatant was aspirated, and the pellet was re-suspended in 1 mL Modified Lacks Media [30] (MLM, Supplementary Table S2). 1:1000 dilutions of the prepared starter aliquots were used to inoculate MLM. Cultures were grown to mid-exponential phase of growth at 37 °C in a 5% CO2 atmosphere. Cultures were centrifuged at 3750 ×g rcf for 10 min, filter sterilized via 0.22 μm PTFE syringe filter (VWR), and 1 mL of the sterile supernatant sealed into 10 mL gas chromatography headspace vials with a PTFE/silicone cap (Sigma-Aldrich, St. Louis, MO). As a control group for headspace analysis, sterile MLM was processed in the same way.
Samples and media were stored at – 20 °C and analyzed within two months of sample preparation. Sterile media control samples were prepared using the same protocol.
2.3. Volatile molecule analysis
Samples were thawed to 4 °C and incubated for 10 min at 37 °C. Volatile compounds in the culture headspace were concentrated using headspace solid-phase microextraction (HS-SPME) and analyzed via two-dimensional gas chromatography time-of-flight mass spectrometry (GC × GC-TOFMS). Details of the SPME fiber, GC × GC-TOFMS system (a Pegasus 4D, LECO Corporation, St. Joseph, MI) and experimental conditions are reported in Table 1. For linear retention index calculation, a linear alkanes (C7-C20, Sigma-Aldrich) solution was analyzed under the same chromatographic conditions.
Table 1.
Summary of HS-SPME-GC × GC-TOFMS instrumental parameters.
| SPME fiber and conditions | |
| Fiber composition, length | Divinybenzene-carboxen-polydimethylsiloxane |
| (DVB/CAR/PDMS), 50/30 pm, 2 cm (Supelco, | |
| Bellefonte, PA) | |
| Headspace exposure time, | 60min, 37°C |
| temperature | |
| Comprehensive two-dimensional gas chromatography (GC × GC) | |
| Sample desorption time, | 180 s, 250°C, splitless |
| temperature, split/splitless | |
| Column 1 (length × internal | Rxi-624Sil (Restek, Bellefonte, PA) |
| diameter × film thickness) | (60 m × 250 μm × 1.4 μm) |
| Column 2 (length × internal | Stabilwax (Restek) |
| diameter × film thickness) | (1m× 250 μm × 0.5 μm) |
| Primary oven temperature ramp | , 5 °C/min ramp, 35 °C, 230 °C |
| starting temperature, final | |
| temperature | |
| Temperature offsets relative to | +5 °C (secondary oven); +25 °C |
| primary oven | (modulator) |
| Modulation period (hot/cold) | 2 s total (alternating 0.5 s/0.5 s hot/cold) |
| Carrier gas (flow rate) | Helium (2 mL/min) |
| Transfer line temperature | 250 °C |
| Time-of-flight mass spectrometry (TOF-MS) | |
| Acquisition range | 30–500 m/z |
| Acquisition rate | 200 spectra/s |
| Ion source temperature | 200 °C |
2.4. Chromatographic alignment and feature identification
Chromatographic data was processed and aligned using the ChromaTOF software Statistical Compare (LECO Corporation). For the alignment of peaks across chromatograms, maximum first- and second-dimension retention time deviations were set at 2 s and 0.1 s, respectively, and the inter-chromatogram spectral match threshold was set at 600 (of 1000). For peak identification, a signal-to-noise (S/N) cutoff was set at 50:1, and resulting peaks were identified by a forward search of the NIST 2011 library. For the identification of low abundance peaks, after initial detection at a S/N threshold of 50:1 or greater in at least one chromatogram, all remaining chromatograms were searched at a S/N threshold of 20:1. A forward score of at least 800 was required for putative compound identification. Features were included for statistical analysis if they were present in 80% of S. pneumoniae samples, or 80% of at least one serotype. Linear retention index filtering (LRI unit tolerance ± 10) was also used when applicable for peak identification in the first dimension.
