Abstract
Introduction: Salmonella enterica subspecies enterica serovar Typhimurium (S. Typhimurium) is one of the major cause of foodborne zoonoses in humans. Poultry acts as a reservoir for S. Typhimurium without showing clinical signs and has become a source of infection to humans. Besides, it also became a source of multidrug-resistant (MDR) strains of S. Typhimurium.
Methods: In the present study, we have isolated 9 S. Typhimurium from 503 samples from environmental sources of poultry wet markets in the Kadapa District of Andhra Pradesh, India. The role of efflux pump activity in antibiotic resistance was evaluated by ethidium bromide cartwheel test and efflux pump inhibition assay.
Results: Eight out of nine isolates were resistant to two or more classes of antibiotics. The efflux pump activity of these isolates by ethidium bromide cartwheel method revealed that 66.6% isolates had shown evidence of pronounced efflux activity. The zone of inhibition (ZOI) of resistant antibiotics for each isolate was estimated in the presence or absence of 25 µg/mL of PAβN. Overall, cephalosporins (cefazolin, cefamandole, and cefaclor), kanamycin, polymyxin-B, piperacillin, and imipenem showed significant increase (≥2 mm) of ZOI, indicating the role of efflux pumps for efflux of these drugs. A maximum of 4 antibiotics among EtBr efflux pump positive isolates and 2 antibiotics among EtBr efflux pump negative isolates showed increased ZOI in the presence of PAβN.
Conclusion: The results indicate that efflux pumps of MDR S. Typhimurium may contribute to resistance for at least one antibiotic, even in EtBr cartwheel test negative isolates.
Keywords: Salmonella, EtBr, efflux, PAβN, ZOI
Introduction
Salmonella is the important genus of Enterobacteriaceae family and consists only two species namely S. enterica and S. bongori.1 The former is subdivided into six subspecies (I, II, IIIa, IIIb, IV, and VI). Salmonella isolates are traditionally typed according to the Kauffmann–White serotyping classification scheme. Specific serovars have been defined by immunologically distinct antigenic combinations of the lipopolysaccharide O-, flagellar H-, and polysaccharide Vi-antigens. Till date, 2673 serotypes have been recognized by the WHO Collaborating Centre for Reference and Research on Salmonella, Pasteur Institute, Paris, France.2
Though typhoid fever caused by S. Typhi is the most important among Salmonella infections in human, non-typhoidal Salmonella organisms are implicated as one of the most common foodborne zoonotic pathogens. The most common manifestation of non-typhoidal salmonellosis is mild to moderate gastroenteritis, consisting of diarrhea, abdominal cramps, vomiting, and fever.3 In a small percentage of cases, septicemia and invasive infections of organs and tissues can occur, leading to diseases such as osteomyelitis, pneumonia, and meningitis.4 Among non-typhoidal Salmonella, S. Typhimurium is the most common cause of food poisoning in many countries including Indian sub-continent.5,6 The disease is mainly transmitted by the food of animal origin especially from poultry and its products. Poultry acts as a reservoir for S. Typhimurium and has become a potential source of this pathogen.
Overuse of antimicrobial agents in the poultry industry has lead to the emergence of multidrug-resistant (MDR) strains. The dissemination of these MDR strains to the environment can occur from poultry farms, slaughterhouses, and poultry wet markets and can become a potential source of infection to humans including those who are vegetarians.
Extrusion of antimicrobial compounds through efflux pumps is one of the mechanisms in antibiotic-resistant strains of foodborne pathogens.7 Efflux pumps are classified into 5 major families namely, small multidrug-resistant family, major facilitator superfamily, adenosine triphosphate-binding cassette superfamily, multidrug and toxic compound extrusion family, and resistance nodulation division superfamily.7,8 Efflux pumps of resistance nodulation division superfamily contribute to intrinsic and acquired resistance for wide variety of antibiotics.7,9 The distribution of efflux-mediated drug resistance among Salmonella isolates is important to understand the mechanisms that occur in a natural selection process for antibiotic resistance.
In the present study, we have isolated MDR phenotypes of S. Typhimurium from the environmental samples of poultry wet markets and determined their efflux activity by ethidium bromide cartwheel test. Then, we have determined the extrusion of various resistant antibiotics in MDR isolates using broad spectrum efflux pump inhibitor.
