Abstract
It was identified that microRNAs were involved in the regulation of chronic neuropathic pain. However, the role of miR-206-3p in neuropathic pain was still unclear. In the current study, the role of miR-206-3p, a type of mature miR-206, in neuropathic pain was investigated. The potential mechanisms were also explored. We found that the expression of miR-206-3p decreased in the dorsal root ganglion (DRG) of chronic constriction sciatic nerve injury (CCI) rats, whereas the While histone deacetylase 4 (HDAC4) level increased. Further exploration showed that administration of a miR-206-3p mimic alleviated neuropathic pain and reduced the level of HDAC4, a predicted target of miR-206-3p. Overexpression of HDAC4 attenuated the effects of miR-206-3p on neuropathic pain. Our data revealed a miR-206-3p-HDAC4 signal that played a potentially important role in CCI-induced neuropathic pain.
Keywords: chronic constriction injury, dorsal root ganglion, histone deacetylase 4, MiR-206-3p, neuropathic pain
Introduction
MicroRNAs (miRNAs) refer to a group of noncoding RNAs whose lengths are 21–25 nt. MiRNAs play a wide range of roles in regulating life activities through posttranscriptional inhibition. MiRNAs are involved in multiple biological processes including cell proliferation, differentiation, and metabolism [13, 19, 29] by modulating specific genes. It has been shown that miRNAs are abnormally expressed in many types of diseases, including heart disease, inflammatory diseases, and especially cancers [20, 21, 30].
MiRNAs also play a vital role in neuronal diseases. MiR-592-5p levels decreased in the hippocampus of hypoxic-ischemic brain damage (HIBD)-induced hippocampal neuronal injury. Administration of a miR-592-5p mimic protects against HIBD-caused hippocampal neuronal injury by targeting PTGDR [32]. The microRNA-302/367 cluster facilitates astrocytes conversion to neurons in adult brains by improving the capability of reprogramming, and miR-302/367-induced neurons improve behavior and repair the neural injury in an Alzheimer’s disease animal model [9, 10]. Intracerebroventricular injection of a miR-126-3p mimic alleviates blood-brain barrier disruption, cerebral edema, and neuronal damage by inhibiting PIK3R2 and the Akt signaling pathway in the vascular endothelium of the brain [38]. The miR-155 level is elevated 9-fold in the hippocampus after controlled cortical impact. The levels of microglial marker Iba1 and neuronal degeneration are increased in miR-155 KO mice. This suggests that miR-155 plays a neuroprotective role in the context of traumatic brain injury [12].
Increasing evidence has indicated that miRNAs play a critical role in neuropathic pain [15]. Peripheral nerve injury causes miR-21 upregulation in the dorsal root ganglion (DRG). Intrathecal injection of a miR-21 inhibitor attenuates mechanical allodynia and thermal hyperalgesia [23]. Both miRNA-124 and miRNA-146a alleviate morphine-induced persistent neuropathic pain by targeting Toll-like receptor signaling [11]. In addition, miR-155 is obviously increased in the spinal cord of chronic constriction sciatic nerve injury (CCI) rats. The suppressor of cytokine signalling 1 (SOCS1) is upregulated in the opposite manner. MiR-155 inhibition attenuates neuropathic pain and neuronal inflammation through directly binding to the 3’-untranslated region of SOCS1 [34]. Our previous data show that miR-96 and miR-183-5p are downregulated in the DRG of CCI rats and that intrathecal administration of miR-96 and miR-183-5p alleviates neuropathic pain through targeting of Nav1.3 and TREK-1, respectively [4, 28].
MiR-206 can be divided into two mature types, miR-206-3p and miR-206-5p, in the rat. Recently, it has been indentified that miR-206 is decreased in the DRG of CCI rats. A miR-206 mimic alleviates neuropathic pain through the MEK/ERK signaling pathway by targeting brain-derived neurotrophic factor [31]. However, the expression profile of miR-206-3p in the DRG with the development of CCI-induced neuropathic pain is still unknown. The role of miR-206-3p in neuropathic pain and the potential mechanisms need to be further explored.
Materials and Methods
Animals
Male Sprague Dawley rats (2 months old) were obtained from the Center of Laboratory Animal Science of Nanchang University. The animals were fed a standard laboratory diet under controlled temperature (20–25°C). All experimental procedures were approved by the Institutional Animal Care and Use Committee of the Medical College of Nanchang University. DRGs ipsilateral to a lesion from lumbar 4 to lumbar 5 (L4-5) were collected at different time points.