2.5. Statistical analysis
Statistical analyses were performed using Python 2.7.1. The Mann Whitney U test with Benjamini-Hochberg correction was used to test for statistical significance [31]. A significance level of p < 0.05 was selected. Principal components analysis (PCA) [32] was used to visualize the variance between samples. For the identification of discriminatory compounds between serotypes, the Random Forest (RF) classification algorithm was employed [33]. 100 iterations of RF were performed, with 1000 decision trees generated per iteration. Compounds were defined as discriminatory if they ranked among the top 10 features, as defined by their mean decrease in accuracy, in 80/100 Random Forest iterations. Receiver Operating Characteristic (ROC) curve analysis [34] was used to visualize the discriminatory performance of the Random Forest Model. Coefficient of variation is calculated by dividing the standard deviation of each analyte by its average peak area and multiplying by 100:. The fold change between classes was calculated by calculating Log2(Class 1/Class 2).
3. Results
3.1. Volatile molecules are produced by S. pneumoniae independent of serotype
In the capsule-switch mutation background we found 181 molecules that were detected in 80% of samples for all seven serotypes (6B, 9V, 14, 15, 18C, 19F, 23F), 13 of which were significantly different (p < 0.05) in abundance between S. pneumoniae culture and sterile media (Fig. 1A). Nine of these molecules were more highly abundant in S. pneumoniae culture and were detected in 93–100% of the S. pneumoniae samples analyzed, while four molecules were more highly abundant in sterile media and detectable in 31 –100% of S. pneumoniae samples (100% of sterile media samples). The profile of these 13 core molecules was found to be highly reproducible among the analyzed S. pneumoniae samples, with an average Spearman’s rank correlation (Spearman’s ρ) between samples of 0.89 (Supplementary Fig. S1). Principal components analysis (PCA) was used to visualize the variance between samples when considering the 13 core molecules among the 54 culture samples and 12 sterile media samples evaluated (Fig. 1B). Here, we observe that majority of the variance explained in these 13 molecules is due to sample type (i.e. S. pneumoniae culture versus sterile media; Fig. 1B). See Supplementary Fig. S2 to visualize the same figure with the S. pneumoniae samples labeled by serotype. Here, we observe that the variance due to serotype is negligible when considering these 13 selected volatile compounds.
Fig. 1.
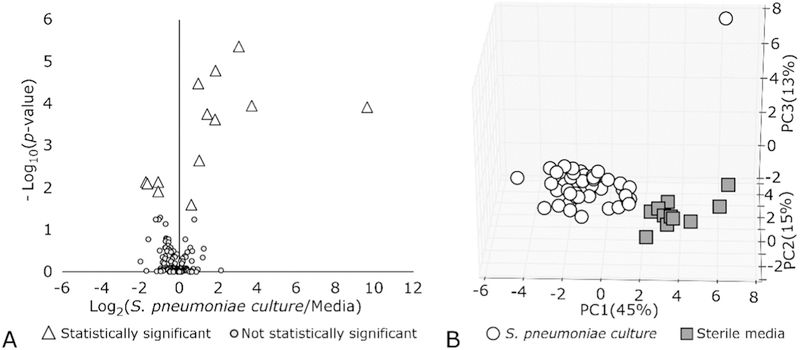
A) Volcano plot showing Log10(p-value) versus Log2(Fold Change) of culture versus sterile media for 181 detected volatile molecules. B) Variance between S. pneumoniae culture and sterile media samples when using the 13 significant molecules, shown via Principal component analysis (PCA).
We were able to assign putative identifications to these 13 molecules based on spectral match scores to the NIST 2011 library (≥800 of 1000) (Table 2). Some of the molecules including benzene, pentanal, heptanal and benzaldehyde, were also confirmed by the use of linear retention indices (within ± 10units). Unfortunately, no retention index data were available for the remaining compounds on the stationary phase used. Eight of the 13 molecules had coefficients of variation of 10% or less, indicating relatively small variability of these metabolites, despite being produced by different serotypes (Table 2). High coefficients of variation, such as 151% for 1-(2,4-dimethyl-furan-3-yl)-ethanone and 93% for the reported halogenated compound were observed for molecules sparsely detected among the S. pneumoniae samples (Table 2).
Table 2.