Materials and methods
Bacterial isolation and its confirmation
A total of 503 samples were randomly collected from environmental sources of poultry wet markets which include meat cutting benches, drainages, weighing machines, and knives in Kadapa District of Andhra Pradesh, India. The enrichment of the samples was done in Rappaport Vassiliadis (RV) media. A loopful of overnight enrichment culture was streaked on hektoen enteric (HE) agar and plates were incubated overnight at 37°C. Typical colonies (maximum three colonies) showing black center with greenish periphery were selected for urease and triple sugar iron (TSI) tests. Molecular confirmation was done by genus-specific (invA)10 and Typhimurium specific (typh)11 PCR. The details of primers are indicated in Table 1. The experiment was carried out with the approval of Institutional Bio-Safety Committee (IBSC), College of Veterinary Science, Sri Venkateswara Veterinary University, Proddatur, Kadapa district, India.
Table 1.
Details of primers used in this study
Antibiotic sensitivity testing for MDR strains
The confirmed isolates of S. Typhimurium were subjected to antibiotic sensitivity testing (Kirby–Bauer disc diffusion method) using 30 antibiotics of different classes. The concentrations of antibiotics used were as per Clinical and Laboratory Standard Institute (CLSI) standards. The lawn cultures were prepared on Mueller Hinton (MH) agar and antibiotic discs were placed with the help of sterile forceps. The plates were incubated at 37°C for overnight. The zone of inhibition around the disc was measured by a scale and interpreted as per CLSI standards. Those isolates which became resistant to two or more classes of antibiotics were considered as MDR strains.
Determination of efflux pump activity by ethidium bromide cartwheel test
The efflux pump activity of MDR S. Typhimurium isolates was determined by ethidium bromide (EtBr) cartwheel test as described by Martins et al (2013)12 with slight modification. Briefly, MH agar plates containing 0 mg/L, 0.5 mg/L, 1 mg/L, 1.5 mg/L, and 2 mg/L of EtBr were prepared on the same day of the experiment. Each MDR S. Typhimurium isolate (approximately 106 cells per mL) was streaked as cartwheel pattern on EtBr plates. The plates were wrapped in aluminum foil to protect from the light and incubated at 37°C for overnight. After incubation, the plates were examined under UV light. The minimum concentration of the EtBr that yielded fluorescence of bacterial colonies was noted.
Determination of antibiotic sensitivity assay in the presence of PAβN
The antibiotic sensitivity assay of MDR isolates for resistant antibiotics was performed in the presence or absence of the efflux pump inhibitor phenylalanine-arginine beta-naphthylamide (PAβN). The minimum inhibitory concentration of PAβN for MDR isolates were determined by broth dilution method. Each isolate was incubated in MH broth containing 25 µg/mL of PAβN for 6 hrs at 37°C. The lawn cultures were prepared on MH agar plates and antibiotic discs were placed and incubated at 37°C for overnight. The zone of inhibition around the antibiotic disc was measured by a ruler. The difference in the zone of inhibition in the presence or absence of PAβN was estimated. Each strain was tested consecutively 3 times to get concurrent values.
Results
Isolation of S. Typhimurium and confirmation
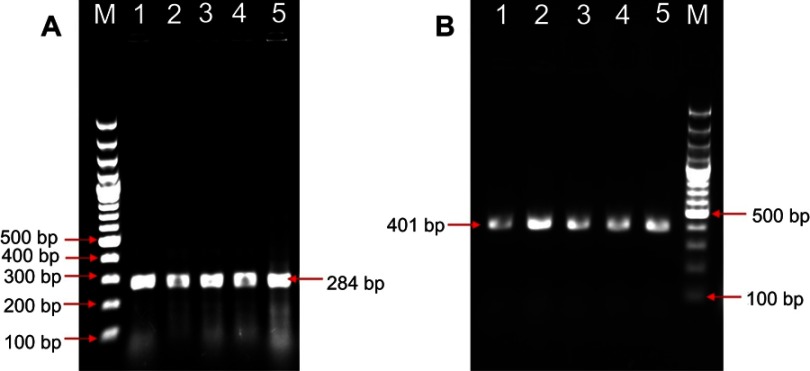
The tentative S. Typhimurium like colonies which were urease negative and showed alkaline slant, acid butt with or without H2S on TSI slants were confirmed by PCR. The genus-specific invA PCR has confirmed with amplicon size of 284 bp, and Typhimurium-specific PCR has confirmed with the size of 401 bp (Figure 1). A total of nine S. Typhimurium isolates has been confirmed by the biochemical and PCR tests.
