Abstract
Luteinizing hormone and human chorionic gonadotropin (hCG) bind to the luteinizing hormone/chorionic gonadotropin receptor (LHCGR). LHCGR is required to maintain corpus luteum function but the mechanisms involved in the regulation of LHCGR in human luteal cells remain incompletely understood. This study aimed to characterize the expression of LHCGR mRNA in primary human luteinized granulosa cells (hLGCs) obtained from patients undergoing in vitro fertilization and to correlate LHCGR expression with the response of hLGCs to hCG by assessing the expression of genes known to be markers of hCG actions. The results show that LHCGR expression is low in freshly isolated cells but recovers rapidly in culture and that hCG maintains LHCGR expression, suggesting a positive feedback loop. The activity of a LHCGR-LUC reporter increased in cells treated with hCG but not with follicle-stimulating hormone. Treatment with hCG also stimulated the expression of genes involved in steroidogenesis in a time-dependent manner. LHCGR promoter expression was found to be regulated by SP1, which we show is highly expressed in hLGCs. Moreover, SP1 inhibition prevented the stimulation of steroidogenic genes and the increase in LHCGR-LUC reporter activity by hCG. Finally, we provide evidence that a complex formed by SP1 and GATA4 may play a role in the maintenance of LHCGR expression. This report reveals the mechanisms involved in the regulation of the LHCGR and provides experimental data demonstrating that the proximal region of the LHCGR promoter is sufficient to drive the expression of this gene in primary hLGCs.
Introduction
Luteinizing hormone (LH) and human chorionic gonadotropin (hCG) bind to the same receptor named luteinizing hormone/chorionic gonadotropin receptor (LHCGR). The expression of this receptor in theca and granulosa cells of ovarian follicles is essential to produce progesterone, androstenedione, and testosterone and consequently also for estradiol production via aromatization of androgens. The LHCGR is also required for ovulation, luteinization, and corpus luteum formation [1]. Luteinizing hormone receptor knockout female mice have small ovaries in which antral follicle formation can be recognized, but preovulatory follicles and corpora lutea are absent [2, 3]. Thus, the LHCGR is needed not only for corpus luteum function but also for follicular maturation and ovulation. The molecular mechanisms involved in the regulation of LHCGR in granulosa cells have been extensively studied [4–6], but these mechanisms remain unexplored in luteal cells, especially in humans.
Whereas low levels of LH are enough to sustain follicle growth and luteal function, ovulation requires high levels of LH that are triggered by the maturation of the preovulatory follicle, which signals its readiness to ovulate by increasing estradiol levels in circulation. This surge of LH causes ovulation, luteinization, and leads to the downregulation of the LHCGR at the protein and mRNA levels in the forming corpus luteum [7–9]. A decrease in Lhcgr mRNA half-life mediated by microRNAs including miR-122 and miR-136 appears to be involved in the rapid drop in Lhcgr mRNA expression associated with the LH surge [9–12]. However, this drop in LHCGR expression is transient and Lhcgr mRNA levels are restored in luteal cells [8, 9, 13, 14]. Thus, LHCGR protein is highly expressed in the human corpus luteum [15]. Recently, it was reported that a progesterone receptor antagonist decreases LHCGR expression in human luteinized granulosa cells (hLGCs)[16], suggesting an autoregulatory mechanism by progesterone. FSH is the main regulator of LHCGR expression in granulosa cells, but FSH is not needed during the luteal phase. Therefore, the factors and mechanisms that control the reactivation of the LHCGR gene in luteal cells are not known, particularly in humans.
In humans, the luteal phase usually lasts 14–16 days, at the end of which progesterone production ceases. Premature luteolysis is recognized clinically as a cause of an abnormally short luteal phase, abnormal vaginal bleeding in the luteal phase, and infertility [17]. Luteal regression occurs in the presence of constant LH pulsatile secretion, suggesting that the responsiveness to LH decreases as the corpus luteum ages [18–20]. In fact, although human luteal cells express LHCGR intensely at the mid-luteal phase, LHCGR is almost undetectable during luteolysis [15, 21]. Similarly, in the rhesus monkey, LHCGR expression increases from the early to mid-luteal phase, remains high until the late luteal phase, and then decreases during the very late luteal phase [22]. The factors causing this loss in LH sensitivity during the late luteal phase are not known. In case of fertilization and implantation, hCG secreted by the syncytiotropho-blast activates the LHCGR and sustains corpus luteum function. Thus, in cynomolgus monkeys, the administration of exponentially increasing levels of hCG prolongs the functional lifespan of the corpus luteum [18]. Therefore, the mechanisms involved in the regulation of LHCGR expression are also essential during early pregnancy in primates.
The aim of this study was to characterize the expression of Lhcgr mRNA in hLGCs obtained from patients undergoing in vitro fertilization and to examine the response of hLGCs to hCG on the expression of several genes known to be markers of LH action in luteal cells. Our findings reveal, for the first time, an autoregulatory effect of hCG on Lhcgr mRNA in primary hLGCs and provide evidence suggesting that SP1 and GATA4 could cooperate to regulate the LHCGR and steroidogenesis in luteal cells.
Material and Methods
Human luteinized granulosa cell culture
Primary hLGCs were obtained from the follicular aspirates of egg donors or women undergoing in vitro fertilization due to male infertility at the University of Illinois at Chicago Fertility Center under Institutional Review Board approval and with the patient’s consent. After controlled ovarian hyperstimulation, patients were administered hCG, underwent transvaginal oocyte retrieval, and follicular aspirates were collected from the ovarian follicles 35 to 36 h after hCG. The hLGCs were isolated from the aspirate milieu after centrifugation at 1000 g for 5 min. An aliquot of cells was processed immediately for RNA isolation as an uncultured ex vivo control group (0d). The rest of the cells were resuspended in 2 ml of serum-free cell culture medium (DMEM/F12, 0.25% BSA, 1 × antibiotic-antimycotic) and applied to a 50% Percoll (Invitrogen) gradient to eliminate contaminating erythrocytes and centrifuged for 20 min at 1000 g. The cellular interphase was collected, pipetted through a 70 μm filter and resuspended in 5 ml of cell culture medium. Cells were cultured for 1, 2, 3, 4, 5 or 6 days in the presence or absence of recombinant hCG (50 ng/ml). This concentration of hCG was chosen because it is widely used in the literature [23–25]. Media and treatment hormones were replaced every two days.
Promoter reporter assays
Promoter reporters were generated by cloning the 174 bp upstream region of the human Lhcgr proximal promoter followed by the firefly luciferase cDNA into pTRIP plasmid [26]. Lentiviruses containing wild-type or SP1-mutant constructs were generated as previously described [27]. All reporter constructs contain an expression cassette for green fluorescence protein to determine infection efficiency, which was greater than 90 percent in all experiments. Empty pTRIP plasmids were used as controls (Luc). To analyze promoter activity, cells were infected with lentiviral constructs for 48 h following treatments as described in the figure legend. Luciferase activity was determined in 50 μl of cell lysate as previously described [27].