Putatively-identified molecules significantly different (p < 0.05) between S. pneumoniae culture and sterile media, independent of serotype.
| Putative Peak Identification | Retention Index (RI)1 | Retention Time 2 (seconds) |
Average MS Similarity4 |
Coefficient of variation5 |
% detection among S. pneumoniae samples6 |
Log2(S. pneumoniae culture/Sterile Media)7 |
|
|---|---|---|---|---|---|---|---|
| Experimental2 | Literature3 | ||||||
| 3-hydroxybutan-2-one * | 779 | NA | 1.6 | 840 | 30.7% | 93% | 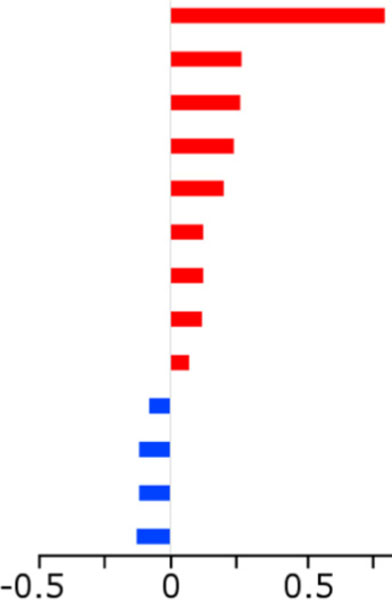 |
| pentane-2,3-dione * | 736 | NA | 1.0 | 920 | 9.3% | 100% | |
| Heptanal | 944 | 943 | 0.9 | 813 | 6.2% | 100% | |
| 2,3-Butanedione | 629 | NA | 1.0 | 963 | 10.0% | 100% | |
| Pentanal | 737 | 736 | 0.8 | 780 | 16.5% | 98% | |
| Benzaldehyde* | 1030 | 1036 | 1.3 | 839 | 5.6% | 100% | |
| (Hetero)-aromatic compound | 949 | NA | 1.1 | <800 | 3.5% | 100% | |
| Benzene* | 687 | 684 | 0.9 | 880 | 16.3% | 98% | |
| 2-methylbutanal | 703 | NA | 0.8 | 860 | 5.0% | 100% | |
| 1-(2,4-dimethyl-furan-3-yl)-ethanone | 1148 | NA | 1.1 | <800 | 151.5% | 30% | |
| Carboxylic acid compound† | 1445 | NA | 0.9 | <800 | 5.5% | 100% | |
| Halogenated compound † | 1558 | NA | 1.1 | <800 | 93.1% | 55% | |
| Ester compound † | 1445 | NA | 1.1 | <800 | 6.6% | 100% | |
NA: not applicable because no RI available from literature.
Retention indices of target analytes compared to values reported in the literature are used to further confirm putative peak identifications.
Retention index of each analyte was calculated based on its first dimension retention time with respect to alkane standards.
Retention index of pure analytical standards were extrapolated from values reported in the literature [35].
Average MS similarity was calculated by ChromaTOF software by comparing the mass spectra of each analyte to the mass spectra of compounds in the NIST 2011 library. Similarity scores are out of 1000.
Coefficient of variation is calculated by dividing the standard deviation of each analyte by its average peak area and multiplying by 100: .
Indicates the percentage of S. pneumoniae samples in which each analyte was detected.
Indicates the Log2(average peak area among S. pneumoniae samples/average peak area among sterile media samples). Red bars indicated analytes higher expressed on average in S. pneumoniae samples. Blue bars indicate analytes higher expressed on average in sterile media samples.
Indicates molecule is statistically different between one or more serotypes.
Compound classification only reported.
3.2. Volatile metabolites discriminate between S. pneumoniae serotypes
We hypothesized that a subset of volatile molecules produced by S. pneumoniae would be produced differentially as a function of serotype. Out of 282 volatile molecules detectable in 80% of at least one serotype, ANOVA analysis identified seven molecules (Fig. 2) that were significantly different (p < 0.05) in relative abundance between serotype 14 and at least one other serotype. Six of these seven molecules (2-methylpropanal, benzene, 2-methylbutanal, 2,3-pentanedione, 3-hydroxybutan-2-one, and benzaldehyde) were significantly different between serotype 14 and every other serotype analyzed (Fig. 2). Significant differences were also observed between other serotype comparisons. Specifically, 2-methylpropanal was significantly different (p < 0.05) between serotypes 19F and 23F (Fig. 2A), 2,3-pentanedione between serotypes 19F and 18C (Fig. 2D); and 2-hydroxy-2-methyl-propanoate between ten serotype-serotype comparisons (Fig. 2G). We also note that five of the seven differentially-expressed molecules (3-hydroxybutan-2-one, 2,3-pentanedione, benzaldehyde, 2-methylbu-tanal, and benzene) are part of the 13 core molecules listed in Table 2, all of which were higher produced in S. pneumoniae culture as compared to sterile media. This suggests that although these molecules are reproducibly produced by every serotype, they are also differentially produced as a function of serotype.
