Abstract
Staphylococcus aureus and other staphylococci continue to cause life-threatening infections in both hospital and community settings. They have become increasingly resistant to antibiotics, especially β-lactams and aminoglycosides, and their infections are now, in many cases, untreatable. Here, we present a non-antibiotic, non-phage method of treating staphylococcal infections by engineering of the highly mobile staphylococcal pathogenicity islands (SaPIs)4. We replaced the SaPIs’ toxin genes with antibacterial cargos to generate antibacterial drones (ABDs) that target the infecting bacteria in the animal host, express their cargo, kill or disarm the bacteria and thus abrogate the infection. As proof of concept, we have constructed ABDs with either a CRISPR-cas9 bactericidal or a CRISPR-dcas9 virulence-blocking module. We show that both ABDs block the development of a murine subcutaneous S. aureus abscess and that the bactericidal module rescues mice given a lethal dose of S. aureus intraperitoneally.
SaPIs are ~15 kb genetic elements that carry tst and other toxic superantigen determinants and are inserted in the staphylococcal chromosomal DNA4. With the help of certain bacteriophages, SaPI DNA is excised from the chromosome, undergoes extensive replication, and becomes packaged in small phage-like particles. These are released upon phage-induced cellular lysis and go on to infect other cells enabling those cells to produce the disease-causing, SaPI-encoded superantigens5–7.
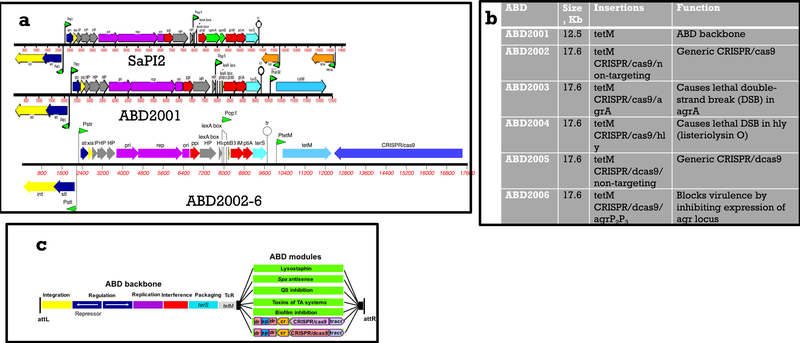
To develop ABDs, we began with a typical SaPI, SaPI2, (Fig. 1a) added tetM, to enable selection for tetracycline (Tc) resistance, and deleted the toxin genes and the two genes, cpmA&B, that cause the formation of small capsids 6. The deletion of cpmA&B resulted in SaPI packaging in full-sized phage particles, thus providing an extra 30 kb of available packaging space, which could be filled with any desired cargo genes. We found that these modifications did not diminish SaPI transfer frequency (data not shown). We then used standard allelic replacement technology to insert generic CRISPR-Cas9 and catalytically inactive CRISPR-dCas9 modules8 with spacers targeting agr, a non-essential global gene regulator of staphylococcal virulence.
Figure 1. (a). Genetic maps of SaPI2 and its ABD derivatives.
Color scheme: yellow – int and xis; dark blue – regulation; gray – hypothetical genes; purple – replication; red – phage interference; green, capsid morphogenesis; turquoise – terminase small subunit (terS); orange – toxin genes; light blue – tetracycline resistance (tetM); medium blue – CRISPR module. (b) ABD constructs. ABD2001 was derived from the prototypical SaPI2 by deleting toxin genes, tst and eta, and the capsid morphogenesis. genes, cpm A&B. ABDs 2002–2006 were derived from ABD2001 by the insertion of the listed genes. (c) ABD scheme. From attL to tetM is the ABD backbone. Black boxes represent the cloning site. Green rectangles represent different modules that have been or will be added to the backbone. dr: direct repeat; sp: spacer; cr: crRNAleader; tracr: tracr RNA.