Total RNA isolation and quantification of gene expression
RNA was isolated using Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. For reverse transcription, we added anchored Oligo DT primers (IDT) to each sample and incubated the samples at 70°C for 5 min. Elongation was performed at 37°C for 1 hour using Moloney Murine Leukemia Virus reverse transcriptase (200U/sample; Thermo Fisher Scientific) in buffer containing 10 mM Tris-HCL pH 8.3, 75 mM KCl, 3 mM MgCl2, 10% Glycerol, 10 mM DTT, 1mM dNTP, and 0.5 U/ul RNase Inhibitor Cocktail (Amresco, Solon, OH, USA). Reverse transcriptase inactivation was performed at 95°C for 5 min, and the resulting cDNA was diluted to a final concentration of 10 ng/μL in RNase and DNase free water. Quantitative real-time PCR (qPCR) was performed using intron-spanning primers. All determinations were performed in duplicate and the number of mRNA copies for each gene was calculated against a standard curve. Primer sequences are shown in table 1. Gene expression was normalized to the expression of RPL19, an internal control. Real-time quantification was performed using an iQcycler Real-Time PCR machine (Bio-Rad, Hercules, CA, USA) using the following protocol: denaturation at 95°C for 2 min followed by 40 cycles of 95°C for 5 sec, 60°C for 10 sec, and 72°C for 40 sec. At the end of each amplification, a melting curve determination was performed for each primer set. PCR products were sequenced to verify the amplification of a single product.
Table 1:
Primer sequences
| Gene Name | Forward Primer | Reverse Primer |
|---|---|---|
| Cyp11a1 | GT GAT GACCT GTTCCGCTTTGC | AAGGTTGAGCATGGGGACGC |
| Cyp19a1 | GCTGGACACCTCTAACACGCT | CAGGTCACCACGTTTCTCTGCT |
| Hsd3b2 | TGCCTT GT GACAGGAGCAGG | TACAGGCGGTGTGGATGACG |
| Lhcgr | TGGAGAAGATGCACAATGGA | GGCAATTAGCCTCTGAATGG |
| Rpl19 | TGIIIIICCGGCATCGAGCCC | GCTGTGGCAAGAAGAAGGTCTGG |
| Sp1 | TTGAGCTTGTCCCTCAGCTGCCACC | ACTGCCTGTGCTGCTACTTCGAGCC |
| Star | GGCTCAGGAAGGACGAAGAACC | ATCACAGCCTGTTGCCTCAGC |
| Fshr | AGCCATTGCTGTGCCTTTGC | ATGCATTCGGCTTAGGGGAGC |
| Vegfa | AGAGATGAGCTTCCTACAGC | GGAACATTTACACGTCTGC |
| Ctgf | GCTAGAGAAGCAGAGCCGCC | GCCGTCAGGGCACTTGAACT |
| Igf2 | AGTCCGAGAGGGACGTGT | TGGACTGCTTCCAGGTGT |
| Sfrp4 | TGGCAACGTATCTCAGCAAA | GGATGGGTGATGAGGACTTG |
| Gata4 | CTCTTGCAATGCGGAAAGAGGGG | CGTTGCTGGAGTTGCTGGAAG |
Protein isolation, immunoprecipitation, and western blotting
Cells were homogenized in ice-cold immunoprecipitation lysis buffer (50 mM Tris-HCl pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA) supplemented with inhibitors of phosphatase activity (1 mM PMSF, 1 mM Na3VO4, 1 mM NaF), and a protease inhibitor cocktail (Sigma, St. Louis, MO, USA). The protein concentration was determined using a BCA assay with BSA as a standard. All samples were denatured and reduced in sample buffer (0.555 mM bis-Tris, 4.44% sodium dodecyl sulfate, 0.333 mM HCl, 30% glycerol, 2.22 mM EDTA, 10% β-mercapto-ethanol, 0.04% bromophenol blue) and heated for 10 min at 90°C. Proteins were separated on a 12% bis-Tris-PAGE gel run for 1.5 h at 120 V. Samples were transferred to nitrocellulose membranes for 2.5 h at 150 mA followed by blocking in 5% milk for 2 h. Next, the membranes were incubated overnight at 4 °C with antibodies against SP1 and ACTB (Abcam, Cambridge, MA, USA). The membranes were then washed three times in Tris-buffered saline + 1% Tween 20 followed by a 2-hour incubation at room temperature with a species-specific secondary antibody conjugated to horseradish peroxidase (Jackson Immunoresearch, West Grove, PA, USA). Protein-antibody bound complexes were visualized by chemiluminescence using horseradish peroxidase conjugated secondary antibodies. Detection was performed using a BioRad ChemiDoc XRS System and the signal intensity was analyzed and quantified with BioRad Image Lab Software. Protein expression was normalized to the expression of ACTB (loading control), for each sample.
Total cell extracts were used to immunoprecipitate SP1 (Abcam) or GATA4 (Cell Signaling, Danvers, MA, USA) using previously described protocols [28]. Western blotting of immunoprecipitated proteins was performed as described above.
Statistics
The number of patient samples used for each experiment is indicated in the corresponding figure legend. Data for continuous variables are presented as mean values ± SEM. Statistical comparisons of mean values between groups were performed with paired t-tests, and multiple comparisons were performed with one-way ANOVA with repeated measures followed by Bonferroni adjustment where appropriate. Differences were considered statistically significant if the P values were less than 0.05.