Fig. 2.
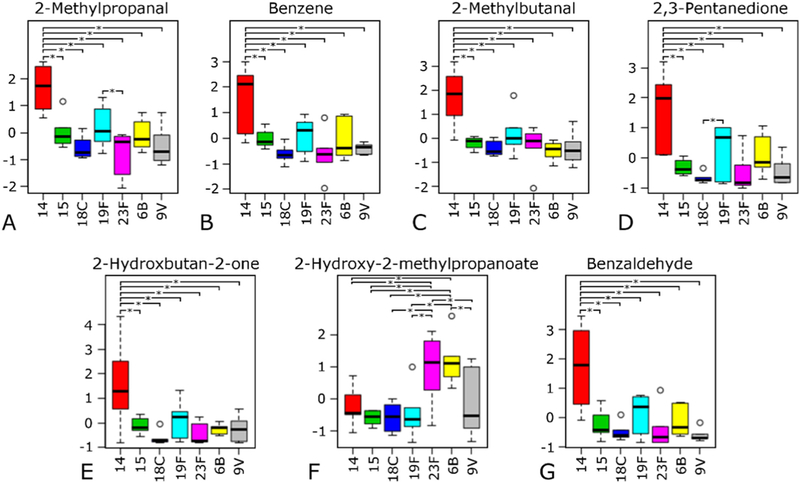
Abundance of seven volatile molecules found to be significantly different in expression between one or more serotypes as indicated by ANOVA analysis.* indicates significant differences in expression between a serotype-serotype comparison.
The ability to discriminate between serotypes using volatile molecules was evaluated using the machine learning algorithm Random Forest. Receiver operating characteristic (ROC) analysis was used to visualize the discriminatory efficacy of the Random Forest model. The highest discriminatory efficacy (AUC = 89%) was achieved in the model that aimed to differentiate serotype 14 from the other serotypes in aggregate (6B, 15, 18C, 19F, 9V and 23F, Fig. 3). The other model comparisons comparing pairs of serotypes achieved AUCs ranging from 36 to 76% (see Supplementary Fig. S3 for the results of each model).
Fig. 3.
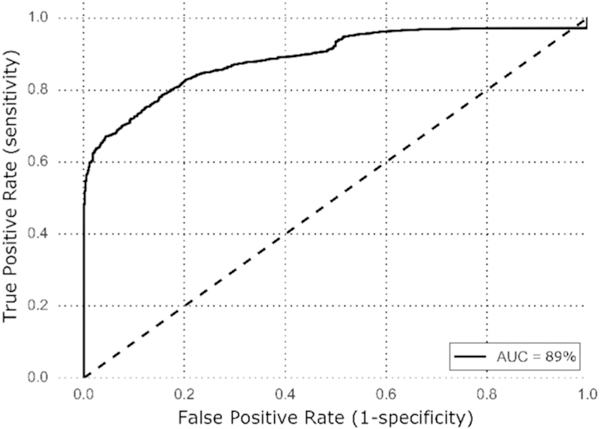
Receiver operating characteristic (ROC) curve showing the dis-criminatory efficacy for differentiating serotype 14 from serotypes 6B, 15, 18C, 19F, 9V and 23F when using 282 molecules detected in 80% of at least one serotype.
Six molecules were found to be important for discriminating serotype 14 from the other serotypes in aggregate (Table 3). Five of the six discriminatory molecules (2-methylbutanal, 2-methylpropanal, benzaldehyde, 2,3-pentanedione, and 3-hydroxybutan-2-one) are part of the seven differentially expressed molecules identified through the ANOVA analysis and all are significantly different (p < 0.05) in abundance between serotype 14 and every other serotype analyzed (Fig. 2). The remaining molecule, propylcyclopropane, although identified as discriminatory, was not significantly different between serotypes.
