Abstract
Background
The microRNA (miRNA) miR-200c-3p is involved in the tumorigenesis and progression of a variety of cancers. However, the underlying regulatory role of miR-200c-3p in prostate cancer (PCa) remains unclear.
Methods
Online databases including Oncomine, Linkedomics and StarBase were used to investigate the clinical significance of miR-200c-3p, along with associated gene targets. PCa tissues and adjacent normal tissues were used for the detection of miR-200c-3p expression. A lentivirus overexpressing miR-200c-3p was constructed and transfected into PC3 and DU145 cells. Cell formation of proliferation, migration, and invasion were determined by cell viability and colony-formation assay, wound healing assay, and Matrigel invasion assay, respectively. Epithelial-mesenchymal transition (EMT)-associated markers were determined by qRT-PCR and Western blot. A luciferase reporter assay was performed to determine the direct relationship of miR-200c-3p and ZEB2. The tumor-suppressive role of miR-200c-3p was further confirmed by a xenograft tumor model and immunohistochemical (IHC) staining.
Results
Online database analyses showed that miR-200c-3p was associated with pathologic T and N stage in PCa, and miR-200c-3p was downregulated in PCa tissues. Overexpression of miR-200c-3p was considered a tumor suppressor and was found to significantly suppress the formation of migration and invasion in PCa cells via repression of E-cadherin-induced EMT. The bioinformatic database indicated that ZEB2 has a significant correlation with miR-200c-3p and was upregulated in PCa tissues. Further, ZEB2 expression was suppressed by the upregulation of miR-200c-3p and was identified as a direct target of miR-200c-3p. In addition, repression of ZEB2 could restore the levels of miR-200c-3p in PCa cells in turn, suggesting a potential negative loop between miR-200c-3p and ZEB2. miR-200c-3p also had an antitumor effect by negatively regulating ZEB2 in a xenograft mouse model.
Conclusions
Taken together, the results of our study demonstrated the novel regulatory loop of miR-200c-3/ZEB2 in PCa progression, providing effective therapeutic strategies for PCa in the future.
Keywords: Prostate cancer (PCa), miR-200c-3p/ZEB2 loop, epithelial-mesenchymal transition (EMT), tumor progression
Introduction
Prostate cancer (PCa) is the most common malignancy and the leading cause of cancer-related mortality in males in Western countries (1). Radical prostatectomy is the most effective treatment in the early stages; however, a certain number of PCa patients eventually experience metastatic cancer progression, which carries a worse prognosis (2). Initial steps in the metastatic process are aided by the phenotype and gene expression changes that occur during the epithelial-mesenchymal transition (EMT) (3). Current studies have identified specific genes, including those of microRNAs (miRNAs), as key regulators of EMT (4,5).
miRNAs are a novel class of small, noncoding single RNAs that regulate the expression of target genes at the posttranscriptional level by binding to the 3’ untranslated region (3’-UTR) of these targets (6). An enormous number of studies indicated that miRNAs play a crucial role in multiple biological processes of human tumors (7-9). The miR-200 family, which includes hsa-miR-200a-3p, hsa-miR-200b-3p, hsa-miR-200c-3p, hsa-miR-141-3p, and hsa-miR-429, is a tumor-suppressive group of miRNAs that play a key role in suppressing EMT (10). The hallmark of EMT is downregulation of the cell adhesion molecule E-cadherin, a key epithelial marker, which is inhibited by promoter suppression by transcription repressors during tumor progression (11). Downregulation of E-cadherin results in loss of epithelial cell characteristics and the adaption of the cells to a mesenchymal phenotype. A number of studies indicate that expression of the miR-200 family members is lost in PCa, thereby resulting in upregulation of targets such as TWIST, Zinc finger E-box-binding homeobox 1 (ZEB1), ZEB2, and SLUG, which drive EMT and tumor progression (12,13). This phenomenon might highlight the potential regulatory mechanism between miR-200 and target genes when metastatic and invasive formation is observed in PCa.
As a member of the miR-200 family, miR-200c-3p is reported to be involved in the progression of various cancers. A study indicated that miR-200c-3p was overexpressed in lung metastases, implicating an inhibitory feedback loop to PI3K-AKT (14). The invasive front in primary human colorectal cancer (CRC) tissues revealed that transfection of miR-200c-3p precursors resulted in enhanced cell proliferation but reduced invasion and migration in CRC cell lines (15). Interestingly, UBQLN1 was identified as a direct functional target of miR-200c-3p involved in irradiation-induced autophagy and radio-resistance, representing a promising therapeutic strategy in breast cancer (16). The effects of miR-200c-3p on ZEB1 and E-cadherin, along with the migration and invasion abilities, were also investigated, to a certain degree, in human PCa DU145 cells (17). However, the research methods were too simple to strongly support the conclusion, and the additional profound mechanism was insufficient. In addition, the regulatory effects of miR-200c-3p and ZEB2 remain unclear in PCa, and more evidence is urgently necessary to determine the expression and regulation of miR-200c-3p/ZEB2 involved in the tumor progression of PCa.
In the present study, we first investigated the regulation of the miR-200c-3p/ZEB2 loop in PCa, and the results indicated that overexpression of miR-200c-3p significantly inhibited cell migration and invasion via ZEB2-induced EMT progression in vitro and in vivo. Furthermore, ZEB2 is identified as the direct target of miR-200c-3p, which also represses the expression of miR-200c-3p in turn. We propose the existence of a novel regulatory loop mediated by miR-200c-3p and ZEB2 in PCa that may be a promising biomarker or potential therapeutic target in the future.
Methods
Clinical specimens and cell culture
Twenty-two pairs of PCa tissues and adjacent normal tissues were obtained from PCa patients who underwent radical proctectomy at the First Affiliated Hospital with Nanjing Medical University, China, from Aug 2013 to Aug 2016. All patients provided signed informed consent, and this study was approved by the Research Ethics Committee of the First Affiliated Hospital of Nanjing Medical University. The human prostate epithelial cell RWPE-1 was obtained from the urological laboratory of the First Affiliated Hospital of Nanjing Medical University. The human PCa cell lines PC3 and DU145 were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and were cultured in F-12K medium and DMEM, respectively, supplemented with 10% fetal bovine serum (FBS; Gibco, Australia) and 1% penicillin-streptomycin in an incubator with humidified 5% CO2 at 37 °C.