Intrathecal catheter implantation
After anesthesia with 10% chloral hydrate, polyethylene catheters (PE-10) were inserted through an incision in the cisterna magna and advanced 7.0–7.5 cm caudally to the level of the lumbar enlargement. Correct position of the catheter was confirmed by hind limb paralysis after the injection of 10 µl of 2% lidocaine.
The chronic constriction injury (CCI) model
After anesthesia with sodium pentobarbital, the sciatic nerve was exposed. Then the sciatic nerve was loosely ligated with sterile 4-0 catgut thread at four consecutive sites. The interval distance of ligation was about 1 mm. The sciatic nerve was not ligated in the rats of the sham group.
Real-time PCR quantification
Total RNA was isolated by using a mirVanaTM miRNA Isolation Kit (Ambion Inc., Carlsbad, CA, USA). A miScript Reverse Transcription Kit (Qiagen, Hilden, Germany) and Applied Biosystems Reverse Transcription Kit were used to synthesize cDNAs of miRNA and mRNA, respectively. Real-time quantitative PCR was performed using an ABI7500 Real-time PCR System (Applied Biosystems, Foster City, CA, USA). The expression the level of miR-206-3p was normalized to U6, while level of Hdac4 was normalized to Gapdh. TaqMan® Fast Advanced Master Mix, miR-206-3p-specific primers, and Hdac4 probes were purchased from Applied Biosystems.
Western blot analysis
Total protein (30 µg) was separated using 8% SDS-polyacrylamide gel electrophoresis. Then the protein was transferred onto a polyvinylidene difluoride membrane. After incubation with primary antibody of anti-HDAC4 (1:500; Abcam, Cambridge, MA, USA) the membrane with protein was incubated with a peroxidase-conjugated secondary antibody (1:10,000; Cell Signaling Technology, Danvers, MA, USA). Immunodetection was performed by the Pierce-enhanced chemiluminescence substrate (Thermo Fisher Scientific, Waltham, MA, USA). The expression level of HDAC4 was normalized to GAPDH.
Luciferase reporter assay
The Full length of 3’UTR of the Hdac4 was cloned into the pmirGLO vector (Promega, Madison, WI, USA), which was designated pmirGLO-HDAC4-wt. The HDAC4-3’UTR-mut1 (site 1 and site 2 mutant), HDAC4-3’UTR-mut2 (site 1 and site 3 mutant), and HDAC4-3’UTR-mut3 (site 2 and site 3 mutant) were constructed by point mutation of HDAC4-3’URT-wt and cloned into the pmirGLO vector, which generated pmirGLO-HDAC4-mut1, pmirGLO-HDAC4-mut2, and pmirGLO-HDAC4-mut1. HEK293T cells were co-transfected with the reporter vectors and a miR-206-3p mimic or miRNA negative control (miRNA-NC) by using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The cells were harvested 48 h later, and luciferase activity was assessed using a Dual Luciferase Reporter Assay System (Promega).
Lentiviral vector package and delivery
The full-length Hdac4, miR-206-3p mimic, or miRNA-NC was subcloned into the GV280 lentiviral vector (GeneChem, Shanghai, China) to construct a lentivirus encoding miR-206-3p (LV-miR-206-3p mimic), lentivirus HDAC4 (LV-HDAC4), and LV-miRNA-NC, respectively. The LV-miR-206-3p vector was constructed by annealing the sense, 5’-CCGGTGGAATGTAAGGAAGTGTGTGGTTCAAGAGACCACACACTTCCTTACTTCCATTTTTG-3’, and antisense, 5’-AATTCAAAAATGGAAGTAAGGAAGTGTGTGGTCTCTTGAACCACACACTTCCTTACATTCCA-3’, oligonucleotides, which formed the double-stranded DNA molecule. Then it was subcloned into the AgeI/EcoRI site of the GV280 lentiviral vector. The sequence of the mature miR-206-3p is as follows: 5’-UGGAAUGUAAGGAAGUGUGUGG-3’. Construction of the LV-miRNA-NC vector was similar to that of the LV-miR-206-3p vector.
Intrathecal drug administration was accomplished using a microinjection syringe connected to the intrathecal catheter. LV-miR-206-3p, LV-miRNA-NC (10 µl), or LV-HDAC4 were delivered at day 5 (d5) after CCI.
Thermal hyperalgesia
The threshold value of thermal hyperalgesia was determined using a thermal paw stimulation system (BME-410C, Tianjin, China), and the data were expressed as thermal withdrawal latency (TWL). The value was determined 3 times for each rat.