Table 3.
Putatively-identified molecules that discriminate serotype 14 from serotypes 6B, 15, 18C, 19F, 9V and 23F.
| Putative Peak Identification | Retention Index (RI)1 | Retention Time 2 (Seconds) |
Average MS Similarity4 |
Log2(Serotype 14/Other Serotypes) |
||
|---|---|---|---|---|---|---|
| Experimental2 | Literature3 | |||||
| 2,3-Pentanedione | 736 | NA | 1.0 | 920 |  |
|
| Benzaldehyde | 1030 | 1036 | 1.3 | 839 | ||
| 2-methylbutanal | 703 | NA | 0.8 | 860 | ||
| 3-hydroxybutan-2-one | 779 | NA | 1.6 | 840 | ||
| Cyclic hydrocarbon compound | 736 | NA | 1.1 | <800 | ||
| 2-methylpropanal | 779 | NA | 0.8 | 903 | ||
NA: not applicable because no RI available.
Retention indices of target analytes compared to values reported in the literature are used to further confirm putative peak identifications.
Retention index of each analyte was calculated based on its first dimension retention time with respect to alkane standards.
Retention index of pure analytical standards were extrapolated from values reported in the literature [35].
Average MS similarity was calculated by ChromaTOF software by comparing the mass spectra of each analyte to the mass spectra of compounds in the NIST 2011 library. Similarity scores are out of 1000.
4. Discussion
To the best of our knowledge, the present study represents the first time the volatile molecule profile of S. pneumoniae serotypes has been evaluated. Here, seven serotypes (6B, 14, 15, 18C, 19F, 9V, and 23F) were analyzed in an isogenic background. We identified 13 core molecules associated with all seven serotypes, and 7 molecules that are differentially-produced as a function of serotype. Serotype 14 was found to have the most distinct volatile profile, and could be dis-criminated from the other 6 serotypes in aggregate (6B, 15, 18C, 19F, 9V, and 23F), with an AUC of 89%.
Previous studies have identified benzaldehyde as a volatile molecule found in the headspace above S. pneumoniae culture when grown on blood agar plates [19], pentanal as significantly greater in S. pneumoniae cultures as compared to sterile media (Brain Heart Infusion broth) [27], and 2-methylpropanal as significantly greater in S. pneumoniae culture as compared to sterile media when grown in tryptic soy broth [16]. In the present study, pentanal was identified as a core volatile molecule that was produced by all seven serotypes, while benzaldehyde and 2-methylpropanal were identified as significantly more abundant in serotype 14 as compared to the six other serotypes (6B, 15, 18C, 19F, 9V and 23F). Benzaldehyde and 2-methylpropanal were also identified as part of a discriminatory panel that could discriminate serotype 14 from the remaining serotypes. Differences in the growth media chosen for bacterial culturing could account for differences in the volatile molecules identified between the present and previous S. pneumoniae studies. In the present study, we used Modified Lacks Media [30] due to its nutrient-limited conditions that are thought to be similar to the growth conditions in the nasopharynx, where S. pneumoniae resides [30]. Previous studies have used nutrient rich media such as BacT/Alert [25,26,28], brain heart infusion broth [27], tryptic soy broth [16], and blood agar [19,36].
In the present study, coefficient of variation was used to evaluate the degree of variation selected analytes exhibited among the S. pneumoniae samples analyzed. For compounds such as 1-(2,4-dimethyl-furan-3-yl)-ethanone, and the unidentified halogenated compound (Table 2), coefficients of variation were found to be quite large: 151% and 93%, respectively. Such high coefficients of variation among the S. pneumoniae samples suggests that despite being significantly different (p < 0.05) in peak area from sterile media, they may not be reliable volatile biomarkers of S. pneumoniae. Both of these analytes were higher expressed in sterile media as compared to the S. pneumoniae samples, and were detected in between 80 and 100% of sterile media samples, with coefficients of variation ranging from 8 to 11% among the sterile media samples analyzed. As these analytes were detected in small percentages of the S. pneumoniae samples (30–55%), it is possible that these compounds may originate in the media and be consumed by this organism. (See Table 2.)