TCGA database and KEGG pathway analysis
TCGA is available from the Cancer Genomics Browser, University of California, Santa Cruz (https://genome-cancer.ucsc.edu/). KEGG is the Kyoto Encyclopedia of Genes and Genomes (https://www.genome.jp/kegg/). The associated integrated databases included Oncomine, Linkedomics and StarBase, which were used to analyze the expression pattern of ZEB2, and the clinical significance of miR-200c-3p in PCa samples. Further potential pathways mediated by miR-200c-3p were identified by the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov) by analyzing the submitted target genes list predicted by StarBase and Linkedomics.
Establishment of miR-200c-3p overexpressing cells
Lentiviruses overexpressing miR-200c-3p and negative control lentivirus were constructed as previously described (18). PC3 and DU145 cells were transduced with the pHAGE-CMV-miR-200c-IzsGreen lentivirus and selected for IzsGreen expression.
RNA isolation and qRT-PCR
Total RNA, including miRNA from 22 pairs of tissues and PCa cells, was extracted using the miRNeasy Mini Kit (Qiagen, Germany). Extracted RNA was prepared for miRNA detection by using a Mir-X™ miRNA First-Strand Synthesis Kit (Takara, Clontech, USA) in a Veriti® 96-Well Thermal Cycler (Applied Biosystems, USA) according to the manufacturer’s protocol. To quantify mRNAs, we used 1 µg of total RNA and synthesized complementary DNA by utilizing a PrimeScript™ RT Master Mix (Takara, Clontech, USA). Real-time quantitative PCR (qRT-PCR) was performed in triplicate in a 96-well plate containing 1 µL of synthesized cDNA by the use of a QuantiNova™ SYBR Green PCR Kit (Qiagen, Germany) on a StepOnePlus™ Real-Time PCR System (Applied Biosystems, USA). The primer sequences used to detect miRNA and mRNA for PCR were as follows: miR-200c-3p reverse: 5'-CACTGGATTGGAGGAGGG-3', forward: 5'-GAGCTTGACCACCGACTC-3'; U6 reverse: 5'-CGCTTCACGAATTTGCGTGTCAT-3', forward: 5'-GCTTCGGCAGCACATATACTAAAAT-3'; ZEB2 reverse: 5'-TTGCAGGACTGCCTTGAT-3', forward: 5'-GAGCTTGACCACCGACTC-3'; GAPDH reverse: 5'-GGCCATCCACAGTCTTCTGAG-3', forward: 5'-CCTGCACCACCAACTGCTTA-3'.
Cell viability assay
The transfected cells were seeded into 96-well plates at a density of 3,000 cells per well for the cell viability assay. The CCK-8 kit was used to determine cell viability according to the manufacturer's protocol (Dojindo Molecular Technologies, Inc., USA). The absorbance was then measured at 450 nm using an Infinite M200 Pro microplate reader (Tecan, USA).
Colony-formation assay
The transfected cell lines were seeded into 6-well plates at a concentration of 600 cells per well. After 14 days of standardized culture, colony counts and rates were calculated after the colonies were fixed with absolute methanol and stained with 0.1% crystal violet.
Wound healing assay
The transfected cell lines were seeded into six-well plates and cultured until they reached 95% confluence. A 200 µL pipette tip was used to generate a linear wound through the center of the well. The cultures were washed with phosphate-buffered saline (PBS) to remove any cell debris and then allowed to migrate for 0 h and 24 h in a humidified 5% CO2 incubator at 37 °C. The wound images for each well were obtained by the use of an inverted microscope (Olympus CKX41, Japan).
Matrigel invasion assay
The BD Matrigel Invasion Chamber (BD Biosciences, USA) was used for the Matrigel invasion assay following the manufacturer’s protocol (19). A total of 2×105 cells were suspended in 100 µL of serum-free medium and seeded on the upper chamber, and 500 µL of medium containing 10% FBS was added to the bottom chamber. After incubation at 37 °C for 36 h, the cells in the top chamber were removed with cotton swabs. The cells on the lower membrane surface were fixed with methanol and stained with 0.1% crystal violet. After three washes with PBS, the cells were counted under a microscope (Olympus CKX41, Japan).
Western blot analysis
The total proteins of tissues and transfected cells were extracted using RIPA buffer (Beyotime, China) with phenylmethylsulfonyl fluoride (PMSF) and quantified by the BCA Protein Assay Kit according to the manufacturer’s instructions (Beyotime, China). Equivalent quantities of protein were separated on 10% SDS-PAGE gels and then transferred to polyvinylidene fluoride (PVDF) membranes. After blocking with 5% dry milk in Tris-buffered saline for 2 h at 20 °C, the PVDF membranes were incubated with rabbit antibodies specific to human ZEB2 (Santa Cruz Biotechnology, USA, 1:1,000), β-Actin, GAPDH (Abcam Inc., USA, 1:5,000), E-cadherin (Bioworld Technology Inc., USA, 1:1,000), N-cadherin (Santa Cruz Biotechnology, CA, USA, 1:1,000) or vimentin (Bioworld Technology Inc., USA, 1:1,000) overnight at 4 °C. The membranes were then incubated with anti-rabbit secondary antibody for 2 h at 20 °C (Thermo Fisher Scientific Inc., USA, 1:5,000). Band signals were detected using a chemiluminescence system (Bio-Rad, USA) and were analyzed using Image Lab Software (Bio-Rad Laboratories Inc, USA).