Mechanical hypersensitivity
To evaluate mechanical hypersensitivity, the mechanical withdrawal threshold (MWT) value was measured by using calibrated von Frey filaments (BME-403, Tianjin, China). The measurement was repeated 3 times with 30 s intervals.
Statistical analysis
Data are shown as the mean ± SD. Comparisons of means between two groups were carried out using a t-test. Statistical comparisons were performed by analysis of variance (ANOVA) with Dunnett’s test for multiple comparisons. A value of P<0.05 was considered to be statistically significant.
Results
MiR-206-3p was decreased in the DRG of CCI rats
The expression profile of miR-206-5p was detected during the development of CCI-caused neuropathic pain by real-time PCR. MiR-206-3p expression was measured at days 2 (d2), 4, 6, and 8 after CCI and in the sham group. The data showed that miR-206-3p was significantly decreased beginning at d2 (Fig. 1A). At d2, the miR-206-3p level was 0.50-fold that of the sham group (P<0.001). MiR-206-3p levels continued to decrease following d4 after CCI (P<0.001) and remained at low levels. At d8, the miR-206-3p level was still lower compared with that of the sham group (P<0.001). At this point, it was 0.25-fold that of the sham group. The data suggested that CCI caused the decrease of miR-206-3p.
Fig. 1.
Expressions profiles of miR-206-3p and HDAC4 in the DRG of CCI rats. A: MiR-206-3p was downregulated. The miR-206-3p level was detected at d2, d4, d6, and d8 after CCI and in the sham group (n=6). B: Hdac4 mRNA was upregulated. Hdac4 mRNA was detected d2, d4, d6, and d8 after CCI and in the sham group (n=6). C: HDAC4 protein was upregulated. HDAC4 protein was measured at d4 and d8 after CCI and in the sham group (n=3). The DRGs in rats of the sham group were collected at d8. Data are as shown as the mean ± SD. *P<0.05 vs. sham group. **P<0.01 vs. sham group. ***P<0.001 vs. sham group.
HDAC4 was increased in the DRG of CCI rats
To assess the expression pattern of HDAC4 in the DRG during neuropathic pain development, Hdac4 mRNA and protein were measured. The Hdac4 mRNA level was detected at d2, d4, d6, and d8 and in the sham group. The results indicated that Hdac4 mRNA did not change at d2. It began to increase significantly at d4 and was 1.63-fold that of the sham group. Hdac4 mRNA remained at high levels at d8 and was 1.71-fold that of the sham group (Fig. 1B). The HDAC4 protein level was measured at d4 and d8 and in the sham group. The results indicated that HDAC4 protein increased both at d4 and d8. The protein levels were 1.71-fold and 1.92-fold those of the sham group at d4 and d8, respectively (Fig. 1C). The data suggested that CCI caused the increase of HDAC4.
HDAC4 was a direct target of miR-206-3p
Sequence alignments of miR-206-3p and its predicted target sites in the 3’-UTR of Hdac4 are shown Fig. 2A, which shows that there are 3 potential regions binding to miR-206-3p in the 3’-UTR of Hdac4. To confirm that the miR-206-3p binds to Hdac4 in 3’UTR region, the luciferase reporter assay was performed. The results indicated that miR-206-3p could repress the luciferase activity with the 3’-UTR of Hdac4 wt. MiR-206-3p also repressed the luciferase activity with the 3’-UTR of Hdac4 mut1, mut2, and mut3 (Fig. 2B). Thus, HDAC4 was a direct target of miR-206-3p. The target sites inclued site 1, site 2 and site 3. In addition, published data show that HDAC4 is a direct target of miR-206 [7].
Fig. 2.
HDAC4 was a direct target of miR-206-3p. A Sequence alignment of miR-206-3p and its target sites in the 3’-UTR of Hdac4 (download from http://www.targetscan.org/vert_71). There were 3 regions (site 1, site 2, and site 3) in the 3’-UTR of Hdac4 which were potential targets of miR-206-3p. B Luciferase activities were measured in the 3’-UTR of Hdac4 WT, Mut1 (site 1 and site 2 mutant), Mut2 (site 1 and site 3 mutant), and Mut3 (site 2 and site 3 mutant) groups. Data are as shown as the mean ± SD (n=3). *P<0.05 vs. miRNA-NC group. **P<0.01 vs. miRNA-NC group.