Results
Characterization of the response of hLGCs to hCG in an extended cell culture
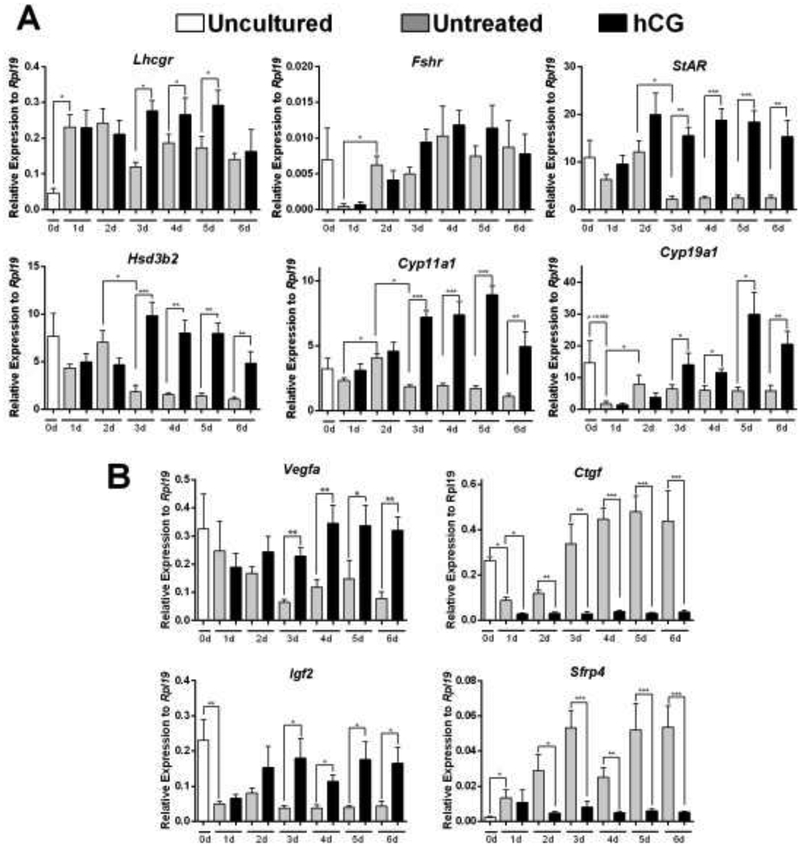
Total RNA was isolated from hLGCs either immediately after follicle aspiration (uncultured ex vivo group, day 0) or following culture in serum-free media for 1, 2, 3, 4, 5 or 6 days in the presence or absence of recombinant hCG. In uncultured ex vivo hLGCs, the mRNA levels of the Lhcgr were low (Figure 1A). However, Lhcgr expression fully recovered after only 1 day in culture reaching peak levels that were maintained for up to 48 h (Figure 1A). This brief increase in expression occurred independent of exposure to hCG. After 3 days in culture, the expression of Lhcgr declined significantly in untreated cells but the expression levels were significantly higher than that of uncultured samples. In contrast, in cells treated with hCG, we did not observe a decrease in Lhcgr mRNA levels after 3 days in culture, rather the receptor remained highly expressed until day 5, and only at this point the Lhcgr mRNA decreased to the levels seen in untreated cells (Figure 1A). We also examined the expression of the FSH receptor during this time-course experiment to determine whether the regulation of LHCGR was unique among gonadotropin receptors. As shown in figure 1, baseline levels of Fshr mRNA were approximately 100 times lower than those of Lhcgr. After 1 day of culture, Fshr expression decreased to almost undetectable levels, but then recovered in a time-dependent manner from day 2 onwards. These changes in Fshr expression were independent from the presence of hCG in the media.
Figure 1: Time-course of gene expression in the presence or absence of hCG.
Human LGCs were obtained after ovarian hyperstimulation from IVF patients at the time of follicle aspiration. Cells were harvested immediately (Uncultured, 0d), or cultured in serum-free media for a period of 1, 2, 3, 4, 5, or 6 days in the presence or absence of recombinant hCG (50 ng/ul). Gene expression was quantified using qPCR. Figure 1A represents LHCGR, FSHR and steroidogenic gene expression, while Figure 1B represents non-steroidogenic gene expression (see Results section). Columns represent the mean ± SEM of six different samples. *P < 0.05, **P < 0.01, ***P < 0.001 vs. C, t-test n=6.
As the main function of the LHCGR in luteal cells is to sustain the expression of steroidogenic enzymes, we next measured the expression of genes involved in steroid synthesis, including StAR, Cyp11a1, and Hsd3b2 (Figure 1A). We observed that, in contrast to Lhcgr expression, culturing cells for one day in the presence or absence of hCG did not significantly affect the mRNA levels for any of these genes. Moreover, in untreated conditions no recovery was observed, and after 3 days the expression of StAR, Cyp11a1, and Hsd3b2 dropped significantly to levels that were similar or lower than those seen in uncultured samples. However, for these genes addition of hCG to the culture medium resulted in a significant increase in their expression. For instance, StAR expression was significantly increased by the second day of culture and remained at this level until the end of the experiment while the expression of Cyp11a1 and Hsd3b2 increased significantly by day 3 and remained at this level until day 6 when a slight decrease was observed.
The human corpus luteum also produces high quantities of estradiol during the luteal phase. Therefore, we examined the expression of aromatase (Cyp19a1), an enzyme essential for estradiol production. The expression of Cyp19a1 decreased significantly between uncultured cells and cells collected after one day of culture (Figure 1A). Cyp19a1 expression recovered on day 2 and remained stable until the end of the study in untreated cells. In the presence of hCG; however, Cyp19a1 expression increased significantly from day 3 until day 6.
Expression profile of non-steroidogenic genes
Although steroidogenesis is the main physiological role of the corpus luteum, several non-steroidogenic genes are known to be crucial for corpus luteum function. Within these genes, vascular endothelial growth factor a (VEGFa), insulin-like growth factor 2 (IGF2), connective tissue growth factor (CTGF), and secreted frizzled-related protein 4 (SFRP4) have been shown to be regulated during luteinization. Therefore, their expression was also examined. The results revealed that Vegfa and Igf2 expression decreases in cultured cells when compared to uncultured cells (Figure 1B). The decrease in Vegfa mRNA levels was progressive while the decrease on Igf2 expression was abrupt and remained low until day 6. The expression of Vegfa and Igf2 was stimulated by hCG from day 3 to day 6 of culture (Figure 1B).
An opposite scenery was observed for Ctgf and Sfrp4. Thus, in untreated cells, the expression of both genes increased gradually in culture from day 1 to day 6. In the case of Ctgf, the expression was high in uncultured cells but fell more than 70 percent after 24 h in culture. The gradual increase in Ctgf and Sfrp4 levels observed in untreated cells was abolished by hCG (Figure 1B).
Activity and regulation of the LHCGR promoter in primary hLGCs
Our results have shown that hCG maintains Lhcgr expression in primary hLGCs, suggesting the presence of a positive feedback loop that directly affects the responsiveness of these cells to hCG. To investigate the mechanisms regulating the expression of the LHCGR gene, we cloned the human LHCGR proximal promoter into a luciferase reporter construct. The proximal region was chosen because the 174 to +1 region of the LHCGR promoter has been shown to display the highest level of expression in granulosa and Leydig cells [4]. We cloned this region into a lentiviral reporter plasmid (LHCGR-LUC) and used a lentiviral system to infect primary hLGCs. The use a lentiviral system to deliver the LHCGR-Luc reported into luteal cells was necessary as primary hLGCs are inherently difficult to transfect by other means. After 24 h of viral infection, cells were treated with hCG for 6, 24, or 48 h then harvested to evaluate luciferase activity. Additionally, samples treated with FSH were included to determine the specificity of our reporter system to hCG. The results showed that gonadotropin treatment had no effect on LHCGR-LUC activity after 6 h of treatment. However, after 24 and 48 h a significant increase in LHCGR-LUC reporter activity was observed in cells treated with hCG but not in those treated with FSH (Figure 2).