In considering our ability to discriminate serotypes based on their volatile profile, serotype 14 was identified with the highest accuracy (AUC = 89%), followed by serotype 15 (AUC = 76%). These results are in line with what would be expected based on the genetic similarities of the serotypes evaluated [37]. For example, hierarchical cluster analysis based on the presence and absence of capsule genes indicates that serotypes 14 and 15 are most similar in genetic composition, while the remaining serotypes (9V, 19F, 6B, 23F, and 18C) form a secondary cluster (Supplementary Fig. S4). Therefore, it is possible that genes unique to serotypes 14 and 15, such as glycosyltransferases wchJ and wchK, wchL, wchM or capsular polysaccharide synthesis protein wchN, may be associated with the production of volatile molecules that are differentially expressed between serotypes. Glycosyltransferases are enzymes that catalyze the transfer of glycosyl residues to an acceptor during the degradation and biosynthesis of polysaccharides, glycoproteins and glycolipids. This class of enzymes have been shown to have a major role in the metabolism of secondary metabolites, and have been linked to the production of flavor-related volatile molecules in plants including strawberries [38], peach [39], and kiwifruit [40]. Future studies could investigate the role of glycosyltransferases in the biosynthesis of S. pneumoniae volatile metabolites.
In the present study, the strains analyzed were isogenic in all areas of their chromosome outside the capsular polysaccharide locus, which determines the serotype. Therefore, we were able to evaluate changes in volatile molecule production due to only this gene cassette. In order to validate these findings, future studies should consider the analysis of wild type strains and measure the production of these molecules in a heterogeneous background.
Supplementary Material
Acknowledgments
Christiaan A Rees was supported by the National Institutes of Health training grant T32 LM012204 (PI: Tor D Tosteson).
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jchromb.2018.08.032.
References
- [1].Catherine Hyams EC, Jonathan M. Cohen, Katie Bax, Brown Jeremy S., The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms, Infect. Immun. 78 (2) (2010) 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Moxon ER, Kroll JS, The role of bacterial polysaccharide capsules as virulence factors, in: Jann K, Jann B (Eds.), Bacterial Capsules, Springer, Berlin Heidelberg, 1990, pp. 65–85. [DOI] [PubMed] [Google Scholar]
- [3].Watson DA, Musher DM, Jacobson JW, Verhoef J, A brief history of the pneumococcus in biomedical research: a panoply of scientific discovery, Clin. Infect. Dis. 17 (1993) 913–924. [DOI] [PubMed] [Google Scholar]
- [4].Weinberger DM, Trzciński K, Lu YJ, Bogaert D, Brandes A, Galagan J, Anderson PW, Malley R, Lipsitch M, Pneumococcal capsular polysaccharide structure predicts serotype prevalence, PLoS Pathog. 5 (6) (2009) e1000476(June 12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wartha F, Beiter K, Albiger B, Fernebro J, Zychlinsky A, Normark S, Henriques-Normark B, Capsule andd-alanylated lipoteichoic acids protect Streptococcus pneumoniae against neutrophil extracellular traps, Cell. Microbiol. 9 (2007) 1162–1171. [DOI] [PubMed] [Google Scholar]
- [6].Nelson AL, Roche AM, Gould JM, Chim K, Ratner AJ, Weiser JN, Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance, Infect. Immun. 75 (1) (2007) 83–90 (January 1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nuorti JP, Whitney CG, Prevention of Pneumococcal Disease Among Infants and Children: Use of 13-valent Pneumococcal Conjugate Vaccine and 23-valent Pneumococcal Polysaccharide Vaccine: Recommendations of the Advisory Committee on Immunization Practices (ACIP), Department of Health and Human Services, Centers for Disease Control and Prevention, 2010. [PubMed] [Google Scholar]