Luciferase reporter assay
For the luciferase reporter assay, non-transfected H293T cells were cultured in 12-well plates and then cotransfected with plasmid containing pEZX/ZEB2-3'-UTR or pEZX/ZEB2-3'-UTR-mutant, together with miR-200c-3p mimics or control (GenePharma Co Ltd, Shanghai, China), by a Lipofectamine 3000 Kit (Thermo Fisher Scientific Inc., USA). Firefly and Renilla luciferase activities were measured 48 h after transfection by the Luc-Pair miR Luciferase Assay (GeneCopoeia, FulenGen, China) according to the manufacturer's instructions.
Xenograft tumor model
The lentivirus-transfected PC3 cell line and control were implanted subcutaneously in the back of Balb/c nude mice (four to five weeks old ordered from the Animal Center of Nanjing Medical University of China) with 2×106 cells per site. The tumors were then dissected and fixed in 4% formalin for immunohistochemical (IHC) analysis. All experimental procedures were approved by the Animal Ethics Committee of Nanjing Medical University.
IHC staining
Tissue sections were immersed in UltraCruz Blocking Reagent (Santa Cruz Biotechnology, CA, USA) and then incubated with primary antibody at 4 °C overnight. The sections were then incubated with HRP-conjugated secondary antibody at room temperature for 1 h, and reactive products were visualized by staining with 3,3'-diaminobenzidine according to the manufacturer’s instructions. The images of PCNA-stained and ZEB2-stained cells were obtained under a magnification of ×200 (Olympus, Japan).
Statistical analysis
Statistical analyses were performed using SPSS 21.0 (IBM, Armonk, NY, USA) and GraphPad Prism 5 (GraphPad Software Inc., California, USA). For TCGA-based analysis, the relationship of miR-200c-3p with PCa pathologic T stage was via the Kruskal-Wallis test, and the Wilcox test was used for the correlation of miR-200c-3p with PCa pathologic N stage. The hazard ratio (HR) for miR-200c-3p in the overall survival of PCa was determined via the Cox regression test. Comparisons between groups were conducted using Student’s t-test. A two-sided P value <0.05 was considered statistically significant.
Results
miR-200c-3p expression is frequently decreased in PCa
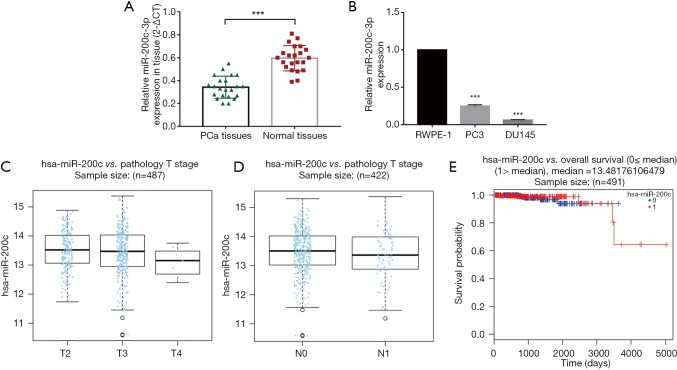
Our result of qRT-PCR showed that the miR-200c-3p expression level was significantly reduced in 22 PCa tissues (P<0.0001) compared to adjacent normal tissues (Figure 1A). The main characteristics of the 22 patients included in this study are mentioned in Table S1. In addition, we found that the expression of miR-200c-3p was strikingly downregulated in the PC3 and DU145 cell lines when compared with that in RWPE-1 (human prostatic epithelial cell line) (Figure 1B). In addition, the bioinformatic database website (http://www.linkedomics.org) indicated that miR-200c-3p expression was positively correlated with pathologic T stage (Figure 1C) and was negatively related to pathologic N stage in PCa samples (Figure 1D) but was not significantly associated with overall survival in PCa patients (Figure 1E). These results strongly indicate the potential tumor-suppressive role of miR-200c-3p in PCa progression.
Figure 1.
miR-200c-3p expression is frequently decreased in PCa. (A) Expression of miR-200c-3p by qRT-PCR in 22 pairs of PCa tissues and normal tissues; (B) expression of miR-200c-3p in PCa cell lines (PC3 and DU145), compared with RWPE-1 cell (human prostatic epithelial cell line); (C) miR-200c-3p was positively correlated with PCa pathologic T stage (the total number of enrolled cases from TCGA database is 487, with a P value of 1.667e-01 by the Kruskal-Wallis test); (D) miR-200c-3p was negatively related to PCa pathologic N stage (the total number of enrolled cases from TCGA database was 422, with a P value of 2.618e-01 by the Wilcox test); (E) miR-200c-3p was not significantly associated with overall survival in PCa (0, low miR-200c-3p group with the sample number of 246; 1, high miR-200c-3p group with the sample number of 245, HR for miR-200c-3p =0.68, with a P value of 6.399e-01 by the Cox regression test). ***, P<0.001. PCa, prostate cancer.
miR-200c-3p suppresses cell proliferation and migration abilities of PCa cell lines in vitro
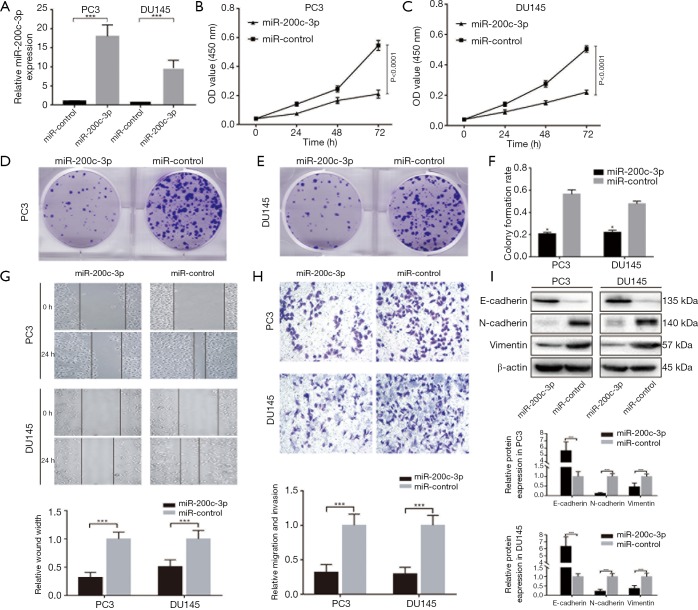
To determine the tumor-suppressive effects of miR-200c-3p in PCa cell lines, we established overexpression of miR-200c-3p in PC3 and DU145 cell lines (Figure 2A). The cell viability assay indicated that overexpression of miR-200c-3p obviously inhibited PCa cell proliferation, with a 63.6% decrease in PC3 cells and a 57.1% decrease in DU145 cells (P<0.0001) (Figure 2B,C). The colony-formation assay consistently supported that miR-200c-3p could reduce the colony rate in PCa cell lines (Figure 2D,E,F). A wound healing assay demonstrated that miR-200c-3p overexpression could significantly reduce the cell migratory capacity of PC3 and DU145 when compared to the control groups (Figure 2G). Moreover, transwell assay data showed that miR-200c-3p overexpression resulted in a significant decrease in the invasive capabilities of PCa cells (Figure 2H). Concerning the positive results induced by miR-200c-3p, we further detected metastasis-associated proteins. Overexpression of miR-200c-3p could upregulate the expression of E-cadherin and downregulate N-cadherin and Vimentin expression, which are all key regulators of EMT progression (Figure 2I). Therefore, miR-200c-3p overexpression significantly repressed the migration and invasion of PCa in vitro.