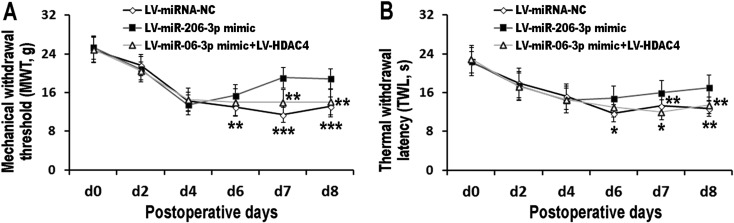
MiR-206-3p alleviated the neuropathic pain
LV-miR-206-3p was intrathecally delivered at d5 before the behaviors test. TWL and MWT were measured to evaluate the effects of miR-206-3p on pain thresholds. They were measured 2 h after each treatment. One day after (at d6) intrathecal injection of LV-miR-206-3p, both TWL and MWT were dramatically increased as compared with scramble-treated CCI rats. Three days after intrathecal injection of LV-miR-206-3p (at d8), TWL and MWT still remained higher. However, intrathecal injection of LV-HDAC4 efficiently blocked the role of LV-miR-206-3p (Fig. 3). The data suggested that miR-206-3p alleviated neuropathic pain through inhibition of HDAC4.
Fig. 3.
MiR-206-3p alleviated the neuropathic pain. MWT (A) and TWL (B) were measured. LV-miRNA-NC, LV-miR-206-3p mimic, and LV-miR-206-3p mimic+LV-HDAC4 were intrathecally injected at d5 after CCI. Data are as shown as the mean ± SD (n=8). *P<0.05 vs. miR-206-3p mimic group. **P<0.01 vs. miR-206-3p mimic group. ***P<0.001 vs. miR-206-3p mimic group.
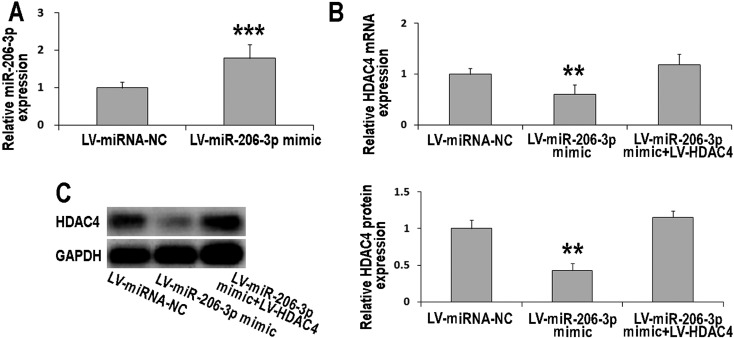
MiR-206-3p decreased the expression of HDAC4
The transfection efficiency was validated by real-time PCR. The data showed that the miR-206-3p mimic could significantly increase the expression levels of miR-206-3p (Fig. 4A). To further confirm the direct target of miR-206-3p, the expression of Hdac4 mRNA and protein were evaluated (Figs. 4B and 4C). Our data indicated that miR-206-3p could suppress CCI-induced HDAC4. However, HDAC4 transfection blocked the suppression role of miR-206-3p. These data confirmed that HDAC4 was a direct target of miR-206-3p. Thus, HDAC4 was involved in the regulation of miR-206-3p in neuropathic pain.
Fig. 4.
MiR-206-3p attenuated the expression of HDAC4. A: The miR-206-3p mimic increased the expression of miR-206-3p in the DRG of the CCI rat. B: MiR-206-3p attenuated the expression of Hdac4 mRNA. HDAC4 eliminated the inhibition role of miR-206-3p. n=6. C: MiR-206-3p blocked the expression of HDAC4 protein. HDAC4 eliminated the inhibition role of miR-206-3p. n=3. The DRGs in each group were collected at d8 after detection of MWT and TWL was finished. Data are as shown as the mean ± SD. **P<0.01 vs. miRNA NC group. ***P<0.001 vs. miRNA NC group.
Discussion
Neuropathic pain arises from injury of peripheral nerves and is caused by many neurological diseases. Patients with diabetes, cancer, and AIDS also experience with neuropathic pain. Several lines of evidence indicate that expression changes of mRNA and protein in the DRG are related to neuropathic pain [5, 33]. It is well known that miRNAs are involved in the regulation of neuropathic pain [1, 3]. In the late phase of neuropathic pain induced by oxaliplatin, miR-15b was robustly increased in the DRG. Overexpression of miR-15b promoted mechanical allodynia through inhibition of the expression of BACE1, which is the direct target of miR-15b [14]. The expression of miR-30b is downregulated in the DRG of spared nerve injury (SNI) rats. By contrast, Nav1.7 is upregulated. MiR-30b overexpression alleviates pain through inhibition of SCN9A transcription. However, miR-30b downregulation markedly increases hypersensitivity to pain in naive rats [27]. Our previous data show that both miR-96 and miR-183-5p are downregulated in the DRG of CCI rats [4, 28]. In the present study, we found that miR-206-3p was decreased in the DRG of CCI rats. Intrathecal miR-206-3p attenuated CCI-induced neuropathic pain. These data indicated that miR-206-3p was involved in the regulation of CCI-induced neuropathic pain.