- [8].Weinberger DM, Malley R, Lipsitch M, Serotype replacement in disease following pneumococcal vaccination: a discussion of the evidence, Lancet 378 (2011) 1962–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Richter SS, Heilmann KP, Dohrn CL, Riahi F, Diekema DJ, Doern GV, Evaluation of pneumococcal serotyping by multiplex PCR and Quellung reactions, J. Clin. Microbiol. 51 (2013) 4193–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Boilot P, Hines EL, Gardner JW, Pitt R, John S, Mitchell J, Morgan DW, Classification of bacteria responsible for ENT and eye infections using the Cyranose system, IEEE Sensors J. 2 (2002) 247–253. [Google Scholar]
- [11].Dutta R, Hines EL, Gardner JW, Boilot P, Bacteria classification using Cyranose 320 electronic nose, Biomed. Eng. Online 1 (2002) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dutta R, Das A, Stocks NG, Morgan D, Stochastic resonance-based electronic nose: a novel way to classify bacteria, Sensors Actuators B Chem. 115 (2006) 17–27. [Google Scholar]
- [13].Lai SY, Deffenderfer OF, Hanson W, Phillips MP, Thaler ER, Identification of upper respiratory bacterial pathogens with the electronic nose, Laryngoscope 112 (2002) 975–979. [DOI] [PubMed] [Google Scholar]
- [14].Lim SH, Mix S, Anikst V, Budvytiene I, Eiden M, Churi Y, Queralto N, Berliner A, Martino RA, Rhodes PA, Bacterial culture detection and identification in blood agar plates with an optoelectronic nose, Analyst 141 (2016) 918–925. [DOI] [PubMed] [Google Scholar]
- [15].Moens M, Smet A, Naudts B, Verhoeven J, Ieven M, Jorens P, Geise H, Blockhuys F, Fast identification of ten clinically important micro-organisms using an electronic nose, Lett. Appl. Microbiol. 42 (2006) 121–126. [DOI] [PubMed] [Google Scholar]
- [16].Filipiak W, Sponring A, Baur MM, Ager C, Filipiak A, Wiesenhofer H, Nagl M, Troppmair J, Amann A, Characterization of volatile metabolites taken up by or released from Streptococcus pneumoniae and Haemophilus influenzae by using GC- MS, Microbiology 158 (2012) 3044–3053. [DOI] [PubMed] [Google Scholar]
- [17].Ishimaru M, Yamada M, Nakagawa I, Sugano S, Analysis of volatile metabolites from cultured bacteria by gas chromatography/atmospheric pressure chemical ionization-mass spectrometry, J. Breath Res. 2 (2008) 037021. [DOI] [PubMed] [Google Scholar]
- [18].Julák J, Procházková-Francisci E, Stránská E, Rosová V, Evaluation of exudates by solid phase microextraction–gas chromatography, J. Microbiol. Methods 52 (2003) 115–122. [DOI] [PubMed] [Google Scholar]
- [19].Preti G, Thaler E, Hanson CW, Troy M, Eades J, Gelperin A, Volatile compounds characteristic of sinus-related bacteria and infected sinus mucus: analysis by solidphase microextraction and gas chromatography-mass spectrometry, J. Chromatogr. B 877 (2009) 2011–2018. [DOI] [PubMed] [Google Scholar]
- [20].Syhre M, Chambers ST, The scent of Mycobacterium tuberculosis, Tuberculosis 88 (2008) 317–323. [DOI] [PubMed] [Google Scholar]
- [21].Syhre M, Manning L, Phuanukoonnon S, Harino P, Chambers ST, The scent of Mycobacterium tuberculosis–part II breath, Tuberculosis 89 (2009) 263–266. [DOI] [PubMed] [Google Scholar]
- [22].Nizio K, Perrault K, Troobnikoff A, Ueland M, Shoma S, Iredell J, Middleton P, Forbes S, In vitro volatile organic compound profiling using GC × GC-TOFMS to differentiate bacteria associated with lung infections: a proof-of-concept study, J. Breath Res. 10 (2016) 026008. [DOI] [PubMed] [Google Scholar]
- [23].Ballabio C, Cristoni S, Puccio G, Kohler M, Sala MR, Brambilla P, Sinues PM-L, Rapid identification of bacteria in blood cultures by mass-spectrometric analysis of volatiles, J. Clin. Pathol. 67 (2014) 743–746. [DOI] [PubMed] [Google Scholar]