Figure 2.
miR-200c-3p suppresses cell proliferation and migration abilities of PCa cell lines in vitro. (A) Established overexpression of miR-200c-3p in PCa cell lines, showing a 17.2-fold increase in miR-200c-3p in PC3 cells and an 8.9-fold increase in miR-200c-3p in DU145 cells; (B,C) overexpression of miR-200c-3p obviously inhibited PCa cell proliferation, with a 63.6% decrease in PC3 cells and a 57.1% decrease in DU145 cells; (D,E) miR-200c-3p reduced colony-formation in PC3 and DU145 cells; (F) miR-200c-3p overexpression resulted in a 61.8% decrease in PC3 colony rate and a 54.2% decrease in DU145 colony rate, (G) miR-200c-3p overexpression significantly reduced the PC3 and DU145 cell migratory capacity (P<0.05); (H) miR-200c-3p overexpression resulted in a significant decrease in the invasive capabilities of PCa cells (P<0.05); (I) Western blot, along with histograms of gray intensity value, indicated that overexpression of miR-200c-3p significantly upregulated E-cadherin protein expression, and downregulated the protein expressions of N-cadherin and vimentin in PC3 and DU145 cells. All experiments were conducted separately three times. *, P<0.05; ***, P<0.001. PCa, prostate cancer.
ZEB2 acts as a direct target of miR-200c-3p in regulating EMT of PCa
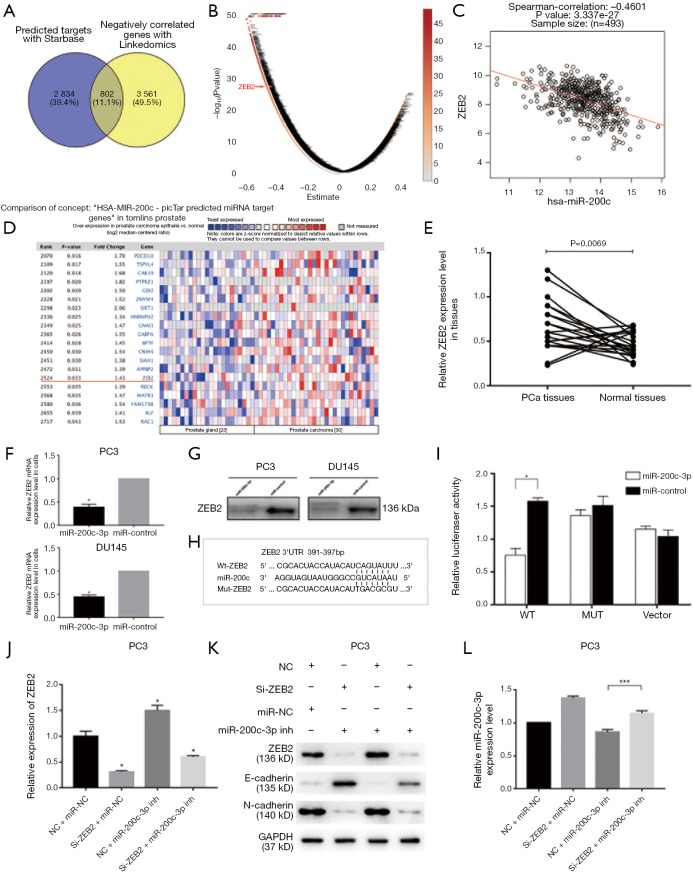
To initially screen the potential targets of miR-200c-3p, we used bioinformatic prediction websites (StarBase, and Linkedomics), and 802 co-targets were identified (Figure 3A). Further pathway analysis of these targets was performed with the DAVID Bioinformatic Resources 6.8 (https://david.ncifcrf.gov/) by submitting a list of these genes. The results showed that 51 signal pathways are predicted to be involved (P<0.05), most of them directly associated with cancers or signal pathways indirectly regulating cancers (Table 1). The potential target ZEB2, which was involved in EMT, was selected through Linkedomics, suggesting a significant negative correlation with miR-200c-3p (Figure 3B,C). Moreover, the bioinformatic database website (https://www.oncomine.org/) was consulted to analyze the ZEB2 level in patients with clinical PCa extracted and integrated from TCGA. The results indicated that ZEB2 has an increased tendency in PCa tissues (fold change: 1.434, P=0.033) compared to normal subjects (Figure 3D). In addition, our results were consistent and indicated that ZEB2 protein expression was significantly upregulated in 22 PCa tissues compared with normal tissues (Figure 3E).
Figure 3.