Histone acetyltransferases (HATs) mediate histone acetylation, which leads to nucleosomal relaxation, and promote transcriptional activation. Conversely, histone deacetylases (HDACs) cause the histone deacetylation and thereby reduce gene transcription.
Eighteen HDACs have been characterized in mammals [35]. These HDACs can be categorized into four distinct classes: class I, class II, class III, and class VI. HDAC4 belongs to class II HDACs. Accumulating evidence suggests that HDACs participate in the regulation of various types of cancers. Many HDAC inhibition drugs have been undergoing clinical trials. Several drugs like abexinostat, belinostat, entinostat, and romidepsin are used as chemotherapeutics [8]. Novel HDAC inhibitors also have greatly advanced the therapeutic potential for a wide range of noncancer indications, including neurodegenerative disease, psychiatric disorders, and inflammatory disease states [22].
Recently, emerging evidence had suggested that the epigenetic modulations of genes were involved in chronic pain. The HDAC1 protein level is increased in the spinal cord of SNI mice. Administration of LG325, a selective HDAC1 inhibitor, significantly attenuates SNI-induced neuropathic pain [26]. Further research shows that LG325 attenuates SNI-induced neuropathic pain through regulation of JNK signaling pathway [25]. HDAC2 expression in the spinal dorsal horn is upregulated during the development of chronic pancreatitis-induced chronic pain. Intrathecal injection of AR-42, a selective HDAC2 inhibitor, obviously alleviated chronic pancreatitis-induced mechanical allodynia by rescuing MOR activity [17].
HDAC4 is involved in several processes that may influence sensory neuron development, regeneration, and pain. HDAC4 plays a unnecessary role in regulation transcriptional of naïve sensory neurons. However, it is essential for appropriate transcriptional responses after injury. HDAC4 knockout decreases thermal hypersensitivity in the inflammatory pain induced by complete Freund’s adjuvant (CFA) [6]. HDAC4 phosphorylation and cytoplasmic redistribution in the dorsal horn neurons were presented in CFA-induced hyperalgesia rats [16]. SGK1 activation-dependent HDAC4 phosphorylation and 14-3-3β promotion of cytoplasmic HDAC4 retention in dorsal horn neurons play crucial roles in neuropathic pain maintenance [18]. In addition, CFA injection upregulates the expressions of HDAC4, HDAC5, HDAC7, and HDAC9 (class II HDAC members) in the spinal dorsal horn, while the levels of HDAC1, HDAC2, and HDAC3 (class I HDAC members) show no significant changes. Pretreated with SAHA, TSA, or LAQ824, inhibitors targeting class I as well as II HDACs, alleviates the CFA-induced thermal hyperalgesia [2]. As a transcriptional repressor, HDAC4 regulates the transcription of genes related to synaptic plasticity in cortical neurons [24]. Because of negative regulation of transcription, HDAC4 modulates its targets indirectly. For example, HDAC4 reduces the level of TRPV1, a neuropathic pain-associated factor, through suppression of another transcription factor [6]. In the present study, we found that chronic constriction sciatic nerve injury increased the expression of HDAC4 in the DRG. This implied that HDAC4 in the DRG participated in CCI-induced neuropathic pain.
Analysis data from TargetScan showed that HDAC4 was a predicted target of miR-206-3p. In the present study, the results of a luciferase reporter assay confirmed that HDAC4 was a direct target of miR-206-3p. Many reports have also provided a link between HDAC4 and miR-206. Overexpression of miR-206 in the muscles of mice decreases endogenous HDAC4 levels, and miR-206 inhibits hypertrophy of myogenic cells by targeting HDAC4 [36]. The dual-luciferase reporter assay indentified that miR-1 and miR-206 directly target the 3’ untranslated region (3’UTR) of paired-box transcription factor (Pax7) and HDAC4. Both miR-1 and miR-206 facilitate skeletal muscle satellite cell myogenic differentiation by targeting Pax7 and HDAC4 [7]. In addition, miR-206 promotes myogenic differentiation by directly targeting HDAC4 [37]. In the current study, we found that overexpression of miR-206-3p suppressed the HDAC4 level. Administration of LV-HDAC4 blocked the inhibition roles of miR-206-3p in chronic pain. Thus, miR-206-3p/HDAC4 signaling in the DRG was related to the CCI-induced neuropathic pain.