- [24].Dolch ME, Janitza S, Boulesteix A-L, Graßmann-Lichtenauer C, Praun S, Denzer W, Schelling G, Schubert S, Gram-negative and-positive bacteria differentiation in blood culture samples by headspace volatile compound analysis, J. Biol. Res. (Thessaloniki) 23 (2016) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Allardyce RA, Langford VS, Hill AL, Murdoch DR, Detection of volatile metabolites produced by bacterial growth in blood culture media by selected ion flow tube mass spectrometry (SIFT-MS), J. Microbiol. Methods 65 (2006) 361–365. [DOI] [PubMed] [Google Scholar]
- [26].Allardyce RA, Hill AL, Murdoch DR, The rapid evaluation of bacterial growth and antibiotic susceptibility in blood cultures by selected ion flow tube mass spectrometry, Diagn. Microbiol. Infect. Dis. 55 (2006) 255–261. [DOI] [PubMed] [Google Scholar]
- [27].Chippendale TW, Gilchrist FJ, Španěl P, Alcock A, Lenney W, Smith D, Quantification by SIFT-MS of volatile compounds emitted by in vitro cultures of S. aureus, S. pneumoniae and H. influenzae isolated from patients with respiratory diseases, Anal. Methods 6 (2014) 2460–2472. [Google Scholar]
- [28].Scotter JM, Allardyce RA, Langford VS, Hill A, Murdoch DR, The rapid evaluation of bacterial growth in blood cultures by selected ion flow tube–mass spectrometry (SIFT-MS) and comparison with the BacT/ALERT automated blood culture system, J. Microbiol. Methods 65 (2006) 628–631. [DOI] [PubMed] [Google Scholar]
- [29].Dimandja J-MD, Peer reviewed: GC × GC, Anal. Chem. 76 (2004) (167 A–174 A). [Google Scholar]
- [30].Hathaway LJ, Brugger SD, Morand B, Bangert M, Rotzetter JU, Hauser C, Graber WA, Gore S, Kadioglu A, Mühlemann K, Capsule type of Streptococcus pneumoniae determines growth phenotype, PLoS Pathog. 8 (3) (2012) e1002574(Mar 8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mann HB, Whitney DR, On a test of whether one of two random variables is stochastically larger than the other, Ann. Math. Stat. (1947) 50–60. [Google Scholar]
- [32].Hotelling H, Analysis of a complex of statistical variables into principal components, J. Educ. Psychol. 24 (1933) 417–441. [Google Scholar]
- [33].Breiman L, Random forests, Mach. Learn. 45 (2001) 5–32. [Google Scholar]
- [34].Lasko TA, Bhagwat JG, Zou KH, Ohno-Machado L, The use of receiver operating characteristic curves in biomedical informatics, J. Biomed. Inform. 38 (2005) 404–415. [DOI] [PubMed] [Google Scholar]
- [35].Schallschmidt K, Becker R, Jung C, Rolff J, Fichtner I, Nehls I, Investigation of cell culture volatilomes using solid phase micro extraction: options and pitfalls exemplified with adenocarcinoma cell lines, J. Chromatogr. B 1006 (2015) 158–166. [DOI] [PubMed] [Google Scholar]
- [36].Jünger M, Vautz W, Kuhns M, Hofmann L, Ulbricht S, Baumbach JI, Quintel M, Perl T, Ion mobility spectrometry for microbial volatile organic compounds: a new identification tool for human pathogenic bacteria, Appl. Microbiol. Biotechnol. 93 (6) (2012) 2603–2614 (March 1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes, PLoS Genet. 2 (3) (2006) e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Song C, Hong X, Zhao S, Liu J, Schulenburg K, Huang F-C, Franz-Oberdorf K, Schwab W, Glucosylation of 4-hydroxy-2, 5-dimethyl-3 (2H)-furanone, the key strawberry flavor compound in strawberry fruit, Plant Physiol. 171 (2016) 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cheng J, Wei G, Zhou H, Gu C, Vimolmangkang S, Liao L, Han Y, Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach, Plant Physiol. 166 (2014) 1044–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Montefiori M, Espley RV, Stevenson D, Cooney J, Datson PM, Saiz A, Atkinson RG, Hellens RP, Allan AC, Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis), Plant J. 65 (2011) 106–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