Figure 1.
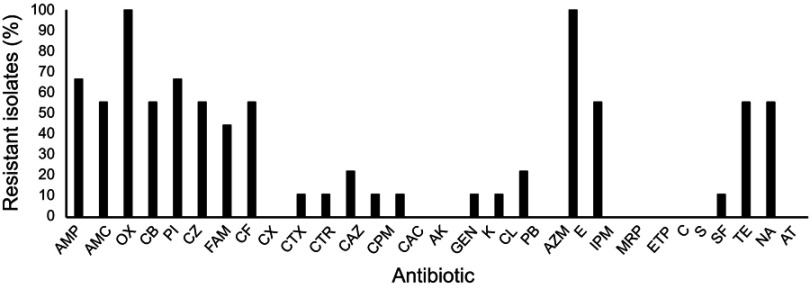
Antibiogram of Salmonella Typhimurium isolates from environmental sources of poultry wet markets. The concentrations of antibiotics used as per clinical and laboratory standards institution (CLSI) standards.
Abbreviations: AMP, ampicillin; AMC, amoxyclav; OX, oxacillin; CB, carbenicillin; PI, piperacillin; CZ, cefazolin; FAM, cefamandole; CF, cefaclor; CX, cefoxitin; CTX, cefotaxime; CTR, ceftriaxone; CAZ, ceftazidime; CPM, cefepime; CAC, ceftazidime/clavulanic acid; AK, Amikacin; GEN, gentamycin; K, kanamycin; CL, colistin; PB, polymyxin B; AZM, azithromycin; E, erythromycin; IPM, imipenem; MRP, meropenem; ETP, ertapenem; C, chloramphenicol; S, streptomycin; SF, sulfisoxazole; TE, tetracycline; NA, nalidixic acid and AT, aztreonam.
Confirmation of phenotypic MDR S. Typhimurium
Antibiogram of S. Typhimurium isolates was performed using 30 antibiotics of various classes which include penicillins, cephalosporins, aminoglycosides, macrolides, polypeptides, carbapenems, and others. Eight out of 9 isolates were found to be resistant to 2 or more classes of antibiotics. However, the resistance pattern within the class varied. The highest resistance was observed among penicillins and cephalosporins. None of the isolates were resistant to Cefoxitin (CX), amikacin (AK), gentamycin (GEN), azithromycin (AZM), meropenem (MRP), ertapenem (ETP), chloramphenicol (C), and aztreonam (AT) (Figure 2).
Figure 2.
PCR detection of Salmonella Typhimurium from field samples. (A) Genus-specific invA PCR for Salmonella produced an amplicon of 284 bp, (B) Typhimurium-specific PCR produced an amplicon of 401 bp. Lane M: 100 bp molecular weight marker; Lane 1–4: Salmonella Typhimurium isolates from field samples, Lane 5: Positive control.