In conclusion, our results showed that miR-206-3p was decreased in DRG of CCI rats. By contrast, HDAC4 was increased. MiR-206-3p directly inhibited the expression of HDAC4. MiR-206-3p increased the TWL and MWT of CCI rats. However, overexpression of HDAC4 abrogated the effects of miR-206-3p. These data demonstrated that miR-206-3p alleviated CCI-induced neuropathic pain through inhibition of HDAC4.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81760213) and Outstanding Young Talent Support Plan of Jiangxi Province (No. 20171BCB23027).
References
- 1.Andersen H.H., Duroux M., Gazerani P.2014. MicroRNAs as modulators and biomarkers of inflammatory and neuropathic pain conditions. Neurobiol. Dis. 71: 159–168. doi: 10.1016/j.nbd.2014.08.003 [DOI] [PubMed] [Google Scholar]
- 2.Bai G., Wei D., Zou S., Ren K., Dubner R.2010. Inhibition of class II histone deacetylases in the spinal cord attenuates inflammatory hyperalgesia. Mol. Pain 6: 51. doi: 10.1186/1744-8069-6-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bali K.K., Hackenberg M., Lubin A., Kuner R., Devor M.2014. Sources of individual variability: miRNAs that predispose to neuropathic pain identified using genome-wide sequencing. Mol. Pain 10: 22. doi: 10.1186/1744-8069-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H.P., Zhou W., Kang L.M., Yan H., Zhang L., Xu B.H., Cai W.H.2014. Intrathecal miR-96 inhibits Nav1.3 expression and alleviates neuropathic pain in rat following chronic construction injury. Neurochem. Res. 39: 76–83. doi: 10.1007/s11064-013-1192-z [DOI] [PubMed] [Google Scholar]
- 5.Chen W., Lu Z.2017. Upregulated TLR3 Promotes Neuropathic Pain by Regulating Autophagy in Rat With L5 Spinal Nerve Ligation Model. Neurochem. Res. 42: 634–643. doi: 10.1007/s11064-016-2119-2 [DOI] [PubMed] [Google Scholar]
- 6.Crow M., Khovanov N., Kelleher J.H., Sharma S., Grant A.D., Bogdanov Y., Wood J.N., McMahon S.B., Denk F.2015. HDAC4 is required for inflammation-associated thermal hypersensitivity. FASEB J. 29: 3370–3378. doi: 10.1096/fj.14-264440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai Y., Wang Y.M., Zhang W.R., Liu X.F., Li X., Ding X.B., Guo H.2016. The role of microRNA-1 and microRNA-206 in the proliferation and differentiation of bovine skeletal muscle satellite cells. In Vitro Cell. Dev. Biol. Anim. 52: 27–34. doi: 10.1007/s11626-015-9953-4 [DOI] [PubMed] [Google Scholar]
- 8.De Souza C., Chatterji B.P.2015. HDAC inhibitors as novel anti-cancer therapeutics. Recent Patents Anticancer Drug Discov. 10: 145–162. doi: 10.2174/1574892810666150317144511 [DOI] [PubMed] [Google Scholar]
- 9.Ghasemi-Kasman M., Baharvand H., Javan M.2017. Enhanced neurogenesis in degenerated hippocampi following pretreatment with miR-302/367 expressing lentiviral vector in mice. Biomed. Pharmacother. 96: 1222–1229. doi: 10.1016/j.biopha.2017.11.094 [DOI] [PubMed] [Google Scholar]
- 10.Ghasemi-Kasman M., Shojaei A., Gol M., Moghadamnia A.A., Baharvand H., Javan M.2018. miR-302/367-induced neurons reduce behavioral impairment in an experimental model of Alzheimer’s disease. Mol. Cell. Neurosci. 86: 50–57. doi: 10.1016/j.mcn.2017.11.012 [DOI] [PubMed] [Google Scholar]