Agr is transcribed from divergent promoters P2 and P3; the P2 operon includes a two-component signaling (TCS) module, where AgrC is the histidine kinase (HK) receptor and AgrA the response regulator (RR). An agr-encoded autoinducing peptide, AIP, is the activating ligand for the TCS and AgrA~P up-regulates both agr promoters. The major effector of virulence regulation is the agrP3 transcript, RNAIII. The AIPs and their cognate HK receptors are highly variable and form 4 distinct agr specificity groups, with the AIPs generally cross-inhibitory between groups 10. The agr RR and promoter region, however, are highly conserved throughout the staphylococci.
Consequently, we have used spacers guide RNAs targeting agrA and agrP2P3 for the ABDs with the CRISPR-Cas9 and dCas9 modules, respectively. As we had earlier found that SaPIs could be transferred to Listeria monocytogenes11, we constructed a third ABD with a CRISPR-Cas9 spacer targeting hly, the gene encoding listeriolysin O. ABDs are labeled in numerical order starting with 2001, which represents the modified parental SaPI2, used for all of the constructs(See Fig. 1b). Targeting spacers are listed after a slash. The existing and potential future ABD constructs are illustrated schematically in Fig. 1c.
To develop a high-titer, phage-free source of ABD particles, we deleted the helper phage small terminase subunit (terS) gene, eliminating the packaging of phage DNA12 without affecting phage or SaPI DNA replication or gene expression. This deletion enabled the production of mitomycin C (MC)-induced lysates with ABD particle titers in the 1010 /ml range. We have tested ABD2003 for toxicity and observed no ill effects (mice were weighed daily for 7 days and observed for signs of toxicity – weight loss, ruffled fur, ataxic gait, sluggishness, etc.) from either intraperitoneal (IP) or intravenous (IV) doses of 1011 particles.
To demonstrate that an ABD could directly kill an infecting staphylococcus, we used ABD2003, containing a CRISPR/cas9 module with a spacer targeting agrA, which was chosen because it is universally conserved among the staphylococci, yet is nonessential. ABD2003 was expected to kill staphylococci by introducing a double-strand break (DSB) in the chromosomal agrA locus. Thus, the agrA-targeting ABD could be used against virtually any Staphylococcus, whereas strains with an agrA deletion could be used as controls, to test for off-target effects, and for ABD2003 production, which requires the absence of the agrA protospacer in the producing strain.
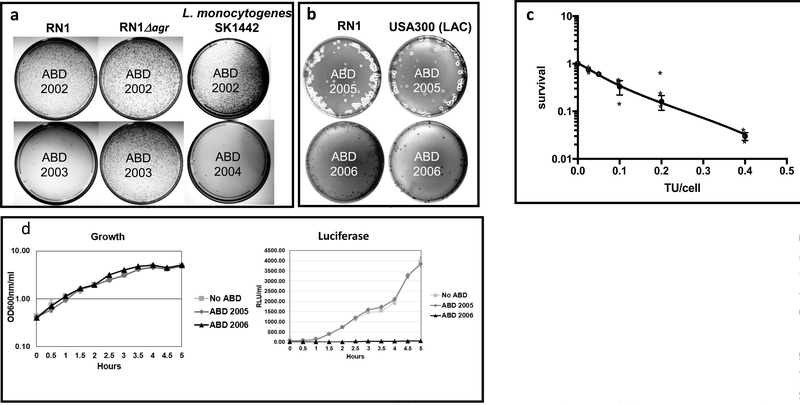
To test for the killing activity of ABDs, we used several S. aureus strains, including RN1, USA300 LAC and other agr+ clinical isolates, with ABD2003. We also tested ABD2004 with L. monocytogenes. We present petri dish photographs (Fig. 2) to show the killing results pictorially: platings were done with 106 particles ml and 109 cells/ml, and 0.1 ml aliquots were plated on Trypticase soy broth (TSB) with Tc, 5μg/ml (Tc5). Colonies seen with the non-targeting spacer (ABD2002) (~104-105/plate) represent ABD-receiving cells; those few seen with the agrA-targeting spacer (ABD2003) were formed by ABD-receiving cells that were not killed by the ABD and represent CRISPR-resistant mutants. Results for these strains are illustrated in Fig. 2a.