Efflux pump activity by EtBr cartwheel method
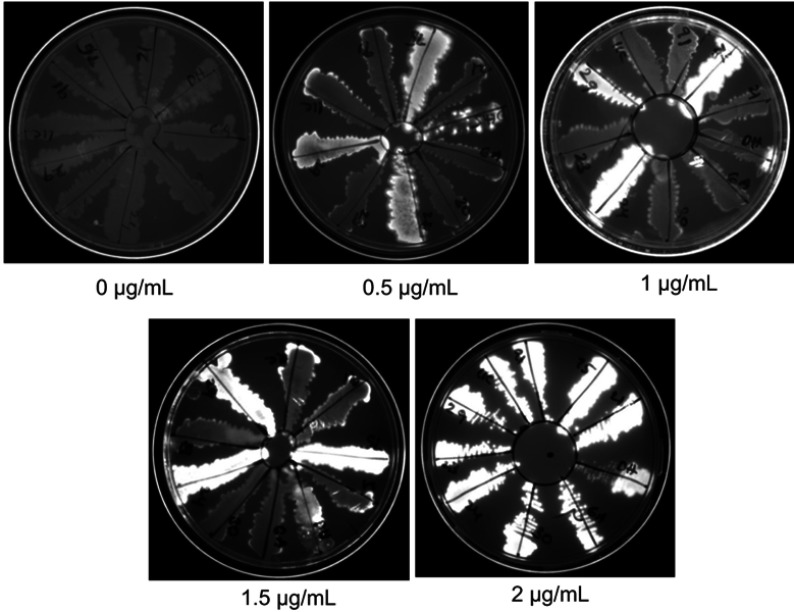
High efflux pump activity is one of the mechanisms of antibiotic resistance. The efflux activity of isolates was determined by the ability of an organism to pump out EtBr out of the cell. The cartwheel test was performed at different concentrations of EtBr. At 1 µg/mL and 1.5 µg/mL of EtBr, the efflux activities of isolates were well distinguished. Out of 9 MDR isolates, 6 isolates did not retain EtBr inside the cells indicating pronounced efflux activity (Figure 3).
Figure 3.
Determination of efflux pump activity by ethidium bromide (EtBr) cartwheel method. Different concentrations of EtBr were added to agar media and a loopful of fresh culture was inoculated. The plates were incubated at 37°C for overnight. The plates were visualized in UV illuminator and documented.
Effect of efflux pump inhibitor-PAβN on antibiogram
The broad spectrum efflux pump inhibitor PAβN was used to assess the efflux activity of 8 MDR and 1 non-MDR isolates of S. Typhimurium. The MIC of PAβN was found to be more than 100 µg/mL. Since PAβN has the effect on membrane integrity and LPS,13 we have used one-fourth of its MIC value (25 µg/mL) in the study. Each MDR strain was tested against antibiotics to which it has shown resistance. The antibiotic sensitivity assay was performed in the presence (Abp) or absence (Ab0) of PAβN. The difference in the zone of inhibition (Abp - Ab0) in the presence and absence of PAβN was measured. The difference of 2 mm or more was considered significant. The difference in the zone of inhibition (ZOI) was found to be significant for antibiotics – cefazolin (2 out of 5 isolates), cefamandole (3 out of 4 isolates), piperacillin (1 out of 6 isolates), cefaclor (3 out of 5 isolates), kanamycin (1 out of 1 isolates), polymyxin-B (2 out of 2 isolates), and imipenem (2 out of 5 isolates) indicating that these drugs were probably effluxed out of the cells (Table 2). Eight out of 9 isolates (88.8%) has shown efflux activity for at least one antibiotic used in the study.
Table 2.
Evaluation of efflux activity by EtBr cartwheel test and PAβN on antibiotic resistance profile of Salmonella Typhimurium isolates. The difference in ZOI was measured by subtracting ZOI of resistant antibiotics for each isolate in the absence of PAβN from the ZOI in the presence of PAβN
| Difference in zone of inhibition (Abp - Ab0) (mm) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Efflux activity by EtBr cart wheel test | Positive | Negative | Positive | Negative | Positive | Positive | Positive | Negative | Positive |
| Resistant antibiotic | ST-21 | ST-75 | ST-30 | ST-29 | ST-6A | ST-23 | ST-91 | ST-24 | ST-11C |
| AMP | 0 | 0 | 0 | 0 | * | 0 | * | 0 | * |
| AMC | 0.66 | * | 0 | 0.66 | * | 1 | * | 0.66 | * |
| OX | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CB | 0 | 0 | 0 | 0 | * | 0 | * | * | * |
| PI | 0 | 0 | 0 | 0 | 3 | 0 | * | * | * |
| CZ | 2.66 | * | 1.66 | 0 | * | 0.66 | * | * | 5 |
| FAM | 11.33 | * | 11.66 | 3 | * | 0 | * | * | * |
| CF | 12.66 | * | 10 | 0 | * | 14 | * | 0 | * |
| CTX | * | 1 | * | * | * | * | * | * | * |
| CTR | * | 0 | * | * | * | * | * | * | * |
| CAZ | 0 | 0 | * | * | * | * | * | * | * |
| CPM | * | 0 | * | * | * | * | * | * | * |
| CAC | * | 0 | * | * | * | * | * | * | * |
| K | * | * | * | * | * | * | * | 3 | * |
| CL | * | 0 | * | * | * | * | * | * | * |
| PB | * | 2 | 5 | * | * | * | * | * | * |
| E | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IPM | 0 | * | 2.66 | 0.66 | * | 0 | * | 6 | * |
| SF | * | 0 | * | * | * | * | * | * | * |
| TE | 0 | * | 0 | 0 | * | 0 | * | 0 | * |
| NA | 0 | * | 0 | 0 | * | 0 | * | 0 | * |
Notes: *Indicates the isolate is sensitive to antibiotic. The significant change in the ZOI is indicated in bold. Abp indicates antibiotic in the presence of PAβN; Ab0 indicates antibiotic in the absence of PAβN.