Figure 2: Effect of gonadotropins on LHCGR promoter activity in primary hLGCs.
Human LGCs were plated for 48 h in serum-free media followed by a 48-hour incubation period with a lentiviral vector carrying a 174 bp region of the human LHCGR proximal promoter. At this point, the cells were either treated with vehicle (C), FSH (50 ng/ml), or hCG (50 ng/ml) for 6, 24, or 48 h and harvested for luciferase reporter assay. *P > 0.05, t-test against specific time-point control, n=6.
SP1 is expressed but not regulated by hCG in primary hLGCs
The promoter region of the LHCGR gene contains two well-characterized SP1 sites (Figure 3A), which have been shown to regulate LHCGR expression in tumor cells, including JAR (human choriocarcinoma) and MCF-7 (human breast adenocarcinoma) cells [29]. Therefore, we first examined the expression of SP1 mRNA and protein in primary hLGCs. The results showed that Sp1 mRNA was detectable in hLGCs but was not regulated by prolonged culture in the presence or absence of hCG (Figure 3B). We detected both the 106 kDa and 95 kDa SP1 protein isoforms using western blot analyses, indicating that SP1 protein is expressed in hLGCs. SP1 protein levels were unaffected by hCG treatment (Figure 3C).
Figure 3:
A) LHCGR promoter - Comparison of human (−174 bp), bovine (−167 bp), and murine (−164 bp) LHCGR promoter regions showing a high degree of conservation between the species or the SP1 sites. B) SP1 mRNA expression in cultured hLGCs. Sp1 mRNA levels were quantified in hLGCs treated as in Figure 1 by qPCR (n=6). C) hLGCs from 3 patients were plated separately and cultured for 48 h in serum-free media to recover LHCGR expression followed by a 48-hour treatment with hCG. Protein expression was assessed by western blot for SP1 (106 kDa and 95 kDa bands) and ACTB as a loading control. Densiometric analysis of protein signal is also shown (ns=not significant, n=3).
SP1 binding sites are required for the activation of the Lhcgr promoter
To determine whether the presence of the −73 and −114 SP1 sites are required for the activation of the LHCGR promoter in primary hLCGs, we infected cells with luciferase reporters carrying either the native LHCGR promoter region or a mutant LHCGR promoter lacking SP1 sites. After plating, cells were treated with the lentivirus for 24 h followed by treatment with hCG, forskolin, or dibutyryl-cAMP (dbcAMP) for an additional 48 h. In cells transduced with the wild-type LHCGR promoter region, treatment with hCG, forskolin, or dbcAMP resulted in a significant increase in luciferase activity (Figure 4). However, in cells transduced with the mutant LHCGR promoter, the stimulatory effect of hCG, forskolin, and dbcAMP was significantly decreased, indicating that the SP1 sites are essential for the full stimulation of LHCGR in primary hLGCs.
Figure 4: Mutation of SP1 sites blunts the activation of the LHCGR promoter.
hLGCs were plated for 24 h in serum-free media followed by a 48-hour incubation with a lentiviral vector carrying a control reporter plasmid, a plasmid containing an intact proximal human LHCGR promoter, or a plasmid containing mutations in the SP1 sites of the LHCGR promoter. Cells were harvested after no treatment (C) or treatment with hCG (50 ng/ml), forskolin (5 μM), or dbcAMP (1 mM) for 48 h. Quantitation of promoter activity was performed using a luciferase reporter assay. *P < 0.05 t-test, n = 6.
SP1, PKA, and AKT participate in the regulation of LHCGR promoter in hLGCs
To investigate the intracellular pathways involved in the activation of the LHCGR promoter, we infected cells with virus carrying the wild-type promoter. Then, cells were treated with hCG for 48 h in the presence or absence of specific inhibitors of protein kinase A (PKA, which is inhibited by H89), extracellular signal-regulated kinases 1 and 2 (ERK1/2, which are inhibited by U0126), serine/threonine kinases (AKT, which is inhibited by MK2206), or a competitive inhibitor of SP1 binding known as Mithramycin A (MTM). We observed that PKA inhibition blunted the increase in promoter activity seen with hCG treatment (Figure 5). There was also a strong tendency for AKT inhibition to prevent hCG-stimulated promoter activity although this inhibition did not reach significance. ERK1/2 inhibition had no effect on LHCGR promoter activity. In contrast, MTM inhibition of SP1 binding not only significantly inhibited hCG-stimulated promoter activity but also decreased promoter activity below the one observed in untreated control sample, suggesting that SP1 is critical for LHCGR expression even in basal conditions. To circumvent the LHCGR, the same experiment was performed with forskolin in place of hCG. In the presence of forskolin a similar pattern of stimulation and inhibition of promoter activity was observed. The only difference was that the inhibition of AKT significantly decreased forskolin stimulation of LHCGR promoter activity.
Figure 5: Effect of LHCGR pathway inhibition on human LHCGR promoter activity.
hLGCs were cultured for 24 h in serum-free media then transfected with the 174 bp human LHCGR proximal promoter region. Cells were left untreated (C) or were treated with hCG (50 ng/ml) or forskolin (5 μM) for 48 h in the presence or absence of H89 (H), U0126 (U), MK2206 (MK) or Mithramycin A (MTM). Paired columns differ significantly from each other by t-test (P < 0.05, n = 6).