Figure 2. (a, b). Plating tests to demonstrate ABD antibacterial activities in vitro. (a) Killing of S. aureus by ABD2003 and of L. monocytogenes by ABD2004.
Suitable dilutions of ABD2002, 2003 or 2004 particle preparations were mixed with RN1, RN1Dagr, or L. monocytogenes SK1442, plated on TSB with 5 μg/ml tetracycline (Tc5), and incubated at 37°C for 48 h. (b) Inhibition of hemolytic activity by ABD2006. Suitable dilutions of ABD2005 or 2006 particle preparations were mixed with RN1 or USA300 LAC, plated on sheep blood agar plates supplemented with Tc5 and incubated at 37°C for 48 h. These experiments have each been done more than 5 times with similar results. (c) Dose-response curve for ABD killing. Data presented in Methods Table 1 are presented as a semi-log plot of surviving cells vs ABD2003 particle dose. Results are averages of the 3 experiments presented in Methods, Table 1. Error bars represent standard errors of the means, where n=3 experiments. (d) Inhibition of agr expression by ABD2006. Strains with integrated ABDs 2001, 2005 and 2006 and a control strain without any ABD were incubated with shaking at 37°C. Two 100 μl samples were withdrawn at each time point and assayed for growth (OD at 600 nM) and Relative Luciferase Units (RLU) in a Molecular Devices luminometer. Graphs show averages of experiments performed in triplicate with error bars representing standard errors, where n=3 experiments.
For L. monocytogenes, strain SK1442, we used ABD2004, which carries CRISPR-Cas9/hly, and observed approximately equivalent cell killing by this ABD, but no killing by ABD2002, which contains the non-targeting CRISPR-Cas9 (See Fig. 2a).
For a more quantitative analysis, cell suspensions were sonicated to disrupt clumps, equal aliquots were mixed with varying numbers of ABD2003 particles and plated on non-selective plates for survivors (which represent cells that were not infected by an ABD particle) and on Tc5 plates for CRISPR-resistant transductants. Methods Table 1 shows the results for ABD2003 (targeting agrA) with RN1. Essentially identical results were obtained with the other strains.
Table 1.
CRISPR/Cas9 killing and mutation frequencies, using ABD2003 with strain RN1
| Sample | TU*/cell (x) | Expected survival (So=e-x) | Observed survival (So) | IP/cell(y) y=ln(1/So) | IP/TU (y/x) | CRISPR-resistant mutants | ||
|---|---|---|---|---|---|---|---|---|
| 1 | %infected | No. | Freq. | |||||
| 1 | .4 | 0.67 | .031±.003 | 3.5±.17 | 8.8±.43 | 97 | 101 | 1.0×10−4 |
| 2 | .2 | 0.82 | .16±.05 | 1.9±.40 | 11±2.1 | 84 | 72 | 8.4×10−5 |
| 3 | .1 | 0.90 | .33±11 | 1.25±.43 | 13±4.3 | 67 | 50 | 7.9×10−5 |
| 4 | .05 | 0.95 | .60±.01 | .52±.006 | 10.3±.33 | 40 | 22 | 6.9×10−5 |
| 5 | 025 | 0.98 | .77±.07 | .28±.087 | 11±3.4 | 23 | 19 | 8.6×10−5 |
| 6 | 0 | 100 | 10.8±.67 | |||||
TU=transduction units; x=TU/cell; Se=expected survival; So= observed survival; IP=infective particles; y=IP/cell
These results confirmed the expected behavior of the three CRISPR modules in vitro and led to pre-clinical testing of ABD2003 in mice. For the in vivo tests of ABD2003, we used 2 different murine infection models with an luminescent agrP3-lux-carrying RN1 derivative13: a subcutaneous (SC) abscess model 14 and an intraperitoneal (IP) lethality model 15.