- 11.Grace P.M., Strand K.A., Galer E.L., Maier S.F., Watkins L.R.2018. MicroRNA-124 and microRNA-146a both attenuate persistent neuropathic pain induced by morphine in male rats. Brain Res. 1692: 9–11. doi: 10.1016/j.brainres.2018.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison E.B., Emanuel K., Lamberty B.G., Morsey B.M., Li M., Kelso M.L., Yelamanchili S.V., Fox H.S.2017. Induction of miR-155 after brain injury promotes type 1 interferon and has a neuroprotective effect. Front. Mol. Neurosci. 10: 228. doi: 10.3389/fnmol.2017.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Z., Li Z., Zhang X., Yin K., Wang W., Xu Z., Li B., Zhang L., Xu J., Sun G., Wang L., Li Q., Huang X., Zhang L., Zhang D., Xu H., Xu Z.2018. MiR-422a regulates cellular metabolism and malignancy by targeting pyruvate dehydrogenase kinase 2 in gastric cancer. Cell Death Dis. 9: 505. doi: 10.1038/s41419-018-0564-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito N., Sakai A., Miyake N., Maruyama M., Iwasaki H., Miyake K., Okada T., Sakamoto A., Suzuki H.2017. miR-15b mediates oxaliplatin-induced chronic neuropathic pain through BACE1 down-regulation. Br. J. Pharmacol. 174: 386–395. doi: 10.1111/bph.13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiangpan P., Qingsheng M., Zhiwen Y., Tao Z.2016. Emerging role of microRNA in neuropathic pain. Curr. Drug Metab. 17: 336–344. doi: 10.2174/1389200216666151015113400 [DOI] [PubMed] [Google Scholar]
- 16.Lai C.Y., Hsieh M.C., Ho Y.C., Chen G.D., Chou D., Ruan T., Lee A.S., Wang H.H., Chau Y.P., Peng H.Y., Lai C.H.2018. GluN2B/CaMKII mediates CFA-induced hyperalgesia via HDAC4-modified spinal COX2 transcription. Neuropharmacology 135: 536–546. doi: 10.1016/j.neuropharm.2018.03.012 [DOI] [PubMed] [Google Scholar]
- 17.Liao Y.H., Wang J., Wei Y.Y., Zhang T., Zhang Y., Zuo Z.F., Teng X.Y., Li Y.Q.2018. Histone deacetylase 2 is involved in µ‑opioid receptor suppression in the spinal dorsal horn in a rat model of chronic pancreatitis pain. Mol. Med. Rep. 17: 2803–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin T.B., Hsieh M.C., Lai C.Y., Cheng J.K., Chau Y.P., Ruan T., Chen G.D., Peng H.Y.2015. Modulation of nerve injury-induced HDAC4 cytoplasmic retention contributes to neuropathic pain in rats. Anesthesiology 123: 199–212. doi: 10.1097/ALN.0000000000000663 [DOI] [PubMed] [Google Scholar]
- 19.Ling X., Yao D., Kang L., Zhou J., Zhou Y., Dong H., Zhang K., Zhang L., Chen H.2017. Involment of RAS/ERK1/2 signaling and MEF2C in miR-155-3p inhibition-triggered cardiomyocyte differentiation of embryonic stem cell. Oncotarget 8: 84403–84416. doi: 10.18632/oncotarget.21218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelosi A., Lunardi C., Fiore P.F., Tinazzi E., Patuzzo G., Argentino G., Moretta F., Puccetti A., Dolcino M.2018. MicroRNA expression profiling in psoriatic arthritis. BioMed Res. Int. 2018: 7305380. doi: 10.1155/2018/7305380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollard J., Burns P.A., Hughes T.A., Ho-Yen C., Jones J.L., Mukherjee G., Omoniyi-Esan G.O., Titloye N.A., Speirs V., Shaaban A.M.2018. Differential Expression of MicroRNAs in Breast Cancers from Four Different Ethnicities. Pathobiology 85: 220–226. doi: 10.1159/000488456 [DOI] [PubMed] [Google Scholar]
- 22.Qiu X., Xiao X., Li N., Li Y.2017. Histone deacetylases inhibitors (HDACis) as novel therapeutic application in various clinical diseases. Prog. Neuropsychopharmacol. Biol. Psychiatry 72: 60–72. doi: 10.1016/j.pnpbp.2016.09.002 [DOI] [PubMed] [Google Scholar]
- 23.Sakai A., Suzuki H.2013. Nerve injury-induced upregulation of miR-21 in the primary sensory neurons contributes to neuropathic pain in rats. Biochem. Biophys. Res. Commun. 435: 176–181. doi: 10.1016/j.bbrc.2013.04.089 [DOI] [PubMed] [Google Scholar]
- 24.Sando R., 3rd, Gounko N., Pieraut S., Liao L., Yates J., 3rd, Maximov A.2012. HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell 151: 821–834. doi: 10.1016/j.cell.2012.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanna M.D., Galeotti N.2018. The HDAC1/c-JUN complex is essential in the promotion of nerve injury-induced neuropathic pain through JNK signaling. Eur. J. Pharmacol. 825: 99–106. doi: 10.1016/j.ejphar.2018.02.034 [DOI] [PubMed] [Google Scholar]