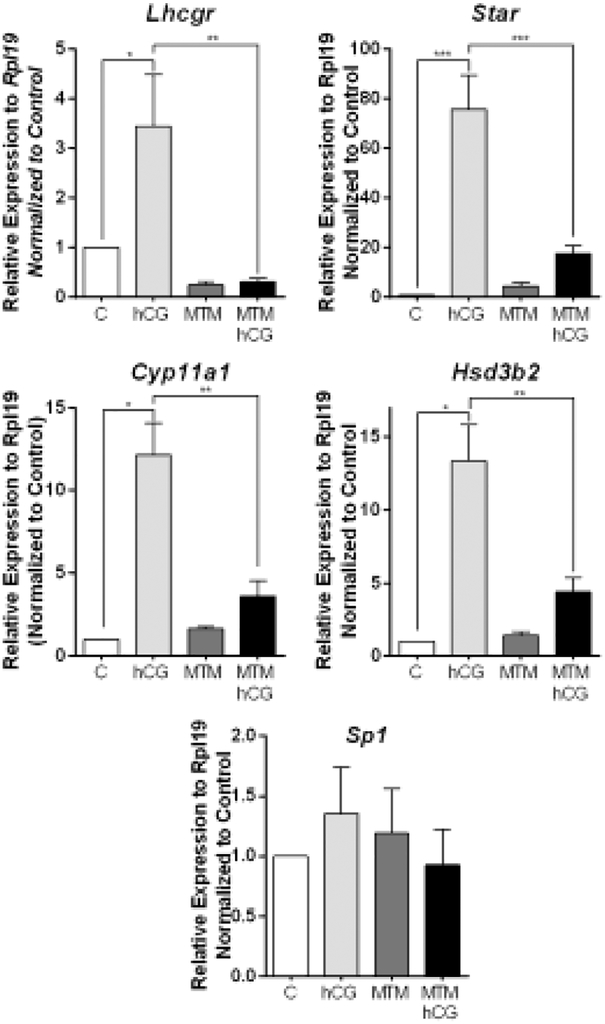
Role of SP1 inhibition on steroidogenesis
Since mutation of the SP1 sites and SP1 inhibition prevent LHCGR promoter activation, we next examined whether inhibition of SP1 translates into a decrease in Lhcgr mRNA levels and whether this affects the response of hLGCs to hCG. For this purpose, we cultured cells with hCG in the presence or absence of the SP1 inhibitor MTM for 48 h. As expected from the time-course experiments, treatment with hCG alone stimulated Lhcgr mRNA expression. In cells treated with MTM alone, Lhcgr mRNA levels were decreased significantly when compared to the control group. In addition, the stimulation of Lhcgr expression by hCG was abolished by MTM co-treatment. Consequently, a significant decrease in the response to hCG was observed in cells exposed to the SP1 inhibitor. Thus, the expression of StAR, Cyp11a1, and Hsd3b2 was also significantly lower in cells treated with MTM and hCG when compared to cells treated with hCG alone (Figure 6). In contrast, the expression of Sp1 was not affected by SP1 inhibition.
Figure 6: Inhibition of SP1 blocks the response of hLGCs to hCG.
Primary hLGCs were plated for 48 h in serum-free media followed by 48 h incubation with hCG, MTM, or hCG+MTM. Quantification of the expression of Lhcgr, Star, Cyp11a1, Hsd3b2, and Sp1 was performed using qPCR. Columns represent the mean ± SEM of six different samples. (*P < 0.05, **P < 0.01, ***P < 0.001 vs. C, one-way ANOVA, Tukey test).
SP1 and GATA4 interaction in primary hLGCs
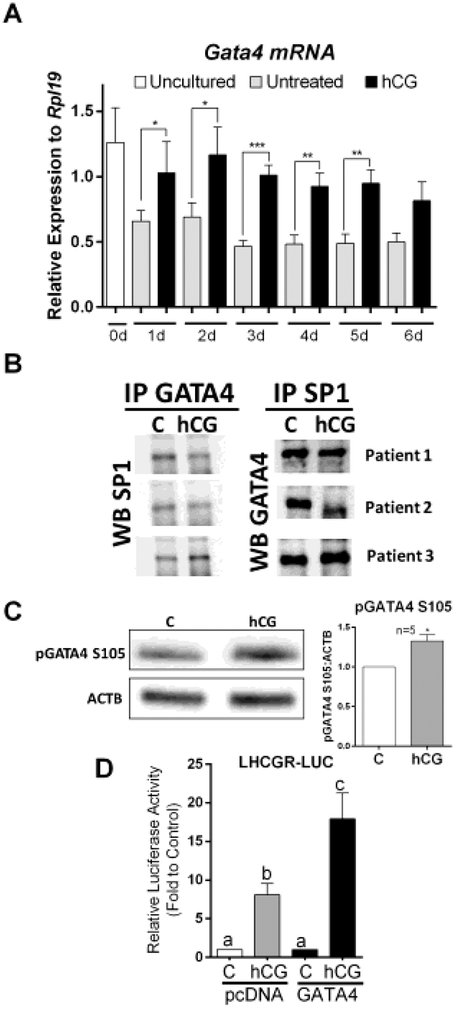
We recently developed GATA4 luteal cell conditional knockout mice [30], which revealed that GATA factors are essential for the expression of LHCGR in mouse luteal cells. Considering these findings, we next examined whether GATA4 may also be involved in the regulation of LHCGR in primary hLGCs. First, we determined the expression and regulation of GATA4 mRNA levels in cells cultured for 6 days in the presence or absence of hCG. As shown in figure 7A, the expression of GATA4 was stimulated by hCG from day 1 to day 5 of culture. On day 6, the expression of GATA4 was also higher in cells treated with hCG than in untreated cells, although the difference did not reach statistical significance (P=0.0563).
Figure 7: GATA4 may play a role in the regulation of the proximal promoter of the LHCGR gene.
A) Cells were treated as indicated in Figure 1. The expression of Gata4 was examined by qPCR. Columns represent the mean ± SEM of six different samples. *P < 0.05, **P < 0.01, ***P < 0.001 vs. C, t-test n=6. B) Cells from three patients were cultured in separate 6-well plates for 48 h and then treated with vehicle or hCG for 1 hour. GATA4 and SP1 were immunoprecipitated. Then western blot was used to detect GATA4 in SP1 precipitates or SP1 in GATA4 precipitates. C) Cells were treated with vehicle or hCG for 1 hour. Western blot for phospho-GATA4 and β-actin (BACT) was performed. The experiment was repeated with 5 different patients, a representative blot is shown, and results quantified and expressed as the ratio of pGATA4/BACT. Columns represent the mean ± SEM, *P < 0.05 vs. C, t-Test, n=5. D) Cells were infected with virus carrying LHCGR reporter and virus carrying an empty cassette or a GATA4 expression cDNA. 48 h later, cells were treated with vehicle or hCG for 48 h. Columns represent the mean ± SEM of six different experiments. Columns with different letters differ significantly (P < 0.05; one-way ANOVA, Tukey test).
Despite the observed stimulation of GATA4 expression by hCG, the effect of GATA4 on LHCGR expression appears to be indirect as no GATA response elements (WGATAR) are present in the LHCGR promoter (Figure 4). Therefore, we tested whether GATA4 interacts with SP1 in primary hLGCs. Immuno-precipitation of GATA4 followed by western blot demonstrated that SP1 was precipitated by the anti-GATA4 antibody. Similarly, GATA4 was also pulled down by the immunoprecipitation of SP1 (Figure 7B). However, no changes were observed between the control and hCG groups, suggesting that the interaction between GATA4 and SP1 is not affected by hCG.
We have previously demonstrated that GATA4 transcriptional activity increases after phosphorylation of the Ser105 residue [31]. Therefore, we next examined whether phosphorylation of GATA4 may be regulated by hCG. As shown in figure 7C, treatment with hCG significantly increased Ser105 phosphorylation of GATA4. These results suggest that even if the interaction of SP1 and GATA4 is not regulated by hCG, the SP1-GATA4 complex may become active upon GATA4 phosphorylation.