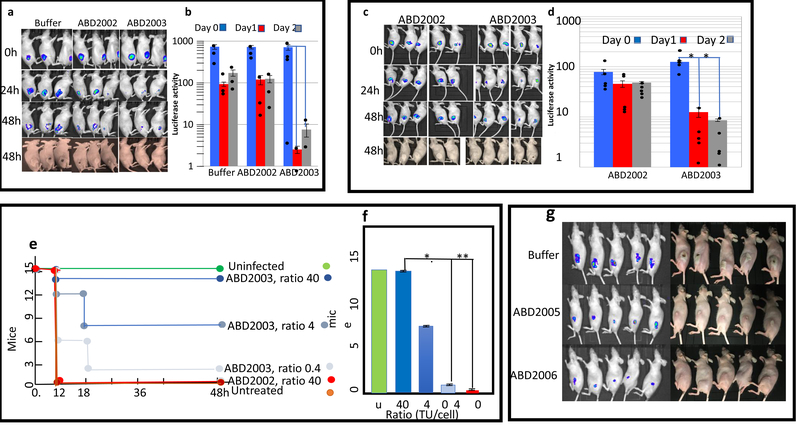
For the SC model, we used hairless mice and a dose of 4×108 organisms (the number of staphylococci in a culture is actually about 3× the number of cfu, because of the natural clumpiness of the organism and we refer to the actual number of organisms rather than the cfu throughout this paper), which uniformly caused a ~2 cm abscess that opened and drained within 72 h. Since there is poor circulation in the subcutaneous space, we needed to deliver the ABD particles to the same site as the bacteria. For this, we injected the bacteria via a cannula into the subcutaneous tissue space, followed immediately by the ABD particles. Here, with an ABD particle:bacteria ratio of 10, abscess formation was completely prevented by ABD2003, and was slightly diminished by ABD2002, which contains CRISPR/cas9 with a non-targeting spacer (we have observed that incoming SaPI or ABD particles cause a slight inhibition of growth (data not shown)) and was unaffected by PBS alone (Fig. 3 (a, b)). Essentially the same result was obtained with an ABD particle:bacteria ratio of 3, while a ratio of 1 reduced the size of the abscess but did not fully block it. Although these experiments showed good efficacy, we recognize that the injection of ABD particles at precisely the same location as the infecting bacteria is a somewhat artificial test. As a more realistic test of therapeutic efficacy, we needed to demonstrate that particles could be administered in one site and reach the infecting bacteria in a second site.
Figure 3. (a,b) Blocking of subcutaneous (SC) murine infections by ABDs.
Bacteria, 4×108, were injected into the SC space of hairless mice through a cannula, followed by 1.2×109 ABD2002 or 2003 particles, through the same cannula. Bacteria were imaged in the in vivo imaging system (IVIS) immediately and after 24 and 48 h. In panel (a) is shown the IVIS images for luciferase activity (Top 3 rows) and photographs of the abscesses of the same mice (bottom row). In panel (b) is shown a quantitative analysis of the luciferase signals. Error bars represent standard errors of the mean, n=3 animals. **P=.01; *P=.05, determined by the one-tailed Fisher’s exact test. Luciferase signals were quantitated using Living Image software (Perkin-Elmer, Inc.). Experiment done once. (c,d) treatment of anbSC murine abscess by IP administration of ABD particles. Mice were injected SC with 4×108 organisms, strain RN1 containing an agrP3::lux fusion, followed immediately by 8.8×109 ABD particles IP. IP treatment with ABD particles at the same dose was repeated 1h later. Mice were imaged in the IVIS immediately afer the first ABD injection, and again after 24 and 48 h. Panel (c) – IVIS images and abscess photographs; panel (d) - quantitative analysis of luciferase signals. Each bar represents the average of the luciferase signals for the five mice in each group at the three time points shown. Error bars represent standard errors of the mean, n=5 animals. Significance was evaluated by the one-tailed Fisher’s exact test; *P=.05. Abscesses at 48 h are the prominent white areas in the mouse flanks – present in all 5 of the ABD2002 mice but in only one of the ABD2003 mice. Difference in abscess formation is significant at the 5% level (P=.0476), one-tailed Fisher’s exact test, n=5 amnimals. Experiment done once. (e, f). Rescue by ABD2003 of mice given a lethal IP dose of staphylococci. Groups of 5 mice were infected IP with 5×109 RN1 cells.. This was followed immediately by IP injection of different numbers of ABD2003 particles. Mice were monitored for 48h. In panel (e) is shown the time course of mouse deaths; In panel (f) is a graphical representation of the final results, in which the groups of mice were pooled and statistics calculated on the pooled groups. Error bars represent standard deviations for each set of 3 pools; *P=.01; **P=.004, calculated by the one-tailed Fisher’s exact test, n=5. (g) Blockage of abscess formation by an agr-inhibiting ABD. Hairless mice were infected with 4×108 RN1 cells through a teflon cannula, followed immediately by 4×109 ABD particles, either ABD2005 (non-targeting) or ABD2006 (targeting the agrP2P3 promoter region), as indicated. Mice were imaged in the IVIS immediately and after 18 and 42 h. The 42h images are shown. Fishers exact test, on the basis of visualization of abscesses, gave a P value of 0.0079 for ABD2006 vs either ABD2005 or untreated, n=5 animals. Experiment done once.