- 26.Sanna M.D., Guandalini L., Romanelli M.N., Galeotti N.2017. The new HDAC1 inhibitor LG325 ameliorates neuropathic pain in a mouse model. Pharmacol. Biochem. Behav. 160: 70–75. doi: 10.1016/j.pbb.2017.08.006 [DOI] [PubMed] [Google Scholar]
- 27.Shao J., Cao J., Wang J., Ren X., Su S., Li M., Li Z., Zhao Q., Zang W.2016. MicroRNA-30b regulates expression of the sodium channel Nav1.7 in nerve injury-induced neuropathic pain in the rat. Mol. Pain 12: 12. doi: 10.1177/1744806916671523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi D.N., Yuan Y.T., Ye D., Kang L.M., Wen J., Chen H.P.2018. MiR-183–5p alleviates chronic constriction injury-Induced neuropathic pain through inhibition of TREK-1. Neurochem. Res. 43: 1143–1149. doi: 10.1007/s11064-018-2529-4 [DOI] [PubMed] [Google Scholar]
- 29.Sohn E.J., Park H.T.2018. MicroRNA Mediated Regulation of Schwann Cell Migration and Proliferation in Peripheral Nerve Injury. BioMed Res. Int. 2018: 8198365. doi: 10.1155/2018/8198365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song Y., Higgins H., Guo J., Harrison K., Schultz E.N., Hales B.J., Moses E.K., Goldblatt J., Pachter N., Zhang G.2018. Clinical significance of circulating microRNAs as markers in detecting and predicting congenital heart defects in children. J. Transl. Med. 16: 42. doi: 10.1186/s12967-018-1411-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun W., Zhang L., Li R.2017. Overexpression of miR-206 ameliorates chronic constriction injury-induced neuropathic pain in rats via the MEK/ERK pathway by targeting brain-derived neurotrophic factor. Neurosci. Lett. 646: 68–74. doi: 10.1016/j.neulet.2016.12.047 [DOI] [PubMed] [Google Scholar]
- 32.Sun L.Q., Guo G.L., Zhang S., Yang L.L.2018. Effects of microRNA-592–5p on hippocampal neuron injury following hypoxic-ischemic brain damage in neonatal mice-involvement of PGD2/DP and PTGDR. Cell Physiol. Cell. Physiol. Biochem. 45: 458–473. doi: 10.1159/000486923 [DOI] [PubMed] [Google Scholar]
- 33.Takasu K., Sakai A., Hanawa H., Shimada T., Suzuki H.2011. Overexpression of GDNF in the uninjured DRG exerts analgesic effects on neuropathic pain following segmental spinal nerve ligation in mice. J. Pain 12: 1130–1139. doi: 10.1016/j.jpain.2011.04.003 [DOI] [PubMed] [Google Scholar]
- 34.Tan Y., Yang J., Xiang K., Tan Q., Guo Q.2015. Suppression of microRNA-155 attenuates neuropathic pain by regulating SOCS1 signalling pathway. Neurochem. Res. 40: 550–560. doi: 10.1007/s11064-014-1500-2 [DOI] [PubMed] [Google Scholar]
- 35.Verdin E., Dequiedt F., Kasler H.G.2003. Class II histone deacetylases: versatile regulators. Trends Genet. 19: 286–293. doi: 10.1016/S0168-9525(03)00073-8 [DOI] [PubMed] [Google Scholar]
- 36.Winbanks C.E., Beyer C., Hagg A., Qian H., Sepulveda P.V., Gregorevic P.2013. miR-206 represses hypertrophy of myogenic cells but not muscle fibers via inhibition of HDAC4. PLoS One 8: e73589. doi: 10.1371/journal.pone.0073589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winbanks C.E., Wang B., Beyer C., Koh P., White L., Kantharidis P., Gregorevic P.2011. TGF-beta regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J. Biol. Chem. 286: 13805–13814. doi: 10.1074/jbc.M110.192625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xi T., Jin F., Zhu Y., Wang J., Tang L., Wang Y., Liebeskind D.S., He Z.2017. MicroRNA-126-3p attenuates blood-brain barrier disruption, cerebral edema and neuronal injury following intracerebral hemorrhage by regulating PIK3R2 and Akt. Biochem. Biophys. Res. Commun. 494: 144–151. doi: 10.1016/j.bbrc.2017.10.064 [DOI] [PubMed] [Google Scholar]