ZEB2 acts as direct target of miR-200c-3p in regulating EMT of PCa. (A) Bioinformatic prediction websites used; (B,C) prediction of miR-200c-3p-associated targets, and the significant correlation of ZEB2 and miR-200c-3p; (D) ZEB2 level in PCa patients extracted and integrated from the TCGA; (E) ZEB2 expression was significantly upregulated in 22 PCa tissues compared with normal tissues; (F) downregulation of ZEB2 was induced by miR-200c-3p overexpression in PCa cell lines by qRT-PCR (P<0.05); (G) downregulation of ZEB2 protein level as determined by Western blot; (H,I) dual luciferase reporter assay of miR-200c-3p and ZEB2 (P<0.05); (J,K) cotransfected miR-200c-3p inhibitor attenuated the ZEB2 mRNA and protein expression repressed by Si-ZEB2 in PC3 cells (P<0.05); (L) downregulation of ZEB2 rescued miR-200c-3p expression under the context of a miR-200c-3p inhibitor in PC3 cells (***P<0.001). All experiments were conducted separately three times. *, P<0.05. PCa, prostate cancer; EMT, epithelial-mesenchymal transition.
Table 1. KEGG analysis of miR-200c-3p with DAVID online tool.
| Category | Term | Gene counts | P |
|---|---|---|---|
| KEGG_PATHWAY | Focal adhesion | 42 | 7.8E-16 |
| KEGG_PATHWAY | Pathways in cancer | 53 | 2.5E-12 |
| KEGG_PATHWAY | PI3K-Akt signaling pathway | 44 | 8.7E-10 |
| KEGG_PATHWAY | Ras signaling pathway | 34 | 1.2E-9 |
| KEGG_PATHWAY | Rap1 signaling pathway | 33 | 1.3E-9 |
| KEGG_PATHWAY | Regulation of actin cytoskeleton | 33 | 1.9E-9 |
| KEGG_PATHWAY | Proteoglycans in cancer | 31 | 5.0E-9 |
| KEGG_PATHWAY | Bacterial invasion of epithelial cells | 18 | 8.6E-8 |
| KEGG_PATHWAY | MicroRNAs in cancer | 23 | 3.7E-7 |
| KEGG_PATHWAY | Platelet activation | 19 | 2.1E-5 |
| KEGG_PATHWAY | Neurotrophin signaling pathway | 18 | 3.2E-5 |
| KEGG_PATHWAY | Endocytosis | 28 | 3.4E-5 |
| KEGG_PATHWAY | Axon guidance | 18 | 5.5E-5 |
| KEGG_PATHWAY | Osteoclast differentiation | 18 | 7.5E-5 |
| KEGG_PATHWAY | Adherens junction | 13 | 9.1E-5 |
| KEGG_PATHWAY | Amoebiasis | 16 | 1.0E-4 |
| KEGG_PATHWAY | ECM-receptor interaction | 14 | 1.7E-4 |
| KEGG_PATHWAY | Renal cell carcinoma | 12 | 1.8E-4 |
| KEGG_PATHWAY | Chronic myeloid leukemia | 12 | 4.5E-4 |
| KEGG_PATHWAY | Small cell lung cancer | 13 | 4.7E-4 |
| KEGG_PATHWAY | ErbB signaling pathway | 13 | 6.5E-4 |
| KEGG_PATHWAY | HTLV-I infection | 24 | 1.1E-3 |
| KEGG_PATHWAY | Leukocyte transendothelial migration | 15 | 1.2E-3 |
| KEGG_PATHWAY | TGF-beta signaling pathway | 12 | 1.2E-3 |
| KEGG_PATHWAY | cGMP-PKG signaling pathway | 18 | 1.7E-3 |
| KEGG_PATHWAY | MAPK signaling pathway | 23 | 2.9E-3 |
| KEGG_PATHWAY | Hippo signaling pathway | 16 | 4.4E-3 |
| KEGG_PATHWAY | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 10 | 5.4E-3 |
| KEGG_PATHWAY | Cholinergic synapse | 13 | 5.7E-3 |
| KEGG_PATHWAY | Prolactin signaling pathway | 10 | 5.9E-3 |
| KEGG_PATHWAY | Sphingolipid signaling pathway | 13 | 1.0E-2 |
| KEGG_PATHWAY | Jak-STAT signaling pathway | 14 | 1.2E-2 |
| KEGG_PATHWAY | Chemokine signaling pathway | 16 | 1.2E-2 |
| KEGG_PATHWAY | Oxytocin signaling pathway | 15 | 1.3E-2 |
| KEGG_PATHWAY | Melanoma | 9 | 1.7E-2 |
| KEGG_PATHWAY | cAMP signaling pathway | 17 | 1.8E-2 |
| KEGG_PATHWAY | Pancreatic secretion | 10 | 2.1E-2 |
| KEGG_PATHWAY | Colorectal cancer | 8 | 2.2E-2 |
| KEGG_PATHWAY | Malaria | 7 | 2.5E-2 |
| KEGG_PATHWAY | Viral myocarditis | 7 | 2.5E-2 |
| KEGG_PATHWAY | Pancreatic cancer | 8 | 2.8E-2 |
| KEGG_PATHWAY | Glioma | 8 | 3.1E-2 |
| KEGG_PATHWAY | Measles | 12 | 3.1E-2 |
| KEGG_PATHWAY | Estrogen signaling pathway | 10 | 3.4E-2 |
| KEGG_PATHWAY | Phosphatidylinositol signaling system | 10 | 3.4E-2 |
| KEGG_PATHWAY | Dorso-ventral axis formation | 5 | 3.5E-2 |
| KEGG_PATHWAY | Glycosaminoglycan biosynthesis—keratan sulfate | 4 | 3.6E-2 |
| KEGG_PATHWAY | Wnt signaling pathway | 12 | 4.1E-2 |
| KEGG_PATHWAY | Lysosome | 11 | 4.6E-2 |
| KEGG_PATHWAY | Vascular smooth muscle contraction | 11 | 4.6E-2 |
| KEGG_PATHWAY | Prostate cancer | 9 | 4.9E-2 |
Preliminarily, qRT-PCR showed that downregulation of ZEB2 was induced by miR-200c-3p overexpression in PCa cell lines (Figure 3F). In addition, the consistent downregulation of ZEB2 protein levels was determined by Western blot (Figure 3G). To further confirm the direct targeting relationship, a dual luciferase reporter assay was performed (Figure 3H,I). Our findings showed that miR-200c-3p significantly reduced the luciferase activity of ZEB2 containing a wild-type (WT) 3'-UTR but did not suppress the activity of ZEB2 containing a mutant 3'-UTR (MUT). Therefore, these results indicated that ZEB2 is the direct target of miR-200c-3p.