As it has been reported that phage can cure a subcutaneous staph infection if administered by intraperitoneal injection 16, we performed a second test of ABD efficacy in the SC model by injecting the ABD particles intraperitoneally. These results, shown in Fig. 3 (c, d), demonstrate that ABD2003 particles can reach and eradicate bacteria in one compartment/tissue/site when administered at a distant site. The final test of ABD2003 was for intraperitoneal lethality. For this test, the mean lethal dose of bacteria was ~5×109 and we used ABD2003 at 3 different ABD particle:cell ratios – 40, 4, and 0.4, with 5 groups of 15 mice each, as shown in Fig. 3 (e, f). Particles were injected at a nearby site immediately following the bacteria. In this experiment, all of 15 untreated mice died within 12 h, as did all of 15 treated with CRISPR/cas9 with a non-targeting spacer (ABD2002). For mice treated with graded doses of ABD2003 particles, survivals were: 14 of 15 at ABD particle:cell ratio 40, 8 of 15 at ratio 4, 2 of 15 at ratio 0.4.
As it was important to determine whether the system would be widely applicable, we tested ABDs and phages on a series of strains in our collection. Here, 100 μl of equivalent dilutions providing about 105 TU of ABD2002 (which carries CRISPR/cas9 with a non-targeting spacer) and ABD2003 (which carries CRISPR/cas9 with an agrA-targeting spacer), were mixed with about 108 cells of each of the 14 listed strains and plated on TSB-Tc5 plates. As can be seen (Table 2), ABD2002 was transduced at very high frequency (~104 colonies/plate) to 11 of the 14 strains, at a somewhat lower frequency for one, and for these12 strains, there were extremely few (usually<10) ABD2003 tetracycline resistant transductants, which is the usual result for killing by ABD2003. The few survivors were probably CRISPR-resistant mutants. The two strains (4850 and 17855) that were transduced at very low frequency by ABD2002 were essentially insensitive to the ABDs. The occurrence of such strains is obviously problematic for a therapeutic agent. Staphylococcal phages (and, therefore, ABDs) adsorb nonspecifically to wall teichoic acid (WTA), resulting in very broad host specificity; however, there is a minor WTA variant that results in resistance to most phages 17, which may be responsible for the observed resistance. This possibility will be investigated in the future.
Table 2.