Finally, we examined whether overexpression of GATA4 contributes to the regulation of the proximal promoter of the LHCGR. For this purpose, we infected cells with virus caring an empty plasmid (pcDNA) or an expression cassette for GATA4. All cells were also infected with virus carrying the LHCGR-LUC reporter. As shown in figure 7D, in cells overexpressing GATA4, hCG was able to increase the activity of the LHCGR-LUC reporter to levels that were significantly higher than those found in cells infected with the empty plasmid.
Discussion
Despite the importance of the LHCGR for normal luteal cell function in humans, the molecular mechanisms controlling the promoter of this receptor has not been investigated in human luteal cells. Here, we described the transcriptional profile of LHCGR in long-term cultures of primary hLGCs and show not only the expression and regulation of the LHCGR in these cells but also the response of these cells to hCG. We also demonstrate for the first time that SP1 is essential for LHCGR expression in primary hLGCs.
LHCGR mRNA expression was low in uncultured hLGCs, which corresponds to approximately 36 h after induction of final follicular maturation by hCG administration. Low LHCGR mRNA expression is most probably due to the action of miRNAs that decrease LHCGR mRNA half-life [32]. We observed that LHCGR expression recovers rapidly, reaching maximum levels after only 1 day of culture, and remains high through day 2 both in the presence or absence of hCG. This finding suggests that the inhibitory effect of these miRNAs quickly vanish, leading to the restoration of LHCGR expression.
High expression of Lhcgr mRNA was maintained for 5 days in hLGCs cultured in the presence of hCG before dropping to levels found in untreated cells. In vehicle-treated cells, Lhcgr mRNA expression decreased as early as day 3 and remained low until the end of the study. These findings support evidence that LHCGR mRNA recovers after ovarian hyperstimulation in primary human granulosa-lutein cells [33]. However, in contrast to previous observations demonstrating that hCG downregulates the Lhcgr mRNA in rats [33], downregulation of Lhcgr after hCG treatment was not observed in hLGCs. Differences in cell culture conditions or species-specific mechanisms may be responsible for these discrepancies. However, considering the importance of LH for luteal function, the stimulatory effect of hCG on Lhcgr observed here appears to be physiologically important.
Interestingly, while Lhcgr expression recovers to its maximum level after only 1 day in culture, the stimulation of gene expression by hCG was delayed until day 2 for StAR or until day 3 for Lhcgr, Cyp19a1, Hsd3b2, Cyp11a1, Vegfa, and Igf2. This observation indicates that although Lhcgr mRNA has recovered there appears to be a significant delay in the recovery of LHCGR protein levels or translocation of the protein to the membrane. However, this hypothesis requires further investigation.
Previously, the regulation of the human LHCGR promoter has only been explored using cell lines or human myometrial smooth muscle cells [29]. Our report provides the first evidence demonstrating that the proximal region of the LHCGR promoter responds to hCG in primary hLGCs. In contrast, FSH did not stimulate the LHCGR promoter at any time point assessed, providing evidence of the specificity of LH/hCG on LHCGR gene activation in luteal cells. This finding is notable as the LHCGR receptor is a classical target of FSH in undifferentiated granulosa cells and leads us to postulate that there are significant differences between the molecular mechanisms controlling LHCGR expression in granulosa and luteal cells.
By inhibiting SP1 binding with MTM, we show that SP1 is required for hCG-induced stimulation of LHCGR, StAR, Cyp11a1, and HSD3B2. Similarly, inhibition of SP1 binding by MTM and deletion of the two SP1 regulatory elements prevented the activation of LHCGR promoter by hCG. These results indicate that SP1 is a critical element driving LHCGR expression in human luteal cells. This conclusion is further supported by the detection of Sp1 mRNA and protein expression in primary hLGCs. However, Sp1 mRNA and protein levels are not regulated by hCG. SP1 transcriptional activity has been shown to occur through posttranslational modifications [34]. In fact, the human SP1 protein has 61 putative phosphorylation sites [34]. Thus, despite constant SP1 levels, it is still possible that its activity is modulated by phosphorylation or protein-protein interactions.
An interesting conclusion of this report is the possibility that GATA4 may interact with SP1 to stimulate LHCGR expression. The proximal promoter of the LHCGR gene lacks canonical GATA response elements. However, the overexpression of GATA4 potentiated hCG activation of a reporter carrying the proximal Lhcgr promoter. Therefore, the role of GATA4 in the regulation of Lhcgr mRNA expression is likely not a result of the direct binding of GATA4 to the LHCGR promoter. Indeed, we observed that GATA4 and SP1 interact in primary hLGCs, supporting an indirect role of GATA4 on LHCGR expression. Although this interaction was not regulated by hCG, we observed that hCG stimulates the expression of GATA4 and its phosphorylation at Ser105, a residue known to increase GATA4 transcriptional activity [31]. Thus, it is possible that SP1 and GATA4 form a complex that increases the transcriptional activity of the LHCGR promoter. Further experiments are needed to confirm this hypothesis.
The development of an extensive vasculature needed for the delivery of progesterone to the circulation is crucial for normal luteal function. We observed that, at least at the mRNA level, hCG strongly stimulated the expression of VEGFa, which is known to participate in luteal angiogenesis [35]. We also observed that hCG stimulated IGF2 expression in luteal cells. We have previously demonstrated that IGF2 is essential for the stimulation of aromatase in granulosa cells [36]; however, the role of IGF2 in the human corpus luteum remains to be examined. We also observed that hCG inhibits the expression of CTGF and sFRP4. CTGF expression has been shown to be downregulated during follicle maturation [37]. Thus, our results suggest that hCG may play a major role in the silencing of this transcription factor in luteal cells, supporting previous reports [38]. In contrast, we show that hCG strongly inhibited the expression of sFRP4; whereas in rats, hCG has been shown to induce sFRP4 expression [39]. This suggests species-specific molecular differences in the regulation of sFRP4 in luteal cells.
The direct targeting of LHCGR activity or expression as a means of fertility management is an area that has not been explored. Dysfunctional LHCGR activity can adversely affect the outcome of IVF treatments by leading to the development of a potentially life-threatening condition called ovarian hyperstimulation syndrome (OHSS), which is a systemic disease resulting from vasoactive products released by the ovaries. The risk of OHSS complications is a significant clinical problem due to the increased demand for IVF treatments [40]. The main culprit leading to OHSS development are an exaggerated response to hCG leading to abnormally high levels of VEGF [41]. The release of VEGF from LGCs to circulation has been shown to increase capillary permeability and fluid retention [42–44]. In fact, inhibition of VEGF actions has been shown to decrease OHSS [45, 46]. Thus, understanding LHCGR regulation could reveal new therapeutic targets to control OHSS.
In conclusion, this report contributes to a better understanding of the mechanisms involved in the regulation of LHCGR in human luteal cells. We have provided the first experimental data suggesting that the proximal 174 bp region of the LHCGR promoter is sufficient to drive the expression of this gene in human luteal cells. We also show that hCG strongly stimulates the expression of its own receptor. Finally, our findings suggest that SP1 and GATA4 are involved in the regulation of LHCGR in human luteal cells.