Abbreviations: AMP, ampicillin; AMC, amoxyclav; OX, oxacillin; CB, carbenicillin; PI, piperacillin; CZ, cefazolin; FAM, cefamandole; CF, cefaclor; CX, cefoxitin; CTX, cefotaxime; CTR, ceftriaxone; CAZ, ceftazidime; CPM, cefepime; CAC, ceftazidime/clavulanic acid; AK, Amikacin; GEN, gentamycin; K, kanamycin; CL, colistin; PB, polymyxin B; AZM, azithromycin; E, erythromycin; IPM, imipenem; MRP, meropenem; ETP, ertapenem; C, chloramphenicol; S, streptomycin; SF, sulfisoxazole; TE, tetracycline; NA, nalidixic acid; AT, aztreonam; ZOI, zone of inhibition.
Discussion
At present, antibiotic resistance is one of the global health problems. Microbes are becoming resistant to last sort antibiotics and thus making the treatment of infectious diseases more difficult. The world health organization stated that “the world may be poised to enter in a post-antibiotic era.”14 Indiscriminate use of antibiotics in the veterinary sector is one of the reasons for the emergence of MDR strains. Being an asymptomatic reservoir, poultry and its products are the major sources of non-typhoidal Salmonella infections in humans.15 Poultry has been considered as a single largest contributor of non-typhoidal salmonellosis in humans.16 The global burden of non-typhoidal salmonellosis was estimated to be around 93.8 million causing 155 000 deaths each year.17 MDR isolates of Salmonella from poultry sources has been well reported around the globe.18–20 MDR Salmonella adapt various mechanisms to resist antibiotics viz., modification of drug target sites, production of drug degrading enzymes, and overexpression of efflux pumps.7,21–23 Efflux pump-mediated drug resistance via RND superfamily was well recognized in gram-negative bacteria.24 Among RND pumps, AcrAB-TolC efflux system is an important contributor for induction of resistance to multiple antibiotics.