ABD and phage host ranges
| S. aureus strains | Genotype | SaPI | agr group | Transduction Frequency* | Phage Sensitivity | ||
|---|---|---|---|---|---|---|---|
| ABD2002 (control) | ABD2003 (killer) | 80α | Phage K | ||||
| RN1 | NCTC8325 wt | SaPIΔ6 | I | H | F | S | R |
| RN450 | RN1 Δ all 3 phages | SaPIΔ6 | I | H | F | S | R |
| RN12134 | RN1 Δagr | SaPIΔ6 | Ι | H | H | S | R |
| Newman | wt | SaPIΔ6 | I | I | F | S | S |
| Newman7B4 | Newman Δ all 4 phages | SaPIΔ6 | I | H | F | S | S |
| RN4282 | wt | SaPI1 | I | H | F | R | S |
| N315 | wt | SaPI2 | II | H | F | R | S |
| RN4850 | wt | IV | L | F | R | S | |
| RN9130 | 502A ΔpT502A | II | H | F | R | R | |
| 17855 | wt | L | <1 | R | R | ||
| RN408 | PS29 wt | H | F | R | R | ||
| RN5006 | wt | H | F | R | R | ||
| RN5007 | wt | H | F | R | R | ||
| RN5934 | wt | H | F | R | R | ||
H-very high frequency; I – intermediate frequency; L – low frequency; F - A very few colonies, probably CRISPR-resistant mutants.
Plating by helper phage 80α and lytic phage K 18, was also variable among these strains, with 12 of the 14 being totally resistant to one or the other or both, as shown in Table 2. Certain strains (such as RN9130, RN408, RN4282, RN5006, RN5007 & RN5934) that were resistant to the two phages were sensitive to transduction and killing by the ABD. This indicates that ABDs will have considerably broader host ranges than individual phages. Aside from WTA variation, most differences in staphylococcal phage sensitivity are well-known to be due mostly to post-adsorption, intracellular blockage 19 which can affect any step in the complex phage life cycle. These would not affect ABD activity, since the expression of only one ABD gene is sufficient, and, presumably, would not affect other types of ABD-mediated cell killing, such as that mediated by bacteriocins, lytic enzymes, etc., nor would they affect ABD-mediated cell inhibition, biofilm formation, or virulence inhibition, because all of these effects depend solely on the expression of the ABD cargo.
A second goal was to develop ABD particles that could block bacterial functionality without directly killing the organism. These would be expected to diminish host-mediated resistance, to avoid the massive release of toxic bacterial contents, to block virulence, or to interfere with biofilms. Here, we utilized the CRISPR-dcas9/agrP2P3-containing ABD (ABD2006) designed to block staphylococcal virulence by inhibiting agr expression, in comparison with ABD2005, which contains CRISPR/dcas9 with a non-targeting spacer. These tests used hemolytic activity, which is up-regulated by agr, as a test for agr function, In Fig. 2b is shown sheep agar blood plates with colonies of RN1 and USA300 containing either ABD2005 or ABD2006, both integrated at the SaPI2 att site. As can be seen, the ABD2005 colonies of either strain show strong hemolytic activity, whereas those containing ABD2006 show none, indicating that ABD2006 blocks agr activity. We also tested for inhibition of agr during growth, using these two RN1 derivatives plus two others – an ABD-negative and one carrying ABD2001, each containing an agrP3-lux reporter. As shown in Fig. 2c, ABD2006 completely inhibited agr expression, by inhibiting expression of the agrP3-lux reporter during growth, whereas strains carrying no ABD, ABD2001, or ABD2005 showed strong and equivalent luciferase induction. All four strains grew indistinguishably. These results indicate that ABD2006 completely inhibited agr expression, whereas ABD2001 and ABD2005 had no effect.
We have previously shown that inhibition of agr expression by a heterologous auto-inducing agr peptide (AIP) could attenuate (but not totally block) a subcutaneous murine abscess in hairless mice 20. Accordingly, we tested the effects of ABD2006 (CRISPR-dcas9/agrP2P3) on subcutaneous abscess formation in the mouse. Using the cannula design as above, we showed that ABD2006 at an ABD:bacteria ratio of 3 completely blocked abscess formation, whereas ABD2005 did not (Fig. 3g), though the abscesses were somewhat smaller than those seen with untreated animals. This effect remains unexplained. Although we did not do a contemporaneous comparison of AIP vs ABD in this model, we suggest that blocking agr expression by the ABD is probably more effective than blocking it by the AIP. Note that in the treated bacteria, which did not develop any abscess, bacteria were still seen at the injection site, as revealed by a persistent and weak luciferase signal, consistent with inhibition of virulence but not loss of viability. The persistent signal is assumed to represent weak residual agr expression.