Highlights.
The expression of the LHCGR gene correlates with the expression of steroidogenic genes and the response of hLGCs to hCG.
The proximal 174 bp region of the LHCGR promoter in enough to activate LHCGR expression in human luteal cells.
hCG strongly stimulates the expression of its own receptor.
SP1 regulates LHCGR expression and the response of luteal cells to hCG.
SP1 and GATA4 cooperate in the regulation of LHCGR in human luteal cells.
Funding
This work was supported by the National Institute of Health grant number R56HD086054 to CS.
Support: NIH grant R56HD086054 (CS)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- [1].Stocco C, Telleria C, Gibori G, The molecular control of corpus luteum formation, function, and regression, Endocr Rev, 28 (2007) 117–149. [DOI] [PubMed] [Google Scholar]
- [2].Lei ZM, Mishra S, Zou W, Xu B, Foltz M, Li X, Rao CV, Targeted disruption of luteinizing hormone/human chorionic gonadotropin receptor gene, Mol Endocrinol, 15 (2001) 184–200. [DOI] [PubMed] [Google Scholar]
- [3].Zhang FP, Poutanen M, Wilbertz J, Huhtaniemi I, Normal prenatal but arrested postnatal sexual development of luteinizing hormone receptor knockout (LuRKO) mice, Mol Endocrinol, 15 (2001) 172–183. [DOI] [PubMed] [Google Scholar]
- [4].Hamalainen T, Poutanen M, Huhtaniemi I, Promoter function of different lengths of the murine luteinizing hormone receptor gene 5’-flanking region in transfected gonadal cells and in transgenic mice, Endocrinology, 142 (2001) 2427–2434. [DOI] [PubMed] [Google Scholar]
- [5].Nakao K, Kishi H, Imai F, Suwa H, Hirakawa T, Minegishi T, TNF-alpha suppressed FSH-induced LH receptor expression through transcriptional regulation in rat granulosa cells, Endocrinology, 156 (2015) 3192–3202. [DOI] [PubMed] [Google Scholar]
- [6].Law NC, Weck J, Kyriss B, Nilson JH, Hunzicker-Dunn M, Lhcgr expression in granulosa cells: roles for PKA-phosphorylated beta-catenin, TCF3, and FOXO1, Mol Endocrinol, 27 (2013) 1295–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].LaPolt PS, Oikawa M, Jia XC, Dargan C, Hsueh AJ, Gonadotropin-induced up- and down-regulation of rat ovarian LH receptor message levels during follicular growth, ovulation and luteinization, Endocrinology, 126 (1990) 3277–3279. [DOI] [PubMed] [Google Scholar]
- [8].Segaloff DL, Wang HY, Richards JS, Hormonal regulation of luteinizing hormone/chorionic gonadotropin receptor mRNA in rat ovarian cells during follicular development and luteinization, Mol Endocrinol, 4 (1990) 1856–1865. [DOI] [PubMed] [Google Scholar]
- [9].Peegel H, Randolph J Jr., Midgley AR, Menon KM, In situ hybridization of luteinizing hormone/human chorionic gonadotropin receptor messenger ribonucleic acid during hormone-induced down-regulation and the subsequent recovery in rat corpus luteum, Endocrinology, 135 (1994) 1044–1051. [DOI] [PubMed] [Google Scholar]
- [10].Lu DL, Peegel H, Mosier SM, Menon KM, Loss of lutropin/human choriogonadotropin receptor messenger ribonucleic acid during ligand-induced down-regulation occurs post transcriptionally, Endocrinology, 132 (1993) 235–240. [DOI] [PubMed] [Google Scholar]
- [11].Menon B, Sinden J, Franzo-Romain M, Botta RB, Menon KM, Regulation of LH receptor mRNA binding protein by miR-122 in rat ovaries, Endocrinology, 154 (2013) 4826–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kitahara Y, Nakamura K, Kogure K, Minegishi T, Role of microRNA-136–3p on the expression of luteinizing hormone-human chorionic gonadotropin receptor mRNA in rat ovaries, Biol Reprod, 89 (2013) 114. [DOI] [PubMed] [Google Scholar]
- [13].Xu F, Stouffer RL, Muller J, Hennebold JD, Wright JW, Bahar A, Leder G, Peters M, Thorne M, Sims M, Wintermantel T, Lindenthal B, Dynamics of the transcriptome in the primate ovulatory follicle, Mol Hum Reprod, 17 (2011) 152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kishi H, Kitahara Y, Imai F, Nakao K, Suwa H, Expression of the gonadotropin receptors during follicular development, Reprod Med Biol, 17 (2018) 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yung Y, Aviel-Ronen S, Maman E, Rubinstein N, Avivi C, Orvieto R, Hourvitz A, Localization of luteinizing hormone receptor protein in the human ovary, Mol Hum Reprod, 20 (2014) 844–849. [DOI] [PubMed] [Google Scholar]
- [16].Yung Y, Maman E, Ophir L, Rubinstein N, Barzilay E, Yerushalmi GM, Hourvitz A, Progesterone antagonist, RU486, represses LHCGR expression and LH/hCG signaling in cultured luteinized human mural granulosa cells, Gynecol Endocrinol, 30 (2014) 42–47. [DOI] [PubMed] [Google Scholar]
- [17].Griesinger G, Meldrum D, Introduction: Management of the luteal phase in assisted reproductive technology, Fertility and sterility, 109 (2018) 747–748. [DOI] [PubMed] [Google Scholar]
- [18].Zeleznik AJ, In vivo responses of the primate corpus luteum to luteinizing hormone and chorionic gonadotropin, Proc Natl Acad Sci U S A, 95 (1998) 11002–11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cameron JL, Stouffer RL, Gonadotropin receptors of the primate corpus luteum. I. Characterization of 125I-labeled human luteinizing hormone and human chorionic gonadotropin binding to luteal membranes from the rhesus monkey, Endocrinology, 110 (1982) 2059–2067. [DOI] [PubMed] [Google Scholar]
- [20].Cameron JL, Stouffer RL, Gonadotropin receptors of the primate corpus luteum. II. Changes in available luteinizing hormone- and chorionic gonadotropin-binding sites in macaque luteal membranes during the nonfertile menstrual cycle, Endocrinology, 110 (1982) 2068–2073. [DOI] [PubMed] [Google Scholar]
- [21].Takao Y, Honda T, Ueda M, Hattori N, Yamada S, Maeda M, Fujiwara H, Mori T, Wimalasena J, Immunohistochemical localization of the LH/HCG receptor in human ovary: HCG enhances cell surface expression of LH/HCG receptor on luteinizing granulosa cells in vitro, Mol Hum Reprod, 3 (1997) 569–578. [DOI] [PubMed] [Google Scholar]