In the present study, we have isolated 9 S. Typhimurium from environmental sources of poultry wet markets. Upon antibiogram analysis, 88.88% of the isolates were found to be phenotypically resistant to 2 or more classes of antibiotics. As mentioned earlier, the expression of efflux pumps is one of the contributing factors of antimicrobial drug resistance in S. Typhimurium. The efflux activity of all nine MDR isolates was evaluated by EtBr cartwheel method. Ethidium bromide (EtBr) is considered as a common substrate of efflux pumps in Enterobacteriaceae.25 EtBr cartwheel method has been used for rapid detection of MDR bacteria associated with efflux pumps.26 EtBr has been used to assess the intrinsic efflux activity in E. coli,27 Staphylococcus aureus,28 Klebsiella pneumoniae, and Pseudomonas aeruginosa.29 In our study, about 66.6% of the isolates had shown prominent efflux activity at 1 µg/mL and 1.5 µg/mL of EtBr. However, the presence of efflux pump activity may not be always associated with antibiotic resistance. Apart from antibiotic resistance, the pumps are physiologically involved in the extrusion of various metabolites, dyes, and chemicals that are toxic to the cells. However, the efflux pumps may play an important role in the emergence of multiple drug resistance among bacteria. Thus, here, we assessed antibiotics of different classes which get extruded via efflux pumps using efflux pump inhibitor in S. Typhimurium isolates. PAβN is a well-studied broad spectrum efflux pump inhibitor. It has been widely used for studying efflux activity in P. aeruginosa,30 S. Typhimurium,30,31 S. Typhi32 and E. coli.30 Each isolate of S. Typhimurium was subjected to antibiotic sensitivity assay (against those antibiotics to which each isolate was found resistant) in the presence or absence of PAβN and the difference between zone of inhibition (ZOI) was measured. The significant difference in ZOI indicates the efflux activity for that particular antibiotic. Overall, 88.8% of the isolates have shown efflux activity for at least one antibiotic. Prominent efflux activity was shown for cefazolin (in 2 out of 5 cefazolin-resistant isolates), cefamandole (in 3 out of 4 cefamandole-resistant isolates), cefaclor (in 3 out of 5 cefaclor-resistant isolates) followed by polymyxin-B (in 2 out of 2 polymyxin-B-resistant isolates), imipenem (2 out of 5 imipenem-resistant isolates), kanamycin (1 out of 1 kanamycin-resistant isolates), and piperacillin (1 out of 6 piperacillin-resistant isolates). Efflux activity for cephalosporins has been well reported in the literature. According to previous reports, various efflux pumps are involved in pumping of cefazolin,8,33 cefamandole,33 polymyxin-B,34 imipenem,35 kanamycin,36 and cefaclor37.
All (except ST-91) EtBr cartwheel test positive isolates (i.e. isolates showed active efflux pumps) have shown efflux of maximum of 3 antibiotics. Although the results of efflux activity tested by EtBr cartwheel method is complimentary with results of EPI test, some of the EtBr negative isolates (isolates ST-24, ST-29, and ST-75) also had efflux activity for at least one antibiotic. On the other hand, isolate ST-91 has shown efflux activity for EtBr but was found to be sensitive to most of the antibiotics. This may be the due difference in the substrate specificity of the efflux pumps for EtBr and antibiotics hence, lack of efflux activity for EtBr may not indicate the total absence of efflux pumps in bacterial strains. Similarly, it may not be true that the same efflux activity will be expected for EtBr and antibiotics.
The efflux pump inhibitor-PAβN did not affect the resistance of quinolones (nalidixic acid) in any of the resistant isolates. In contrast to our findings, Li et al (2015)31 reported that the resistance to nalidixic acid was inhibited by PAβN at 20 µg/mL in S. Typhimurium. However, the high level of nalidixic acid resistance may also be associated with a plasmid (qnr)38 or mutations in the quinolone resistance-determining region of gyrA gene.39
Overall, our results indicate that environmental sources of wet poultry markets are the potential contributor of zoonotic MDR S. Typhimurium. Efflux activity for different antibiotics varied among MDR isolates of S. Typhimurium. The efflux activity for cephalosporins was more common as compared to other class of antibiotics in MDR isolates. Efflux pumps of MDR S. Typhimurium contribute to resistance for at least one antibiotic even if EtBr cartwheel test shows negative. Isolates of EtBr cartwheel test negative should not be neglected as these strains too have multiple drug resistance. Therefore, use of EtBr cartwheel test and EPI assay simultaneously may help to identify the MDR isolates more efficiently than using the individual test.
Acknowledgments
The corresponding author is thankful to the Science and Engineering Research Board (SERB), Department of Science and Technology (DST) for providing Postdoctoral Fellowship (PDF/2016/003206) and funds to carry out the research. The authors are thankful to the Dean, Sri Venkateswara Veterinary University, Tirupati, for providing necessary facilities.