Inhibition of miR-200c-3p expression partially rescues the Si-ZEB2-induced suppression of EMT
We performed rescue experiments to test whether miR-200c-3p inhibition could abrogate the inhibition of EMT by Si-ZEB2 (GenePharma Co., Ltd, Shanghai, China). In parallel, we cotransfected miR-200c-3p inhibitor (GenePharma Co., Ltd, Shanghai, China) to attenuate ZEB2 mRNA and protein expression repressed by Si-ZEB2 in the PC3 cell line (Figure 3J,K). The protein expression patterns of E-cadherin and N-cadherin were also observed. Transfection of miR-200c-3p inhibitor partially but significantly reversed the suppression of EMT by Si-ZEB2 (P<0.05). Moreover, downregulation of ZEB2 by Si-ZEB2 was found to rescue the expression of miR-200c-3p in the context of the miR-200c-3p inhibitor, suggesting the existence of the miR-200c-3p/ZEB2 loop (Figure 3L). Thus, we confirmed that ZEB2 is a key mediator of miR-200c-3p in tumor suppression in PCa.
Tumor suppressor role of miR-200c-3p in vivo
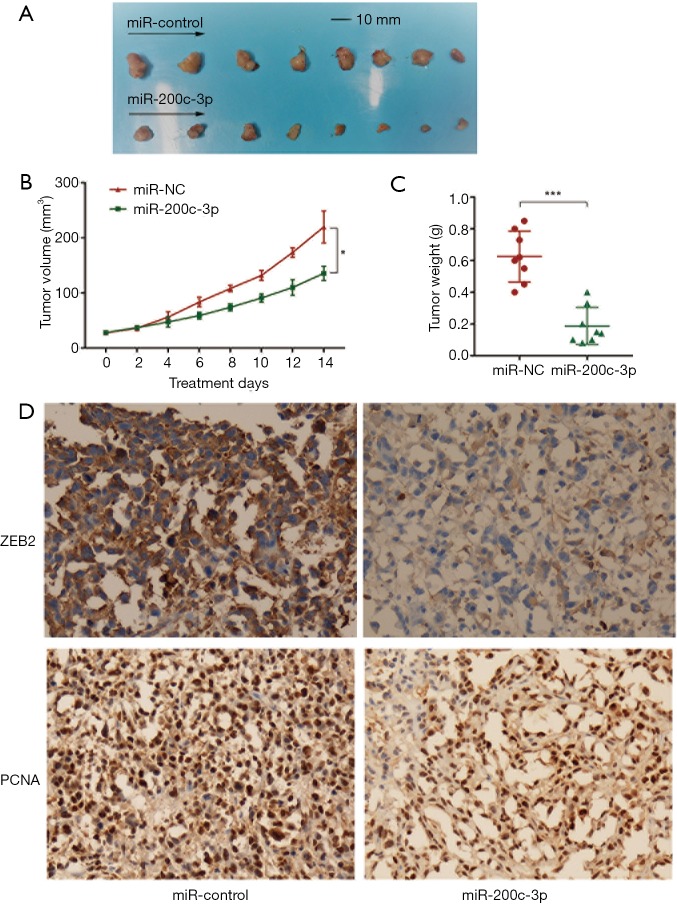
To evaluate the effects of miR-200c-3p overexpression on tumor growth in vivo, the transfected PC3 cells were implanted subcutaneously into the back of nude mice, and the tumor volumes were measured after injection (eight nude mice were included in the miR-200c-3p overexpression group, and eight were included in the miR-200c-3p control group). After two weeks of treatment with miR-200c-3p, the mice were humanely euthanized, and tumor xenografts were removed for further tests. Tumor xenografts were weighed; an obvious weight reduction was found in the miR-200c-3p-treated group, and the size of each tumor xenograft is shown in Figure 4A,B,C. To revalidate the expression level of ZEB2, tumor xenografts were analyzed by immunohistochemistry (IHC) (Figure 4D), and the results revealed that ZEB2 was downregulated in tumor xenografts treated with miR-200c-3p (P<0.05). IHC was also used to further confirm the proliferation indicator (PCNA), which suggested a consistent reduction by miR-200c-3p-treated xenografts (P<0.05). In summary, a tumor xenograft experiment in vivo reconfirmed the tumor suppressor role of miR-200c-3p in PCa.
Figure 4.
Tumor-suppressive role of miR-200c-3p in vivo. (A) The size of each tumor xenograft between two groups (eight tumor xenografts from miR-200c-3p overexpression group, and eight from miR-200c-3p control group); (B) tumor volumes of two groups by treatment days; (C) tumor weight between two groups (***P<0.001); (D) immunohistochemistry of ZEB2 (×400) and PCNA (×400) in tumor xenografts between two groups (P<0.05).
Discussion
This study investigated the role of miR-200c-3p in regulating prostate carcinoma migration. We indicated a novel regulatory loop of miR-200c-3p/ZEB2 in PCa progression. To determine the crucial gene in the regulating loop, we referred to Oncomine (https://www.oncomine.org/) and found that ZEB2 expression was increased in prostate adenocarcinoma compared with noncancerous prostate tissues. We also used bioinformatics and a dual luciferase reporter assay to confirm the relationship between miR-200c-3p and ZEB2. We conducted in vitro and in vivo studies to examine the function of the miR-200c-3p/ZEB2 regulatory axis in PCa. Therefore, targeting the miR-200c-3p/ZEB2 regulatory loop may be a novel strategy for the treatment of PCa.
miRNA regulation is highly associated with multiple biological processes, such as differentiation, proliferation, migration, survival and invasion. Subsequently, deregulation of certain miRNA levels has been found to be associated with tumor progression, metastasis, and recurrence (20). miR-646 was reported to directly target FOXK1 and regulate Akt/mTOR signaling after FOXK1, which resulted in inhibition of proliferation and EMT-induced metastasis in human gastric cancer (21). Moreover, miR-485 was found to inhibit metastasis and EMT by targeting Flot2 and suppress the activity of PI3K/AKT/mTOR signaling in lung adenocarcinoma cells (22). In addition, miR-138 overexpression could inhibit metastasis of breast cancer cells and downregulate vimentin expression and upregulate E-cadherin expression, which are involved in EMT (23).