These data demonstrate excellent efficacy for CRISPR-carrying ABDs in two mouse infection models. However, the ABD system is not without potential problems, including concomitant packaging of unwanted host genes, induction of or recombination with resident SaPIs or plasmids carrying virulence or resistance genes, the occurrence of resistance to the ABDs and stable establishment of only a fraction of incoming ABDs carrying nonlethal cargo.
Resistance to ABDs can be caused by entry exclusion or post entry interference involving either DNA degradation or gene expression. Most S. aureus phages adsorb nonspecifically to the common wall teichoic acid (WTA). A minority of strains (ST395) have a variant WTA that is not recognized by typical phages. To enable ABDs to target ST395 strains, we would use a helper phage with an ST395 receptor17. Resistance due to DNA degradation could be circumvented by modifying problematic restriction site(s) or using different SaPI backbones.
CRISPR resistance can affect either the CRISPR itself, e.g., loss of spacer, or the host cell e.g., unlinked host gene(s) or loss of protospacer. In a sample of 15 mutants, we found both types – 5 had retained both the spacer and the protospacer and were therefore due to mutations in unlinked host gene(s) and the other 10 had simply lost the spacer. Resistance would be circumvented by including in the ABD a non-CRISPR module with a similar mode of action. Therefore, at least 2 different antibacterial modules would be included in any ABD destined for clinical use.
A different problem arises for ABDs that must be functional for an extended period of time. These would have to be equipped with a replicon or with an efficient integration mechanism, and would be subject to resistance based on interference with replication or integration, in addition to the above.
Nevertheless, the ABD system appears to have great potential as a therapeutic modality and to have decided advantages over existing antibacterial strategies. The addition of extra antibacterial modules should circumvent the resistance problem; because of their specificity, the ABDs will have no impact on the microbiome. Though formally akin to phage therapy, they are predicted to have major advantages over phage therapy: i) efficiency: A therapeutic phage generally functions by killing its bacterial target – which means utilizing the entire metabolic machinery of the bacterial cell. An entering ABD molecule needs only to express its antibacterial gene(s). Bacteria have developed a very wide variety of phage resistance mechanisms, affecting virtually every step in the phage reproductive cycle. Most of these mechanisms would not affect an ABD. ii) versatility: Phage genomes are not greatly expandable – for example, coliphage λ with more than about 3 kb of added DNA 21 becomes defective22 – whereas ABD genomes can accommodate >30 kb of added DNA, over twice their genome size, without functional detriment. This, for example, would enable the insertion of additional killing modules to cover for CRISPR resistance; iii) broader host range than any individual phage; iv) non-killing alternatives: Spacers that block cell growth, cell division, virulence gene expression, biofilm formation, etc. can readily be added to dcas9, and genes with these properties can be incorporated independently of CRISPRs; v) trans activity: Genetic systems can be incorporated that encode secretable inhibitors, such as quorum sensing inhibitors, lysins, toxins, etc., which can act in an infection site on bacteria that have escaped infection by the ABD.
We also suggest that the ABD system is by far the most effective delivery system for CRISPR and other explicit antibacterial functions. It is substantially more effective than the phagemids described by Bikard, et al, 23 and by Citorik, et al 24 which are based on rolling circle and other plasmids and phages, some of which are notoriously unstable and none of which can be transferred at high enough frequency to cure an infection.
The results described in this paper point the way to further development of the system, using other types of antibacterial modules directed against bacterial viability, growth, and biofilm formation and directed against Gram-negative as well as other Gram-positive pathogens.
Supplementary Material
Acknowledgements.
This work was supported by NIH grant R01-AI22159 to RPN and by a grant-in-aid from NYU School of Medicine. The animal experiments described in this paper were conducted in conformity with all relevant ethical regulations, under IACUC protocol #160722–01, approved by the NYUSOM IACUC on 7/21/16.