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Brenner FW, Villar RG, Angulo FJ, Tauxe R, Swaminathan B. Salmonella nomenclature. J Clin Microbiol. 2000;38:2465–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Issenhuth-Jeanjean S, Roggentin P, Mikoleit M, et al. Supplement 2008–2010 (no. 48) to the White–Kauffmann–Le minor scheme. Res Microbiol. 2014;165:526–530. doi: 10.1016/j.resmic.2014.07.004 [DOI] [PubMed] [Google Scholar]
- 3.Griffin AJ, McSorley SJ. Development of protective immunity to Salmonella, a mucosal pathogen with a systemic agenda. Mucosal Immunol. 2011;4:371–382. doi: 10.1038/mi.2011.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen JI, Bartlett JA, Corey GR. Extra-intestinal manifestations of Salmonella infections. Medicine (Baltimore). 1987;66:349–388. doi: 10.1097/00005792-198709000-00003 [DOI] [PubMed] [Google Scholar]
- 5.Greig JD, Ravel A. Analysis of foodborne outbreak data reported internationally for source attribution. Int J Food Microbiol. 2009;130:77–87. doi: 10.1016/j.ijfoodmicro.2008.12.031 [DOI] [PubMed] [Google Scholar]
- 6.Bakshi CS, Singh VP, Malik M, Singh RK, Sharma B. 55 kb plasmid and virulence-associated genes are positively correlated with Salmonella Enteritidis pathogenicity in mice and chickens. Vet Res Commun. 2003;27:425–432. [DOI] [PubMed] [Google Scholar]
- 7.Martins M, McCusker M, Amaral L, Fanning S. Mechanisms of antibiotic resistance in Salmonella: efflux pumps, genetics, quorum sensing and biofilm formation. Lett Drug Des Discov. 2011;8(2):114–123. doi: 10.2174/157018011794183770 [DOI] [Google Scholar]
- 8.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2009;69(12):1555–1623. doi: 10.2165/11317030-000000000-00000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikaido H. How do exported proteins and antibiotics bypass the periplasm in gram-negative bacterial cells? Trends Microbiol. 2000;8(11):481–483. [DOI] [PubMed] [Google Scholar]
- 10.Galan JE, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella invasion gene invA: homology of invA to members of a new protein family. J Bacteriol. 1992;174:4338–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarez J, Sota M, Vivanco A, et al. Development of a multiplex PCR technique for detection and epidemiological typing of Salmonella in human clinical samples. J Clin Microbiol. 2004;42:1734–1738. doi: 10.1128/JCM.42.4.1734-1738.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martins M, McCusker MP, Viveiros M, et al. A simple method for assessment of MDR bacteria for over-expressed efflux pumps. Open Microbiol J. 2013;7:72–82. doi: 10.2174/1874285801307010072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun J, Deng Z, Yan A. Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem Biophys Res Commun. 2014;2014(453):254–267. doi: 10.1016/j.bbrc.2014.05.090 [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization. Antimicrobial resistance: global report on surveillance. World Health Organization; 2014.
- 15.Pesingi PK, Kumawat M, Behera P, et al. Protein-l-isoaspartyl methyltransferase (PIMT) is required for survival of Salmonella Typhimurium at 42° C and contributes to the virulence in poultry. Front Microbiol. 2017;8:361. doi: 10.3389/fmicb.2017.00361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikanatha NM, Sandt CH, Localio AR, et al. Multidrug-resistant Salmonella isolates from retail chicken meat compared with human clinical isolates. Foodborne Pathog Dis. 2010;7:929–934. doi: 10.1089/fpd.2009.0499 [DOI] [PubMed] [Google Scholar]
- 17.Majowicz SE, Musto J, Scallan E, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–889. doi: 10.1086/650733 [DOI] [PubMed] [Google Scholar]
- 18.European food safety authority and European centre for disease prevention and control. EU summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2013. EFSA J. 2015;13(2):4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National antimicrobial resistance monitoring system: the 2012–2013 integrated NARMS report; 2015. Available from: http://www.fda.gov/downloads/AnimalVeterinary/SafetyHealth/AntimicrobialResistance. Accessed April 24, 2019.