- [22].Bogan RL, Murphy MJ, Stouffer RL, Hennebold JD, Systematic determination of differential gene expression in the primate corpus luteum during the luteal phase of the menstrual cycle, Mol Endocrinol, 22 (2008) 1260–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Martinez JC, Stouffer RL, Hazzard TM, Patton PE, Molskness TA, Insulin-like growth factors-1 and −2, but not hypoxia, synergize with gonadotropin hormone to promote vascular endothelial growth factor-A secretion by monkey granulosa cells from preovulatory follicles, Biol Reprod, 68 (2003) 1112–1118. [DOI] [PubMed] [Google Scholar]
- [24].Moravek MB, Shang M, Menon B, Menon K, HCG-mediated activation of mTORC1 signaling plays a crucial role in steroidogenesis in human granulosa lutein cells, Endocrine, 54 (2016) 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Palaniappan M, Menon KM, Human chorionic gonadotropin stimulates theca-interstitial cell proliferation and cell cycle regulatory proteins by a cAMP-dependent activation of AKT/mTORC1 signaling pathway, Mol Endocrinol, 24 (2010) 1782–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Stove V, Smits K, Naessens E, Plum J, Verhasselt B, Multiple gene knock-down by a single lentiviral vector expressing an array of short hairpin RNAs, Electronic Journal of Biotechnology, 9 (2006) 572–579. [Google Scholar]
- [27].Zhou P, Baumgarten SC, Wu Y, Bennett J, Winston N, Hirshfeld-Cytron J, Stocco C, IGF-I signaling is essential for FSH stimulation of AKT and steroidogenic genes in granulosa cells, Mol Endocrinol, 27 (2013) 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wu YG, Bennett J, Talla D, Stocco C, Testosterone, not 5alpha-dihydrotestosterone, stimulates LRH-1 leading to FSH-independent expression of Cyp19 and P450scc in granulosa cells, Mol Endocrinol, 25 (2011) 656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dufau ML, Liao M, Zhang Y, Participation of signaling pathways in the derepression of luteinizing hormone receptor transcription, Mol Cell Endocrinol, 314 (2010) 221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Convissar SM, Bennett J, Baumgarten SC, Lydon JP, DeMayo FJ, Stocco C, GATA4 and GATA6 knockdown during luteinization inhibits progesterone production and gonadotropin responsiveness in the corpus luteum of female mice, Biol Reprod, 93 (2015) 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kwintkiewicz J, Cai Z, Stocco C, Follicle-stimulating hormone-induced activation of Gata4 contributes in the up-regulation of Cyp19 expression in rat granulosa cells, Mol Endocrinol, 21 (2007) 933–947. [DOI] [PubMed] [Google Scholar]
- [32].Menon B, Gulappa T, Menon KM, miR-122 regulates LH receptor expression by activating sterol response element binding protein in rat ovaries, Endocrinology, 156 (2015) 3370–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Menon KM, Menon B, Regulation of luteinizing hormone receptor expression by an RNA binding protein: role of ERK signaling, The Indian journal of medical research, 140 Suppl (2014) S112–119. [PMC free article] [PubMed] [Google Scholar]
- [34].Tan NY, Khachigian LM, Sp1 phosphorylation and its regulation of gene transcription, Mol Cell Biol, 29 (2009) 2483–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fraser HM, Bell J, Wilson H, Taylor PD, Morgan K, Anderson RA, Duncan WC, Localization and quantification of cyclic changes in the expression of endocrine gland vascular endothelial growth factor in the human corpus luteum, J Clin Endocrinol Metab, 90 (2005) 427–434. [DOI] [PubMed] [Google Scholar]
- [36].Baumgarten SC, Convissar SM, Zamah AM, Fierro MA, Winston NJ, Scoccia B, Stocco C, FSH regulates IGF-2 expression in human granulosa cells in an AKT-Dependent Manner, J Clin Endocrinol Metab, 100 (2015) E1046–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Slee RB, Hillier SG, Largue P, Harlow CR, Miele G, Clinton M, Differentiation-dependent expression of connective tissue growth factor and lysyl oxidase messenger ribonucleic acids in rat granulosa cells, Endocrinology, 142 (2001) 1082–1089. [DOI] [PubMed] [Google Scholar]
- [38].Phan B, Rakenius A, Pietrowski D, Bettendorf H, Keck C, Herr D, hCG-dependent regulation of angiogenic factors in human granulosa lutein cells, Mol Reprod Dev, 73 (2006) 878–884. [DOI] [PubMed] [Google Scholar]
- [39].Hsieh M, Mulders SM, Friis RR, Dharmarajan A, Richards JS, Expression and localization of secreted frizzled-related protein-4 in the rodent ovary: evidence for selective up-regulation in luteinized granulosa cells, Endocrinology, 144 (2003) 4597–4606. [DOI] [PubMed] [Google Scholar]
- [40].Sunderam S, Kissin DM, Crawford SB, Folger SG, Jamieson DJ, Warner L, Barfield WD, Assisted Reproductive Technology Surveillance - United States 2014, MMWR Surveill Summ, 66 (2017) 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nelson SM, Prevention and management of ovarian hyperstimulation syndrome, Thromb Res, 151 Suppl 1 (2017) S61–S64. [DOI] [PubMed] [Google Scholar]
- [42].Al-Shawaf T, Grudzinskas JG, Prevention and treatment of ovarian hyperstimulation syndrome, Best practice & research. Clin Obstet Gynecol, 17 (2003) 249–261. [DOI] [PubMed] [Google Scholar]
- [43].Naredi N, Talwar P, Sandeep K, VEGF antagonist for the prevention of ovarian hyperstimulation syndrome: Current status, Med J Armed Forces India, 70 (2014) 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pietrowski D, Szabo L, Sator M, Just A, Egarter C, Ovarian hyperstimulation syndrome is correlated with a reduction of soluble VEGF receptor protein level and a higher amount of VEGF-A, Hum Reprod, 27 (2012) 196–199. [DOI] [PubMed] [Google Scholar]
- [45].Soares SR, Gomez R, Simon C, Garcia-Velasco JA, Pellicer A, Targeting the vascular endothelial growth factor system to prevent ovarian hyperstimulation syndrome, Hum Reprod Update, 14 (2008) 321–333. [DOI] [PubMed] [Google Scholar]
- [46].Cenksoy C, Cenksoy PO, Erdem O, Sancak B, Gursoy R, A potential novel strategy, inhibition of vasopressin-induced VEGF secretion by relcovaptan, for decreasing the incidence of ovarian hyperstimulation syndrome in the hyperstimulated rat model, Eur J Obstet Gynecol Reprod Biol, 174 (2014) 86–90 [DOI] [PubMed] [Google Scholar]