EMT is an important process during tumor development by which epithelial cells acquire mesenchymal, fibroblast-like properties and show reduced intercellular adhesion and increased motility (24). A recent study has revealed that the loss of miR-200c-3p contributes to the acquisition of EMT phenotype and drug resistance in cancer stem cells (25). As an important regulator of EMT, miR-200c-3p was previously studied in breast cancer, gastric cancer and some other epithelial cancers (15,26-28). In addition, the expression of miR-200c-3p and miR-141-3p in prostate biopsy tissue was inversely correlated with methylation in promoter CpG sites closest to the miR-200c-3p/miR-141-3p loci, indicating that they are under epigenetic regulation in PCa cells (29). Therefore, the molecular mechanisms of miR-200c-3p in PCa progression require further investigation.
Recent studies have demonstrated that transcription repressors, including ZEB, are strongly correlated with the induction of EMT phenotype (30-32). To gain further insight into the association between ZEB2 and miR-200c-3p during EMT progression in PCa, we overexpressed miR-200c-3p in PC3 and DU145 cells. As expected, overexpression of miR-200c-3p led to a significant decrease in ZEB2 expression and an increase in the expression of the epithelial cell marker E-cadherin in both PC3 and DU145 cells. These findings indicated a partial reversal of the EMT signature upon miR-200c-3p overexpression. Subsequently, in the xenograft model, our findings were consistent with the effects in vitro. Taken together, the results of both in vitro and in vivo studies strongly suggest a tumor-suppressive role for miR-200c-3p in PCa pathogenesis.
Bioinformatics analysis demonstrated that ZEB2 is a potential direct target of miR-200c-3p in PCa, with a significant P value of negative correlation. The zinc-finger structures of ZEB2 are highly conserved and allow DNA binding at E-boxes present within the promoter regions of target genes, such as E-cadherin. It was revealed that as an oncogene, ZEB2 is expressed and is a strong suppressor of E-cadherin expression as well as an inducer of N-cadherin and vimentin expression in some cancers (33-35). In our study, the dual luciferase reporter assay confirmed that miR-200c-3p directly targets ZEB2. Therefore, miR-200c-3p was considered to maintain the epithelial phenotype and control the EMT process by targeting ZEB2 in PCa. Moreover, downregulation of ZEB2 by Si-ZEB2 was found to rescue the expression of miR-200c-3p under the context of a miR-200c-3p inhibitor, suggesting the existence of the miR-200c-3p/ZEB2 loop in PCa.
Conclusions
In conclusion, we found that miR-200c-3p could reduce EMT through targeting the expression of ZEB2. Moreover, ZEB2 could also repress the expression of miR-200c-3p in turn. Further study of the pleiotropic functions of miR-200c-3p/ZEB2 may help in understanding PCa metastasis. Based on our novel findings, we believe that targeting miR-200c-3p/ZEB2 could be promising for designing strategies for the treatment of invasive and metastatic PCa.
Table S1. Main characteristics of 22 included patients in this study.
| Characteristics | Patients (n=22) |
|---|---|
| Age (year), median ± SD | 59.6±5.4 |
| Positive surgical margins, % | 18.2 |
| Median pre-surgery PSA concentration (ng/mL) (IQR range) | 8.9 (6.4, 15.7) |
| Pathologic Gleason sum, n (%) | |
| 6 | 5 (22.7) |
| 7 | 15 (68.2) |
| ≥8 | 2 (9.1) |
| Pathologic stage, n (%) | |
| T2 | 6 (27.3) |
| T3a | 12 (54.5) |
| T3b/N1 | 4 (18.2) |
PSA, prostate specific antigen.
Acknowledgements
We would like to thank Animal Ethics Committee of Nanjing Medical University.
Funding: This work was supported by the National Natural Science Foundation of China (Grant no. 81570676, 81770751, 81100532, and 81470981), the Science and Education Health Project of Jiangsu Province for Important Talent (grant number ZDRCA2016009), the “333 High Level Talents Project” in Jiangsu Province (grant number BRA2017532), the Standardized Diagnosis and Treatment Research Program of Key Diseases in Jiangsu Province, China (grant number BE2016791), the Postgraduate Research & Practice Innovation Program of Jiangsu Province (grant number KYCX17_1253) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, JX10231801).