- 20.Asif M, Rahman H, Qasim M, et al. Molecular detection and antimicrobial resistance profile of zoonotic Salmonella Enteritidis isolated from broiler chickens in Kohat, Pakistan. J Chin Med Assoc. 2017;80:303–306. doi: 10.1016/j.jcma.2016.11.007 [DOI] [PubMed] [Google Scholar]
- 21.Giraud E, Baucheron S, Cloeckaert A. Resistance to fluoroquinolones in Salmonella: emerging mechanisms and resistance prevention strategies. Microb Infec. 2006;8(7):1937–1944. doi: 10.1016/j.micinf.2005.12.025 [DOI] [PubMed] [Google Scholar]
- 22.Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agent. 2005;25(5):358–373. doi: 10.1016/j.ijantimicag.2005.02.006 [DOI] [PubMed] [Google Scholar]
- 23.Giraud E, Cloeckaert A, Kerboeuf D, Chaslus-Dancla E. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother. 2000;44(5):1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poole K. Efflux pumps as antimicrobial resistance mechanisms. Ann Med. 2007;39(3):162–176. doi: 10.1080/07853890701195262 [DOI] [PubMed] [Google Scholar]
- 25.Schumacher A, Trittler R, Bohnert JA, et al. Intracellular accumulation of linezolid in Escherichia coli, Citrobacter freundii and Enterobacter aerogenes: role of enhanced efflux pump activity and inactivation. Antimicrob Agents Chemother. 2006;59(6):1261–1264. doi: 10.1093/jac/dkl380 [DOI] [PubMed] [Google Scholar]
- 26.Martins M, Santos B, Martins A, et al. An instrument-free method for the demonstration of efflux pump activity of bacteria. In vivo. 2006;20(5):657–664. [PubMed] [Google Scholar]
- 27.Amaral L, Martins A, Spengler G, Molnar J. Efflux pumps of gram-negative bacteria: what they do, how they do it, with what and how to deal with them. Front Pharmacol. 2014;4:168. doi: 10.3389/fphar.2013.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costa SS, Falcão C, Viveiros M, et al. Exploring the contribution of efflux on the resistance to fluoroquinolones in clinical isolates of Staphylococcus aureus. BMC Microbiol. 2011;11(1):241. doi: 10.1186/1471-2180-11-241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rana T, Kaur N, Farooq U, et al. Efflux as an arising cause of drug resistance in Punjab-India. IJBPAS. 2015;4(9):5967–5979. [Google Scholar]
- 30.Kourtesi C, Ball AR, Huang YY, et al. Suppl 1: microbial efflux systems and inhibitors: approaches to drug discovery and the challenge of clinical implementation. Open Microbiol. 2013;7:34–52. doi: 10.2174/1874285801307010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li XZ, Plésiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in gram-negative bacteria. Clin Microbiol Rev. 2015;28(2):337–418. doi: 10.1128/CMR.00117-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith AM, Govender N, Keddy KH. Quinolone-resistant Salmonella Typhi in South Africa, 2003–2007. Epidemiol Infect. 2010;138(1):86–90. doi: 10.1017/S0950268809990331 [DOI] [PubMed] [Google Scholar]
- 33.Nagano K, Nikaido H. Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci. 2009;106(14):5854–5858. doi: 10.1073/pnas.0901695106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikaido H, Pagès JM. Broad-specificity efflux pumps and their role in multidrug resistance of gram-negative bacteria. FEMS Microbiol Rev. 2012;36(2):340–363. doi: 10.1111/j.1574-6976.2011.00290.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidhu K, Talbot M, Van Mil K, et al. Treatment with sub-inhibitory kanamycin induces adaptive resistance to aminoglycoside antibiotics via the AcrD multidrug efflux pump in Escherichia coli K-12. J Exp Microbiol Immunol. 2012;16:11–16. [Google Scholar]
- 37.Moreira MAS, Souza ECD, Moraes CAD. Multidrug efflux systems in gram-negative bacteria. Braz J Microbiol. 2004;35(1–2):19–28. doi: 10.1590/S1517-83822004000100003 [DOI] [Google Scholar]
- 38.Tran JH, Jacoby GA. Mechanism of plasmid-mediated quinolone resistance. Proc Natl Acad Sci. 2002;99(8):5638–5642. doi: 10.1073/pnas.082092899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacoby GA. Mechanisms of resistance to quinolones. Clin Infect Dis. 2005;41(Supplement_2):S120–S126. doi: 10.1086/428052 [DOI] [PubMed] [Google Scholar]