Footnotes
Competing financial interests statement. A patent application has been filed by NYU with Drs. Ram, Ross and Novick as inventors, on the basis of results presented in this paper.
References
- 1.Kennedy AD et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: Recent clonal expansion and diversification. Proc Natl Acad Sci U S A (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King MD et al. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann Intern Med 144, 309–317 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Gould IM et al. New insights into meticillin-resistant Staphylococcus aureus (MRSA) pathogenesis, treatment and resistance. Int J Antimicrob Agents 39, 96–104, doi: 10.1016/j.ijantimicag.2011.09.028 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Lindsay JA, Ruzin A, Ross HF, Kurepina N & Novick RP The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29, 527–543 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Novick RP, R. & Ruzin A Molecular Genetics of SaPI1-a Mobile Pathogenicity Island in Staphylococcus aureus. submitted (2000). [DOI] [PubMed] [Google Scholar]
- 6.Tallent SM, Langston TB, Moran RG & Christie GE Transducing Particles of Staphylococcus aureus Pathogenicity Island SaPI1 Are Comprised of Helper Phage-Encoded Proteins. J Bacteriol 189, 7520–7524 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tormo-Mas MA, Shrestha A, Mir I, Campoy S, Lasa I, Barbé J, Novick RP, Christie GE,, Penadés JR Moonlighting phage proteins de-repress staphylococcal pathogenicity islands. Nature 465, 779–782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, La Russa M & Qi LS CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem 85, 227–264, doi: 10.1146/annurev-biochem-060815-014607 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Novick RP & Geisinger E Quorum sensing in staphylococci. Annu Rev Genet 42, 541–564 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Ji G, Beavis RC & Novick RP Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92, 12055–12059 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J & Novick RP Phage-mediated intergeneric transfer of toxin genes. Science 323, 139–141 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Ram G, Penades JR, Brown S & Novick RP Pathogenicity Island-Directed Transfer of Unlinked Chromosomal Virulence Genes. Molecular cell, doi: 10.1016/j.molcel.2014.11.011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright JS 3rd, Lyon GJ, George EA, Muir TW & Novick RP Hydrophobic interactions drive ligand-receptor recognition for activation and inhibition of staphylococcal quorum sensing. Proc Natl Acad Sci U S A 101, 16168–16173 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barg N, Bunce C, Wheeler L, Reed G & Musser J Murine Model of Cutaneous Infection with Gram-Positive Cocci. Infect. and Immun. 60, 2636–2640 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brouillette E et al. DNA immunization against the clumping factor A (ClfA) of Staphylococcus aureus. Vaccine 20, 2348–2357 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Ryan EM, Gorman SP, Donnelly RF & Gilmore BF Recent advances in bacteriophage therapy: how delivery routes, formulation, concentration and timing influence the success of phage therapy. J Pharm Pharmacol 63, 1253–1264, doi: 10.1111/j.2042-7158.2011.01324.x (2011). [DOI] [PubMed] [Google Scholar]
- 17.Winstel V et al. Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat Commun 4, 2345–54, doi: 10.1038/ncomms3345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Flaherty S et al. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J Bacteriol 186, 2862–2871 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labrie SJ, Samson JE & Moineau S Bacteriophage resistance mechanisms. Nat Rev Microbiol 8, 317–327, doi: 10.1038/nrmicro2315 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Mayville P et al. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. USA 96, 1218–1223 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weil J, Cunningham R, Martin R 3rd, Mitchell E & Bolling B Characteristics of lambda p4, a lambda derivative containing 9 per cent excess DNA. Virology 50, 373–380 (1972). [DOI] [PubMed] [Google Scholar]
- 22.Frischauf AM, Lehrach H, Poustka A & Murray N Lambda replacement vectors carrying polylinker sequences. J Mol Biol 170, 827–842 (1983). [DOI] [PubMed] [Google Scholar]
- 23.Citorik RJ, Mimee M & Lu TK Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol 32, 1141–1145, doi: 10.1038/nbt.3011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bikard D et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol 32, 1146–1150, doi: 10.1038/nbt.3043 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.