Ethical Statement: The study was approved by the Research Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (IRB number: 2016-SR-029) and written informed consent was obtained from all patients.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013;63:11-30. 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- 2.Amaral TM, Macedo D, Fernandes I, et al. Castration-resistant prostate cancer: mechanisms, targets, and treatment. Prostate Cancer 2012;2012:327253. 10.1155/2012/327253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banyard J, Chung I, Wilson AM, et al. Regulation of epithelial plasticity by miR-424 and miR-200 in a new prostate cancer metastasis model. Sci Rep 2013;3:3151. 10.1038/srep03151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Ma L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev 2012;31:653-62. 10.1007/s10555-012-9368-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullock MD, Sayan AE, Packham GK, et al. MicroRNAs: critical regulators of epithelial to mesenchymal (EMT) and mesenchymal to epithelial transition (MET) in cancer progression. Biol Cell 2012;104:3-12. 10.1111/boc.201100115 [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang D, Shen Y, Wang M, et al. Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. Eur J Cancer Prev 2013;22:540-8. 10.1097/CEJ.0b013e32835f3be9 [DOI] [PubMed] [Google Scholar]
- 8.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008;451:147-52. 10.1038/nature06487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White NM, Yousef GM. MicroRNAs: exploring a new dimension in the pathogenesis of kidney cancer. BMC Med 2010;8:65. 10.1186/1741-7015-8-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng X, Wang Z, Fillmore R, et al. MiR-200, a new star miRNA in human cancer. Cancer Lett 2014;344:166-73. 10.1016/j.canlet.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009;119:1420-8. 10.1172/JCI39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mongroo PS, Rustgi AK. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol Ther 2010;10:219-22. 10.4161/cbt.10.3.12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu YN, Yin JJ, Abou-Kheir W, et al. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene 2013;32:296-306. 10.1038/onc.2012.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berlanga P, Munoz L, Piqueras M, et al. miR-200c and phospho-AKT as prognostic factors and mediators of osteosarcoma progression and lung metastasis. Mol Oncol 2016;10:1043-53. 10.1016/j.molonc.2016.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hur K, Toiyama Y, Takahashi M, et al. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut 2013;62:1315-26. 10.1136/gutjnl-2011-301846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Q, Liu T, Yuan Y, et al. MiR-200c inhibits autophagy and enhances radiosensitivity in breast cancer cells by targeting UBQLN1. Int J Cancer 2015;136:1003-12. 10.1002/ijc.29065 [DOI] [PubMed] [Google Scholar]
- 17.Shi R, Xiao H, Yang T, et al. Effects of miR-200c on the migration and invasion abilities of human prostate cancer Du145 cells and the corresponding mechanism. Front Med 2014;8:456-63. 10.1007/s11684-014-0353-z [DOI] [PubMed] [Google Scholar]
- 18.Chen D, Zhang Y, Wang J, et al. MicroRNA-200c overexpression inhibits tumorigenicity and metastasis of CD117+CD44+ ovarian cancer stem cells by regulating epithelial-mesenchymal transition. J Ovarian Res 2013;6:50. 10.1186/1757-2215-6-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moilanen JM, Kokkonen N, Loffek S, et al. Collagen XVII expression correlates with the invasion and metastasis of colorectal cancer. Hum Pathol 2015;46:434-42. 10.1016/j.humpath.2014.11.020 [DOI] [PubMed] [Google Scholar]
- 20.Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol 2014;24:R762-76. 10.1016/j.cub.2014.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang P, Tang WM, Zhang H, et al. MiR-646 inhibited cell proliferation and EMT-induced metastasis by targeting FOXK1 in gastric cancer. Br J Cancer 2017;117:525-34. 10.1038/bjc.2017.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mou X, Liu S. MiR-485 inhibits metastasis and EMT of lung adenocarcinoma by targeting Flot2. Biochem Biophys Res Commun 2016;477:521-6. 10.1016/j.bbrc.2016.04.043 [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Liu D, Feng Z, et al. MicroRNA-138 modulates metastasis and EMT in breast cancer cells by targeting vimentin. Biomed Pharmacother 2016;77:135-41. 10.1016/j.biopha.2015.12.018 [DOI] [PubMed] [Google Scholar]
- 24.Larue L, Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3' kinase/AKT pathways. Oncogene 2005;24:7443-54. 10.1038/sj.onc.1209091 [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Li Y, Ahmad A, et al. Targeting miRNAs involved in cancer stem cell and EMT regulation: An emerging concept in overcoming drug resistance. Drug Resist Updat 2010;13:109-18. 10.1016/j.drup.2010.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vrba L, Jensen TJ, Garbe JC, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One 2010;5:e8697. 10.1371/journal.pone.0008697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Li G, Chen Y, et al. Metformin Inhibits Tumorigenesis and Tumor Growth of Breast Cancer Cells by Upregulating miR-200c but Downregulating AKT2 Expression. J Cancer 2017;8:1849-64. 10.7150/jca.19858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui FB, Liu Q, Li RT, et al. Enhancement of radiotherapy efficacy by miR-200c-loaded gelatinase-stimuli PEG-Pep-PCL nanoparticles in gastric cancer cells. Int J Nanomedicine 2014;9:2345-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch SM, O'Neill KM, McKenna MM, et al. Regulation of miR-200c and miR-141 by Methylation in Prostate Cancer. Prostate 2016;76:1146-59. 10.1002/pros.23201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mooney SM, Talebian V, Jolly MK, et al. The GRHL2/ZEB Feedback Loop-A Key Axis in the Regulation of EMT in Breast Cancer. J Cell Biochem 2017;118:2559-70. 10.1002/jcb.25974 [DOI] [PubMed] [Google Scholar]
- 31.Figiel S, Vasseur C, Bruyere F, et al. Clinical significance of epithelial-mesenchymal transition markers in prostate cancer. Hum Pathol 2017;61:26-32. 10.1016/j.humpath.2016.10.013 [DOI] [PubMed] [Google Scholar]
- 32.Graham TR, Zhau HE, Odero-Marah VA, et al. Insulin-like growth factor-I-dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res 2008;68:2479-88. 10.1158/0008-5472.CAN-07-2559 [DOI] [PubMed] [Google Scholar]
- 33.Ahmad A, Sarkar SH, Bitar B, et al. Garcinol regulates EMT and Wnt signaling pathways in vitro and in vivo, leading to anticancer activity against breast cancer cells. Mol Cancer Ther 2012;11:2193-201. 10.1158/1535-7163.MCT-12-0232-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacob S, Nayak S, Fernandes G, et al. Androgen receptor as a regulator of ZEB2 expression and its implications in epithelial-to-mesenchymal transition in prostate cancer. Endocr Relat Cancer 2014;21:473-86. 10.1530/ERC-13-0514 [DOI] [PubMed] [Google Scholar]
- 35.Sun DK, Wang JM, Zhang P, et al. MicroRNA-138 Regulates Metastatic Potential of Bladder Cancer Through ZEB2. Cell Physiol Biochem 2015;37:2366-74. 10.1159/000438590 [DOI] [PubMed] [Google Scholar]